Abstract
Regulating growth and the cell cycle in response to environmental fluctuations is important for all organisms in order to maintain viability. Two major pathways for translational regulation are found in higher eukaryotes: the Tor signaling pathway and those operating through the eIF2α kinases. Studies from several organisms indicate that the two pathways are interlinked, in that Tor complex 1 (TORC1) negatively regulates the Gcn2 kinase. Furthermore, inactivation of TORC1 may be required for activation of Gcn2 in response to stress. Here, we use the model organism Schizosaccharomyces pombe to investigate this crosstalk further. We find that the relationship is more complex than previously thought. First, in response to UV irradiation and oxidative stress, Gcn2 is fully activated in the presence of TORC1 signaling. Second, during amino-acid starvation, activation of Gcn2 is dependent on Tor2 activity, and Gcn2 is required for timely inactivation of the Tor pathway. Our data show that the crosstalk between the two pathways varies with the actual stress applied.
Keywords: S. pombe, Tor, Gcn2, oxidative stress, starvation, UV irradiation
Introduction
All living organisms are from time to time exposed to stress that can be potentially harmful to the cellular and/or genetic integrity of the organism. Similarly, all organisms have the ability to activate different signaling pathways that make them better equipped to deal with particular stresses. Some of these pathways are targeting the translation machinery in order to change the composition of the proteome, which again changes the general metabolism as well as the regulation of growth and the cell cycle. The target of rapamycin (TOR) kinase is the key player in such a pathway, integrating environmental signals to regulate protein synthesis and other metabolic processes. The TOR kinases form an evolutionarily conserved family of large proteins, and they are members of the phosphatidylinositol 3-kinase (PI3-kinase)-related family.1 The mammalian TOR (mTOR, also referred to as mechanistic TOR) can perform its tasks when incorporated into either of 2 distinct multiprotein complexes, mTOR complex 1 (mTORC1) or mTOR complex 2 (mTORC2). The 2 complexes have clearly separate functions, even if the effector kinase mTOR is the same in both.2 The responses of the TOR pathway to nutrient fluctuations have been well studied. When nutritional conditions are good, the activity of mTOR as a part of mTORC1 is high.3,4 Elevated mTOR activity keeps translation rates high by phosphorylating 2 important translation regulators, the S6 kinase (S6K) and the eIF4E-binding protein (4E-BP1).5,6
Unlike higher eukaryotes, fission yeast has 2 Tor kinases, namely Tor1 and Tor2, of which Tor2 is essential for growth.7 Like mTOR, the fission yeast Tor kinases form large multiprotein complexes with other proteins. Tor2 is the major Tor kinase in TORC1, and Tor1 has mainly been found to associate with TORC2 components. However, Tor1 has also been implied as part of TORC1 in the regulation of mitotic entry in response to nutrient stress.8 The Tor complexes in fission yeast harbor many homologs of proteins found in the mammalian complexes,9 and the upstream regulatory pathways are evolutionarily conserved,10,11 which makes Schizosaccharomyces pombe an excellent model organism for studies of TOR functions. This is further supported by the quite recent finding in fission yeast of a mechanism similar to downstream signaling of mTORC1.12 It was shown that the ribosomal protein S6 (Rps6), an S6K target, was phosphorylated in a Tor2-dependent manner in the presence of adequate nitrogen supply.
Another important way to regulate translation in response to nutrient limitation and other stresses is through phosphorylation of the serine52 of eIF2α, a subunit of the translation initiation factor 2.13 This phosphorylation leads to a general downregulation of translation that is accompanied by enhanced translation of specific mRNAs encoding proteins that are thought to be required for stress responses.14-16 In fission yeast there are 3 known eIF2α kinases, Gcn2, Hri1, and Hri2.17 The kinases share sequence and structural features in their catalytic domains, but have unique flanking regulatory domains, allowing each to respond to distinct stress conditions. Hri2 is the primary responder to heat shock, arsenite, and cadmium, while Gcn2 is the main kinase induced upon nutrient downshift and also after exposure to H2O2, methyl methane sulfonate (MMS), and short-wavelength (254nm) UV light (UVC).17-20 Hri1 was recently shown to respond to nitrogen depletion.21
In S. cerevisiae, a link between inhibition of TOR signaling and activation of Gcn2 to phosphorylate eIF2α has been demonstrated.22 Treating budding yeast with rapamycin, an inhibitor of the TOR kinases, leads to removal of an inhibitory phosphorylation on serine 577 of Gcn2, activating it to phosphorylate eIF2α. However, this phosphorylation site in Gcn2 is not conserved in fission yeast or in mammalian cells. Nonetheless, in a recent study23 it was shown that a link between Tor signaling and eIF2α phosphorylation does exist in fission yeast.
Here, we have investigated the crosstalk between the Tor pathway and eIF2α phosphorylation in fission yeast. We find that the relationship depends on the particular type of stress employed. The two pathways seem to interact in a complex network where signaling from one pathway can affect the signaling of the other and vice versa.
Results
Fission yeast Tor proteins are not involved in the Gcn2-dependent, UVC-induced phosphorylation of eIF2α
We have demonstrated that irradiating fission yeast cells with UVC leads to a Gcn2-dependent phosphorylation of eIF2α and downregulation of translation.18 How Gcn2 is activated in response to such stress is, however, not known. Since Tor signaling was found to be involved in negative regulation of Gcn2 under some conditions in both budding and fission yeast, we set out to explore whether the fission yeast Tor kinases are involved also in regulation of the Gcn2-eIF2α-pathway after UVC irradiation.
