Abstract
Surface-bound polypeptides and proteins are increasingly used to functionalize inorganic interfaces such as electrodes, but their structural characterization is exceedingly difficult with standard technologies. In this paper, we report the first two-dimensional sum-frequency generation (2D SFG) spectra of a peptide monolayer, which is collected by adding a mid-IR pulse shaper to a standard femtosecond SFG spectrometer. On a gold surface, standard FTIR spectroscopy is inconclusive about the peptide structure because of solvation-induced frequency shifts, but the 2D lineshapes, anharmonic shifts, and lifetimes obtained from 2D SFG reveal that the peptide is largely α-helical and upright. Random coil residues are also observed, which do not themselves appear in SFG spectra due to their isotropic structural distribution, but which still absorb infrared light and so can be detected by cross-peaks in 2D SFG spectra. We discuss these results in the context of peptide design. Because of the similar way in which the spectra are collected, these 2D SFG spectra can be directly compared to 2D IR spectra, thereby enabling structural interpretations of surface-bound peptides and biomolecules based on the well-studied structure/2D IR spectra relationships established from soluble proteins.
Introduction
Bio-functionalized inorganic interfaces play a role in a wide range of emerging materials and analytical technologies. Peptides tethered to gold surfaces have recently been used as the basis for a variety of sensors, ranging from the detection of enzymes and cell metabolites1,2 to metal ions.3 Peptide-functionalized interfaces also find use in the design and creation of biocompatible surfaces, 4 and play a role in biofouling and non-specific cell adsorption in medical and other devices. 5,6
As for all proteins, the sequence and structure of the protein is critical for the function of the interface. Many principles have been established for designing the folded structure of soluble peptides, but design principles are not so clear when it comes to surface-bound peptide structures. Most surface attachment strategies rely on thiol grafting through a cysteine residue.7 Also important are hydrophobic contacts and amino acids with planar side groups such as arginine.8 Peptides with these amino acids bind to gold even without cysteine.9,10 Binding to gold can cause structural distortions by either inducing non-natural curvatures or by disrupting side chain contacts that are necessary for the native fold.11–13
Currently, these surface forces are not well understood, underscoring the need to experimentally assess peptide structures adsorbed to surfaces. Methods capable of assessing both structure and dynamics are highly desirable. Imaging techniques and scanning-probe microscopies14,15 provide information about packing and morphology from which structure can be inferred. Secondary structure information can be gleaned from ATR-FTIR,16 vibrational sum frequency generation (SFG) spectroscopy,17–21 and tip-enhanced vibrational spectroscopies22–24, which can all achieve monolayer or sub-monolayer sensitivity. When applicable, chiral SFG spectroscopy is particularly sensitive to peptide secondary structure.25,26 However, these spectroscopies are all one-dimensional techniques that have few observables from which one can deduce structure or dynamics. For a solvent-free peptide on a gold surface, like those studied here, not even the amide-I vibrational frequency is a reliable measure of secondary structure, as we show below.
In the bulk, there are many multidimensional vibrational techniques for obtaining very detailed structures and dynamics of proteins and other biomolecules. We and several other research groups are extending these new multidimensional spectroscopies to interfaces.27–33 2D IR spectroscopy is a very useful technique for probing peptide and protein structures in solution. In a 2D IR experiment, a pair of ultrafast mid-IR pulses pumps a vibrational transition, and a probe pulse monitors the vibrations of the system some time later. The resulting 2D spectrum provides information about structure through cross-peaks between coupled vibrational modes and about environment through 2D lineshapes. Time-resolved dynamics probe chemical exchange and energy transfer.34,35 Our approach is a technique called 2D SFG spectroscopy that combines the information content of 2D IR spectroscopy with the surface sensitivity of SFG spectroscopy. By using phase-sensitive heterodyne detection (HD) and femtosecond pump pulses,36–38 we can directly compare 2D IR and 2D SFG (and thus solution and surface) spectra.29,39,40 By utilizing a mid-IR pulse shaper we turn a typical broadband SFG spectrometer into an instrument capable of performing either 2D or pump/probe experiments.
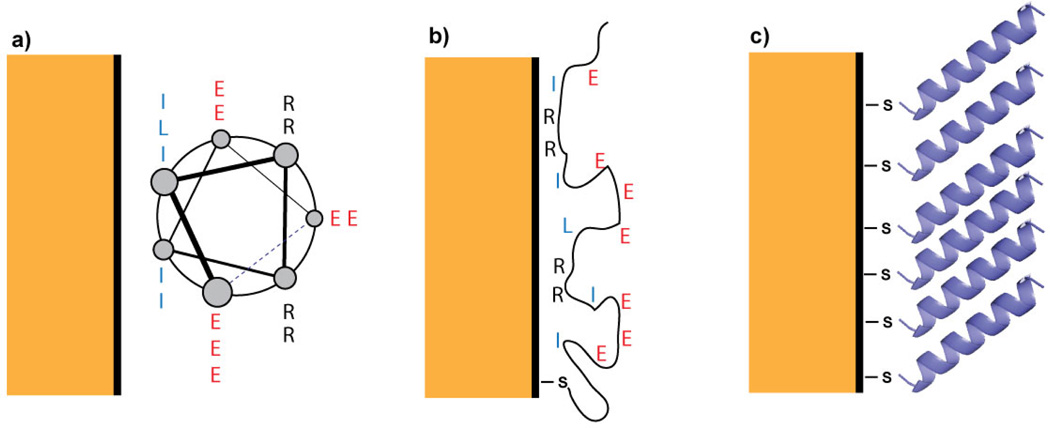
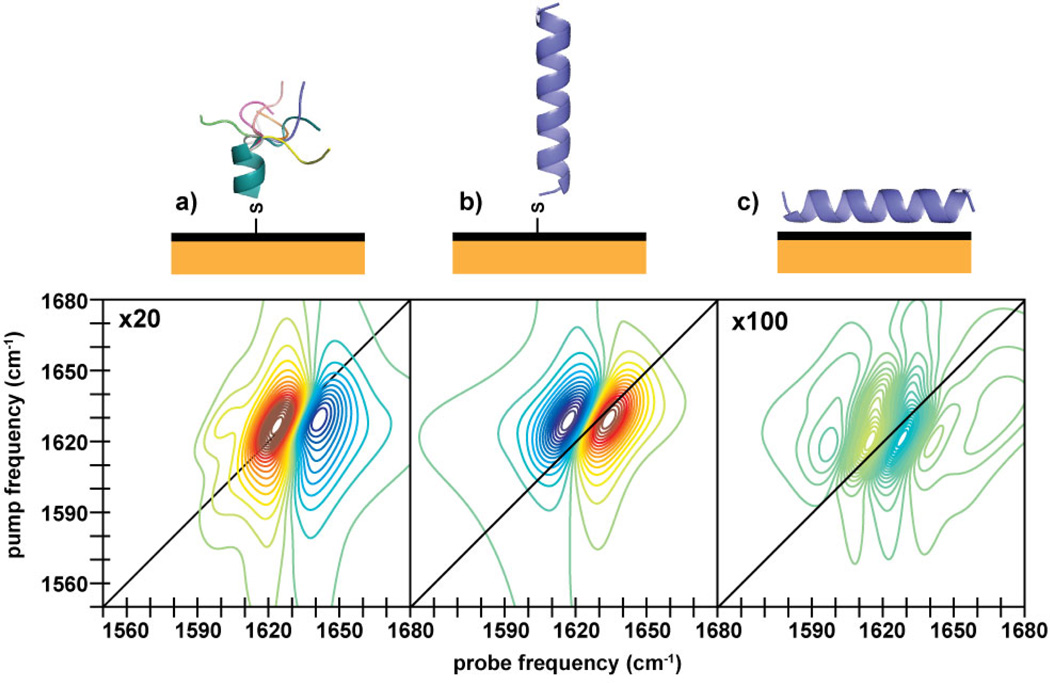
In this paper, we apply this emerging technique to a peptide monolayer on gold, with the goal of determining the peptide’s conformation and orientation. The peptide, Succ–YEIRRIEEELRRIEEEIGGC–CONH2 (hereafter called AHP), bears a Cys residue near the C-terminus which provides a thiol group to mediate attachment to the gold surface. In solution, AHP adopts a mostly α-helical conformation in which the hydrophobic Ile and Leu residues are clustered in a “stripe” that runs along one side; the remaining helical surface is hydrophilic (see Figure 1a). It is well-established that cysteine-containing peptides preferentially bind through the cysteine thiol,13,14 but even with the thiol attachment, AHP may interact with the gold surface in one of three ways. First, the peptide may lie down to bury the hydrophobic surface of the helix at the gold surface (Figure 1a); second, the peptide may denature to maximize favorable side chain-surface interactions (Figure 1b); or third, the Cys side chain thiolate may make a single-point contact, causing the helix to project away from the surface (Figure 1c). A combination of interaction modes is possible as well.
