Abstract
Development of lipid-based adjuvant formulations to enhance the immunogenicity of recombinant vaccine antigens is a focus of modern vaccine research. Characterizing interactions between vaccine antigens and formulation excipients is important for establishing compatibility between the different components and optimizing vaccine stability and potency. Cryogenic transmission electron microscopy (TEM) is a highly informative analytical technique that may elucidate various aspects of protein- and lipid-based structures, including morphology, size, shape, and phase structure, while avoiding artifacts associated with staining-based TEM. In this work, cryogenic TEM is employed to characterize a recombinant tuberculosis vaccine antigen, an anionic liposome formulation, and antigen–liposome interactions. By performing three-dimensional tomographic reconstruction analysis, the formation of a population of protein-containing flattened liposomes, not present in the control samples, was detected. It is shown that cryogenic TEM provides unique information regarding antigen–liposome interactions not detectable by light-scattering-based methods. Employing a suite of complementary analytical techniques is important to fully characterize interactions between vaccine components.
Keywords: vaccine adjuvant, cryo-TEM, antigen-adjuvant interactions, vaccine physical characterization, vaccine formulation morphology, 3D tomographic reconstruction
Introduction
ID93 is a recombinant fusion protein of 93 kDa that is being developed as a prophylactic and therapeutic tuberculosis vaccine antigen candidate.1,2 When formulated with the Toll-like receptor (TLR)-4 ligand synthetic glucopyranosyl lipid adjuvant (GLA), ID93 induces T helper type 1 immune responses in mice, guinea pigs, and non-human primates.1,3 Phase I clinical trials designed to evaluate the safety and immunogenicity of ID93 with an oil-in-water emulsion formulation containing GLA (GLA-SE) are ongoing.4,5
Liposomes represent an alternative method of formulation of TLR4 ligands such as GLA. Unlike oil-in-water emulsions, liposomes do not contain an oily core component but rather consist of an aqueous core surrounded by a phospholipid bilayer. As such, liposomes appear to be less reactogenic than oil-in-water emulsions while maintaining potent immunogenic responses. For instance, GlaxoSmithKline’s liposomal formulation of the naturally derived TLR4 ligand monophosphoryl lipid A (MPL®) and the Quillaja saponaria-derived saponin QS21 demonstrated greater immunogenicity and less reactogenicity than an oil-in-water emulsion formulation of the same compounds when combined with a recombinant malaria vaccine;6 this vaccine is now in Phase III clinical trials in African children. We have developed an anionic liposomal formulation containing GLA (GLA-LS) and demonstrated protective immune responses in mice when combined with ID93,7 although T helper type 1 immunogenicity markers were not induced to the same magnitude as with GLA-SE. Interestingly, in contrast to GLA-SE, there appeared to be little association of ID93 with GLA-LS. In fact, changing the liposome charge from anionic to cationic increased the ID93 association but not the immunogenicity or protective responses to the protein.7
In order to achieve a greater understanding of the interactions between anionic liposomes and ID93, we carried out cryogenic transmission electron microscopy (cryo-TEM) analysis of each of the vaccine components alone as well as after mixing. Cryo-TEM is a high-resolution imaging technique that avoids interference from staining artifacts associated with traditional TEM approaches.8 Since sample preparation maintains the aqueous environment of the sample using a flash-freeze approach, cryo-TEM is particularly suitable for aqueous formulations such as protein solutions and liposomes. Here, we present cryo-TEM-based analysis of ID93 and anionic liposomes, complemented by dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA).
Materials and methods
ID93 production and purification
ID93 consists of a fusion construct of Rv3619-Rv1813-Rv3620-Rv2608 and was expressed in Escherichia coli as previously described.1 The antigen was purified by a series of column chromatographies (DEAE, Q Sepharose, and Phenyl Sepharose) and a reducing agent (2-mercaptoethanol) was added before a final diafiltration step.
Liposome formulation
1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol (DPPG) were purchased from Avanti Polar Lipids Inc. (Alabaster, AL, USA). Cholesterol was obtained from JT Baker (Avantor Performance Materials, Inc., Center Valley, PA, USA) and phosphate-buffered saline (PBS) at pH 7.2 was purchased from Invitrogen (Life Technologies, Carlsbad, CA, USA). GLA-LS was manufactured by combining 3.6 g DPPC, 1.0 g cholesterol, and 0.4 g DPPG in 20 mL of chloroform:methanol:water (65:35:8), which was then evaporated using a BUCHI Rotovapor R-114 (BUCHI, Flawil, Switzerland). The dried components were rehydrated in 100 mL PBS (pH 7.2), then sonicated in a VWR 75D (VWR International, Radnor, PA, USA) or Crest Powersonic CP230D (Crest Ultrasonics Corp, Trenton, NJ, USA) sonicating water bath at room temperature until all the solids were removed from the sides of the glass container. The mixture was then transferred to the Microfluidics M110P (Microfluidics, West-wood, MA, USA) for high-pressure homogenization at 10,000 psi for 12 passes. The stock liposome formulation contained 18 mg/mL DPPC, 2 mg/mL DPPG, and 5 mg/mL cholesterol, and was stored at 5°C. Liposomes were diluted 2.5-fold in PBS prior to cryo-TEM experiments.