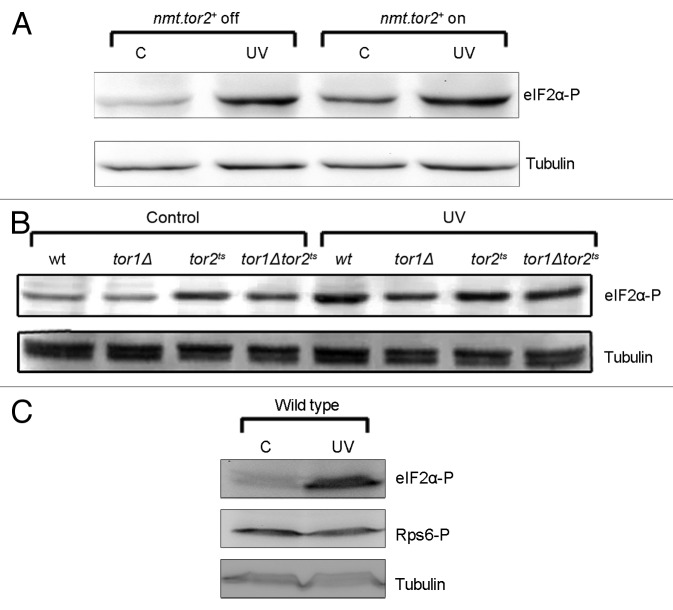
If TORC1 can inhibit the Gcn2-eIF2α pathway after UVC irradiation, a high Tor2 activity would be expected to reduce the Gcn2-dependent phosphorylation of eIF2α. To examine the effect of high Tor2 activity on the UVC-induced phosphorylation of eIF2α, we used a strain in which the tor2 gene was overexpressed and driven by an inducible nmt1 promoter. The strain was grown for 26 h without thiamine to induce the expression of Tor2. The cells were then irradiated with UVC and compared with irradiated cells grown in the presence of thiamine, with the nmt1 promoter turned off. Samples were collected immediately after UVC irradiation and the level of eIF2α phosphorylation was assessed by immunoblotting, using an antibody specific for the phosphorylated form of eIF2α. UVC irradiation induced phosphorylation of eIF2α regardless of whether Tor2 was overexpressed or not (Fig. 1A). Surprisingly, eIF2α phosphorylation was also induced in the non-irradiated samples overexpressing Tor2 (Fig. 1A), although not to the same extent as after UVC irradiation. We conclude that the overproduction of Tor2 leads to phosphorylation of eIF2α, indicating that Tor2 can promote eIF2α phosphorylation.
Figure 1. Tor signaling is not involved in UVC-induced eIF2α phosphorylation. For all experiments, the cells were growing exponentially in EMM at 25 °C, except where noted. (A) nmt.tor2+ cells were cultured without thiamine (nmt.tor2+ on) for 26 h to induce expression of Tor2 (right) or in the presence of thiamine (nmt.tor2+ off; (left) before exposure to UVC. Samples were collected immediately after UVC irradiation. Unirradiated cells were used as control (lanes marked C). (B) Wild-type (wt), tor1∆, tor2ts, and tor1∆tor2ts cells were UVC-irradiated or left unirradiated and samples collected immediately after irradiation. (C) Wild-type cells treated as in (B) to show phosphorylation levels of Rps6. In all panels cell extracts were run on SDS-PAGE and subjected to immunoblotting in order to detect Ser52-phosphorylation of eIF2α and/or phosphorylation of Rps6. Tubulin was used as loading control.
To further test whether the Tor proteins regulate the UVC-induced phosphorylation of eIF2α, we performed a similar experiment in a strain lacking Tor2 activity. Since Tor2 is an essential protein, we used a temperature-sensitive (ts) mutant grown at restrictive temperature. In this strain, eIF2α was phosphorylated in the unirradiated control cells, in agreement with earlier data,23 and phosphorylation was further induced upon UVC irradiation (Fig. 1B). Furthermore, UVC-induced phosphorylation of eIF2α was also seen in cells lacking Tor1 activity (Fig. 1B). To address the possibility that both Tor1 and Tor2 are involved in UVC-induced phosphorylation of eIF2α, we constructed a tor1∆tor2ts double mutant. Also in this strain, where both Tor proteins are nonfunctional, UVC-induced phosphorylation of eIF2α was observed (Fig. 1B). We conclude that the activities of Tor2 and Tor1 are not essential for the induction of eIF2α phosphorylation in response to UVC irradiation.
Since Tor2 inactivation in itself leads to eIF2α phosphorylation, we investigated whether Tor2 activity is reduced in response to UVC irradiation, which could, in turn, result in the UVC-induced eIF2α phosphorylation that we have observed.18 To this end, we made use of an antibody recognizing the phosphorylated form of Rps6, a substrate in the Tor signaling pathway. The phosphorylation status of Rps6 was not changed in a wild-type strain after UVC irradiation (Fig. 1C), indicating that the Tor2 signaling pathway is not affected by UVC irradiation.
We conclude that the Tor proteins are not involved in the regulation of UVC-induced phosphorylation of eIF2α in fission yeast, implying that Gcn2 can be fully activated without a change in Tor activity. However, we find that both overexpression and inactivation of Tor2 lead to phosphorylation of eIF2α in unirradiated control cells, arguing that Tor2 is involved in regulating eIF2α kinase activity.
Rps6 as a readout of Tor2 activity
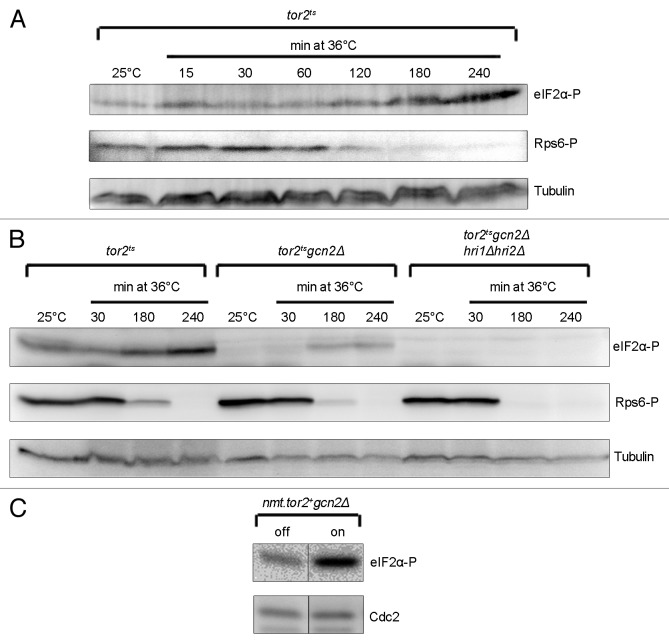
Our assay for measuring Tor2 activity relies on monitoring the phosphorylation of Rps6. Phosphorylation of this substrate has been shown, under some circumstances and to a marginal extent, to depend on the activity of Tor1.24 In our hands, Rps6 phosphorylation was not detectable in a tor2ts mutant incubated at the restrictive temperature (Fig. 2A), confirming that Rps6 phosphorylation is mainly a measure of Tor2 activity. Furthermore, Rps6 is not a direct substrate of the Tor proteins and its dephosphorylation most likely depends on both reduced Tor activity and the action of phosphatase(s). This may introduce a time lag from actual Tor2 inactivation to a reduction in Rps6 phosphorylation, which would pose a difficulty when comparing the kinetics of Tor2 inactivation and eIF2α phosphorylation. However, when the tor2ts mutant was shifted to the restrictive temperature, reduced phosphorylation of Rps6 coincided with increased phosphorylation of eIF2α (Fig. 2A). Since the 2 events coincide under a condition where phosphorylation of eIF2α is most likely a response to Tor2 inactivation (ref. 23; Fig. 2A), Rps6 phosphorylation appears to be a good measure of Tor2 activity and was used as such throughout this work.