Figure 1.
Schematics of possible surface structures of AHP, which is helical in solution. The helical peptide may (a) lie flat on the surface to bury its hydrophobic stripe, (b) denature to maximize sidechain contacts, or (c) stand upright. The G, Y, and sometimes C residues are omitted to highlight the non-thiol sidechains expected to interact most strongly with the gold surface.
As we show below, the new observables from 2D SFG and comparison to 2D IR spectra of the soluble peptide provide direct measures of secondary structure and lead us to conclude that the peptide is helical and standing upright. Moreover, we study the dynamics of the peptide and vibrational energy transfer along its backbone, from which we learn that there are also disordered structural elements that are invisible in conventional SFG spectra but appear as cross-peaks in 2D SFG spectra. Our work establishes that HD 2D SFG spectroscopy is a viable and helpful probe of surface-bound peptide structures, and our results provide insight into design principles for peptides at interfaces. Figure 1: Schematics of possible surface structures of AHP, which is helical in solution. The helical peptide may (a) lie flat on the surface to bury its hydrophobic stripe, (b) denature to maximize sidechain contacts, or (c) stand upright. The G, Y, and sometimes C residues are omitted to highlight the non-thiol sidechains expected to interact most strongly with the gold surface.
Methods
Sample Preparation
We prepared peptide monolayers on gold using a 20-residue cysteine-containing peptide with sequence Succ–YEIRRIEEELRRIEEEIGGC–CONH2. The peptide contains a succinyl capping group at the N-terminus and a primary amide capping group at the C-terminus. Substrates were prepared via electron-beam evaporation of a 20 nm adhesion layer of Ti on a polished silicon (111) substrate, followed by a 100 nm film of gold. The sample deposition rate was 1 Å/s with a chamber base pressure less than 2×10−6 Torr. Gold samples were immersed in 0.1 mM solutions of the peptide in 100 mM acetic acid buffer in H2O adjusted with KOH to pH 5, which is close to the peptide’s isoelectric point and gave the best monolayer formation. Samples were left to soak overnight, and were then rinsed with fresh H2O and dried under a stream of nitrogen before measurement. Samples were characterized by reflection FTIR and surface coverage was estimated by x-ray photoelectron spectroscopy, which gave 3.12 nm2 per molecule (see Supporting Information and Discussion, below). At other pH values, peptides either aggregated or did not attach at appreciable coverages on the gold surface. 2D SFG spectra were identical for peptides deposited from D2O and from H2O.
2D IR and circular dichroism (CD) spectra of bulk samples were obtained both in MES buffer in D2O at pH 5, matching the deposition pH for the surface samples, and in phosphate buffer in D2O at pH 7. We use the 2D IR and CD spectra at pH 7 for our analysis because phosphate buffer at pH 7 provided a cleaner CD spectrum and more consistent fitting to extract the degree of helicity, but as shown in the Supporting Information, spectra at pH 5 showed no qualitative differences from those at pH 7.
Two-Dimensional Infrared and Sum Frequency Generation Spectroscopies
Our 2D IR and 2D SFG apparati were similar to those we have described previously.29,41 Both experiments utilized mid-IR pulses generated by optical parametric amplification and difference frequency generation from the output of a regeneratively-amplified Ti:sapphire laser. A germanium mid-IR pulse shaper was used to produce pump pulse pairs and scan the requisite t1 time (0 to 1500 fs for 2D SFG and 0 to 3200 fs for 2D IR, in 50 fs steps, with a rotating frame frequency of 1450 cm−1). For 2D IR experiments, the pump pulses were focused onto a sample sandwiched between two CaF2 windows along with an infrared probe pulse, which was detected on an MCT array. For 2D SFG experiments, the pump pulses were focused onto peptide monolayer samples along with the mid-IR probe and visible upconversion pulse. The upconversion pulse was temporally overlapped with the mid-IR probe for all waiting times in order to create a large non-resonant background signal. This non-resonant signal was used as a local oscillator to heterodyne the 2D SFG signal; the local oscillator was removed during data processing by Fourier filtering (see Supporting Information). The resulting sum-frequency signal was then dispersed in a polychromator and detected on a thermoelectrically-cooled CCD. The frequency range of the 2D IR spectra along the probe dimension was determined by the width of the mid-IR array detector, which spanned 1545–1670 cm−1. The larger number of pixels in the CCD detector used for SFG measurements enabled us to acquire 2D SFG spectra across the entire bandwidth of the infrared pulse. Further details of the spectrometer setup are provided in the Supporting Information, but we point out that the pulse shaper was used to produce identical pump pulses for both 2D IR and 2D SFG experiments, which enables a one-to-one comparison between the two datasets.
