Abstract
Background
The aim was to analyse trends in clinically relevant resistance to first-line antiretroviral drugs in Spain, applying the Stanford algorithm, and to compare these results with reported Transmitted Drug Resistance (TDR) defined by the 2009 update of the WHO SDRM list.
Methods
We analysed 2781 sequences from ARV naive patients of the CoRIS cohort (Spain) between 2007–2011. Using the Stanford algorithm “Low-level resistance”, “Intermediate resistance” and “High-level resistance” categories were considered as “Resistant”.
Results
70% of the TDR found using the WHO list were relevant for first-line treatment according to the Stanford algorithm. A total of 188 patients showed clinically relevant resistance to first-line ARVs [6.8% (95%Confidence Interval: 5.8–7.7)], and 221 harbored TDR using the WHO list [7.9% (6.9–9.0)]. Differences were due to a lower prevalence in clinically relevant resistance for NRTIs [2.3% (1.8–2.9) vs. 3.6% (2.9–4.3) by the WHO list] and PIs [0.8% (0.4–1.1) vs. 1.7% (1.2–2.2)], while it was higher for NNRTIs [4.6% (3.8–5.3) vs. 3.7% (3.0–4.7)]. While TDR remained stable throughout the study period, clinically relevant resistance to first line drugs showed a significant trend to a decline (p = 0.02).
Conclusions
Prevalence of clinically relevant resistance to first line ARVs in Spain is decreasing, and lower than the one expected looking at TDR using the WHO list. Resistance to first-line PIs falls below 1%, so the recommendation of screening for TDR in the protease gene should be questioned in our setting. Cost-effectiveness studies need to be carried out to inform evidence-based recommendations.
Introduction
HIV drug resistance due to transmitted mutations in the reverse transcriptase (RT) and protease (Pro) regions has been associated with a higher risk of virological failure to first line antiretroviral therapy (ART) [1], having a greater impact for initial regimens containing a non-nucleoside reverse transcriptase inhibitor (NNRTI) [2]. Testing for transmitted drug resistance (TDR) in newly diagnosed patients with HIV is strongly recommended by treatment guidelines [3]–[6], as it has shown to be cost-effective, in terms of gain in quality-adjusted life year (QALYs) when drug resistance prevalence is over 1–5% [7].
The Spanish cohort of naïve HIV infected individuals (CoRIS) offers relevant information about the current epidemiological profile of HIV infection [8], [9], and is an excellent scenario to characterise the prevalence of TDR over time in Spain. Two previous analyses of viral sequences in CoRIS were carried out for the periods 2004–2008 and 2007–2010, and have been published elsewhere [10], [11]. For these two previous studies, we used the 2009 update World Health Organization (WHO) comprehensive list of mutations [12], which has been also widely used for TDR evaluation [13]–[16]
WHO mutations list has overcome the major limitation of TDR studies across the world, thus providing high levels of standardization into these studies. However, it defines TDR to the different classes of antiretroviral drugs based on the presence of at least one mutation related to drug resistance, while first line treatment drugs that are currently approved by many of the latest updates of clinical guidelines, often include compounds, for which more than one mutation is necessary in order to reduce in vivo drug activity (e.g, abacavir, and boosted Protease Inhibitors). Further approaches, such as the Stanford HIV Drug Resistance Database algorithm, calculate the effective resistance given the combination of mutations present in a particular strain, allowing analyzing clinically relevant resistance to specific antiretroviral drugs (ARVs) and regimens, providing a complementary and invaluable input for informing clinical recommendations.
The objective of this study was to analyse clinically relevant resistance to drugs included in recommended first-line regimens in Spain (CoRIS) from 2007 to 2011, using the Stanford algorithm, and compare it to TDR, defined by the WHO list of mutations.
Patients and Methods
CoRIS is an open, multicentre, prospective cohort of HIV-positive, antiretroviral-naïve subjects over 13 years of age, including both seroprevalent and seroconverter patients. Subjects are recruited and followed up in 31 HIV units from 13 of the 17 Autonomous Communities of Spain. Ethics approval was obtained from participating sites and a written informed consent was obtained from every patient included in the study. Detailed descriptions of the cohort have been previously published [8], [9]. As part of the cohort data collection process, which began in 2004, sites are asked every year to provide a FASTA viral sequence, encoding the HIV protease and RT obtained at the time of inclusion, available from routine resistance testing. As of October 2011, 23 sites from 10 Autonomous Communities were collaborating in the collection of FASTA sequences. In the cohort-coordinating centre, these sequences are linked to clinical and epidemiological data of the patient. Further, the cohort coordinating centre double checks that dates of FASTA sample collection are previous to initiation of ART. Study period for this analysis was from January 2007 to October 2011 and only naïve patients with availability of a FASTA sequence of medium or high quality were included. Sequence data may be available to qualified researchers upon request.
Clinically relevant drug resistance to the first line antiretrovirals (ARVs) included in recommended first-line regimens in Spain as of current guidelines [6] [abacavir, emtricitabine, lamivudine and tenofovir, (Nucleoside Analogue Reverse Transcriptase Inhibitors, NRTIs), efavirenz and nevirapine (Non-Nucleoside Analogue Reverse Transcriptase Inhibitors, NNRTIs), atazanavir, darunavir and lopinavir (Protease Inhibitors, PIs)] was evaluated using the Stanford Database algorithm [17]. For analysis, Stanford HIVdb “Low-level resistance”, “Intermediate resistance” and “High-level resistance” categories were considered as “Resistant”, while “Susceptible” and “Potential Low Level Resistance” were pooled into Susceptible. For subsequent genotypic sensitivity score (GSS) analysis, “Susceptible” and “Potential Low Level Resistance” were scored as 1, “Low Level Resistance” and “Intermediate Resistance” were pooled into Intermediate and scored as 0.5, and “High Level Resistance” was scored as 0. GSS was calculated adding individual resistance scores for each first-line drug in the regimen.