Cryo-TEM
The samples were prepared and imaged individually. ID93 (0.5 mg/mL stock concentration in 20 mM Tris buffer, pH 8) was diluted 1:10 with dilution buffer (20 mM Tris, pH 8) or mixed 1:1 with the liposome sample (7.2 mg/mL DPPC, 0.8 mg/mL DPPG, 2 mg/mL cholesterol in PBS, pH 7.2) at room temperature immediately prior to vitrification. Samples were preserved by vitrification and supported by holey carbon films on 400-mesh copper grids. Vitrified samples were prepared by applying a 3-μL drop of sample suspension to a cleaned grid, blotting away with filter paper, and immediately plunging into liquid ethane. Grids were stored under liquid nitrogen until transferred to the electron microscope for imaging. Electron microscopy was performed using an FEI Tecnai T12 electron microscope (FEI, Hillsboro, OR, USA), operating at 120 keV equipped with an FEI Eagle 4k × 4k CCD camera (FEI). Vitreous ice grids were transferred into the electron microscope using a cryostage that maintains the grids at a temperature below −170°C. Images of each grid were acquired at multiple scales to assess the overall distribution of the specimen. After identifying potentially suitable target areas for imaging at lower magnifications, pairs of high magnification images were acquired using the Leginon multi-scale imaging application (http://ami.scripps.edu/redmine/projects/leginon/wiki; National Resource for Automated Molecular Microscopy, The Scripps Research Institute, La Jolla, CA, USA)9 at nominal magnifications of 110,000× (0.10 nm/pixel), 52,000× (0.21 nm/pixel), and 21,000× (0.50 nm/pixel). The images were acquired at a nominal underfocus of −1 μm (110,000×), −4 μm (52,000×), and −5 μm (21,000×) and electron doses of ~10–39 e/Å2. Three-dimensional (3D) tomography tilt series were collected for the sample preserved in vitreous ice. Single axis tilt series of images were acquired at angles every 2° from −60° to 60° using a dose per image of ~1–4 e/Å2 at each increment. The images were aligned and used to reconstruct a 3D volume using IMOD (http://bio3d.colorado.edu/imod/; Boulder Laboratory for 3-D Electron Microscopy of Cells, University of Colorado, Boulder, CO, USA).10 Chimera software (https://www.cgl.ucsf.edu/chimera/; Resource for Biocomputing, Visualization, and Informatics, University of California, San Francisco, CA, USA) was used to create rendered volumes of the 3D structures of the sample presented as single frames and as part of the movie loop that shows the structure rotating through a range of angles, as shown in Video S1.11
The 52,000× magnification images were used in the particle counting analysis. These images have a field of view of 0.8 μm × 0.8 μm and a pixel size of 0.208 nm. The images in the experiment were first pre-screened for quality and then randomly ordered for the analysis. Seven hundred and one particles in the images were picked by a human operator. General particle selection was based on the following criteria: particles were not selected if they were over the carbon film surrounding the hole or touching the edge of the carbon film. The following are the criteria for the particle categories: spherical liposome – liposome appeared spherical or mostly round in shape with an uninterrupted bilayer; flattened liposome – liposome appeared flattened so that it had a rectangular shape or appeared as two parallel rod shapes.
Negative-stain TEM
The ID93 sample was imaged at full concentration (0.5 mg/mL) and also at a 1:10 dilution made with the dilution buffer (20 mM Tris, pH 8). Samples were prepared over continuous carbon nitrocellulose supported 400-mesh copper grids. Samples were prepared by applying 3 μL of sample suspension to a cleaned grid, wicking most of the sample with filter paper, and immediately staining with uranyl formate. Electron microscopy was performed using methods as described above. Negative stain grids were transferred into the electron microscope using a room temperature stage. Images of each grid were acquired at multiple scales to assess the overall distribution of the specimen. After identifying potentially suitable target areas for imaging at lower magnifications, pairs of high magnification images were acquired at nominal magnifications of 110,000× (0.10 nm/pixel), 52,000× (0.21 nm/pixel), and 21,000× (0.50 nm/pixel). The images were acquired at a nominal underfocus of −1.5 μm (110,000×), −4 μm (52,000×), and −5 μm (21,000×) and electron doses of ~10–45 e/Å2.