Two-Exciton Hamiltonian Model for 2D SFG Spectroscopy
Spectra were simulated using the transition dipole coupling model and a two-exciton Hamiltonian, as described in more detail in the supporting information. Briefly, peptide structures were loaded from a protein databank (PDB) file or generated via a Ramanchandran plot, as described. A transition dipole and Raman polarizability were assigned to each amide-I local mode. Transition dipole coupling was used to calculate couplings between all pairs of local amide-I modes. The resulting Hamiltonian was diagonalized to yield the eigenstates of the system,34 whose coefficients were used to calculate the transition dipoles and Raman tensors connecting every pair of zero-, one-, and two-exciton normal modes by taking appropriate linear combinations of the local mode transition dipoles and Raman tensors. These responses were rotated to the laboratory frame, mapped into a two-dimensional stick spectrum, and convoluted with a two-dimensional lineshape consistent with model amide-I chromophor N-methyl-acetamide (NMA)42 to give the final simulated spectra. Helices were constructed using ϕ=−60°,ψ=−45°. For simulations involving frayed ends, dihedral angles were varied over standard regions of the Ramachandran plot to generate 2000 different structures. A HD 2D SFG spectrum was calculated for each, which were summed to generate the spectrum for the ensemble average. Computer code is freely available from our website to calculate 2D SFG spectra.43 We did not calculate spectra for the full-length peptide because it was too computationally expensive for a laptop computer. The helix absorption frequency depends on helix length. We set the local mode frequencies to the experimental helix signal with a 20 cm−1 diagonal frequency shift for random coil regions.
Results
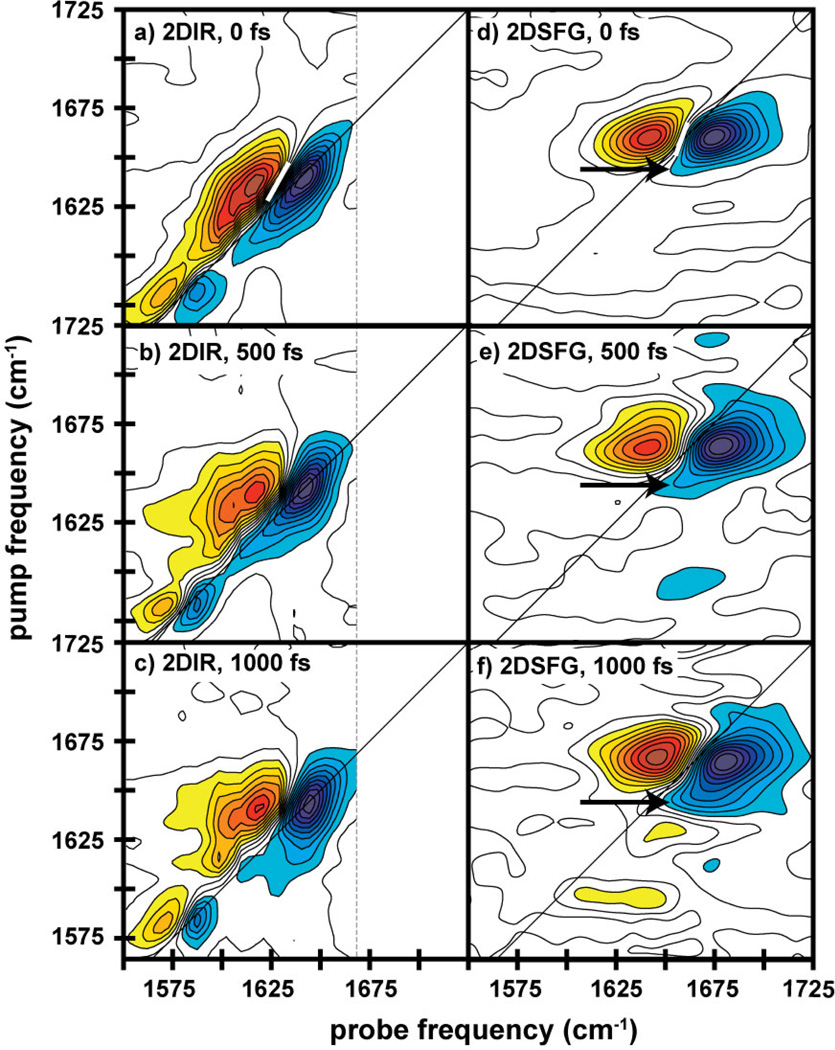
In Figures 2a and 2d, we present 2D IR spectra of AHP in buffer and HD 2D SFG spectra of dehydrated peptide on gold collected with a t2=0 fs waiting time. In the 2D IR spectra, we assign the primary pair of peaks at 1642 cm−1 to the amide-I band, which consists mostly of the backbone carbonyl stretch. 2D IR spectra always exhibit pairs of out-of-phase peaks.34 The negative (blue) peak on the diagonal is the fundamental transition, and the positive (red) peak is the corresponding sequence band. The separation between these two peaks reflects the anharmonic shift of the vibrational mode. These two peaks are elongated along the diagonal and have a prominent shoulder on the low-frequency edge and a smaller one on the high-frequency side. Circular dichroism (CD) spectra estimate the peptide is 50% helical and monomeric in solution (see Supporting Information). Thus, we assign the 1642 cm−1 peaks to the α-helix and the shoulders to random coil. These assignments match frequencies expected for the amide-I band.44 Protonated arginine side chains contribute peaks at 1584 and 1615 cm−1.44
Figure 2.
Two-dimensional infrared (a–c) and 2D SFG (d–f) spectra of peptide AHP, at waiting times of (a,d) t2=0, (b,e) 500, and (c,f) 1000 fs. Contours are unfilled between ±20% to emphasize the peaks under discussion. The frequency range of the probe dimension in the 2D IR and 2D SFG spectra are determined by the width of the mid-IR array detector and CCD camera, respectively. The white bar in panels (a) and (d) indicate the nodal line at t2 = 0.
HD 2D SFG spectra can be interpreted like 2D IR spectra since the signal is heterodyned by the nonresonant background and the infrared pulse sequence is exactly the same.29 The two spectra share similarities, but differ in many respects. First, there is a very large difference in the peak frequency. The primary pair of peaks in the 2D SFG spectra is near 1660 cm−1. Second, there are no side chain features. Third, the anharmonic shift appears larger in the 2D SFG spectrum than in the 2D IR spectra. And fourth, the spectral features are narrower along the diagonal in the 2D SFG than the 2D IR spectrum. We discuss these features in the Discussion.
