Abstract
The spatiotemporal manipulations of gene expression by the Cre recombinase (Cre) of bacteriophage P1 has become an essential asset to understanding mammalian genetics. Accumulating evidence suggests that Cre activity can, in addition to excising targeted loxP sites, induce cytotoxic effects, including abnormal cell cycle progression, genomic instability, and apoptosis, which can accelerate cancer progression. It is speculated that these defects are caused by Cre-induced DNA damage at off-target sites. Here we report the formation of tetraploid keratinocytes in the epidermis of keratin 5 and/or keratin 14 promoter-driven Cre (KRT5- and KRT14-Cre) expressing mouse skin. Biochemical analyses and flow cytometry demonstrated that Cre expression also induces DNA damage, genomic instability, and tetraploidy in HCT116 cells, and live-cell imaging revealed an extension of the G2 cell cycle phase followed by defective or skipping of mitosis as cause for the tetraploidy. Since tetraploidy eventually leads to aneuploidy, a hallmark of cancer, our findings highlight the importance of distinguishing non-specific cytopathic effects from specific Cre/loxP-driven genetic manipulations when using Cre-mediated gene deletions.
Keywords: Cre recombinase, cytokinesis failure, bypass of mitosis, tetraploidy, aneuploidy, cancer, DNA damage, transgenic mice, homologous recombination, apoptosis
Introduction
Cre recombinase of the bacteriophage P1 is a member of the phage λ integrase family of site-specific recombinases. It efficiently catalyzes the recombination of two 34-base pairs (bp)-long target sites called loxP (locus of crossover in phage P1), which consist of two 13-bp inverted repeats separated by a directional 8-bp core sequence.1-4 The ability to excise genomic regions flanked with loxP sites (called “floxed allele”) allows for controlled ablation of genes in vivo by using transgenic mouse lines with tissue-specific Cre expression. Examples include the keratin 5 and keratin 14 promoter-driven Cre transgenes (KRT5- and KRT14-Cre), which ablate floxed alleles in keratin 5/14-expressing cells such as epidermal keratinocytes.5 Cre expression can also be controlled in a temporal manner by using ligands such as tamoxifen, which activates a Cre recombinase fused with a mutated ligand-binding domain of the human estrogen receptor (Cre-ERT2).6-9 Together, these tools enable the inducible and conditional manipulation of genes, thereby circumventing lethal or severe developmental defects resulting from their systemic ablation.10-13 Thus, Cre-mediated site-specific recombination became an invaluable tool to manipulate the mammalian genome in a controlled manner.11,14-16
A caveat of Cre is its toxicity, which develops in a dose-dependent manner even in the absence of loxP target sites and was shown to result in growth arrest, chromosomal abnormalities, and apoptosis.17-22 By an unknown mechanism, Cre was shown to accelerate cancer progression.23 Since a lysine-173 to arginine substituion (R173K) in Cre abrogated both its nuclease activity and cytotoxicity, DNA damage and/or unfaithful recombination of genomic DNA are thought to cause Cre toxicity.18-21 Although wild-type mammalian genomes lack perfect loxP sequences, so-called pseudo-loxP sites have been identified, which occur, for instance, in the mouse genome 1.2× per megabase.24 These imperfect sites serve as substrates for Cre resulting in DNA recombinations, nicks and double-stranded DNA breaks (DSBs).24-26 Consequently, Cre-expressing cells were shown to accumulate diverse chromosomal abnormalities ranging from chromatid breaks, dicentric chromosomes, sister chromatid exchange, to ring-shaped chromosomes.18,27 How these abnormalities occur is unclear. Since balanced chromosome exchanges are rare, Cre-mediated unfaithful recombination of cryptic loxP sites can be excluded as major cause for the abnormalities. A much more plausible cause is DNA damage, which can result both in growth arrest and cell death.28 Surprisingly, however, this has not been experimentally addressed so far. Furthermore, Cre toxicity was noted after extended periods of Cre activity, but not immediately after Cre activation. Therefore, it can only be speculated which of the observed defects are directly or indirectly caused by Cre activity.
For cancer studies conducted in Cre transgenic mice it is of upmost importance to know whether Cre can indeed induce DNA damage, which, in turn, can cause tetraploidy,29,30 aneuploidy,31-33 and eventually cancer.34,35 We made the serendipitous observation that wild-type mice carrying the KRT5- and/or KRT14-Cre transgene showed increased binucleation (tetraploidy) and apoptosis in epidermal keratinocytes. In cultured cells, we observed that induced Cre expression evokes a persistent DNA damage response, which leads to tetraploidy by bypassing mitosis or cytokinesis failure. Our findings show that persistent DNA damage is responsible for the Cre-induced genomic instability and emphasize the importance of including Cre-expressing control mice in cancer studies to avoid misinterpretations and incorrect conclusions.
Results
Cre expression induces tetraploidy in vivo
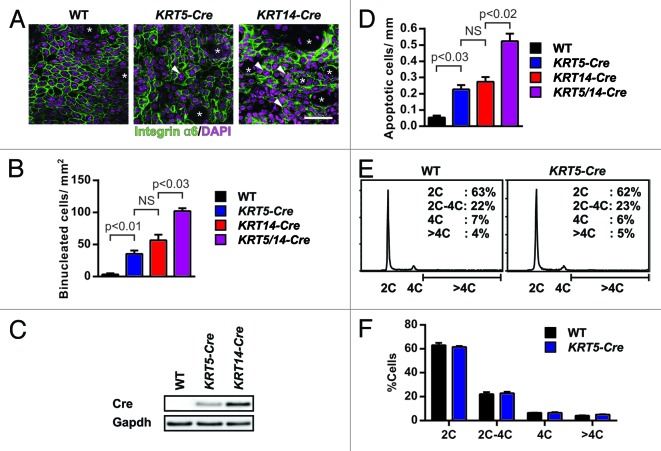
Wild-type mice carrying a KRT5- or KRT14-Cre transgene have no apparent phenotype and are frequently used to delete genes in epidermis and hair follicles.5,36-38 When we immunostained epidermal whole mounts from 3-d-old pups for the expression of the α6 integrin subunit, which outlines basal keratinocytes, we observed that the epidermis of Cre-expressing mice contained binucleated cells (Fig. 1A). While binucleated basal keratinocytes were extremely rare in wild-type epidermis (3.3 ± 1.7 cells per mm2), their numbers were increased 10.6-fold in KRT5-Cre (35.5 ± 5.2 cells per mm2), 18.7-fold in KRT14-Cre (56.7 ± 8.6 cells per mm2), and 30.7-fold in KRT5/KRT14-Cre epidermis (102.1 ± 4.4 cells per mm2) (Fig. 1B). The increasing abundance of tetraploid cells in KRT5-Cre and KRT14-Cre epidermis correlated with increasing Cre expression levels, indicating a Cre dosage effect (Fig. 1C).