Alternatively, transmitted drug resistance (TDR) associated mutations were evaluated following the WHO surveillance drug resistance mutation list updated in 2009 by Bennett and colleagues [12].
The sample was described using proportion or median (interquartile range) for categorical and continuous variables, respectively; and bivariate analysis was performed using Chi Square or Kruskall-Wallis test as appropriate. Resistance mutations were described using prevalence, and the corresponding confidence intervals were calculated with an analytically derived variance estimator. Their linear trend over the study period was analysed using chi-square test for trend. Significance level was 5%. All the analyses were conducted using Stata software (V.11.1, Stata Corporation, College Station, Texas, USA).
Results
A total of 23 out of the 28 centres in CoRIS contributed FASTA sequences from 2007 to 2011. These centres provided 7,351 patients, 92.2% of the CoRIS cohort, and a complete naïve sequence was submitted for 2,781 (37.8%). Compared to the total cohort, those with an available FASTA sequence were younger (median age 33.9 vs. 34.9), more frequently males (88.3% vs. 83.9%), infected through sex between men (69.2% vs. 58.9%), with a higher educational level (63.5% vs. 56.2%), originating from Spain compared to other regions (70.1% vs. 68.0%), and recruited in earlier stages of HIV disease progression (CD4 count at recruitment 424 vs. 323 cells/mm3). All differences were statistically significant at p<0.01.
A description of the study population by year in which sample for sequencing was obtained is shown in Table 1. An increase in the proportion of men who have sex with men (MSM), of subjects with a higher education, and of patients at earlier CDC stages and higher CD4 counts was observed over time. Median time from cohort entry to resistance study was -1 day (IQR: -21 to 13).
Table 1. Baseline characteristics of the study population.
| 2007 | 2008 | 2009 | 2010 | 2011 | Total | |||||||||
| (N = 484) | (N = 582) | (N = 596) | (N = 756) | (N = 363) | (N = 2,781) | p | ||||||||
| n | (%) | n | % | n | % | n | % | n | % | n | % | |||
| Sex (Male) | 436 | 90.1 | 481 | 82.6 | 529 | 88.8 | 683 | 90.3 | 326 | 89.8 | 2,455 | 88.3 | <0.01 | |
| Age*, Median (IQR) | 33.8 (28.3–41.0) | 34.3(28.2–41.5) | 33.2(28.0–39.7) | 34.5(28.5–41.6) | 33.8(27.3–42.3) | 33.9 (28.2–41.0) | 0.37 | |||||||
| Mode of transmission | IDU | 35 | 7.2 | 47 | 8.1 | 27 | 4.5 | 38 | 5.0 | 14 | 3.9 | 161 | 5.8 | |
| MSM | 321 | 66.3 | 347 | 59.6 | 429 | 72.0 | 550 | 72.8 | 278 | 76.6 | 1,925 | 69.2 | <0.01 | |
| Heterosexual | 119 | 24.6 | 178 | 30.6 | 126 | 21.1 | 140 | 18.5 | 63 | 17.4 | 626 | 22.5 | ||
| Other/NA | 9 | 1.9 | 10 | 1. 7 | 14 | 2.4 | 28 | 3.7 | 8 | 2.2 | 69 | 2.5 | ||
| Country of origin | Spain | 331 | 68.4 | 408 | 70.1 | 435 | 73.0 | 513 | 67.9 | 262 | 72.2 | 1,949 | 70.1 | |
| Africa | 22 | 4.5 | 23 | 4.0 | 27 | 4.5 | 23 | 3.0 | 7 | 1.9 | 102 | 3.7 | 0.07 | |
| Latin America | 106 | 21.9 | 108 | 18.6 | 106 | 17.8 | 174 | 23.0 | 67 | 18.5 | 561 | 20.2 | ||
| Other/unknown | 25 | 5.2 | 43 | 7.4 | 28 | 4.7 | 46 | 6.1 | 27 | 7.4 | 169 | 6.1 | ||
| Educational level | Lower | 127 | 26.2 | 175 | 30.1 | 148 | 24.8 | 191 | 25.3 | 88 | 24.2 | 729 | 26.2 | |
| Higher | 299 | 61.8 | 341 | 58.6 | 380 | 63.7 | 503 | 66.5 | 244 | 67.2 | 1,767 | 63.5 | <0.05 | |
| Unknown | 58 | 12.0 | 66 | 11.3 | 68 | 11.4 | 62 | 8.2 | 31 | 8.5 | 285 | 10.3 | ||
| Viral Load* (log c/ml) | N | 349 | 429 | 440 | 622 | 291 | 2,131 | 0.11 | ||||||
| Median (IQR) | 4.5 (4.0–5.1) | 4.6 (4.0–5.0) | 4.6 (4.0–5.1) | 4.6 (4.1–5.1) | 4.7 (4.2–5.2) | 4.6 (4.1–5.1) | ||||||||
| CD4 count * (cells/mm3) | N | 416 | 501 | 503 | 699 | 330 | 2,449 | <0.05 | ||||||
| Median (IQR) | 392 (237–604) | 371 (223– 559) | 396 (251– 593 | 413 (276–580) | 432(260–606) | 399 (251–588) | ||||||||
| Duration of the infection* | Recent infection | 43 | 8.9 | 47 | 8.1 | 51 | 8.6 | 70 | 9.3 | 34 | 9.4 | 245 | 8.8 | |
| Chronic infecion | 112 | 23.1 | 121 | 20.8 | 138 | 23.1 | 169 | 22.3 | 76 | 20.9 | 616 | 22.2 | 0.96 | |
| Not evaluable | 329 | 68.0 | 414 | 71.1 | 407 | 68.3 | 517 | 68.4 | 253 | 69.7 | 1,920 | 69.0 | ||
| CDC stage* | P/A | 402 | 83.6 | 492 | 84.8 | 537 | 90.2 | 658 | 87.2 | 326 | 89.8 | 2,415 | 87.1 | |
| B | 40 | 8.3 | 38 | 6.6 | 29 | 4.9 | 53 | 7.0 | 23 | 6.3 | 183 | 6.6 | <0.05 | |
| C | 39 | 8.1 | 50 | 8.6 | 29 | 4.9 | 44 | 5.8 | 14 | 3.9 | 176 | 6.3 | ||
| History of delayed diagnosis | Yes | 149 | 30.8 | 205 | 35.2 | 169 | 28.4 | 218 | 28.8 | 114 | 31.4 | 855 | 30.7 | |
| No | 248 | 51.2 | 277 | 47.6 | 325 | 54.5 | 439 | 58.1 | 192 | 53.0 | 1,481 | 53.3 | <0.05 | |
| Not evaluable | 87 | 18.0 | 100 | 17.2 | 102 | 17.1 | 99 | 13.1 | 57 | 15.7 | 445 | 16.0 | ||
*At the time of the resistance testing; IDU: injecting drug users; MSM: men who have sex with men; P/A: Primoinfection or CDC stage A; NA: Not available; IQR: interquartile range.