Also presented in Figure 2 are 2D IR and 2D SFG spectra measured at t2=500 and 1000 fs. These waiting time experiments measure the dynamics inherent to the system. In the 2D IR spectra, the nodal line between the out-of-phase peaks becomes more vertical at longer waiting times, as structural and environmental dynamics cause the frequencies of the amide-I backbone to evolve. In addition, the spectra narrow along the diagonal at longer waiting times, which is illustrated in Figure 3d, due to a decrease in intensity at the lower-frequency edge of the peak (arrow). At t2=0 fs, the diagonal linewidth of the 2D IR spectrum is 26 cm−1, but by 1 ps it is 21 cm−1. We attribute this narrowing to the short lifetimes of random coil modes.45 Thus, the lifetimes are consistent with our assignment of the lower frequency modes to disordered peptide residues, most likely due to frayed ends.45,46 Other secondary structures can absorb at this frequency, such as small beta-sheets, but we do not observe the corresponding cross-peaks nor higher-frequency modes.47–49 We also note that at 1 ps, cross-peaks begin to form between the amide-I mode and the arginine sidechain at 1615 cm−1 due to energy transfer with the backbone.50
Figure 3.
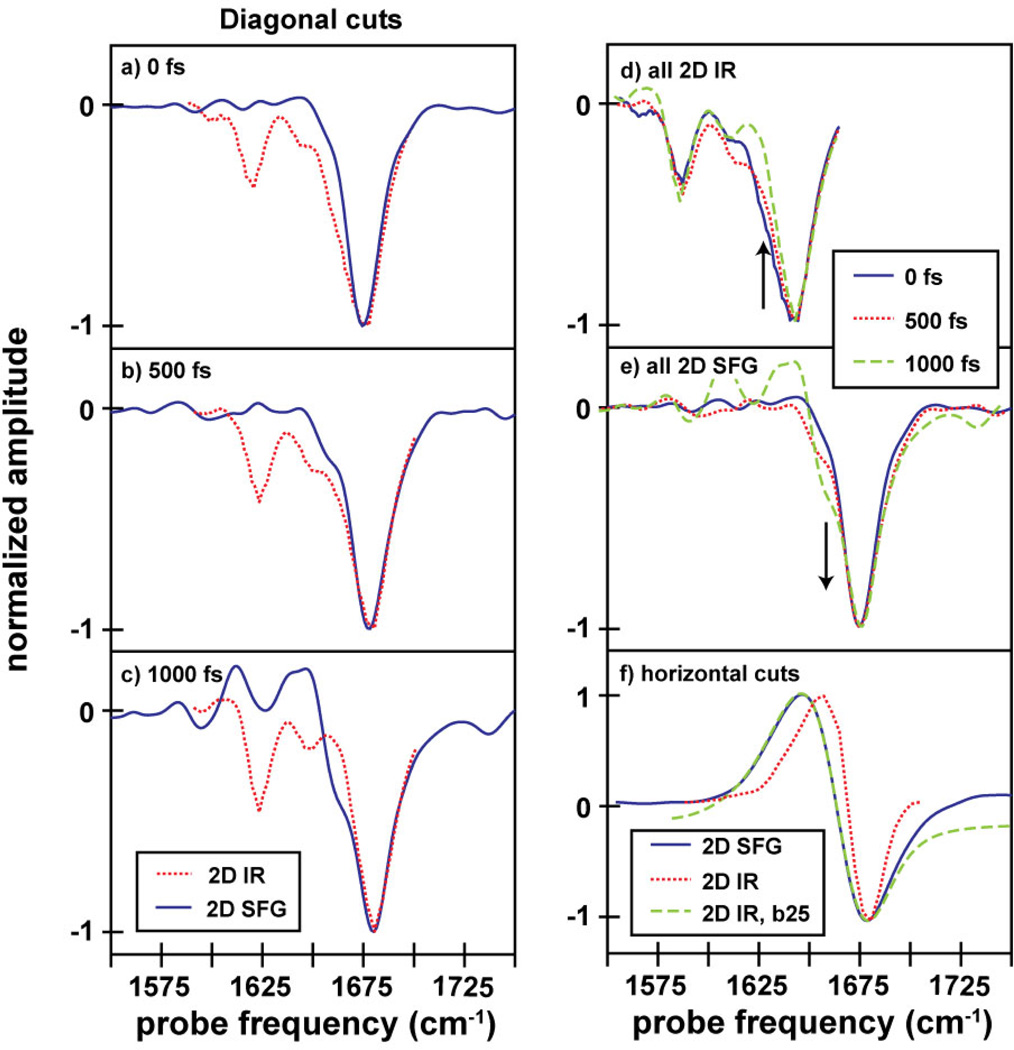
Comparison of various cuts through the 2D IR and 2D SFG spectra. In (a–c), we give a comparison between diagonal cuts through the fundamental peaks in the 2D IR and 2D SFG spectra (a) t2=0, (b) 500, and (c) 1000 fs. In (d) and (e), we show the evolution of these cuts as a function of waiting time. In (f), we comapare a horizontal cut through the 2D SFG spectrum at ωpump = 1660 cm−1 with a cut through the 2D IR spectrum at ωpump = 1642 cm−1, both with and without broadening by a 25 cm−1 Lorentzian (labeled b25). In all spectra comparing cuts from the 2D IR spectra with cuts from the 2D SFG spectra, the 2D IR cuts have been shifted along the probe axis so that the fundamental lines up with that from the 2D SFG spectrum to enable direct linewidth and lineshape comparisons.
In contrast, little spectral diffusion occurs in the 2D SFG spectra, since the nodal line changes very little with waiting time. This indicates that the dehydrated peptide’s environment is less dynamic on the surface than in solution. Moreover, the diagonal linewidth of the 2D SFG spectra broadens with waiting time instead of decreasing as in the 2D IR spectra (Figure 3). The broadening is caused by a shoulder on the low-frequency side the main peak (arrow). In the overlapped spectra, one sees that the shoulder in the 2D SFG matches the shoulder in the 2D IR spectrum (arrow). For reasons that we discuss below, we assign this feature to vibrational energy flow from disordered to ordered structures. We also note that the linewidths of the 2D SFG spectrum at t2=0 match the 2D IR linewidth at t2=1 ps when the random coil features have disappeared.