Figure 1. Cre induces tetraploidy in vivo. (A) Integrin α6 immunostainings (green, DAPI counterstaining in purple) of epidermal whole mounts of wild-type mice without Cre (WT) or carrying the KRT5- or KRT14-Cre transgene, imaged onto the basal surface of the epidermis. Arrowheads indicate binucleated cells. Infundibula are indicated by asterisks (*). Bar: 50 µm. (B) Binucleated cells in epidermal whole mounts of the indicated genotype from 3-d-old pups (2–5 animals per genotype); columns indicate mean + s.e.m. (C) Immunoblot for Cre and Gapdh in skin lysates of wild-type mice without Cre (WT) or with the indicated Cre transgene. (D) Quantification of absolute numbers of apoptotic keratinocytes per mm epidermis of skin cryosections from 3-d-old pups (3 animals per genotype), columns indicate mean + s.e.m. (E) Representative flow cytometry analysis of propidium iodide (PI)-stained primary keratinocytes isolated from 3-d-old pups. Percentage of cells with different ploidy levels, C denotes haploid DNA complement. (F) Quantification of the percentage of cells in different phases of cell cycle. Columns indicate mean+s.e.m of 3 independent experiments.
Since tetraploid cells have an increased propensity to undergo apoptosis,34 we also quantified the number of apoptotic keratinocytes in the epidermis of 3-d-old pups by immunostaining for cleaved caspase 3. The experiments revealed that wild-type epidermis contained 0.05 ± 0.01 cleaved caspase 3-positive cells per mm. In Cre-expressing epidermis, the number of apoptotic cells increased with the Cre gene dosage: KRT5-Cre epidermis contained 4.4-fold, KRT14-Cre epidermis 5.3-fold, and KRT5/14-Cre double transgenic epidermis 10.3-fold more apoptotic cells per mm than control epidermis (Fig. 1D).
To test whether the binucleated cell population is also visible in freshly isolated keratinocyte populations from 3-d-old KRT5-Cre pups, we examined their DNA content by flow cytometry. The cell cycle distribution of keratinocytes was not changed between KRT5-Cre mice and wild-type cell populations (Fig. 1E and F). These findings indicate that flow cytometry is not suitable to detect very low abundant ploidy defects in KRT5-Cre keratinocytes.
Cre expression increases DNA content in cells
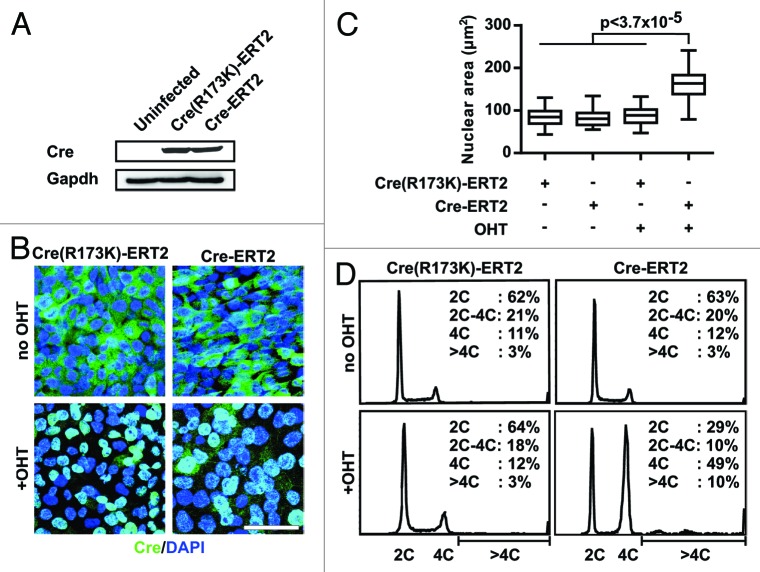
Since the abundance of ploidy defects in Cre expressing keratinocytes is low, we decided to further analyze the mechanism(s) underlying Cre-induced tetraploidy in vitro using HCT116 cells overexpressing Cre recombinase. HCT116 cells are chromosomally stable, have a normal karyotype and spindle checkpoint, and, with the exception of nucleotide mismatch repair, all other DNA damage response mechanisms operate normally.39-42 We infected HCT116 cells with a retroviral construct expressing 4-hydroxytamoxifen (OHT)-inducible wild-type Cre recombinase (Cre-ERT2) or endonuclease-deficient Cre recombinase (CreR173K-ERT2).18 Both constructs were expressed at comparable levels (Fig. 2A). In the absence of OHT induction proliferation rates, nuclear sizes and cell cycle profiles of infected cells were comparable to parental cells (data not shown). In these cells, wild-type as well as mutant Cre proteins were retained in the cytoplasm, while treatment with OHT triggered a rapid nuclear translocation of wild-type and mutant Cre proteins (Fig. 2B). Strikingly, within 24 h after induction of Cre-ERT2, the cells enlarged and increased the size of their nuclei by 1.9-fold, suggesting that also their DNA content increased (Fig. 2B and C). Importantly, OHT-induced expression of CreR173K-ERT2 had no effect indicating that Cre induces cell changes via its catalytic activity (Fig. 2B and C).