Figure 2. Gcn2 is not the sole kinase activated upon Tor2 inactivation. (Aand B) tor2ts, and gcn2∆ and gcn2∆ hri1∆ hri2∆ in a tor2ts background were incubated at the permissive (25 °C) or the restrictive (36 °C) temperature and samples collected at the indicated time points. (C) nmt.tor2+gcn2∆ cells were cultured without thiamine (on) for 26 h to induce expression of Tor2 (right) or in the presence of thiamine (off; left) (the figure is a composite image deriving from different parts of the same blot). Immunoblotting was performed as in Figure 1, except for the use of Cdc2 as a loading control in (C).
Different kinases are activated upon Tor2 inactivation and overexpression
Gcn2 has been shown to be the major kinase responsible for phosphorylation of eIF2α upon Tor2 inactivation.23 Here we confirmed this result in a strain where gcn2 had been deleted in the tor2ts background (Fig. 2B). However, in a tor2ts gcn2∆ strain the phosphorylation of eIF2α after inactivation of Tor2 was not abolished, only reduced (Fig. 2B). In a strain where all 3 eIF2α kinase genes were deleted the phosphorylation of eIF2α was completely abolished (Fig. 2B), suggesting that in addition to Gcn2, Hri1 and/or Hri2 contribute to eIF2α phosphorylation when Tor2 is inactivated. We also explored whether Gcn2 is the main kinase responsible for phosphorylation of eIF2α when Tor2 is overexpressed. For this purpose, we used a strain where gcn2 had been deleted in the nmt.tor2+ background. The strain was cultivated for 26 h without thiamine to induce the expression of Tor2. The phosphorylation of eIF2α upon high Tor2 expression was similar in the gcn2+ and gcn2Δ strains (compare Fig. 1A to Fig. 2C), showing that Gcn2 is not responsible for eIF2α phosphorylation in these cells.
We conclude that Gcn2 plays a major role in phosphorylation of eIF2α when Tor2 is inactivated, but it is not responsible for the phosphorylation when Tor2 is overexpressed.
Tor2 is required for phosphorylation of eIF2α upon amino-acid starvation
Gcn2 is activated by amino-acid starvation,17 and Tor2 is involved in sensing the levels of amino acids.23 This prompted us to investigate whether Tor2 affects the Gcn2-dependent phosphorylation of eIF2α during amino-acid starvation.
Cells auxotrophic for leucine were grown in minimal medium (EMM) supplemented with leucine. To induce starvation, the cells were washed and resuspended in EMM with no leucine, and samples were collected at different time points afterwards. In wild-type (gcn2+) cells phosphorylation of eIF2α occurred at 60 min after withdrawal of leucine (Fig. 3A). In a similarly treated gcn2∆ mutant eIF2α phosphorylation did not occur (Fig. 3A), demonstrating that Gcn2 is the sole kinase responsible for the phosphorylation response. In the same samples, we measured the phosphorylation of Rps6 to study the kinetics of Tor2 activity. In wild-type cells at the time of eIF2α phosphorylation (60 min) the level of Rps6 phosphorylation was the same as in non-starved cells, and it was not reduced until 1 h later (Fig. 3A), indicating that Tor2 activity is not reduced at the time of Gcn2 activation. Thus, in leucine-starved cells, the activation of Gcn2 is not coupled to Tor2 inactivation. Interestingly, the decrease in phosphorylated Rps6 seen in wild-type cells was not detectable within the time range of the experiment in the gcn2∆ mutant (Fig. 3A), suggesting that phosphorylation of eIF2α or a yet unknown function of Gcn2 is required for downregulation of the Tor2–Rps6 pathway in response to leucine starvation.
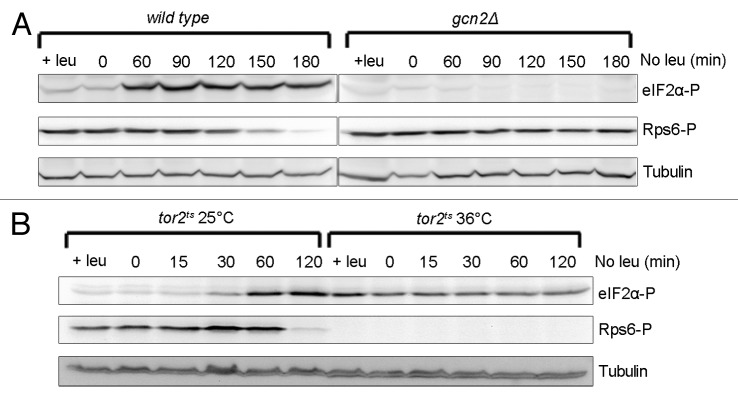
Figure 3. Phosphorylation of eIF2α in response to leucine starvation is dependent on both Gcn2 and Tor2. Leucine-auxotroph cells were grown in the presence of leucine, washed 3 times by filtering with medium lacking leucine, resuspended in leucine-free medium (no leu) and samples collected at the indicated time points. Cells grown in the presence of leucine (+leu) were used as control. (A) Wild type and gcn2∆ (the figure is a composite image deriving from different parts of the same blot). (B) tor2ts cells were cultured at the restrictive temperature (36 °C) for 4 h or at the permissive temperature (25 °C) before shift to medium without leucine. Cultures were kept at their respective temperatures after medium shift. Immunoblotting was performed as in Figure 1.
To investigate the effect of loss of Tor2 activity on the leucine-starvation response, we incubated a tor2ts leu1–32 strain at the restrictive temperature for 4 h before removing the leucine. The culture was kept at the restrictive temperature, and samples were collected at different time points. Incubation at the restrictive temperature induced phosphorylation of eIF2α due to Tor2 inactivation, but a further increase in phosphorylation as a response to leucine starvation was not observed (Fig. 3B). In the same strain grown at permissive temperature, eIF2α was phosphorylated to the same extent and followed the same kinetics as in wild-type cells (Fig. 3B). We conclude that the phosphorylation of eIF2α in response to leucine starvation is absolutely dependent on both Gcn2 and Tor2.