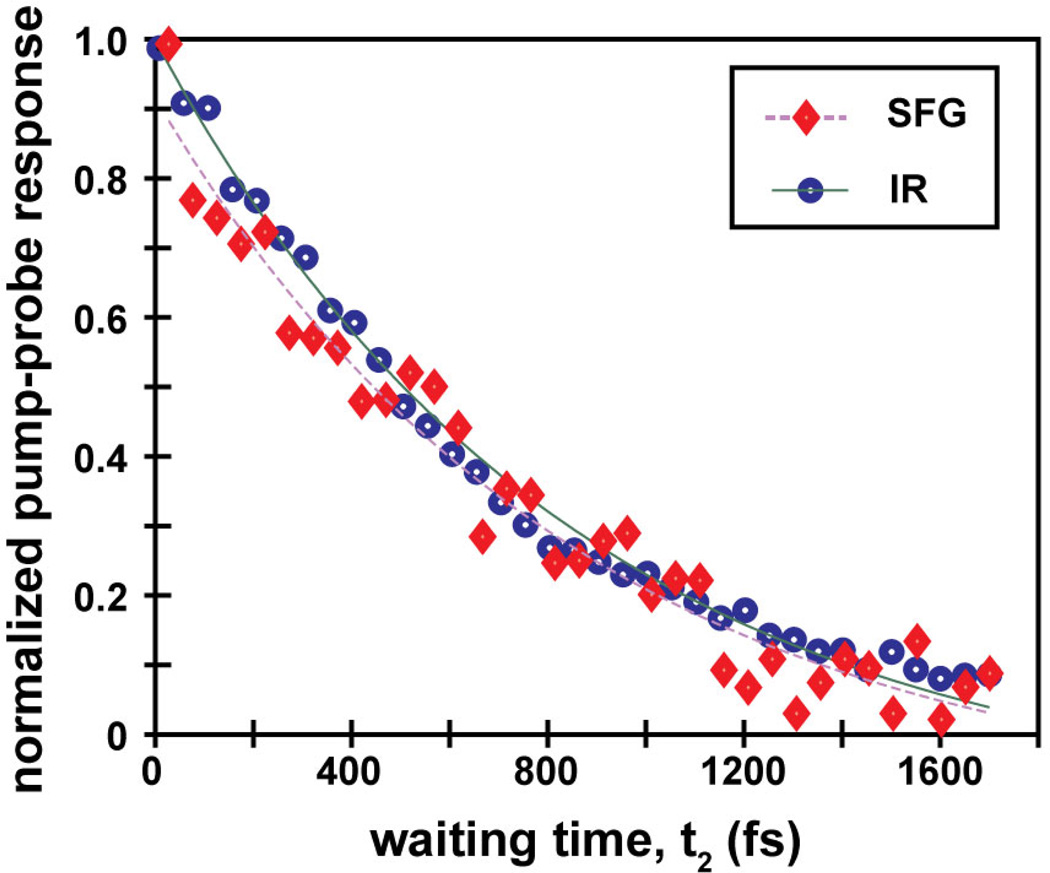
To compare the vibrational lifetimes associated with the main peaks, we measure pump-probe kinetics by setting t1=0 fs and scanning t2, shown in Figure 4. The fundamental peak decays with a time-constant of 850 and 870 fs for 2D IR and 2D SFG measurements, respectively. Thus, the overall vibrational relaxation rate is the same for the soluble and surface bound peptides. Figure 4: Overlaid pump-probe traces of AHP on gold and in D2O with fitted exponential decays.
Figure 4.
Overlaid pump-probe traces of AHP on gold and in D2O with fitted exponential decays.
Discussion
Amide-I frequencies are often used as indicators of secondary structure in peptides and proteins. In solution, AHP’s CD spectrum (see SI) and amide-I frequency at 1642 cm−1 indicate the peptide is largely helical. On gold, obtaining a CD spectrum is prohibitively difficult,51 and the frequency as measured by FTIR-RAS is now 1660 cm−1. Hydrophobic membrane peptides can absorb as high as 1660 cm−1 as can dehydrated random coils and turns.16,52,53 Interpretation of a standard SFG spectrum based on frequency alone has the same difficulties as the FTIR response. Chiral SFG responses, which are sensitive to secondary structure in a similar manner as CD spectroscopy, can lend insight into secondary structure,25,26 but are difficult to measure for samples on gold due to suppression of s-polarized responses.54 Thus, frequency alone is insufficient for definitive secondary structure assignment, because solvation causes similar frequency shifts in this region.
2D IR and 2D SFG measurements provide additional observables indicative of secondary structure, including anharmonic shifts, lineshapes, vibrational lifetimes, phases, and intensities. These observables depend on the extent of vibrational excitonic delocalizations, which depends strongly on the secondary structure. We start by investigating the anharmonic shift of the principle amide-I normal mode. Amide-I modes of single amino acids and random coils have anharmonic shifts of 14 cm−1, which is the frequency difference between the fundamental (v=0→1) and sequence band (v=1→2) transitions.55,56 Well-ordered secondary structures have smaller anharmonic shifts because their vibrational modes are delocalized over multiple residues. α-helices have anharmonic shifts of 5 to 10 cm−1 because the amide-I vibrational mode extends over 3–4 amino acids, whereas the random coils have larger anharmonic shifts because the structural disorder prevents delocalization. 34,57–59 Accurately fitting the spectra to obtain the anharmonic shift is difficult because the peaks are separated by less than their linewidths. Instead, we use the 2D IR spectrum as a reference since we know that the peptide is α-helical in solution and exhibits an anharmonic shift that is typical of α-helices34,57–59. In Figure 3, we compare horizontal slices from the 2D IR and 2D SFG spectra passing through the principle amide-I peak. To make a valid comparison, we must take into account the visible laser pulse in the 2D SFG experiments, which acts as a window function;38 we convolute the cut through the 2D IR spectrum with a 25 cm−1 Lorentzian, which corresponds to the bandwidth of our visible pulse. Figure 3 shows a comparison of the resulting cuts. The separation of the positive and negative peaks is almost perfectly matched between the 2D SFG spectrum and the broadened 2D IR spectrum. Thus, the anharmonic shift is consistent with AHP adopting α-helical structure on the gold surface.
2D lineshapes are another useful indicator of secondary structure. The 2D IR and 2D SFG spectra collected here utilize a photon echo pulse sequence that removes inhomogeneous broadening along the anti-diagonal of the spectra. The asymmetry of the 2D lineshape is thus a measure of the homogeneous and inhomogeneous contributions to the vibrational dynamics. Random coils and turns are very inhomogeneous, and their spectra appear elongated along the diagonal with nodal lines tilted at 45°. In contrast, excitonic delocalization in regular secondary structures creates homogeneous broadening, which causes round 2D lineshapes and nodal lines that tend toward the vertical (0°). In our 2D SFG spectra, at the 0 fs waiting time the diagonal cut (shown in Figure 3) has a linewidth of approximately 23 cm−1 and nodal line tilted 20°(white line, Figure 2d). The 2D linewidths of soluble helices are usually about 21 cm−1 and have a nodal line tilt of 28° or less58(see Supplemental). Thus, the 2D lineshape is consistent with a delocalized excitonic state such as an α-helix.
Finally, vibrational lifetimes can also be indicators of secondary structure.45 The fact that the peptide has the same lifetime in the bulk and on the gold surface argues that they have similar structures. Thus, based on the anharmonic shift, nodal line, lifetimes, and homogeneous 2D line-shape, we conclude that AHP adopts a helical structure on the gold surface. The conditions under which peptides adhere to surfaces are often constrained to a small range of conditions. For the peptide studied here, pH 5 was used because other conditions either caused surface aggregation or little binding. The ability to directly compare 2D SFG spectra to 2D IR spectra is thus particularly useful, because the experimental conditions for 2D IR spectra can be widely varied and we already know and understand the spectral signatures of alpha-helices, random coils and other secondary structures.