Figure 2. Cre activity induces increased DNA content in HCT116 cells. (A) Immunoblot for Cre and Gapdh in HCT116 cell lysate either left uninfected or infected with Cre-ERT2 or CreR173K-ERT2 containing retrovirus. (B) Immunofluorescence staining for Cre (green) and nuclei (blue) in CreR173K-ERT2 or Cre-ERT2 expressing HCT116 cells non-induced or induced with 500 nM OHT for 24 h. Bar: 50 µm. (C) Quantification of the nuclear areas in a box-and-whisker plot (between 80 and 100 cells were evaluated for each condition in 3 independent experiments). (D) Flow cytometry analysis of PI-stained cells. Cells and treatment are as in (B). Percentage of cells with different ploidy levels, (C) denotes haploid DNA complement.
Next we analyzed the cellular DNA content by flow cytometry before and after OHT treatment for 24 h. Before OHT treatment, 63 ± 0.5% cells were in G0/G1 phase with 2C DNA content (C = chromosome complement), 20 ± 0.9% were in S phase with 2C to 4C DNA content, and 12 ± 0.8% were in G2/M phase with 4C DNA content. Only 3 ± 0.2% had >4C DNA content. OHT induction of the endonuclease-deficient CreR173K-ERT2 did not change the distribution of DNA content (Fig. 2D). In contrast, OHT treatment of cells containing Cre-ERT2 increased the percentage of cells with 4C to 49 ± 0.6% or > 4C to 10 ± 0.4% DNA content and concomitantly decrease cell numbers with 2C or 2C-4C DNA content (Fig. 2D). These results indicate that the nuclear activity of endonuclease-proficient Cre has 2 immediate consequences: first, Cre increases the percentage of cells with 4C DNA content indicative for a G2/M arrest; second, Cre increases the percentage of cells with > 4C DNA content indicative for polyploidy.
Cre induces DNA damage leading to G2/M phase defects
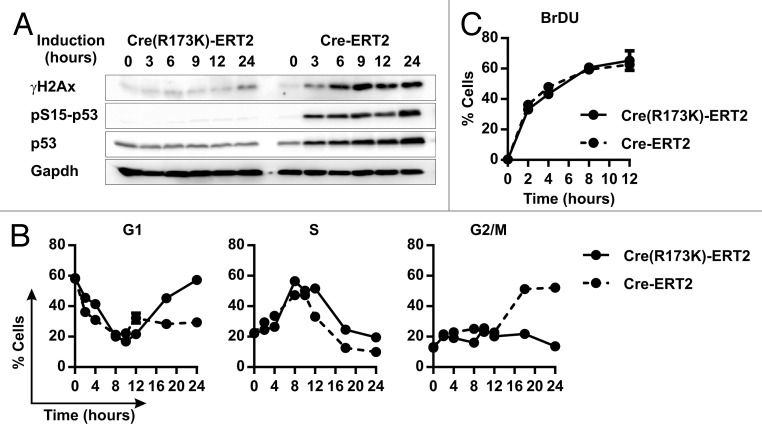
Since a persistent DNA damage can increase the percentage of cells with 4C and higher DNA content,28,29 we determined the expression levels of the DNA damage marker phosphoSer139-histone H2Ax (γH2Ax) and the DNA damage response markers phosphoSer15-p53 and total p53. Cells expressing CreR173K-ERT2 exhibited low basal levels of γH2Ax, and total p53 and lacked detectable levels of phosphoSer15-p53 during 24 h of OHT treatment (Fig. 3A). In contrast, cells expressing Cre-ERT2 upregulated γH2Ax, phospho-Ser15-p53 and total p53 levels within 3 h after OHT treatment, which further rose at later time points (Fig. 3A). These findings prove that Cre activity induces DNA damage and a DNA damage response.
Figure 3. Cre-induced DNA damage results in G2/M arrest. (A) Immunoblot for indicated proteins of CreR173K-ERT2 or Cre-ERT2 expressing HCT116 cells untreated or treated with 500 nM OHT for indicated time points. (B) Flow cytometry analysis of PI-stained cells. Cells and treatment were as in (A). Percentage of cells in different phases of cell cycle at indicated time points. Data are displayed as mean ± s.d. of 3 independent experiments. (C) Flow cytometry analysis of BrdU and PI-stained cells. Cells and treatment were as in (A). Quantification of the percentage of BrdU-positive cells at indicated time points. Data are displayed as mean ± s.d. of 3 independent experiments.
To test if Cre expression arrests the cell cycle at specific phases, we plated the cells at low density to achieve synchronous entry into S phase and treated them with OHT. OHT treated cells expressing Cre-ERT2 entered S phase at similar rates as cells expressing CreR173K-ERT2 (Fig. 3B). However, unlike CreR173K-ERT2-expressing cells, the Cre-ERT2-expressing cells accumulated in G2/M phase, which resulted in a reduction of cells in the G1 phase at later time points (Fig. 3B). Normal S-phase entry and progression of Cre-ERT2-expressing cells were confirmed with the 5-bromo-2'-deoxyuridine (BrdU) incorporation assay, which showed a similar increase of BrdU-positive cells in Cre-ERT2 and CreR173K-ERT populations at all time-points analyzed (Fig. 3C). Taken together, these results indicate that Cre-endonuclease activity interferes with mitotic progression, while entry into and progression through the S phase are normal.