Phosphorylation of eIF2α in response to nitrogen starvation coincides with Tor2 inactivation
Amino-acid starvation and nitrogen depletion are thought to be quite similar stress types, since depletion of nitrogen eventually will result in amino-acid starvation. Thus, one would expect cells to respond in a similar way upon withdrawal of nitrogen sources and amino acids. However, the response to leucine starvation (above) is quite different from that previously reported for nitrogen starvation.12,23 Therefore, we re-examined the relationship between Tor2 inactivation and eIF2α phosphorylation upon nitrogen depletion to ascertain that the observed differences are due to the different treatments rather than experimental or strain differences. Wild-type cells cultured in EMM with NH4Cl as a nitrogen source were harvested by filtration, washed 3 times with EMM with or without NH4Cl, resuspended in the same medium, and samples were collected at different time points after the medium change. Phosphorylation of Rps6 was dramatically reduced at 60 min after withdrawal of the nitrogen source, due to reduced Tor2 activity, and this coincided with a small and transient increase in eIF2α phosphorylation (Fig. 4A). Thus, a reduction in Tor2 activity upon nitrogen starvation correlates with a weak and transient phosphorylation of eIF2α. These results are consistent with those previously reported12,23 and confirm that the different responses after leucine and nitrogen starvation does not derive from experimental or strain differences.
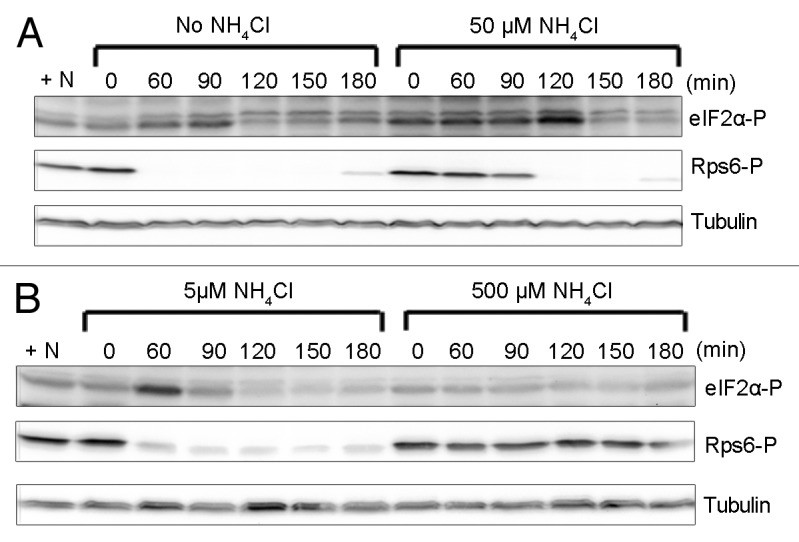
Figure 4. Inactivation of Tor2 in response to nitrogen starvation coincides with a small and transient increase in eIF2α phosphorylation. (A and B) Wild-type cells were harvested on filter, washed 3 times with equal volumes of medium lacking or supplemented with 5, 50, or 500 µM NH4Cl and resuspended in washing media. Samples were collected at the indicated time points. Cells washed and resuspended in standard minimal medium were used as control (+N). Immunoblotting was performed as in Figure 1.
We also investigated whether a gradual depletion of the nitrogen source, a scenario that is more likely in nature, has a more severe effect on eIF2α phosphorylation than a sudden depletion. To gradually deplete the cells of their nitrogen source, we performed the experiment as described above, but instead of washing and resuspending the cells in EMM without NH4Cl, we used EMM containing 5, 50, or 500 µM NH4Cl. In the cultures shifted to medium with the lowest concentrations of ammonium chloride, phosphorylation of Rps6 was reduced within 60 min (5 µM) or 120 min (50 µM), and this coincided with a weak and transient increase in eIF2α phosphorylation (Fig. 4A and B). No change in phosphorylation of either protein occurred upon a shift to medium with 500 µM NH4Cl (Fig. 4B). These data suggest that there is no significant difference in the response whether the nitrogen source is depleted rapidly or gradually. We also note that as little as 500 µM of NH4Cl in the growth medium was enough to keep Tor2 signaling high, suggesting that standard minimal medium (94 mM NH4Cl)25 contains nitrogen source in vast excess.
Gcn2 activation in response to oxidative stress occurs in the presence of active Tor2
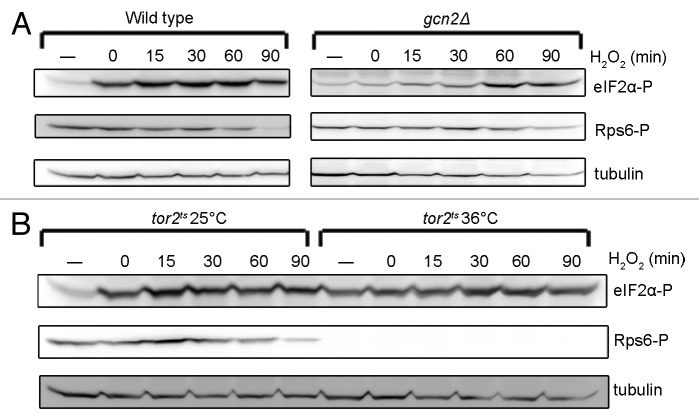
The Gcn2 and the Tor signaling pathways both respond to oxidative stress. Treatment with high concentrations of H2O2 (1–5 mM) leads to Gcn2-dependent phosphorylation of eIF2α17,19,20 and to inactivation of Tor2.12 However, the 2 events have never been investigated in the same study, and therefore a potential crosstalk between the pathways has not been revealed. We set out to investigate a potential interaction by examining the phosphorylation kinetics of eIF2α and Rps6. If Tor2 inactivation is a prerequisite for Gcn2 activation in response to oxidative stress, a tight correlation between reduced Tor2 activity and phosphorylation of eIF2α is expected. Wild-type cells were treated with 5 mM H2O2 for different periods of time, and the phosphorylation of eIF2α and inactivation of Tor2 were measured by immunoblotting using the phosphospecific eIF2α and Rps6 antibodies. A rapid and robust phosphorylation of eIF2α was observed after treatment with H2O2 (Fig. 5A), as described previously.17,19,20 Oxidative stress also led to a reduction in Rps6 phosphorylation (Fig. 5A). However, this response occurred at least an hour after the increase in eIF2α phosphorylation. Taken together, these data suggest that phosphorylation of eIF2α upon oxidative stress is not dependent on inactivation of Tor2.