Transmitted Drug Resistance (TDR) according to the WHO list
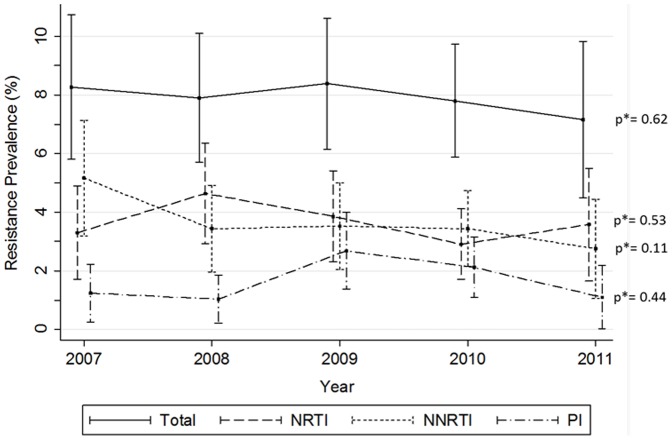
A total of 221 patients had at least one mutation among those contained in the WHO surveillance list, giving a prevalence of TDR of 7.9% (95%Confidence Interval: 6.9–9.0). Regarding mutations affecting specific ARV classes, similar prevalence of TDR was found for nucleoside reverse transcriptase inhibitors (NRTIs) and non-NRTIs (NNRTIs), being 3.6% (2.9–4.3) for NRTIs and 3.7% (3.0–4.7) for NNRTIs. TDR to protease inhibitors (PIs) was less prevalent, of 1.7% (1.2–2.2). Figure 1 shows prevalence of TDR for each ARV class throughout the study period. TDR to NNRTIs decreased from 5.2% in 2007 to 2.8% in 2011, although the trend was not statistically significant. TDR to more than one class of ARV was uncommon; 1.0% (0.6–1.3) and 0.1% (0.0–0.2) of the subjects showed TDR to a minimum of two or three ARV classes, respectively. TDR to two ARV classes decreased from 1.5% in 2007 to 0.3% in 2011, but again was not statistically significant.
Figure 1. Prevalence of TDR (WHO SDRM estimates) by antiretroviral class between 2007 and 2011.
NRTI: nucleoside reverse transcriptase inhibitors; NNRTI: non-nucleoside reverse transcriptase inhibitors; PI: protease inhibitors. * p value for Chi-square test for trend.
Table 2 shows prevalence of specific mutations in the WHO surveillance list. The main mutation related to TDR to NNRTI was K103N/S, which had a prevalence of 2.8% (2.2–3.4). Three mutations were identified as being mainly responsible for TDR to NRTIs: T215 revertants (C/D/E/N/I/V/S) (1.3%; 0.9–1.8), M41L (1.2%; 0.8–1.5) and K219EQNR (1.0%; 0.7–1.4). For PIs, the mutation mainly responsible for TDR was M46IL (0.9%; 0.6–1.3). Of interest, a single mutation (singleton) was responsible for TDR to NRTIs, NNRTIs or PIs in 55.5%, 85.3% and 93.8% of the cases.
Table 2. Prevalence of mutations from the WHO transmitted drug resistance surveillance list.