Cre expression inhibits mitosis and causes cytokinesis failure and tetraploidy
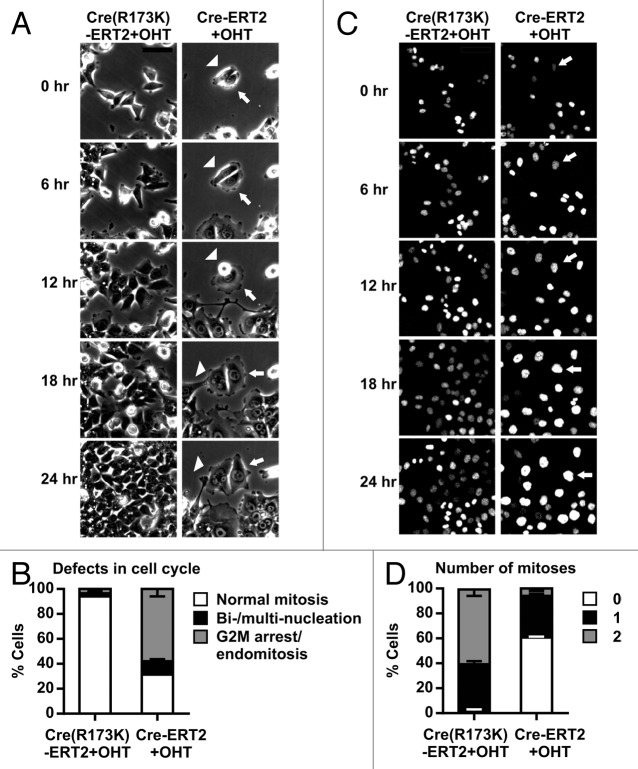
To explore the mechanism leading to Cre-induced increase in DNA content or ploidy, we analyzed asynchronously growing cells expressing Cre by phase contrast time-lapse imaging. After treating cells for 24 h with OHT, 94 ± 2% of CreR173K-ERT2-expressing cells divided at least once, with each division giving rise to 2 daughter cells, each cell with a single nucleus (Fig. 4A and B; Video S1). In the same period, only 31 ± 3% of Cre-ERT2 underwent an apparently normal cell division. Another 11 ± 3% of the Cre-ERT expressing cells entered mitosis and progressed through anaphase (as seen by nuclear envelope breakdown and reformation), but became bi- or multinucleated after cytokinesis failure (Fig. 4A and B, arrowheads; Video S2). The majority (58 ± 6%) of Cre-ERT2 expressing cells failed to enter mitosis, evidenced by a lack of nuclear envelope breakdown (Fig. 4A and B, see arrows; Video S2).
Figure 4. Cre activity results in mitotic defects. (A) Phase contrast images of CreR173K-ERT2 (Video S1) or Cre-ERT2 (Video S2) expressing HCT116 cells treated with OHT for indicated times. Images were taken every 10 min for 24 h. Selected stills are shown. Arrowhead indicates a multinucleated cell after defective mitosis. Arrow indicates a cell failed to enter mitosis. Bar: 100 µm. (B) Relative quantification of mitotic defects of cells shown in (A). Data are displayed as mean+s.d. of 3 independent experiments. (C) GFP-images of CreR173K-ERT2 (Video S3) or Cre-ERT2 (Video S4) expressing HCT116-H2B-GFP cells treated with OHT for the indicated time points. Images were taken every 10 min for 24 h. Selected stills are shown. Arrow indicates a nucleus of a cell that failed to enter mitosis. Bar: 100 µm. (D) Relative quantification of mitoses per cell. Cells were treated and imaged as in (C). Error bars indicate s.e.m. of 3 independent experiments.
These findings were confirmed with Cre-ERT2 or CreR173K-ERT2 expressing HCT116 cells stably transduced with the chromosome marker histone H2B fused to green fluorescent protein (H2B-GFP). Almost all (98 ± 2%) OHT-treated CreR173K-ERT2/H2B-GFP-expressing cells divided normally and displayed normal nuclear size as judged from GFP fluorescence, with 38 ± 3% of the cells dividing once and 60 ± 5% dividing twice (Fig. 4C and D; Video S3). In contrast, only 39 ± 1% of OHT-treated Cre-ERT2/H2B–GFP expressing cells completed mitosis. The remaining 61 ± 1% cells increased their size without entering mitosis (Fig. 4C and D, arrows; Video S4). The cell size increase indicates that the cells were not terminally arrested in G2, but rather skipped mitosis and re-entered the cell cycle as tetraploid G1 cells.
To exclude fusion of 2 diploid cells as cause for tetraploidy,43,44 we carefully re-examined all movies but did not observe cell–cell fusion events. Taken together, our findings indicate that Cre activity interferes predominantly with entry into mitosis and to a lesser extent completion of mitosis.
Cre-induced DNA damage results in G2/M arrest and bypass of mitosis
To examine whether Cre-induced failure to enter mitosis is due to G2/M arrest and/or bypass of mitosis, we followed the expression of doxycycline-inducible GFP-tagged cyclin B1 (cyclin B1-GFP) in CreR173K-ERT2 and Cre-ERT2 cells, respectively. Addition of doxycycline induces cyclin B1-GFP upregulation in S phase, translocation into the nucleus at prophase and degradation at the onset of anaphase.45-47 This was observed in 93 ± 1% OHT/doxycycline-treated, CreR173K-ERT2-expressing HCT116 cells (Fig. 5A and B, see numbers; Video S5). Furthermore, the mean duration of mitosis (time from nuclear import of cyclin B1-GFP at prophase until its destruction at the onset of anaphase) in these cells was 44 ± 6.6 min (n = 40 cells). In contrast, normal cyclin B1-GFP kinetics was observed in only 41 ± 6% of OHT/doxycycline-treated, Cre-ERT2-expressing cells. 50 ± 6% of OHT/doxycycline-treated, Cre-ERT2-expressing cells retained cyclin B1-GFP in the cytoplasm for at least 10–12 h (Fig. 5A, see arrows; Video S6), followed by degradation of cyclin B1-GFP without nuclear accumulation in 9 ± 1% of the cells, indicating bypass of mitosis/endomitosis (Fig. 5A, see arrowheads; Video S6). These findings confirm that Cre-induced DNA damage can lead to G2/M arrest that is resolved by a bypass of mitosis, finally resulting in tetraploidy.