Figure 5. Oxidative stress leads to rapid phosphorylation of eIF2α and precedes inactivation of Tor2. (A) Wild-type and gcn2∆ cells were treated with H2O2 at a final concentration of 5 mM. Samples were collected at the indicated time points and untreated cells (−) were used as control. (B) tor2ts cells were cultured at restrictive temperature (36 °C) for 4 h or permissive temperature (25 °C) and treated as in (A), then kept at their respective temperatures. Immunoblotting was performed as in Figure 1.
Since the reduction in phosphorylated Rps6 upon oxidative stress is slow compared with the observed increase in phosphorylation of eIF2α, we asked whether Tor2 activity is required for eIF2α phosphorylation. A culture of the tor2ts strain incubated at the restrictive temperature for 4 h was treated with 5 mM H2O2, further incubated at the restrictive temperature, and samples collected at different time points. Exposure to H2O2 led to an increase in eIF2α phosphorylation beyond that induced by Tor2 inactivation, although slightly less than that observed in the control at permissive temperature (Fig. 5B). This indicates that Tor2 signaling is not required for eIF2α phosphorylation in response to oxidative stress.
We also investigated whether Gcn2 affects Tor2 signaling in response to oxidative stress. A strain lacking gcn2 was treated with H2O2 as described above, and the phosphorylation of eIF2α and Rps6 was measured by immunoblotting. In the absence of Gcn2, the early and robust phosphorylation of eIF2α, observed in wild-type cells, was absent (Fig. 5A). Tor2 activity was reduced, but 30 min later than what was observed in gcn2+ cells (Fig. 5A). Some phosphorylation of eIF2α could be detected at later time points (Fig. 5A), probably due to the activation of Hri2.20
In summary, we find that under oxidative stress phosphorylation of eIF2α and the activity of Tor2 are not coupled. Neither the presence nor the inactivation of Tor2 is required for eIF2α phosphorylation, and Gcn2-dependent phosphorylation of eIF2α is not required for Tor2 inactivation.
Inhibition of Tor by rapamycin coincides with phosphorylation of eIF2α
To explore how phosphorylation of eIF2α is affected by yet another treatment repressing Tor activity, we exposed fission yeast cells to rapamycin, a well-known inhibitor of Tor proteins.26-28 Both Tor proteins in fission yeast are inhibited by rapamycin,12,29,30 although rapamycin-treatment of fission yeast cells does not result in the same phenotype as mutating tor1 and tor2.7,31
Wild-type cells were treated with rapamycin, and the phosphorylation status of eIF2α was compared with that found in untreated cells. After 15–30 min of rapamycin treatment, eIF2α became phosphorylated, and this was closely followed by a drop in Tor2 activity (Fig. 6). These results, taken together with the fact that inactivation of Tor2 is sufficient to induce eIF2α phosphorylation, indicate that phosphorylation during rapamycin treatment is a result of reduced Tor2 activity.
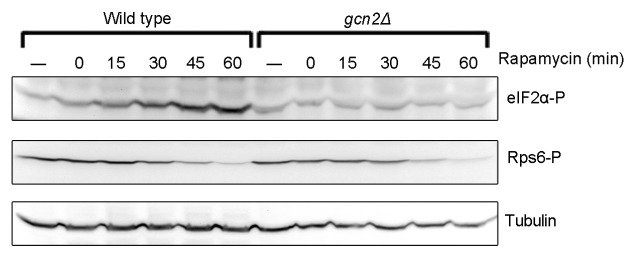
Figure 6. eIF2α is phosphorylated in a Gcn2-dependent manner in response to rapamycin treatment. Wild-type and gcn2∆ cells in exponential phase grown in YES at 25 °C were treated with rapamycin at a final concentration of 200 nM. Samples were collected at the indicated time points and cells treated with solvent only (−) were used as control. Immunoblotting was performed as in Figure 1.
In a gcn2∆ mutant strain the rapamycin-induced phosphorylation of eIF2α was abolished (Fig. 6), showing that Gcn2 is responsible for the drug-induced phosphorylation in wild-type cells.
These results suggest that rapamycin treatment of fission yeast results in a phenotype that shares features with, but is not equivalent to, that of the temperature-sensitive tor2 mutant. Both treatments induce a weak eIF2α phosphorylation, but, unlike in rapamycin, phosphorylation is not completely dependent on Gcn2 in the tor2ts strain (compare Fig. 6 to Fig. 2B). Also, after rapamycin treatment the phosphorylation of eIF2α seems to follow Tor2 inactivation more closely in time than it does in the tor2ts (compare Fig. 6 to Fig. 2A).
Discussion
We have investigated the crosstalk between 2 major pathways involved in translation regulation: the Tor signaling network and the phosphorylation of the translation-initiation factor eIF2α. We show that the Tor pathway can affect eIF2α phosphorylation under some conditions, consistent with earlier reports.22,23 However, the interplay between the 2 pathways is more complex than previously proposed and depends on the particular stress employed.
In S. cerevisiae, the activity of Gcn2 can be inhibited by phosphorylation on serine 577. It has been demonstrated that both TOR kinases can inhibit dephosphorylation of this serine 577, TOR1 (Tor2 in S. pombe) being the main contributor. Furthermore, inactivation of the TOR kinases by rapamycin leads to activation of a phosphatase that removes the inhibitory phosphate on serine 577 of Gcn2.22 The only known mechanism for direct activation of Gcn2 is the binding of uncharged tRNAs, which accumulate during amino-acid starvation.32 Under conditions that lead to dephosphorylation of serine 577, the activation of Gcn2 is still dependent on binding of uncharged tRNAs. However, removal of the inhibitory phosphorylation increases the affinity of Gcn2 for uncharged tRNAs.33 Therefore, it has been proposed that under types of stress where the levels of uncharged tRNAs do not rise, activation of Gcn2 is dependent on dephosphorylation of serine 577.33 A similar mechanism of Tor activity (Tor2) being inhibitory to Gcn2 has also been shown in S. pombe.23
Our observations presented here, together with previously published results,12,23 demonstrate that reducing Tor2 signaling by 3 different strategies: nitrogen starvation, a temperature-sensitive tor2 mutant, and exposure to rapamycin, all lead to phosphorylation of eIF2α in fission yeast. Under these conditions, a reduction in Tor2 activity and eIF2α phosphorylation occur at about the same time, suggesting a causal relationship. Interestingly, serine 577 is not conserved in S. pombe or in mammalian cells, suggesting that a potential regulatory role of Tor2 over Gcn2 activity is not fully conserved.