| NRTI mutations | NNRTI mutations | PI mutations | ||||||||
| Mutation | N | (Pv, %) | Mutation | n | (Pv, %) | Mutation | n | (Pv, %) | ||
| M41L | 32 | (1.15) | L100I | 2 | (0.07) | L24I | 2 | (0.07) | ||
| K65R | 1 | (0.04) | K101EP | 8 | (0.29) | D30N | 1 | (0.04) | ||
| D67EGN | 23 | (0.83) | K103N/S | 77 | (2.77) | V32I | 2 | (0.07) | ||
| T69D | 5 | (0.18) | Y181CIV | 9 | (0.32) | M46IL | 26 | (0.93) | ||
| K70ER | 5 | (0.18) | Y188CHL | 5 | (0.18) | I47AV | 2 | (0.07) | ||
| L74IV | 3 | (0.11) | G190AES | 11 | (0.40) | F53LY | 2 | (0.07) | ||
| F77L | 2 | (0.07) | P225H | 3 | (0.11) | I54ALMSTV | 2 | (0.07) | ||
| Y115F | 2 | (0.07) | M230L | 1 | (0.04) | V82ACFLMST | 5 | (0.18) | ||
| M184IV | 13 | (0.47) | N83D | 1 | (0.04) | |||||
| L210W | 12 | (0.43) | I85V | 1 | (0.04) | |||||
| T215REV* | 37 | (1.33) | N88DS | 2 | (0.07) | |||||
| T215YF | 2 | (0.07) | L90M | 10 | (0.36) | |||||
| K219EQNR | 29 | (1.04) | ||||||||
| 1 mut. | 56 | (2.01) | 1 mut. | 87 | (3.13) | 1 mut. | 45 | (1.62) | ||
| 2 mut. | 32 | (1.15) | 2 mut. | 13 | (0.47) | 2 mut. | 2 | (0.07) | ||
| ≥3 mut. | 13 | (0.47) | ≥3 mut. | 2 | (0.07) | ≥3 mut. | 1 | (0.04) | ||
| Prevalence (95%CI) | 3.6 | (2.9–4.3) | Prevalence (95%CI) | 3.7 | (3.0–4.4) | Prevalence (95%CI) | 1.7 | (1.2–2.2) | ||
NRTI: nucleoside reverse transcriptase inhibitors; NNRTI: non-nucleoside reverse transcriptase inhibitors; PI: protease inhibitors; Pv: prevalence; mut.:mutation. Amino acide abbreviations: A, alanine; C, cysteine; D, aspartate; E, glutamate; F, phenylalanine; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M methionine; N, asparagine; P, proline; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; W, tryptophan; Y, tyrosine. * = C/D/E/N/I/V/S.
Clinically relevant resistance to first-line drugs according to the Stanford Algorithm
When investigating clinically relevant resistance to ARV drugs that make part of recommended initial regimens, the number of patients that showed any resistance was 188, for a total prevalence of 6.8% (5.8–7.7). Prevalence of clinically relevant resistance to any first line ARV was lower than TDR for NRTIs [2.3% (1.8–2.9)] and PIs [0.8% (0.4–1.1)], while it increased in the case of NNRTIs to reach 4.6% (3.8–5.3). In fact, only 70.1% of patients harboring TDR actually showed any clinically relevant resistance to first-line drugs. Percentages for NRTIs and PIs were 65.3% and 35.4%, respectively. For NRTIs, the mutations responsible for having TDR, but without clinically relevant resistance to first line drugs, were present mainly as singletons: a Thymidine Associated Mutation (TAM) alone (M41L, n = 8; T69D, n = 1; K70R, n = 1; L210W, n = 4; K219N/Q, n = 7), a T215 revertant alone (D/N/S n = 7), a F77L alone (n = 2), or a combination of D67N+K219Q (n = 4), or D67N+T69D (n = 1). For PIs, mutations responsible for these discordances were always detected as singletons (L24I, n = 1; D30N, n = 1; M46I/L, n = 24; F53Y, n = 1, V82L, n = 2 and N88D, n = 2). On the other hand, the increase in clinically relevant resistance for NNRTIs was driven mainly by the combination of two or more mutations that are not present in the WHO list (V90I+V108I+H221Y, n = 1; A98G+V179E, n = 1; V108I+E138A, n = 3; V108I+V179E, n = 2; E138A+V179D, n = 2; E138A+V179D+Y188H, n = 1; V179D+ V106I, n = 1), and in 7 cases by the presence of a A98G singleton.
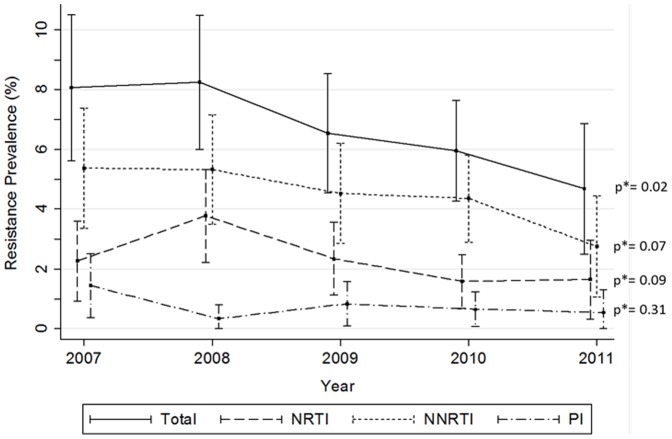
Figure 2 shows the prevalence of clinically relevant resistance to first line ARV drugs by ARV class. A significant decrease (p = 0.02) was found from 2007 (8.1%) to 2011 (4.7%), explained by a borderline statistical significance decrease in both NRTIs and NNRTI. No trend was found in case of PIs. Primary drug resistance to two or three ARV classes was low, of 0.8% (0.5–1.2) and of 0.04% (0.0–0.1), respectively.
Figure 2. Prevalence of clinically relevant resistance to first line drugs (Stanford HIVdb) by antiretroviral class between 2007 and 2011.
NRTI: nucleoside reverse transcriptase inhibitors (abacavir, emtricitabine, lamivudine and tenofovir); NNRTI: non-nucleoside reverse transcriptase inhibitors (efavirenz and nevirapine); PI: protease inhibitors (atazanavir, darunavir and lopinavir). * p value for Chi-square test for trend.
For individual first line NRTIs, clinically relevant resistance was 2.2% (1.7–2.8) for abacavir, 0.7% (0.4–1.0) for emtricitabine and lamivudine, and 1.6% (1.1–2.1) for tenofovir; for first line NNRTIs, resistance was observed in 4.0% (3.3–4.8) for efavirenz and 4.6% (3.8–5.3) for nevirapine; finally, for PIs, resistance was observed in 0.7% (0.4–1.0) for atazanavir, 0.1% (0.0–0.3) for darunavir, and 0.3% (0.1–0.5) for lopinavir.