DLS
Particle size was determined using the Malvern Instruments (Malvern, UK) Zetasizer Nano-S or Nano-ZS via DLS. ID93 (0.5 mg/mL) was diluted 1:10 in 20 mM Tris buffer (pH 8) in a 1.5-mL polystyrene disposable cuvette (this dilution was the same as that used for the electron microscopy imaging). The default values supplied by Malvern for refractive index, absorption, and viscosity of protein and water were selected; the data were analyzed by the manufacturer’s supplied protein analysis model, consisting of a standard regularized non-negative least squares analysis where the regularization is determined from an “L curve”. Liposomes were measured after dilution in PBS (1:5) or water (1:100). ID93–liposome mixture was prepared at the same concentrations as for the electron microscopy imaging described above. For liposome-containing samples, the default values supplied by Malvern for refractive index, absorption, and viscosity of polystyrene and water were selected; the data were analyzed by the manufacturer’s supplied general purpose (normal resolution) model. All DLS measurements were performed three times with each cuvette.
NTA
Particle size distributions were evaluated for the ID93–liposome mixture as well as each component alone (at the same concentration) using a NanoSight LM10 nanosizer with a 40 mW 410 nm laser (Malvern Instruments) and a Hamamatsu Orca Flash 2.8 CMOS camera (Hamamatsu Photonics KK, Hamamatsu, Japan). Before and after each measurement the sample cell was rinsed with three cell volumes of Millipore Milli-Q (Billerica, MA, USA) ultrapure water. Samples were diluted 1:106 or 1:105 to obtain a final particle concentration of approximately 5 × 108 particles/mL. Appropriate dilution factors were determined empirically. Samples were diluted in ultrapure Milli-Q water in two or three steps. Each formulation was diluted four times, independently, to account for dilution error. One milliliter of diluted formulation was loaded into a 1-mL syringe and infused into the sample chamber at 15 μL/minute. Four consecutive 60-second videos were recorded for each dilution. Shutter and gain settings were optimized for each sample. The camera histogram gating was adjusted to maximize sensitivity. Data analysis was performed in NanoSight NTA 2.3 (Malvern Instruments) in batch mode.
Isoelectric point (pI) determination
Theoretical calculation of the pI of ID93 was performed using the web-based tool at isoelectric.ovh.org and represented the average pI value calculated by various distinct algorithms. The pI of ID93 was determined via zeta potential measurement using the Malvern Instruments MPT-2 multipurpose titrator in conjunction with the Zetasizer Nano-ZS. ID93 (0.5 mg/mL) was diluted 1:20 in distilled water for a total of 10 mL of volume in a disposable sample container. Titrants 0.5 M HCl and 0.5 M NaOH were used. The start and end pH values were 9.0 and 3.0, respectively, with an increment of 0.5 pH units and a target pH tolerance of 0.5. Three zeta potential measurements were obtained at each pH value.
Results and discussion
Liposome particle size and stability
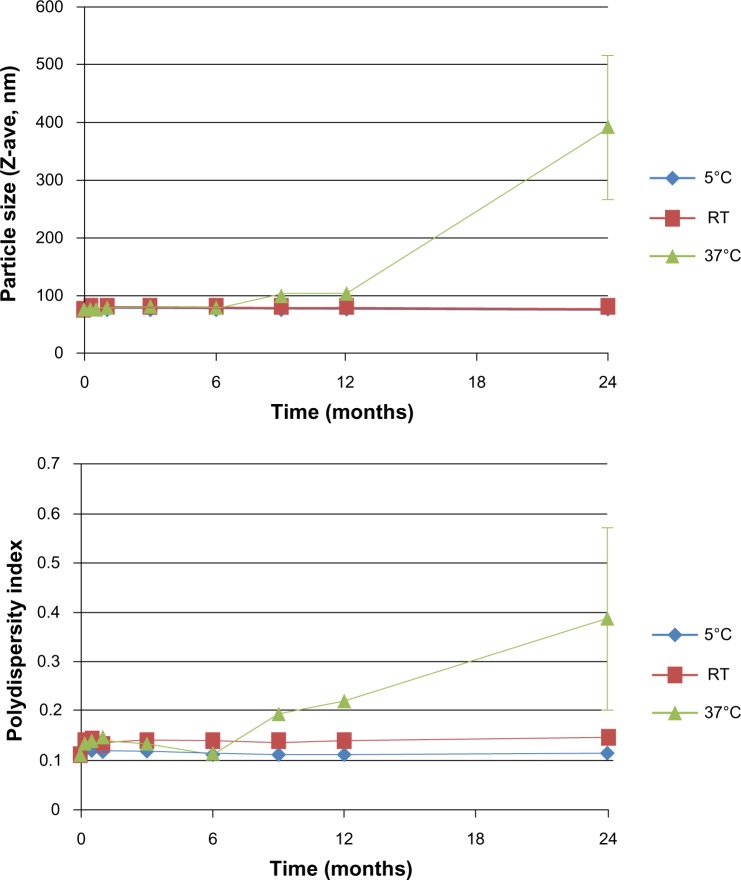
The intensity-based average size (Z-ave) of the liposomes after 100-fold dilution in water is 75.9 nm with polydispersity index (PDI) of 0.111, as measured by DLS. The size and PDI show little change for at least 2 years when stored at 5°C or ambient temperature (Figure 1); at 37°C, size growth is evident after 9 months; at 60°C, the liposome visual appearance indicated substantial sedimentation after 2 weeks. As reported previously, the liposomes are negatively charged, as expected due to the presence of the anionic DPPG.7 Theoretical prediction of the pI of ID93 indicates a value of ~4.7, which was confirmed by zeta potential measurement at different pH values (Figure S1), indicating that the protein is negatively charged at neutral pH. This charge repulsion may explain why little association was detected between protein and liposome when analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) following size exclusion chromatography (SEC).7 However, in order to investigate whether any interactions were present between protein and liposome not detectable by SEC/SDS-PAGE, we subjected the ID93–liposome mixture to cryo-TEM analysis and compared the images to those obtained from each component alone as well as a negative stain TEM of ID93.