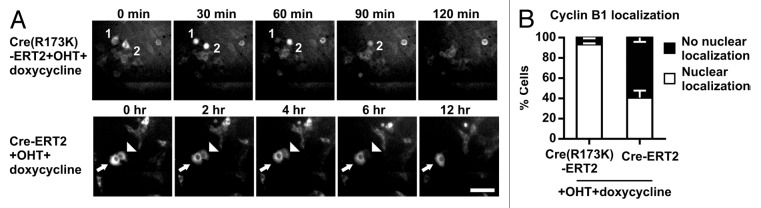
Figure 5. Cre induces G2/M arrest and a bypass of mitosis in HCT116 cells. (A) Time-lapse imaging of CreR173K-ERT2 (Video S5) or Cre-ERT2 (Video S6) expressing HCT116-cyclin B1-GFP cells treated with OHT and doxycycline. GFP images were taken every 10 min for 24 h. Selected stills are shown. Numbers indicate cells with normal kinetics of cyclin B1-GFP and arrow indicates a cell positive for cytosolic cyclin B1-GFP for more than 12 h. Arrowhead indicates a cell with cytosolic cyclin B1-GFP degradation and absent localization in the nucleus. Bar: 50 µm. (B) Relative quantification of OHT- and doxycycline-treated cells showing degradation of cyclin B1-GFP with or without prior nuclear localization. Error bars indicate s.e.m. of 3 independent experiments. Cells and treatment were as in (A).
Discussion
The caveats associated with Cre transgene expression are well-documented, including proliferation defects, cytotoxicity, chromosomal abnormalities, and accelerated cancer progression. We report that Cre transgene expression in the epidermis of normal mice induces tetraploidy and apoptosis. To our knowledge this is the first report showing that Cre can cause tetraploidy in the absence of floxed alleles in vivo. Complementing in vitro studies revealed that tetraploidy development is due to a bypass of mitosis or a failure to complete mitosis. These findings highlight potential problems that may arise when the Cre/loxP system is used to study DNA damage, DNA repair, and cancer development.
Mechanisms of Cre-induced genomic instability
Previous in vitro analyses of Cre-induced cytotoxicity were designed as end-point studies after an extended period of Cre activity.18,21 Although these studies showed that Cre can cause polyploidy, further analysis exclusively focused on chromosomal aberrations and aneuploidy without addressing how Cre activity caused polyploidy. Furthermore, the mechanism(s) of Cre-induced chromosomal defects was also not entirely clear. Our live cell analysis allowed for the first time to directly observe the effect of Cre activity on cell cycle progression.
Activation of Cre for 24 h induced a swift and persistent DNA damage response in HCT116 cells without leading to cell cycle arrest or apoptosis, which are usually triggered by p53 upon extensive DNA damage.48 This finding is in line with previous observations showing that low and persistent DNA damage is unable to efficiently induce cell cycle arrest,49,50 but instead induced polyploidy by endomitosis29 or cytokinesis defects.30 Interestingly, the limited DNA damage caused by Cre was also associated with high rates of tetraploidy/polyploidy due to either endomitosis or cytokinesis failure. Defective resolutions of mitosis followed by polyploidy is a well-known starting point to aneuploidy and extensive chromosomal rearrangements, which can cause a plethora of secondary effects, including cell death.51
How can our in vitro results explain the in vivo cytotoxicity of Cre? Both KRT5- and KRT14-Cre mice are phenotypically normal,5,37 and flow cytometry analysis of primary keratinocytes failed to detect cells with abnormal ploidy. However, careful analysis revealed a very small number of binucleated (tetraploid/polyploid) or apoptotic keratinocytes in wild-type epidermis lacking a Cre transgene. The number of tetraploid and apoptotic cells increased concomitantly with the Cre activity.
Interestingly, previous in vivo studies reported that Cre-mediated cytotoxicity is associated with cell death19,27 that increases in a Cre dose-dependent manner.52 This “side effect” of Cre-mediated cytotoxicity has even been exploited to target and ablate specific cell populations such as mast cells and basophilic neutrophils in vivo.53,54 How Cre toxicity achieves cell death at the molecular level is not known. It has been proposed that p53 activity is responsible for Cre-induced cell death.53 However, we found that deletion of p53 in epidermis of Trp53-floxed KRT5-Cre mice did not abolish epidermal apoptosis (unpublished results). Since Cre-induced DNA damage was insufficient to induce cell death while being permissive for the development of abnormal ploidy, we favor the possibility that the observed cytotoxicity in vivo is secondary to polyploidy: Dividing polyploid cells are prone for increased apoptosis, and even viable aneuploid daughter cells resulting from such polyploid mitoses obtain unpredictable chromosome complements that either lead to cell death or transformation and cancer.51,55,56 Unfortunately, the chromosme complement was not examined in previous studies on Cre cytotoxicity, leaving the question unanswered, whether genomic instability triggered Cre-induced cell death in these cells.
Cre-induced tetraploidy and carcinogenesis
Our data link Cre activity with increased tetraploidy even in absence of noticeable phenotypes. Since tetraploidy is an unstable intermediate to aneuploidy and efficiently promotes tumorigenesis in vivo,35,40,51,57,58 Cre might have an oncogenic off-target function in vivo even in absence of floxed alleles. The absence of oncogenic effects in our mice may simply be due to the rapid movement of tetraploid cells to the upper epidermal layers, where they cornify and eventually slough. In line with this hypothesis, we observed binucleated cells in the post-mitotic suprabasal layers of the epidermis indicating that terminal differentiation and shedding may indeed suppress transformation and growth of cells with an abnormal genome (unpublished observation). However, if the oncogenic cells are growth promoted before they are lost, they may induce tumors. Such a situation could have caused an increased malignant progression in mice homozygous for a KRT5-Cre transgene and exposed to the 2-stage carcinogenesis protocol.23 Although the ploidy was not analyzed in this study, our finding of increased tetraploidy in Cre-positive epidermis provides a mechanistic explanation for the observed oncogenic function of Cre in these mice.
Dealing with Cre-dependent cytotoxicity
Our as well as published results from other laboratories emphasize the problems with the Cre/loxP system in cancer or gene manipulation studies addressing DNA damage repair, cell cycle regulation, apoptosis, and/or mitosis. The possibility of combinatorial effects of targeted gene modification and illegitimate Cre activity requires appropriate controls to accurately interpret results of gene-targeting studies. Appropriate controls are wild-type or heterozygous floxed mice carrying the Cre transgene. When such controls are not included, it will be impossible to discriminate the Cre effects from the impact of the targeted mutation. This can severely compromise the validity of the experimental results.