Therapeutic barrier to resistance of first-line drug regimens
Table 3 shows resistance when combining these drugs into first line recommended regimens. Overall, 188 patients (6.8%) showed resistance to at least one regimen (GSS ≤3). Most affected regimens were those containing NNRTIs with patients showing resistance between 5.6% and 6.2%, depending on the specific combination of drugs. Resistance to PI based regimens was less common, between 2.2% and 2.7%. Of note, only 0.7% to 0.9% of the PI containing regimens showed two or less fully active drugs.
Table 3. Therapeutic barrier of first line ARV regimens using Stanford Algorithm.
| ARV regimen | Genotypic Sensitivity Score | |||||
| <3 | ≤2 | |||||
| n | (%) | n | (%) | |||
| EFV | TDF | FTC/3TC | 155 | (5.6) | 94 | (3.4) |
| ABC | FTC/3TC | 157 | (5.7) | 95 | (3.4) | |
| NVP | TDF | FTC/3TC | 169 | (6.1) | 109 | (3.9) |
| ABC | FTC/3TC | 171 | (6.2) | 109 | (3.9) | |
| LPV | TDF | FTC/3TC | 64 | (2.3) | 20 | (0.7) |
| ABC | FTC/3TC | 68 | (2.5) | 20 | (0.7) | |
| ATZ | TDF | FTC/3TC | 72 | (2.6) | 25 | (0.9) |
| ABC | FTC/3TC | 75 | (2.7) | 26 | (0.9) | |
| DRV | TDF | FTC/3TC | 61 | (2.2) | 19 | (0.7) |
| ABC | FTC/3TC | 65 | (2.3) | 19 | (0.7) | |
* Threshold of the genotypic sensitivity score (GSS) used to consider the strain as resistant to a given ARV combination. TDF: tenofovir; ABC: abacavir; 3TC: lamivudine; FTC: emtricitabine; EFV: efavirenz; NVP: nevirapine; LPV: lopinavir; DRV: darunavir; ATZ: atazanavir.
Discussion
Our study shows that, in Spain for the period 2007–2011, the prevalence of clinically relevant resistance to approved first line ARV drugs analyzed using the Stanford HIV interpretation system is lower than the one we would expect when looking at the Transmitted Drug Resistance (TDR) defined in the WHO mutations list. Moreover, a significant trend to a reduction in resistance was found, driven by a decrease in resistance to the first line NNRTIs efavirenz and nevirapine, as well as to the NRTIs abacavir, emtricitabine, lamivudine, and tenofovir, while resistance to the PIs atazanavir, darunavir and lopinavir, remained stable and very low throughout the whole period. In contrast, no significant trend in the TDR defined by WHO list has been observed. Resistance to more than one antiretroviral class was very uncommon by year 2011.
Many studies have evaluated trends in TDR transmission across Europe and the US [10], [11], [13]–[16], [18]–[21]. The use of the 2009 update of the WHO surveillance recommendations for the estimation of TDR has brought uniformity to all TDR surveys, and has had the great benefit of making all these studies comparable. It also has proved extremely useful for characterizing the epidemiology of TDR and carrying out population-based surveillance. However, its correlation with resistance to the different ARVs is not always straightforward, and further analysis of these mutations such as the one proposed by the Stanford HIV Resistance Database, can provide a complementary and useful input to clinicians, public health professionals and policy makers to develop evidence-based clinical guidelines.
In our study, TDR was of 7.9%, higher than clinically relevant resistance to first line drugs (prevalence of 6.8%). This shows that effective resistance to first line regimens in Spain would lie below the one we would expect when looking at the list of resistance mutations surveillance. In fact, up to 30% of patients harboring TDR did not actually show clinically relevant resistance, being this percentage higher in the case of PIs (65%) and NRTIs (34%). Specifically looking at clinical resistance to first line drugs is therefore relevant and can have important implications.
Clinically relevant resistance to first line PIs remained below 1% for all years except 2007, and was mostly of intermediate level, showing that transmitted resistance to PIs is not threatening effectiveness of PI-based first line ART in Spain. In agreement with what was previously suggested by an Irish report [14], our study greatly supports the hypothesis that there is no need from the clinical point of view to screen for PI resistance in our setting, though having the protease sequence may be useful for epidemiologic studies tracing TDR. A recent study has reported a misinterpretation of resistance to PIs, with an increase in the level of transmitted resistance to this class of antiretrovirals when using the WHO list [22]. In our study we have found that focusing only on TDR may also overestimate the impact of circulating resistances on the clinical effectiveness of first-line PI-containing regimens. In fact, 93.8% of the patients with TDR to PIs harboured a single mutation (singleton) in the protease. For classes including drugs with a high genetic barrier to resistance, such as PIs [23], the presence of just one mutation should not preclude its further use for first line regimens. Specific studies on clinically relevant mutations would be needed for informing this decision.
In a similar way, we have also shown this effect for NRTIs, with a prevalence of TDR of 3.6%, and a prevalence of clinically relevant resistance to first line drugs of 2.3%. Again, singletons were responsible for these differences (55.5% of the patients with TDR to NRTIs harboured a single mutation). Abacavir and tenofovir are the two drugs in the NRTI class with the best resistance profile, being the least affected drugs by single mutations. In fact most of the discordant cases showing TDR showed resistance to zidovudine or stavudine, two NRTIs that are no longer included in Spanish and most international recommendations for first line treatment. In contrast, the NNRTI class, represented for first line with two low genetic barrier drugs, showed higher clinically relevant resistance than TDR. This difference was mainly driven by intermediate resistance calls for efavirenz and/or nevirapine by a combination of two or more mutations that are not included in the WHO list, or by the presence of A98G alone.