The above structural conclusion comes primarily from an analysis of the 2D SFG spectrum collected at t2=0, but spectra at longer waiting times reveal that the peptide also contains random coil residues. Isotropic distributions of chromophores are SFG-inactive and do not appear in 1D SFG spectra or as diagonal peaks in 2D SFG spectra. For example, the Arg side chains are absent in the 2D SFG spectra, indicating that they are not aligned. The fact that the 2D IR spectrum at t2= 1ps, in which the random coil has disappeared,45 matches the linewidth of the 2D SFG spectrum at t2=0, is consistent with the standard assumption that SFG spectra do not contain random coil signals.60 However, random coil residues can be observed in 2D SFG spectra by coupling or energy transfer to SFG active modes, as we show below.
To illustrate this point, we performed the simulation shown in Figure 5a. The simulation consists of a 7-residue α-helix with a 5-residue frayed end. In the simulated spectra the helix gives rise to a doublet at 1650 cm−1 but the frayed end does not contribute along the diagonal nor does it create a signal in 1D SFG spectra (see SI), because it forms a nearly isotropic structural distribution and the ensemble averaged SFG signals cancel. However, the frayed ends still absorb light from the pump pulses at 1620 cm−1, which creates cross-peaks to the SFG active helix modes. The cross-peaks only appear below the diagonal because random coils can absorb IR radiation but not emit SFG signals, so peaks only appear where ordered modes are probed.61 If the SFG inactive mode were higher in frequency, then the cross-peaks would appear above the diagonal. In our simulations, the cross-peaks are generated purely by coupling. They would be enhanced by energy transfer from the SFG inactive to SFG active modes.
Figure 5.
Simulated HD 2D SFG spectra of (a) a 7-residue helix followed by a 5-residue random coil, (b) a 20-residue helix perpendicular to the surface, and (c) a 20-residue helix lying flat on the surface.
With this simulation in mind, we turn back to the experiments. A shoulder grows in on the low-frequency edge of the fundamental transition at t2=500 fs and 1 ps at the same relative frequency as the random coil mode that disappears from the corresponding 2D IR spectra (Figures 3e and 3f, arrow). This feature is due to a cross-peak below the diagonal which becomes more prominent at longer waiting times and does not have a corresponding diagonal peak. The only ways for cross-peaks to grow in in 2D IR and 2D SFG spectra are by energy transfer or chemical exchange. Chemical exchange is usually caused by solvation dynamics, which our dehydrated peptide should not experience. Therefore, we conclude that these cross-peaks arise from energy transfer from the random coil region to the ordered helix modes. In essence, disordered secondary structures that cannot be observed by typical SFG measurements can still interact with the pump pulses, and can be detected via 2D SFG if they transfer energy (or are otherwise coupled) to ordered modes. Thus, we learn from the dynamical measurements that AHP is primarily helical, but, as in solution, does have some disordered residues on the gold surface.
In Figures 1b through 1c, we posited three possible binding motifs for AHP on gold. Our 2D SFG spectra show that the dehydrated surface-bound peptide largely maintains its helical secondary structure. Interactions between the gold surface and the arginines and hydrophobic residues thus are not strong enough to significantly denature the peptide, which rules out the first structural scenario (Figure 1b). To distinguish between the other two binding modes, we simulated the spectra shown in Figures 5b and 5c for a helix standing up and lying down on the surface. The spectrum of the vertical helix is similar to that observed in our experiment, while that of the horizontal helix has a much different lineshape that is 100-times weaker and would probably be unmeasurable with our current signal-to-noise. Like the spectrum simulated for the vertical helix, simulated spectra for tilted helices are dominated by the helix A-mode and resemble the experimental spectrum until tilt angles of more than 70°. Knowing that we have an alpha helix also enables a rough measure of the tilt from a ratio of the amide-I and amide-II intensities in the FTIR spectra (see Supporting Information), giving a tilt of approximately 48°, which is consistent with the observed 2D SFG lineshape. Thus, we conclude that the helix is upright, if somewhat tilted. The orientation reveals that even though the gold surface is hydrophobic and may serve to mirror the charge distribution of the peptide, attraction of the sidechains to the surface is not strong enough to destabilize the peptide’s intrinsic helical propensity.13,14
It is interesting to compare the upright nature of the helix to the peptide surface coverage as measured by x-ray photoelectron spectroscopy (XPS). According to XPS, there are 3.21×1013 molecules/cm2, which corresponds to an area of approximately 3.12 nm2 per peptide. A 20-residue perfect helix is expected to have a length of 30 Å (5.4 Å per 3.6 residue turn) and a diameter of approximately 10 Å including sidechains;62,63 for a helix standing up these dimensions correspond to an area per molecule of approximately 0.75 nm2, while for a helix lying down these dimensions correspond to an area per molecule of 3 nm2. While these estimates are very rough, they suggest that there should be enough space on the gold surface for all the peptides to lie down flat, if that is what was preferred. The fact that they do not, suggests either that there is an intrinsic property to each helix that causes it to stand upright or there are peptide/peptide interactions inducing upright monolayers. Additional concentration and/or mutational studies might be able to uncover the origins of this structural observation.
Conclusions
In this paper, we have demonstrated the first HD 2D SFG spectrum of a peptide monolayer, obtained simply by adding a mid-IR pulse shaper to a typical broadband, ultrafast SFG spectrometer. Heterodyne detection enabled a direct comparison to 2D IR spectra of the same peptide in solution. By measuring the anharmonic shift, 2D lineshapes, nodal line tilts, and lifetimes, we concluded that the peptide maintains its helical structure when adsorbed to gold, despite the presence of many residues whose sidechains’ affinities for gold might be expected to disrupt the peptide’s structure. While other factors, such as solvation environment,64 may also affect these characteristics of 2D spectra, the substantial similarities between the 2D IR and 2D SFG spectra indicate that the structure does not change significantly upon surface attachment. Moreover, we found that the peptide still has structurally disordered components, which typical SFG experiments cannot probe. Because 2D SFG can pump disordered modes, disordered modes can still be detected through coupling and energy transfer to ordered modes, such as the helix modes measured here. 2D SFG spectroscopy is also amenable to larger proteins and can provide site-specific or domain-specific resolution when used in conjuction with isotope labeling.65 Moreover, since a single monolayer is being measured, ligand binding studies can be done as well as voltage dependent measurements that might be important for sensors.66 Thus, we think that heterodyne detected 2D SFG is poised to become a powerful technique for investigating molecular structures and dynamics at interfaces in the host of interfacial systems already being studied with SFG spectroscopy and beyond.