Numerous reports have shown that Cre expression can cause toxicity in absence of a “floxed allele”, regardless whether its expression is ubiquitous, tissue-specific and/or ligand-inducible.17,20,22,23,27,52-54,59,60 Therefore, minimizing the exposure of cells to Cre recombinase is clearly a sensible strategy. Several measures have been developed to minimize Cre cytotoxicity; in vitro studies utilize self-deleting Cre vectors that flank Cre with loxP sites to combine gene ablation with Cre elimination,19,21,61 and in vivo studies employ ligand-activated Cre transgenic lines, in which the ligand is applied for a limited time to avoid extended Cre expression, and at low dose to avoid ligand-induced side effects.27,59,62 Although this requires an increased experimental burden, Cre cytotoxicity can be minimized or even overcome.
Materials and Methods
Ethics statement
Mice were kept and bred according to German animal welfare laws at the animal facilities of the Max Planck Institute of Biochemistry.
Antibodies and reagents
The antibodies used were: Gapdh, Cre, γH2Ax (Merck Millipore), phospho-p53 (Ser15), total p53 and cleaved caspase 3 (Cell Signaling Technology/NEB), integrin α6 and fluorescein isothiocyanate (FITC) conjugated anti-BrdU antibody (Becton Dickinson). All reagents were of cell culture grade (Sigma or Life Technologies).
Cell lines and culture
The cell lines HCT116 (ATCC No. CCL-247), HCT116-H2B-GFP and doxycycline inducible HCT116-cyclin B1-GFP were kindly provided by Zuzana Storchova (Max Planck Institute of Biochemistry) and maintained at 37 °C with 5% CO2 atmosphere in Dulbecco modified Eagle medium containing 10% fetal bovine serum, 100 U penicillin and 100 µg/ml streptomycin. For induction of cyclin B1-GFP, cells were cultured in the presence of 1 µg/ml doxycycline.
Retroviral constructs
To generate the CreR173K-ERT2 expression construct the Cre-ERT2 cDNA was mutated with the Quickchange kit (Agilent Technologies). Successful mutagenesis was confirmed by sequencing. Wild-type and mutant CreR173K-ERT2 cDNAs were cloned into pCLMFG-MCS63 to generate virus-containing supernatants for infecting the cell lines.
Live-cell imaging
Freshly cultured cell were seeded sparsely on a 6 channel μ-slide (Ibidi) 24 h before the experiment. The slide was placed onto a sample stage with an incubator chamber (EMBLEM) maintained at 37 °C, 40% humidity, in an atmosphere with 5% CO2. Imaging was performed using a Zeiss Axio Observer Z1 microscope equipped with a Plan Neofluar 10× or 20× air objective. Metamorph 7.1 software (Molecular Devices) was used to control the microscope. Movies were evaluated using the ImageJ 1.42I software (Schneider, Rasband et al. 2012).
Flow cytometry
For measurement of DNA content, cells were cultured in the presence or absence of 500 nM OHT for indicated time points, collected, fixed, stained with propidium iodide. For BrdU incorporation assays, cells were cultured in presence of 100 µM BrdU for indicated time points and fixed immediately in 70% ethanol. Nuclei were isolated from fixed cells and stained with FITC-conjugated anti-BrdU antibody. Cells and nuclei analyzed by flow cytometry on a FACSCalibur instrument (Becton Dickinson) as described.64 Cell cycle profiles and BrdU indices were analyzed using FlowJo (Tree Star).
Generation of epidermal whole mounts
Three-day-old pups were sacrificed and skinned. Muscle and fatty layer were scraped off, followed by careful mechanical separation of dermis and epidermis. Epidermis was fixed for 10 min in 4% PFA/PBS and kept in PBS with 0.05% NaN3 at 4 °C until use.
Immunofluorescence and imaging
Skin cryosections were prepared and embedded as described.65 Cells grown on glass coverslips were fixed in 3.7% PFA/PBS, permeabilized with 0.1% Triton X-100/PBS and blocked in 5% BSA/PBS. Primary antibodies were diluted in blocking solution and applied overnight at 4 °C. After washing with PBS, appropriate secondary antibodies were diluted in blocking solution and applied for 1 h at room temperature. After washing and DAPI staining, slides were mounted in Elvanol. Pictures were taken with a TCS SP5 AOBS Confocal Laser Scanning Microscope (Leica).
For determination of binucleation, epidermal whole mount preparations were immunostained for integrin α6 and counterstained with DAPI. Stacks of pictures were taken as above to account for waves and wrinkles in the epidermis. Suprabasal cells (negative for integrin α6) were excluded from analysis, as were outer root sheath keratinocytes of infundibula due to their altered spatial orientation compared with interfollicular keratinocytes. Cells were scored as bi-/multinucleated basal cells when there was no intervening integrin-signal between their nuclei; furthermore, nuclei within a cell were aligned and deformed each other. If in doubt, binucleation was either confirmed by examination of adjacent z-layers, or the cells were not scored as binucleated. Apoptosis was measured by staining skin cryosections from 3-d-old pups for cleaved caspase 3 (clCasp3), integrin α6 and DAPI. Ten or more sections of at least 1 cm length per animal were used.
Western blotting
Lysates from cells were separated on a polyacrylamide gel and transferred onto a PVDF membrane (Millipore). Membrane blocking and antibody dilution were performed with Tris-buffered saline (TBS), pH 7.6, supplemented with 5% (w/v) skim milk powder, and 0.1% Tween 20 (Serva). Subsequently, membranes were incubated for 1 h at room temperature or overnight at 4 °C with antibodies. Appropriate HRP-coupled secondary antibodies (BioRad) were applied for 1 h at room temperature followed by enhanced chemiluminescence (ECL) detection (Merck Millipore).