We show here that Tor2 is not inhibiting eIF2α phosphorylation under a number of conditions, and these results challenge a straightforward model for Tor2-dependent Gcn2 regulation. First, we show that Gcn2 can be fully activated by UVC and oxidative stress without any detectable change in Tor2 activity. Second, under leucine starvation the phosphorylation of eIF2α is dependent on maintained Tor2 activity rather than on Tor2 inactivation. Third, both Tor2 overexpression and inactivation bring about eIF2α phosphorylation. Fourth, we find that eIF2α phosphorylation and Tor2 inactivation does not follow a consistent pattern, since both can precede the other, depending on the type of stress employed. Finally, we find that under leucine starvation the Gcn2 pathway seems to regulate Tor2 activity.
Gcn2 is activated in response to UVC and oxidative stress, conditions where uncharged tRNAs are unlikely to accumulate. Under these stresses, we find that Tor signaling is either left unchanged (UVC), or its downregulation does not precede the phosphorylation of eIF2α (H2O2), suggesting that the mechanism proposed for S. cerevisiae does not apply to S. pombe.
In response to leucine starvation, the activation of Gcn2 occurred without any detectable change in Tor2 activity. This is consistent with data from S. cerevisiae, showing that Gcn2 activation in response to histidine starvation is not dependent on dephosphorylation of ser577.22 More surprisingly, we find that instead of being inhibitory to Gcn2 activity, Tor2 signaling is required for starvation-induced phosphorylation of eIF2α. This seems paradoxical, since Tor2 is known to promote translation and therefore seems an unlikely candidate to induce eIF2α phosphorylation. Furthermore, it has been reported that high Tor activity suppresses eIF2α phosphorylation under amino acid-replete conditions.23 We propose that the major mechanism whereby Tor2 affects eIF2α phosphorylation in response to amino-acid starvation is through its effects on maintaining optimal translation levels. We suggest that ongoing translation might be a prerequisite for accumulation of uncharged tRNAs and, thus, the activation of Gcn2.
Surprisingly, nitrogen starvation, a type of stress that is thought to lead to amino-acid starvation, elicited quite a different response from leucine starvation. Gcn2 was activated by leucine starvation in a Tor2-dependent manner, and Tor2 inactivation was detected 60 min after the increase in eIF2α phosphorylation, whereas after nitrogen starvation Tor2 was quickly inactivated, but only a transient and weak induction of eIF2α phosphorylation could be observed. These results indicate that regulation of the 2 pathways depends upon the type of starvation that the cells are subjected to. Similarly, in mammalian cells, the mTOR pathway is regulated by different upstream elements in response to different environmental stimuli. The canonical pathway of mTORC1 regulation by insulin goes via the tuberous sclerosis complex 1 and 2 (TSC1/TSC2-complex), which inhibits an mTOR activator named Ras homolog enriched in brain (Rheb).4,34,35 From studies in mammalian cells, it has been suggested that mTORC1 is not regulated through this pathway in response to amino acid deprivation, since in TSC2-knockout cells mTOR activity is still reduced.36,37 This finding suggests the existence of an alternative or additional pathway. It has also been suggested that the alternative pathway for mTORC1 inhibition involves Gcn2.38 Interestingly, and in agreement with this model, we observed that in a gcn2 deletion mutant inactivation of Tor2 does not occur under leucine starvation. In further support of this model, it was recently shown that leucine deprivation reduces mTOR/S6K1 signaling in mammalian cells in a GCN2-dependent manner.39
The observation that overexpressing Tor2 can promote phosphorylation of eIF2α suggests that Tor2 activity is not exclusively inhibitory to the eIF2α kinases. However, it should be noted that an increase in Rps6 phosphorylation, which is our measure of Tor2 activity, could not be detected upon overexpression of Tor2. This might result from the fact that Rps6 phosphorylation is not a direct readout of Tor2 activity (see above). Alternatively, we cannot exclude the possibility that the overall activity of Tor2 is not elevated in an overproducer, and that phosphorylation of eIF2α is a response to stress caused by overexpression per se, and not a response specific to Tor2 activity.
Our finding that eIF2α is phosphorylated in response to rapamycin treatment is in disagreement with some previously published results. In contrast to this work and Valbuena et al.,23 2 other publications12,40 report that treatment of fission yeast with rapamycin does not bring about phosphorylation of eIF2α. The only obvious way their and our experimental procedures differ is the growth media employed. In the 2 experiments where rapamycin treatment leads to induction of eIF2α phosphorylation, the cells were cultured in YES, as opposed to EMM, in the other 2 papers. Since Tor2 activity and translation levels are likely to be higher in rich medium, inhibiting Tor by rapamycin in YES might bring about a more pronounced starvation-mimicking response than in EMM, including detectable changes in eIF2α phosphorylation.
A lot remains to be elucidated about how the pathways of eIF2α kinases and Tor signaling interact with each other in response to a number of different stresses. The current work demonstrates that there is no single mechanism for how the 2 pathways interact that applies to all types of stress. We conclude that rather than operating in a linear pathway, the 2 mechanisms cooperate in a complex signaling network regulating translation.
Materials and Methods
Fission yeast strains and media
All strains used were derived from the Schizosaccharomyces pombe L972h- strain and are listed in Table 1. The growth conditions and media were as described in reference 25. The cells were grown in liquid Edinburgh minimal medium (EMM) (Cat. #2005, Sunrise Science products http://www.sunrisescience.com/pages/ystmedia_sp_emm.html) containing the required supplements, or in yeast extract (YES) (Cat. #2009, Sunrise Science products http://www.sunrisescience.com/pages/ystmedia_sp_yes.html), at 25 °C, to a cell density of 3–5 × 106 cells/ml (OD595 0.15–0.3). For Tor2 overexpression, cells cultured in EMM + 5 µg/ml thiamine (Cat. #T-1270, Sigma-Aldrich http://www.sigmaaldrich.com/catalog/search?interface=All&term=T-1270&lang=en®ion=NO&focus=product&N=0+220003048+219853206+219853286&mode=match%20partialmax) were harvested by filtering, washed 3 times culture volume with EMM lacking thiamine, and resuspended in EMM lacking thiamine to induce expression from the nmt promoter for 26 h before experiments were performed and samples collected. For Tor2-inactivation experiments, cells were grown in EMM at 25 °C, and shifted to 36 °C for 4 h, except where indicated.