Supplementary Material
Acknowledgement
The authors thank the National Science Foundatio (NSF) and National Institutes of Health (NIH) for funding through the following grants. J.E.L. was supported by the NSF Graduate Research Fellowship Program (DGE-0717123) and the University ofWisconsin Materials Research Science and Engineering Center (UW-MRSEC, DMR-1121288). D.R.S. was supported by the UW-MRSEC and by a NIH Molecular Biophysics Training Grant (T32-GM08293). J-J.H. and M.T.Z. received funding from the National Science Foundation through single-investigator grant CHE-1266422. Y.J. and P.G. were supported by the UW-MRSEC. A.L.S. was supported by NIH grant DK079895, and J.R.S. and S.H.G. received funding through NIH R01 GM061238.
Footnotes
Supporting Information Available
CD and FTIR data and further experimental and calculation details.
This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Chen Y, Cruz-Chu ER, Woodard JC, Gartia MR, Schulten K, Liu L. ACS Nano. 2012;6:8847–8856. doi: 10.1021/nn3027408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin DS, Liu Y, Gao Y, Kwa T, Matharu Z, Revsin A. Analytical Chemistry. 2013;85:220–227. doi: 10.1021/ac302547p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gooding JJ, Hibbert DB, Yang W. Sensors. 2001;1:75–90. [Google Scholar]
- 4.Chaikoff EL, Wang HS, Wingert TM, Stephens S, Dluhy RA. MRS Proceedings. 1995;414:17. [Google Scholar]
- 5.Bolduc OR, Correia-Ledo D, Masson JF. Langmuir. 2012;28:22–26. doi: 10.1021/la203493v. [DOI] [PubMed] [Google Scholar]
- 6.Boulos SP, Prigozhin MB, Liu Y, Wirth AJ, Boppart SA, Gruebele M, Murphy CJ. Current Physical Chemistry. 2013;3:128–135. doi: 10.2174/1877946811303020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu Y, Das A, Hecht MH, Scoles G. Langmuir. 2005;21:9103–9109. doi: 10.1021/la046857h. [DOI] [PubMed] [Google Scholar]
- 8.Feng J, Pandey RB, Berry RJ, Farmer BL, Naik RR, Heinz H. Soft Matter. 2011;7:2113–2120. [Google Scholar]
- 9.Tamerler C, Duman M, Oren EE, Gungormus M, Xiong X, Kacar T, Parviz BA, Sarikaya M. Small. 2006;2:1372–1378. doi: 10.1002/smll.200600070. [DOI] [PubMed] [Google Scholar]
- 10.Verde ACV, Acres J, Maranas JK. Biomacromolecules. 2009;10:2118–2128. doi: 10.1021/bm9002464. [DOI] [PubMed] [Google Scholar]
- 11.Peele BR, Krauland EM, Wittrup KD, Belcher AM. Langmuir. 2005;21:6929–6933. doi: 10.1021/la050261s. [DOI] [PubMed] [Google Scholar]
- 12.Heinz H, Farmer BL, Pandey RB, Slocik JM, Patnaik SS, Pachter R, Naik RR. Journal of the American Chemical Society. 2009;131:9704–9714. doi: 10.1021/ja900531f. [DOI] [PubMed] [Google Scholar]
- 13.Nowinski AK, Sun F, White AD, Keefe AJ, Jiang S. Journal of the American Chemical Society. 2012;134:6000–6005. doi: 10.1021/ja3006868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa K, Morita T, Kimura S. Journal of Physical Chemistry B. 2004;108:15090–15095. [Google Scholar]
- 15.Raigoza AF, Webb LJ. Journal of the American Chemical Society. 2012;134:19354–19357. doi: 10.1021/ja309632m. [DOI] [PubMed] [Google Scholar]
- 16.Manor J, Feldblum ES, Zanni MT, Arkin IT. Journal of Physical Chemistry Letters. 2012;3:939–944. doi: 10.1021/jz300150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim G, Gurau MC, Lim SM, Cremer PS. Journal of Physical Chemistry B. 2003;107:1403–1409. [Google Scholar]
- 18.Fu L, Ma G, Yan ECY. Journal of the American Chemical Society. 2010;132:5405–5412. doi: 10.1021/ja909546b. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, Wang J, Boughton AP, Kristalyn CB, Chen Z. Journal of the American Chemical Society. 2007;129:1420–1427. doi: 10.1021/ja067446l. [DOI] [PubMed] [Google Scholar]
- 20.Walter SR, Geiger FM. Journal of Physical Chemistry Letters. 2010;1:9–15. [Google Scholar]
- 21.Roeters SJ, van Dijk CN, Torres-Knoop A, Backus EHG, Campen RK, Bonn M, Woutersen S. Journal of Physical Chemistry A. 2013;117:6311–6322. doi: 10.1021/jp401159r. [DOI] [PubMed] [Google Scholar]
- 22.Xu XG, Rang M, Craig IM, Raschke MB. Journal of Physical Chemistry Letters. 2012;3:1836–1841. doi: 10.1021/jz300463d. [DOI] [PubMed] [Google Scholar]
- 23.Kurouski D, Postiglione T, Deckert-Gaudig T, Deckert V, Lednev IK. Analyst. 2013;138:1665–1673. doi: 10.1039/c2an36478f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pozzi EA, Sonntag MD, Jiang N, Klingsporn JM, Hersam MC, Duyne RPV. ACS. 2013;7:885–888. doi: 10.1021/nn400560t. [DOI] [PubMed] [Google Scholar]
- 25.Fu L, Liu J, Yan ECY. Journal of the American Chemical Society. 2011;133:8094–8097. doi: 10.1021/ja201575e. [DOI] [PubMed] [Google Scholar]
- 26.Fu L, Xiao D, Wang Z, Batista VS, Yan ECY. Journal of the American Chemical Society. 2013;135:3592–3598. doi: 10.1021/ja3119527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong W, Laaser JE, Paoprasert P, Franking RA, Hamers RJ, Gopalan P, Zanni MT. Journal of the American Chemical Society. 2009;131:18040–18041. doi: 10.1021/ja908479r. [DOI] [PubMed] [Google Scholar]
- 28.Donaldson PM, Hamm P. Angewandte Chemie. 2012;52:634–638. doi: 10.1002/anie.201204973. [DOI] [PubMed] [Google Scholar]
- 29.Xiong W, Laaser JE, Mehlenbacher RD, Zanni MT. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20902–20907. doi: 10.1073/pnas.1115055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh PC, Nihonyanagi S, Yamaguchi S, Tahara T. Journal of Chemical Physics. 2012;137:094706. doi: 10.1063/1.4747828. [DOI] [PubMed] [Google Scholar]