Statistics
All data are given as a mean value with standard error of the mean (s.e.m.) or standard deviation (s.d.) as indicated. Statistical significance was tested with a 2-tailed Student t test.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Christian Kuffer, Suzana Storchova, and members of the Fässler lab for cell lines, suggestions and help with microscopy and Roy Zent for discussions and carefully reading the manuscript. We are grateful to Marc Schmidt-Supprian for valuable discussion and suggestions. This work was supported by the Tiroler Landestiftung and the Max Planck Society.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/27271
References
- 1.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–86. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 2.Sternberg N, Hamilton D, Hoess R. Bacteriophage P1 site-specific recombination. II. Recombination between loxP and the bacterial chromosome. J Mol Biol. 1981;150:487–507. doi: 10.1016/0022-2836(81)90376-4. [DOI] [PubMed] [Google Scholar]
- 3.Argos P, Landy A, Abremski K, Egan JB, Haggard-Ljungquist E, Hoess RH, Kahn ML, Kalionis B, Narayana SV, Pierson LS, 3rd, et al. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–40. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–70. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez A, Page A, Gandarillas A, Zanet J, Pibre S, Vidal M, Tusell L, Genesca A, Whitaker DA, Melton DW, et al. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–7. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- 6.Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch JL, Chambon P, Metzger D. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc Natl Acad Sci U S A. 1997;94:14559–63. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–6. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 8.Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci U S A. 1996;93:10887–90. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–7. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Metzger D, Feil R. Engineering the mouse genome by site-specific recombination. Curr Opin Biotechnol. 1999;10:470–6. doi: 10.1016/S0958-1669(99)00012-9. [DOI] [PubMed] [Google Scholar]
- 11.Rajewsky K, Gu H, Kühn R, Betz UA, Müller W, Roes J, Schwenk F. Conditional gene targeting. J Clin Invest. 1996;98:600–3. doi: 10.1172/JCI118828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossant J, McMahon A. “Cre”-ating mouse mutants-a meeting review on conditional mouse genetics. Genes Dev. 1999;13:142–5. doi: 10.1101/gad.13.2.142. [DOI] [PubMed] [Google Scholar]
- 13.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–92. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 14.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–55. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 15.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. doi: 10.1002/(SICI)1526-968X(200002)26:2<99::AID-GENE1>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 16.Schmidt-Supprian M, Rajewsky K. Vagaries of conditional gene targeting. Nat Immunol. 2007;8:665–8. doi: 10.1038/ni0707-665. [DOI] [PubMed] [Google Scholar]
- 17.Jeannotte L, Aubin J, Bourque S, Lemieux M, Montaron S, Provencher St-Pierre A. Unsuspected effects of a lung-specific Cre deleter mouse line. Genesis. 2011;49:152–9. doi: 10.1002/dvg.20720. [DOI] [PubMed] [Google Scholar]
- 18.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, Berns A, Jonkers J. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001;98:9209–14. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer A, Brandon EP, Kootstra N, Gage FH, Verma IM. Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc Natl Acad Sci U S A. 2001;98:11450–5. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci U S A. 2000;97:13702–7. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–43. doi: 10.1016/S1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 22.Thanos A, Morizane Y, Murakami Y, Giani A, Mantopoulos D, Kayama M, Roh MI, Michaud N, Pawlyk B, Sandberg M, et al. Evidence for baseline retinal pigment epithelium pathology in the Trp1-Cre mouse. Am J Pathol. 2012;180:1917–27. doi: 10.1016/j.ajpath.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan EL, Peace BE, Toney K, Kader SA, Pathrose P, Collins MH, Waltz SE. Homozygous K5Cre transgenic mice have wavy hair and accelerated malignant progression in a murine model of skin carcinogenesis. Mol Carcinog. 2007;46:49–59. doi: 10.1002/mc.20192. [DOI] [PubMed] [Google Scholar]
- 24.Semprini S, Troup TJ, Kotelevtseva N, King K, Davis JR, Mullins LJ, Chapman KE, Dunbar DR, Mullins JJ. Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques. Nucleic Acids Res. 2007;35:1402–10. doi: 10.1093/nar/gkl1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abremski K, Wierzbicki A, Frommer B, Hoess RH. Bacteriophage P1 Cre-loxP site-specific recombination. Site-specific DNA topoisomerase activity of the Cre recombination protein. J Biol Chem. 1986;261:391–6. [PubMed] [Google Scholar]
- 26.Thyagarajan B, Guimarães MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/S0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 27.Higashi AY, Ikawa T, Muramatsu M, Economides AN, Niwa A, Okuda T, Murphy AJ, Rojas J, Heike T, Nakahata T, et al. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J Immunol. 2009;182:5633–40. doi: 10.4049/jimmunol.0802413. [DOI] [PubMed] [Google Scholar]
- 28.Su TT. Cellular responses to DNA damage: one signal, multiple choices. Annu Rev Genet. 2006;40:187–208. doi: 10.1146/annurev.genet.40.110405.090428. [DOI] [PubMed] [Google Scholar]
- 29.Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichijima Y, Yoshioka K, Yoshioka Y, Shinohe K, Fujimori H, Unno J, Takagi M, Goto H, Inagaki M, Mizutani S, et al. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS One. 2010;5:e8821. doi: 10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vitale I, Galluzzi L, Senovilla L, Criollo A, Jemaà M, Castedo M, Kroemer G. Illicit survival of cancer cells during polyploidization and depolyploidization. Cell Death Differ. 2011;18:1403–13. doi: 10.1038/cdd.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121:3859–66. doi: 10.1242/jcs.039537. [DOI] [PubMed] [Google Scholar]