Table 1. Strains used in this study.
| Strain | Genotype of the strain | Source |
|---|---|---|
| 214 | leu1–32 h- | P Nurse |
| 489 | cdc10-M17 | P Nurse |
| 1136 | SPBC 36B7.09 gcn2::ura4+ leu1–32 ura4-D18 h- | R Wek |
| 1138 | gcn2::ura4+ cdc10M-17 ura4-D18 h- | This study |
| 1257 | tor1::kanMX6 h- | S Moreno |
| 1258 | kanMX6-P3nmt.tor2+ h+ | S Moreno |
| 1259 | tor2–51:ura4+ ura4-D18 h+ | S Moreno |
| 1279 | tor2–51:ura4+ gcn2::ura4+ ura4-D18 h+ | This study |
| 1305 | tor2–51:ura4+ ura4-D18 leu1–32 | This study |
| 1384 | tor1::kanMX6 tor2ts:ura4+ ura4-D18 leu1–32 | This study |
| 1812 | gcn2::hphMX6 tor2–51:ura4+ ura4-D18 hri1::ura4+ hri2::ura4+ leu1–32 h- | This study |
Leucine starvation
Leucine-auxotroph cells were grown in the presence of leucine in the appropriate minimal medium and washed 3 times, by filtering, with medium lacking leucine. The cells were then resuspended in washing medium, and samples were collected for immunoblot analyses at different time points after leucine removal.
Nitrogen starvation
Nitrogen starvation experiments were performed by harvesting on filters cells exponentially growing in EMM. The filters were washed 3 times with equal volumes of EMM without NH4Cl (Cat. #2023, Sunrise Science products http://www.sunrisescience.com/pages/ystmedia_sp_emm.html) or with reduced concentrations of NH4Cl and resuspended in EMM containing the same concentrations of NH4Cl as in the washing medium. Samples were collected by centrifugation for immunoblot analyses at different time points after change of medium.
Oxidative stress
Cells grown in EMM were treated with 5 mM H2O2 (Cat. #216763, Sigma-Aldrich http://www.sigmaaldrich.com/catalog/search?interface=All&term=216763&lang=en®ion=NO&focus=product&N=0+220003048+219853206+219853286&mode=match%20partialmax), and samples collected at different time points after addition of the oxidative agent for immunoblot analyses.
Rapamycin
Cells growing exponentially in YES medium were exposed to 200 nM of rapamycin (Cat. #553210, EMD Millipore Corporation http://www.millipore.com/search.do?q=553210#0:0) and samples were collected for immunoblot analyses at different time points after addition of the drug.
UVC irradiation
Cells suspended in a thin layer (3 mm) of rapidly stirred liquid medium were irradiated at 20–25 °C with 254-nm UV light. The dose was measured with a radiometer (UV Products), and a dose of 1100 J/m2 was given at an incident dose rate of approximately 250 J/m2/min. This dose results in >90% survival when log-phase cells are irradiated and ca 20% survival when cells arrested in G1 are irradiated.
Immunoblots
Samples for immunoblotting were made by the trichloroacetic acid protein extraction method.41 A total of 100–200 µg protein extracts were run on 12.5% SDS-PAGE, transferred to a PVDF membrane (Cat. #IPVH00010, EMD Millipore Corporation http://www.millipore.com/catalogue/item/ipvh00010) and probed with the following antibodies: anti-phosphorylated eIF2α (Cat. #44-728G, Life Technologies http://www.lifetechnologies.com/order/catalog/product/44728G?ICID=search-44728g) 1:3000; anti-α-tubulin (Cat. #T-5168 Sigma-Aldrich, http://www.sigmaaldrich.com/catalog/search?interface=All&term=T-5168&lang=en®ion=NO&focus=product&N=0+220003048+219853206+219853286&mode=match%20partialmax) 1:30 000; and for the phosphorylation of Rps6, anti-Phospho-(Ser/Thr) Akt Substrate (Cat. #9611, Cell Signaling Technology, http://www.cellsignal.com/products/9611.html) 1:2000.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank S Moreno, P Nurse, R Wek and A Nakashima for kindly providing strains, MO Haugli, L Lindbergsengen, and I Løbersli for technical assistance and S Lopez-Aviles for useful discussions. This work was supported by The Research Council of Norway, the Norwegian Cancer Society, the South-Eastern Norwegian Regional Health Authority and Radiumhospitalets Legater.
Glossary
Abbreviations:
- TOR
target of rapamycin
- TORC1
Tor complex 1
- ,TORC2
Tor complex 2
- mTOR
mammalian TOR
- PI3-kinase
phosphatidylinositol 3-kinase
- S6K
S6 kinase
- 4E-BP1
eIF4E-binding protein
- Rps6
ribosomal protein S6
- MMS
methyl methane sulfonate
- UVC
ultraviolet light
- ts
temperature-sensitive
- EMM
minimal medium
- TSC1/TSC2-complex
tuberous sclerosis complex 1 and 2
- Rheb
Ras homolog enriched in brain
- YES
yeast extract
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27270
References
- 1.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 2.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Proud CG. Regulation of mammalian translation factors by nutrients. Eur J Biochem. 2002;269:5338–49. doi: 10.1046/j.1432-1033.2002.03292.x. [DOI] [PubMed] [Google Scholar]
- 4.Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, Yonezawa K. Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene. 2006;25:6361–72. doi: 10.1038/sj.onc.1209882. [DOI] [PubMed] [Google Scholar]
- 5.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A. 1998;95:1432–7. doi: 10.1073/pnas.95.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–87. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weisman R, Choder M. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J Biol Chem. 2001;276:7027–32. doi: 10.1074/jbc.M010446200. [DOI] [PubMed] [Google Scholar]
- 8.Hartmuth S, Petersen J. Fission yeast Tor1 functions as part of TORC1 to control mitotic entry through the stress MAPK pathway following nutrient stress. J Cell Sci. 2009;122:1737–46. doi: 10.1242/jcs.049387. [DOI] [PubMed] [Google Scholar]
- 9.Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27:3154–64. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uritani M, Hidaka H, Hotta Y, Ueno M, Ushimaru T, Toda T. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells. 2006;11:1367–79. doi: 10.1111/j.1365-2443.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 11.Weisman R, Roitburg I, Schonbrun M, Harari R, Kupiec M. Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics. 2007;175:1153–62. doi: 10.1534/genetics.106.064170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakashima A, Sato T, Tamanoi F. Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J Cell Sci. 2010;123:777–86. doi: 10.1242/jcs.060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimball SR. Eukaryotic initiation factor eIF2. Int J Biochem Cell Biol. 1999;31:25–9. doi: 10.1016/S1357-2725(98)00128-9. [DOI] [PubMed] [Google Scholar]