- 31.Bredenbeck J, Ghosh A, Nienhuys HK, Bonn M. Accounts of Chemical Research. 2009;42:1332–1342. doi: 10.1021/ar900016c. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Piatkowski L, Bakker HJ, Bonn M. Nature Chemistry. 2011;3:888–893. doi: 10.1038/nchem.1158. [DOI] [PubMed] [Google Scholar]
- 33.Rosenfeld DE, Gengeliczki Z, Smith BJ, Stack TDP, Fayer MD. Science. 2011;334:634–639. doi: 10.1126/science.1211350. [DOI] [PubMed] [Google Scholar]
- 34.Hamm P, Zanni MT. Concepts and Methods of 2D Infrared Spectroscopy. 1st ed. Cambridge University Press; 2011. p. 296. [Google Scholar]
- 35.Fayer MD. Ultrafast Infrared Vibrational Spectroscopy. CRC Press; 2013. [Google Scholar]
- 36.Stiopkin IV, Jayathilake HD, Bordenyuk AN, Benderskii AV. Journal of the American Chemical Society. 2008;130:2271–2275. doi: 10.1021/ja076708w. [DOI] [PubMed] [Google Scholar]
- 37.Nihonyanagi S, Yamaguchi S, Tahara T. Journal of Chemical Physics. 2009;130:204704. doi: 10.1063/1.3135147. [DOI] [PubMed] [Google Scholar]
- 38.Laaser JE, Xiong W, Zanni MT. Journal of Physical Chemistry B. 2011;115:2536–2546. doi: 10.1021/jp200757x. [DOI] [PubMed] [Google Scholar]
- 39.Ni Y, Gruenbaum SM, Skinner JL. Proceedings of the National Academy of Sciences. 2013;110:1992–1998. doi: 10.1073/pnas.1222017110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang C, Jansen TLC. Journal of Physical Chemistry B. 2013;117:6937–6945. doi: 10.1021/jp403111j. [DOI] [PubMed] [Google Scholar]
- 41.Middleton CT, Woys AM, Mukherjee S, Zanni MT. Methods. 2010;52:12–22. doi: 10.1016/j.ymeth.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skoff DR, Laaser JE, Mukherjee SS, Middleton CT, Zanni MT. Chemical Physics. 2012 doi: 10.1016/j.chemphys.2012.08.019. (in press - doi:10.1016/j.chemphys.2012.08.019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. http://zanni.chem.wisc.edu/.
- 44.Barth A. Progress in Biophysics and Molecular Biology. 2000;74:141–173. doi: 10.1016/s0079-6107(00)00021-3. [DOI] [PubMed] [Google Scholar]
- 45.Middleton CT, Buchanan LE, Dunkelberger EB, Zanni MT. Journal of Physical Chemistry Letters. 2011;2:2357–2361. doi: 10.1021/jz201024m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silva RAGD, Kubelka J, Bour P, Decatur SM, Keiderling TA. Proceedings of the National Academy of Sciences. 2000;97:8318–8323. doi: 10.1073/pnas.140161997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheatum CM, Tokmakoff A, Knoester J. Journal of Chemical Physics. 2004;120:8201. doi: 10.1063/1.1689637. [DOI] [PubMed] [Google Scholar]
- 48.Demirdöven N, Cheatum CM, Chung HS, Khalil M, Knoester J, Tokmakoff A. Journal of the American Chemical Society. 2004;126:7981–7990. doi: 10.1021/ja049811j. [DOI] [PubMed] [Google Scholar]
- 49.Hahn S, Kim SS, Lee C, Cho M. Journal of Chemical Physics. 2005;123:084905. doi: 10.1063/1.1997151. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh A, Tucker MJ, Hochstrasser RM. The Journal of Physical Chemistry A. 2011;115:9731–9738. doi: 10.1021/jp201794n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gallardo IF, Webb LJ. Langmuir. 2012;28:3510–3515. doi: 10.1021/la204927q. [DOI] [PubMed] [Google Scholar]
- 52.Laird DJ, Mulvihill MM, Whiles Lillig JA. Biophysical Chemistry. 2009;145:72–78. doi: 10.1016/j.bpc.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Boncheva M, Vogel H. Biophysical Journal. 1997;73:1056–1072. doi: 10.1016/S0006-3495(97)78138-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cecchet F, Lis D, Guthmuller J, Champagne B, Caudano Y, Silien C, Mani AA, Thiry PA, Peremans A. ChemPhysChem. 2010;11:607–615. doi: 10.1002/cphc.200900733. [DOI] [PubMed] [Google Scholar]
- 55.DeFlores LP, Ganim Z, Ackley SF, Chung HS, Tokmakoff A. The Journal of Physical Chemistry B. 2006;110:18973–18980. doi: 10.1021/jp0603334. [DOI] [PubMed] [Google Scholar]
- 56.Zanni MT, Asplund MC, Hochstrasser RM. Journal of Chemical Physics. 2001;114:4579–4590. [Google Scholar]
- 57.Wang J, Hochstrasser RM. Chemical Physics. 2004;297:195–219. [Google Scholar]
- 58.Grechko M, Zanni MT. The Journal of Chemical Physics. 2012;137:184202. doi: 10.1063/1.4764861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao J, Wang J. The Journal of Chemical Physics. 2012;136:214112. doi: 10.1063/1.4725181. [DOI] [PubMed] [Google Scholar]
- 60.Ding B, Soblosky L, Nguyen K, Geng J, Yu X, Ramamoorthy A, Chen Z. Scientific Reports. 2013;3:1854. doi: 10.1038/srep01854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Laaser JE, Zanni MT. Journal of Physical Chemistry A. 2013;117:5875–5890. doi: 10.1021/jp307721y. [DOI] [PubMed] [Google Scholar]
- 62.Pauling L, Corey RB. Nature. 1953;171:59–61. doi: 10.1038/171059a0. [DOI] [PubMed] [Google Scholar]
- 63.Hristova K, Wimley WC, Mishra VK, Anantharamiah G, Segrest JP, White SH. Journal of Molecular Biology. 1999;290:99–117. doi: 10.1006/jmbi.1999.2840. [DOI] [PubMed] [Google Scholar]
- 64.Woys AM, Lin YS, Reddy AS, Xiong W, de Pablo JJ, Skinner JL, Zanni MT. Journal of the American Chemical Society. 2010;132:2832. doi: 10.1021/ja9101776. [DOI] [PubMed] [Google Scholar]
- 65.Moran SD, Decatur SM, Zanni MT. Journal of the American Chemical Society. 2012;134:18410–18416. doi: 10.1021/ja307898g. [DOI] [PubMed] [Google Scholar]
- 66.Jiang X, Engelhard M, Ataka K, Heberle J. Journal of the American Chemical Society. 2010;132:10808–10815. doi: 10.1021/ja102295g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.