Evaluating resistance to specific drugs also allows evaluating the global therapeutic barrier of a regimen, measuring the global robustness to resistance of a certain combination rather than resistance to a certain drug. This may be of extreme importance for evaluating clinical resistance to first line regimens, establishing recommendations on how resistance testing should be performed on newly diagnosed patients, and updating treatment guidelines. In our study, first line NNRTI containing regimens showed the lowest therapeutic barrier, as 5.6% to 6.2% of the newly diagnosed patients from CoRIS, showed any degree of resistance to at least one of the drugs in the regimen. In contrast, the therapeutic barrier to first line PI-based regimens was much higher, with between 2.2% and 2.7% showing any degree of resistance, and only 0.7% to 0.9% of the patients with two or less active drugs in the regimen, most of them being fully resistant to emtricitabine or lamivudine. To our knowledge, this is the first study to estimate the therapeutic barrier of first line regimens, and our results reinforce the previously mentioned notion that, in our setting, a reverse transcriptase-only approach for resistance screening in naïve patients is a strategy that should be considered. Cost-effectiveness studies evaluating the different strategies are urgently needed to better inform drug resistance testing recommendations in our setting. Some other issues, related to the epidemiological value of having the protease sequence information, should be considered and modelled in this analysis.
Our study has several limitations. First, we have studied a subset of patients included in CoRIS, as a FASTA sequence was only available for 37.8% of the patients. Restricting to years after 2007, when the recommendation to screen for resistance in all patients was made, minimises the possibility of selective testing of patients with higher probability of resistance. Higher testing in younger males, MSM of Spanish origin and higher education could reflect clinicians' preferences and practices and higher agency of these patients. However, we cannot rule out that selection bias persists at a certain scale, resulting in an infra-estimation of resistance rates. Second, our sequencing data have been obtained through population sequencing and we have not investigated the presence of minor populations. As minor variants are known to have a greater impact on resistance to NNRTIs in naïve patients [24], it is very unlikely that using this technology our main observations could have affected resistance to the other classes, in particular for the PI class. Finally, higher time since HIV infection can limit our ability to find resistance mutations that were present at infection. Unfortunately, we did not have information on the date of infection for most of the patients, although higher CD4 counts, lower percentage of patients in CDC C stage and a slightly higher percentage of recent infections in the latter calendar years could indicate an earlier HIV diagnosis. The fact that we have not found a decreasing trend in TDR (as of WHO list) indicates that this bias, if existing, is probably of small magnitude.
In summary, clinically relevant resistance to ART first line regimens in Spain estimated using the Stanford HIV resistance database algorithm has been found to be lower than the one we expected when looking at the WHO surveillance mutations list, and with a significant trend to a decline through the period 2007–2011. As PI-based first line regimens have shown the highest therapeutic barrier and primary resistance to first-line PIs has been found to be below 1%, we believe that there is a need to question the recommendation of screening for TDR in the protease gene in our setting, and that cost-effectiveness studies need to be carried out to inform evidence-based recommendations.
Acknowledgments
This study would not have been possible without the collaboration of all the patients, physicians, nurses, and data managers who have taken part in the project.
The members of Cohorte de la Red de Investigación en Sida (CoRIS) are:
Steering Committee: Juan Berenguer, Julia del Amo, Federico García, Félix Gutiérrez, Pablo Labarga, Santiago Moreno y María Ángeles Muñoz-Fernández. Field work, data management and analysis: Paz Sobrino Vegas, Victoria Hernando Sebastián, Belén Alejos Ferreras, Débora Álvarez, Susana Monge, Inma Jarrín, Santiago Pérez Cachafeiro. BioBank: M Ángeles Muñoz-Fernández, Isabel García-Merino, Coral Gómez Rico, Jorge Gallego de la Fuente y Almudena García Torre. Participating Centres: Hospital Central de Asturias (Oviedo): Víctor Asensi Álvarez, Eulalia Valle and José AntonioCartón, Santiago Melón. Hospital Universitario de Canarias (Santa Cruz de Tenerife): Juan Luis Gómez Sirvent, Patricia Rodríguez Fortúnez, María Remedios Alemán Valls, María del Mar Alonso Socas, María Inmaculada Hernández Hernández, Felicitas Díaz-Flores, Dácil García Rosado y Ricardo Pelazas González. Hospital Carlos III (Madrid): Vicente Soriano, Pablo Labarga, Pablo Barreiro, Francisco Blanco, Luz Martín Carbonero, Eugenia Vispo, Carmen Solera, Carmen de Mendoza, Ana Treviño, Eva Poveda, Gustavo Manuzza, Lourdes