- 14.Hinnebusch AG. Gene-specific translational control of the yeast GCN4 gene by phosphorylation of eukaryotic initiation factor 2. Mol Microbiol. 1993;10:215–23. doi: 10.1111/j.1365-2958.1993.tb01947.x. [DOI] [PubMed] [Google Scholar]
- 15.Gerlitz G, Jagus R, Elroy-Stein O. Phosphorylation of initiation factor-2 alpha is required for activation of internal translation initiation during cell differentiation. Eur J Biochem. 2002;269:2810–9. doi: 10.1046/j.1432-1033.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- 16.Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics. 2009;38:328–41. doi: 10.1152/physiolgenomics.90396.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhan K, Narasimhan J, Wek RC. Differential activation of eIF2 kinases in response to cellular stresses in Schizosaccharomyces pombe. Genetics. 2004;168:1867–75. doi: 10.1534/genetics.104.031443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tvegård T, Soltani H, Skjølberg HC, Krohn M, Nilssen EA, Kearsey SE, Grallert B, Boye E. A novel checkpoint mechanism regulating the G1/S transition. Genes Dev. 2007;21:649–54. doi: 10.1101/gad.421807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krohn M, Skjølberg HC, Soltani H, Grallert B, Boye E. The G1-S checkpoint in fission yeast is not a general DNA damage checkpoint. J Cell Sci. 2008;121:4047–54. doi: 10.1242/jcs.035428. [DOI] [PubMed] [Google Scholar]
- 20.Berlanga JJ, Rivero D, Martín R, Herrero S, Moreno S, de Haro C. Role of mitogen-activated protein kinase Sty1 in regulation of eukaryotic initiation factor 2alpha kinases in response to environmental stress in Schizosaccharomyces pombe. Eukaryot Cell. 2010;9:194–207. doi: 10.1128/EC.00185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martín R, Berlanga JJ, de Haro C. New roles of the fission yeast eIF2α kinases Hri1 and Gcn2 in response to nutritional stress. J Cell Sci. 2013;126:3010–20. doi: 10.1242/jcs.118067. [DOI] [PubMed] [Google Scholar]
- 22.Cherkasova VA, Hinnebusch AG. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003;17:859–72. doi: 10.1101/gad.1069003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valbuena N, Rozalén AE, Moreno S. Fission yeast TORC1 prevents eIF2α phosphorylation in response to nitrogen and amino acids via Gcn2 kinase. J Cell Sci. 2012;125:5955–9. doi: 10.1242/jcs.105395. [DOI] [PubMed] [Google Scholar]
- 24.Du W, Hálová L, Kirkham S, Atkin J, Petersen J. TORC2 and the AGC kinase Gad8 regulate phosphorylation of the ribosomal protein S6 in fission yeast. Biol Open. 2012;1:884–8. doi: 10.1242/bio.20122022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-L. [DOI] [PubMed] [Google Scholar]
- 26.Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–9. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- 27.Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–8. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- 28.Lorenz MC, Heitman J. TOR mutations confer rapamycin resistance by preventing interaction with FKBP12-rapamycin. J Biol Chem. 1995;270:27531–7. doi: 10.1074/jbc.270.46.27531. [DOI] [PubMed] [Google Scholar]
- 29.Weisman R, Finkelstein S, Choder M. Rapamycin blocks sexual development in fission yeast through inhibition of the cellular function of an FKBP12 homolog. J Biol Chem. 2001;276:24736–42. doi: 10.1074/jbc.M102090200. [DOI] [PubMed] [Google Scholar]
- 30.Weisman R, Roitburg I, Nahari T, Kupiec M. Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics. 2005;169:539–50. doi: 10.1534/genetics.104.034983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schonbrun M, Laor D, López-Maury L, Bähler J, Kupiec M, Weisman R. TOR complex 2 controls gene silencing, telomere length maintenance, and survival under DNA-damaging conditions. Mol Cell Biol. 2009;29:4584–94. doi: 10.1128/MCB.01879-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wek SA, Zhu S, Wek RC. The histidyl-tRNA synthetase-related sequence in the eIF-2 alpha protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15:4497–506. doi: 10.1128/mcb.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia-Barrio M, Dong J, Cherkasova VA, Zhang X, Zhang F, Ufano S, Lai R, Qin J, Hinnebusch AG. Serine 577 is phosphorylated and negatively affects the tRNA binding and eIF2alpha kinase activities of GCN2. J Biol Chem. 2002;277:30675–83. doi: 10.1074/jbc.M203187200. [DOI] [PubMed] [Google Scholar]
- 34.Kwiatkowski DJ, Manning BD. Tuberous sclerosis: a GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14 Spec No. 2:R251–8. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- 35.Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell. 2003;11:1457–66. doi: 10.1016/S1097-2765(03)00220-X. [DOI] [PubMed] [Google Scholar]
- 36.Roccio M, Bos JL, Zwartkruis FJ. Regulation of the small GTPase Rheb by amino acids. Oncogene. 2006;25:657–64. doi: 10.1038/sj.onc.1209106. [DOI] [PubMed] [Google Scholar]
- 37.Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. J Biol Chem. 2005;280:18717–27. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- 38.Yan L, Lamb RF. Amino acid sensing and regulation of mTORC1. Semin Cell Dev Biol. 2012;23:621–5. doi: 10.1016/j.semcdb.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Xiao F, Huang Z, Li H, Yu J, Wang C, Chen S, Meng Q, Cheng Y, Gao X, Li J, et al. Leucine deprivation increases hepatic insulin sensitivity via GCN2/mTOR/S6K1 and AMPK pathways. Diabetes. 2011;60:746–56. doi: 10.2337/db10-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen J, Nurse P. TOR signalling regulates mitotic commitment through the stress MAP kinase pathway and the Polo and Cdc2 kinases. Nat Cell Biol. 2007;9:1263–72. doi: 10.1038/ncb1646. [DOI] [PubMed] [Google Scholar]
- 41.Caspari T, Dahlen M, Kanter-Smoler G, Lindsay HD, Hofmann K, Papadimitriou K, Sunnerhagen P, Carr AM. Characterization of Schizosaccharomyces pombe Hus1: a PCNA-related protein that associates with Rad1 and Rad9. Mol Cell Biol. 2000;20:1254–62. doi: 10.1128/MCB.20.4.1254-1262.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]