- 33.Lv L, Zhang T, Yi Q, Huang Y, Wang Z, Hou H, Zhang H, Zheng W, Hao Q, Guo Z, et al. Tetraploid cells from cytokinesis failure induce aneuploidy and spontaneous transformation of mouse ovarian surface epithelial cells. Cell Cycle. 2012;11:2864–75. doi: 10.4161/cc.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 35.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–7. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 36.Brakebusch C, Grose R, Quondamatteo F, Ramirez A, Jorcano JL, Pirro A, Svensson M, Herken R, Sasaki T, Timpl R, et al. Skin and hair follicle integrity is crucially dependent on beta 1 integrin expression on keratinocytes. EMBO J. 2000;19:3990–4003. doi: 10.1093/emboj/19.15.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hafner M, Wenk J, Nenci A, Pasparakis M, Scharffetter-Kochanek K, Smyth N, Peters T, Kess D, Holtkötter O, Shephard P, et al. Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis. 2004;38:176–81. doi: 10.1002/gene.20016. [DOI] [PubMed] [Google Scholar]
- 38.Lorenz K, Grashoff C, Torka R, Sakai T, Langbein L, Bloch W, Aumailley M, Fässler R. Integrin-linked kinase is required for epidermal and hair follicle morphogenesis. J Cell Biol. 2007;177:501–13. doi: 10.1083/jcb.200608125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brattain MG, Fine WD, Khaled FM, Thompson J, Brattain DE. Heterogeneity of malignant cells from a human colonic carcinoma. Cancer Res. 1981;41:1751–6. [PubMed] [Google Scholar]
- 40.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 41.Roschke AV, Stover K, Tonon G, Schäffer AA, Kirsch IR. Stable karyotypes in epithelial cancer cell lines despite high rates of ongoing structural and numerical chromosomal instability. Neoplasia. 2002;4:19–31. doi: 10.1038/sj.neo.7900197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tighe A, Johnson VL, Albertella M, Taylor SS. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep. 2001;2:609–14. doi: 10.1093/embo-reports/kve127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duelli D, Lazebnik Y. Cell fusion: a hidden enemy? Cancer Cell. 2003;3:445–8. doi: 10.1016/S1535-6108(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 44.Ogle BM, Cascalho M, Platt JL. Biological implications of cell fusion. Nat Rev Mol Cell Biol. 2005;6:567–75. doi: 10.1038/nrm1678. [DOI] [PubMed] [Google Scholar]
- 45.Chang DC, Xu N, Luo KQ. Degradation of cyclin B is required for the onset of anaphase in Mammalian cells. J Biol Chem. 2003;278:37865–73. doi: 10.1074/jbc.M306376200. [DOI] [PubMed] [Google Scholar]
- 46.Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–7. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- 47.Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–9. doi: 10.1016/S0960-9822(99)80308-X. [DOI] [PubMed] [Google Scholar]
- 48.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–8. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deckbar D, Birraux J, Krempler A, Tchouandong L, Beucher A, Walker S, Stiff T, Jeggo P, Löbrich M. Chromosome breakage after G2 checkpoint release. J Cell Biol. 2007;176:749–55. doi: 10.1083/jcb.200612047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deckbar D, Stiff T, Koch B, Reis C, Löbrich M, Jeggo PA. The limitations of the G1-S checkpoint. Cancer Res. 2010;70:4412–21. doi: 10.1158/0008-5472.CAN-09-3198. [DOI] [PubMed] [Google Scholar]
- 51.Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 52.Naiche LA, Papaioannou VE. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 2007;45:768–75. doi: 10.1002/dvg.20353. [DOI] [PubMed] [Google Scholar]
- 53.Feyerabend TB, Weiser A, Tietz A, Stassen M, Harris N, Kopf M, Radermacher P, Möller P, Benoist C, Mathis D, et al. Cre-mediated cell ablation contests mast cell contribution in models of antibody- and T cell-mediated autoimmunity. Immunity. 2011;35:832–44. doi: 10.1016/j.immuni.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 54.Ohnmacht C, Schwartz C, Panzer M, Schiedewitz I, Naumann R, Voehringer D. Basophils orchestrate chronic allergic dermatitis and protective immunity against helminths. Immunity. 2010;33:364–74. doi: 10.1016/j.immuni.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 55.Vitale I, Galluzzi L, Castedo M, Kroemer G. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12:385–92. doi: 10.1038/nrm3115. [DOI] [PubMed] [Google Scholar]
- 56.Gisselsson D, Håkanson U, Stoller P, Marti D, Jin Y, Rosengren AH, Stewénius Y, Kahl F, Panagopoulos I. When the genome plays dice: circumvention of the spindle assembly checkpoint and near-random chromosome segregation in multipolar cancer cell mitoses. PLoS One. 2008;3:e1871. doi: 10.1371/journal.pone.0001871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 58.Shi Q, King RW. Chromosome nondisjunction yields tetraploid rather than aneuploid cells in human cell lines. Nature. 2005;437:1038–42. doi: 10.1038/nature03958. [DOI] [PubMed] [Google Scholar]
- 59.Huh WJ, Mysorekar IU, Mills JC. Inducible activation of Cre recombinase in adult mice causes gastric epithelial atrophy, metaplasia, and regenerative changes in the absence of “floxed” alleles. Am J Physiol Gastrointest Liver Physiol. 2010;299:G368–80. doi: 10.1152/ajpgi.00021.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee JY, Ristow M, Lin X, White MF, Magnuson MA, Hennighausen L. RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. J Biol Chem. 2006;281:2649–53. doi: 10.1074/jbc.M512373200. [DOI] [PubMed] [Google Scholar]
- 61.Mähönen AJ, Airenne KJ, Lind MM, Lesch HP, Ylä-Herttuala S. Optimized self-excising Cre-expression cassette for mammalian cells. Biochem Biophys Res Commun. 2004;320:366–71. doi: 10.1016/j.bbrc.2004.05.175. [DOI] [PubMed] [Google Scholar]
- 62.Heidmann D, Lehner CF. Reduction of Cre recombinase toxicity in proliferating Drosophila cells by estrogen-dependent activity regulation. Dev Genes Evol. 2001;211:458–65. doi: 10.1007/s004270100167. [DOI] [PubMed] [Google Scholar]
- 63.Moik DV, Janbandhu VC, Fässler R. Loss of migfilin expression has no overt consequences on murine development and homeostasis. J Cell Sci. 2011;124:414–21. doi: 10.1242/jcs.075960. [DOI] [PubMed] [Google Scholar]
- 64.Janbandhu VC, Singh AK, Mukherji A, Kumar V. p65 Negatively regulates transcription of the cyclin E gene. J Biol Chem. 2010;285:17453–64. doi: 10.1074/jbc.M109.058974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lange A, Wickström SA, Jakobson M, Zent R, Sainio K, Fässler R. Integrin-linked kinase is an adaptor with essential functions during mouse development. Nature. 2009;461:1002–6. doi: 10.1038/nature08468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.