Anta. Hospital Clinic – IDIBAPS, universidad de Barcelona (Barcelona): José M. Miró, Christian Manzardo, Laura Zamora, Iñaki Pérez, Ma Teresa García, Carmen Ligero, José Luis Blanco, Felipe García-Alcaide, Esteban Martínez, Josep Mallolas, José M. Gatell. Hospital Doce de Octubre (Madrid): Rafael Rubio, Federico Pulido, Silvana Fiorante, Jara Llenas, Violeta Rodríguez, Mariano Matarranz, Rafael Delgado. Hospital Donostia (San Sebastián): José Antonio Iribarren, Julio Arrizabalaga, María José Aramburu, Xabier Camino, Francisco Rodríguez-Arrondo, Miguel Ángel von Wichmann, Lidia Pascual Tomé, Miguel Ángel Goenaga, Ma Jesús Bustinduy, Harkaitz Azkune Galparsoro. Hospital General Universitario de Elche (Elche): Félix Gutiérrez, Mar Masiá, Sergio Padilla, Cristina López, Juan Carlso Rodríguez, Andrés Navarro, Fernando Montolio, Catalina Robledano García, Rafael Pascual. Hospital Germans Trías i Pujol (Badalona): Bonaventura Clotet, Cristina Tural, Lidia Ruiz, Cristina Miranda, Roberto Muga, Jordi Tor, Arantza Sanvisens. Hospital Gregorio Marañón (Madrid): Juan Berenguer, Juan Carlos López Bernaldo de Quirós, Pilar Miralles, Jaime Cosín Ochaíta, Matilde Sánchez Conde, Isabel Gutiérrez Cuellar, Margarita Ramírez Schacke, Belén Padilla Ortega, Paloma Gijón Vidaurreta. Hospital Universitario La Paz (Madrid): Juan González García, Ignacio Bernardino de la Serna, José Ramón Arribas López, María Luisa Montes Ramírez, Jose Ma Peña, Blanca Arribas, Juan Miguel Castro, Francisco Javier Zamora Vargas, Ignacio Pérez Valero, Miriam Estébanez, Silvia García Bujalance, Natalia Stella y Jesús Mingorance. Hospital Universitario La Fé (Valencia): José López Aldeguer, Juan Córdoba Cortijo, José Miguel Molina, Marta Montero Alonso, Marino Blanes Juliá, Miguel Salavert Lletí, José Lacruz Rodrigo, Eva Calabuig, Sandra Cuéllar. Hospital de la Princesa (Madrid): Ignacio de los Santos, Jesús Sanz Sanz, Ana Salas Aparicio, Cristina Sarriá Cepeda. Hospital San Pedro-CIBIR (Logroño): José Antonio Oteo, José Ramón Blanco, Valvanera Ibarra, Luis Metola, Mercedes Sanz, Laura Pérez-Martínez. Hospital Universitario Mutua de Terrassa (Terrassa): David Dalmau, Angels Jaén Manzanera, Mireia Cairó Llobell, Daniel Irigoyen Puig, Laura Ibáñez, Queralt Jordano Montañez, Mariona Xercavins Valls, Javier Martinez-Lacasa, Pablo Velli, Roser Font. Hospital de Navarra (Pamplona): María Rivero, Itziar Casado, Jorge Díaz, Javier Uriz, Jesús Repáraz, Carmen Irigoyen, María Jesús Arraiza. Hospital Parc Taulí (Sabadell): Ferrán Segura, María José Amengual, Eva Penelo, Gemma Navarro, Montserrat Sala, Manuel Cervantes, Valentín Pineda. Hospital Ramón y Cajal (Madrid): Santiago Moreno, José Luis Casado, Fernando Dronda, Ana Moreno, María Jesús Pérez Elías, Dolores López, Carolina Gutiérrez, Beatriz Hernández, María Pumares, Paloma Martí. Hospital San Cecilio (Granada): Federico García García, José Hernández Quero, Alejandro Peña Monje, Leopoldo Muñoz Medina, Jorge Parra Ruiz, Vicente Guillot, Marta Alvarez y Natalia Chueca. Centro Sanitario Sandoval (Madrid): Jorge Del Romero Guerrero, Carmen Rodríguez Martín, Teresa Puerta López, Juan Carlos Carrió Montiel, Paloma Raposo,Mar Vera García y Juan Ballesteros Martín. Hospital Son Espases (Palma de Mallorca): Melchor Riera, Javier Murillas, Maria Peñaranda, Maria Leyes, Ma Angels Ribas, Antoni Campins, Concepcion Villalonga, Carmen Vidal. Hospital Universitario de Valme (Sevilla): Juan Antonio Pineda, Eva Recio Sánchez, Fernando Lozano de León, Juan Macías, José Carlos Palomares, Manuel Parra, Jesús Gómez-Mateos. Hospital Virgen de la Victoria (Málaga): Jesús Santos González, Manuel Márquez Solero, Isabel Viciana Ramos, Rosario Palacios Muñoz. Hospital Universitario Virgen del Rocío (Sevilla): Pompeyo Viciana, Manuel Leal, Luis Fernando López-Cortés, Mónica Trastoy, Pilar Pérez Romero. Hospital Universitario de Santiago (Santiago): Antonio Antela López, Arturo Prieto, Elena Losada, Antonio Aguilera Ortiz. Centro Colaborador: Centro Nacional de Microbiología (Majadahonda): Lucía Pérez Álvarez, Miguel Thomson Okatsu, Elena Delgado Blanco, Yolanda Vega Rocha.
Funding Statement
This work was supported by the Instituto de Salud Carlos III through the Red Temática de Investigación Cooperativa en Sida [ISCIII-RETIC RD06/006 and RD12/0017], and through Grant n° PI12/01053. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wittkop L, Günthard HF, de Wolf F, Dunn D, Cozzi-Lepri A, et al. (2011) Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis 11: 363–371. [DOI] [PubMed] [Google Scholar]
- 2. Kuritzkes DR, Lalama CM, Ribaudo HJ, Marcial M, Meyer WA 3rd, et al. (2008) Preexisting resistance to nonnucleoside reverse-transcriptase inhibitors predicts virologic failure of an efavirenz-based regimen in treatment-naive HIV-1-infected subjects. J Infect Dis 197: 867–870. [DOI] [PubMed] [Google Scholar]
- 3.European AIDS clinical Society. EACS Guidelines v6.1. Available: http://www.europeanaidsclinicalsociety.org. Accessed: 12 Nov 2012.
- 4.Office of AIDS Research Advisory Council (OARAC). Bethesda (US). Guidelines for the Use of Antiretroviral Agents in HIV-1-Infected Adults and Adolescents. Available: http://www.aidsinfo.nih.gov/. Accessed: 28 Mar 2012.
- 5. Thompson MA, Aberg JA, Hoy JF, Telenti A, Benson C, et al. (2012) Antiretroviral Treatment of Adult HIV Infection: 2012 Recommendations of the International Antiviral Society–USA Panel. JAMA 308: 387–402. [DOI] [PubMed] [Google Scholar]
- 6.Grupo de Estudio de SIDA. Documento de consenso de Gesida/Plan Nacional sobre el Sida respecto al tratamiento antirretroviral en adultos infectados por el virus de la inmunodeficiencia humana (Actualización enero 2013). Available: http://www.gesida-seimc.org. Accessed: 6 Mar 2013. [DOI] [PubMed]
- 7. Sax PE, Islam R, Walensky RP, Losina E, Weinstein MC, et al. (2005) Should resistance testing be performed for treatment-naive HIV-infected patients? A cost-effectiveness analysis. Clin Infect Dis 41: 1316–1323. [DOI] [PubMed] [Google Scholar]
- 8. Caro-Murillo AM, Castilla J, Pérez-Hoyos S, Miró JM, Podzamczer D, et al. (2007) Spanish cohort of naïve human immunodeficiency virus infected patients (CoRIS): rationale, organization and initial results. Enferm Infecc Microbiol Clin 25: 23–31. [DOI] [PubMed] [Google Scholar]
- 9. Sobrino-Vegas P, Gutiérrez F, Berenguer J, Labarga P, García F, et al. (2011) The cohort of the spanish hiv research network (coris) and its associated biobank; organizational issues, main findings and losses to follow-up. Enferm Infecc Microbiol Clin 29: 645–653. [DOI] [PubMed] [Google Scholar]
- 10. García F, Pérez-Cachafeiro S, Guillot V, Alvarez M, Pérez-Romero P, et al. (2011) Transmission of HIV drug resistance and non-B subtype distribution in the Spanish cohort of antiretroviral treatment naïve HIV-infected individuals (CoRIS). Antivir Res 91: 150–153. [DOI] [PubMed] [Google Scholar]
- 11. Monge S, Guillot V, Alvarez M, Peña A, Viciana P, et al. (2012) Analysis of transmitted drug resistance in Spain in the years 2007–2010 documents a decline in mutations to the non-nucleoside drug class,. Clin Microbiol Infect 18: E485–490. [DOI] [PubMed] [Google Scholar]
- 12. Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, et al. (2009) Drug resistance mutations for surveillance of transmitted human immunodeficiency virus type 1 drug-resistance: 2009 update. PloS One 4: e4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karlsson A, Björkman P, Bratt G, Ekvall H, Gisslén M, et al. (2012) Low prevalence of transmitted drug resistance in patients newlydiagnosed with HIV-1 infection in Sweden 2003–2010. PLoS One 7: e33484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Gascun CF, Waters A, Regan C, O'Halloran J, Farrell G, et al. (2012) Documented Prevalence of HIV Type 1 Antiretroviral Transmitted Drug Resistance in Ireland from 2004 to 2008. AIDS Res Hum Retroviruses 28: 276–281. [DOI] [PubMed] [Google Scholar]
- 15. Dolling D, Sabin C, Delpech V, Smit E, Pozniak A, et al. (2012) Time trends in drug resistant HIV-1 infections in the United Kingdom up to 2009: multicentre observational study. BMJ 345: e5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Descamps D, Chaix ML, Montes B, Pakianather S, Charpentier C, et al. (2010) Increasing prevalence of transmitted drug resistance mutations and non-B subtype circulation in antiretroviral-naive chronically HIV-infected patients from 2001 to 2006/2007 in France. J Antimicrob Chemother 65: 2620–2627. [DOI] [PubMed] [Google Scholar]
- 17. Liu TF, Shafer RW (2006) Web Resources for HIV type 1 Genotypic-Resistance Test Interpretation. Clin Infect Dis 42: 1608–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bartmeyer B, Kuecherer C, Houareau C, Werning J, Keeren K, et al. (2010) Prevalence of transmitted drug resistance and impact of transmitted resistance on treatment success in the German HIV-1 Seroconverter Cohort. PLoS One 5: e12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vercauteren J, Wensing AM, van de Vijver DA, Albert J, Balotta C, et al. (2009) Transmission of drug-resistant human immunodeficiency virus type 1 is stabilizing in Europe. J Infect Dis 200: 1503–1508. [DOI] [PubMed] [Google Scholar]
- 20. Hurt CB, McCoy SI, Kuruc J, Nelson JA, Kerkau M, et al. (2009) Transmitted antiretroviral drug resistance among acute and recent human immunodeficiency virus infections in North Carolina from 1998 to 2007. Antivir Ther 14: 673–678. [PMC free article] [PubMed] [Google Scholar]
- 21. Price MA, Wallis CL, Lakhi S, Karita E, Kamali A, et al. (2011) Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS Res Hum Retroviruses 27: 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frentz D, van de Vijver DA, Boucher CA Albert J (2011) Estimates of HIV transmitted drug resistance can be inflated due to natural sequence polymorphisms. J Acquir Immune Defic Syndr 58: e135–137. [DOI] [PubMed] [Google Scholar]
- 23. Beerenwinkel N, Däumer M, Sing T, Rahnenfuhrer J, Lengauer T, et al. (2005) Estimating HIV evolutionary pathways and the genetic barrier to drug resistance. J Infect Dis 191: 1953–1960. [DOI] [PubMed] [Google Scholar]
- 24. Li JZ, Paredes R, Ribaudo HJ, Kozal MJ, Svarovskaia ES, et al. (2013) Impact of minority nonnucleoside reverse transcriptase inhibitor resistance mutations on resistance genotype after virologic failure. J Infect Dis 207: 893–897. [DOI] [PMC free article] [PubMed] [Google Scholar]