Figure 1.
Liposome size stability. Liposome particle size and polydispersity index as measured by dynamic light scattering indicates an initial size of 75.9 nm and polydispersity of 0.111, with minimal change over 2 years when stored at 5°C or room temperature. When stored at 37°C, the liposomes grew in size and polydispersity after 6 months. Error bars represent standard deviations of three separate measurements of the same batch of liposomes employed in the electron microscopy studies.
Abbreviation: RT, room temperature.
Negative-stain TEM of ID93
Negative-stain TEM was employed in order to obtain maximum contrast and determine whether the protein would have good potential to be visible, in the presence of liposomes, by cryo-TEM. The protein was present in various configurations, including small linear particles ~7 nm in length, medium linear particles ~15 nm in length, tripod-shaped particles ~10 nm in length, and clusters of particles that had lengths typically in the range of 25–50 nm, although some were in the range 50–120 nm (Figure 2). Based on reported results in the literature from proteins containing from 58 to 760 residues, Wilkins et al developed equations to relate protein hydrodynamic radius with the number of amino acid residues they contain, with different constants for folded proteins and highly denatured proteins.12 Although ID93 contains slightly more residues (891) than the range reported, the equations are useful to give an approximate range of the expected size of ID93. The hydrodynamic radius range for folded or denatured proteins using these equations is ~3.4–10.6 nm, which agrees well with the diameters of the smaller structures evident in the TEM images. The presence of tripod-shaped particles, which may represent dimers or trimers, as well as the larger aggregates is consistent with SDS-PAGE and multiangle light scattering data from various batches of ID93 manufactured in our lab (data not shown).
Figure 2.
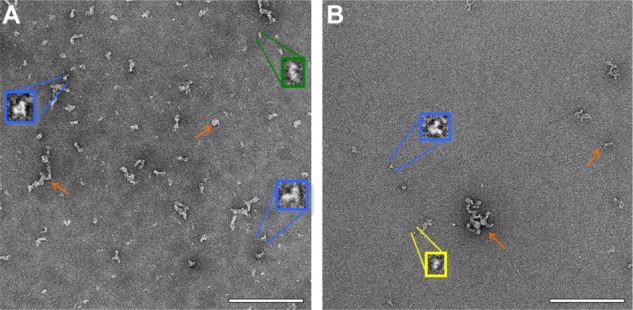
Negative stain TEM of ID93. Negative stain TEM images of ID93 in 20 mM Tris buffer at different protein concentrations indicate the presence of various structures, including small linear particles (yellow), medium linear particles (green), tripod-shaped particles (blue), and particle clusters (orange arrows). The samples were imaged at 52,000× magnification with insets shown at larger scale. (A) 0.5 mg/mL ID93. (B) 0.05 mg/mL ID93. Scale bar =200 nm.
Abbreviation: TEM, transmission electron microscopy.
Cryo-TEM of ID93
Protein particles apparent in unstained cryo-TEM images were mostly consistent with the negative stain TEM images, and included small linear particles of ~7 nm length, medium linear particles of ~15 nm length, tripod-shaped particles of ~10 nm length, and very small particles that appear as black dots (Figure 3). These are likely an end-on view of the linear particles; a long particle viewed end on will appear much denser than a long particle viewed edge on. Also apparent were clusters of particles that had lengths typically in the range of 25–50 nm or were very large and had a mat-like appearance. The very large mat-like clusters were not evident in the negative stain preparation.
Figure 3.
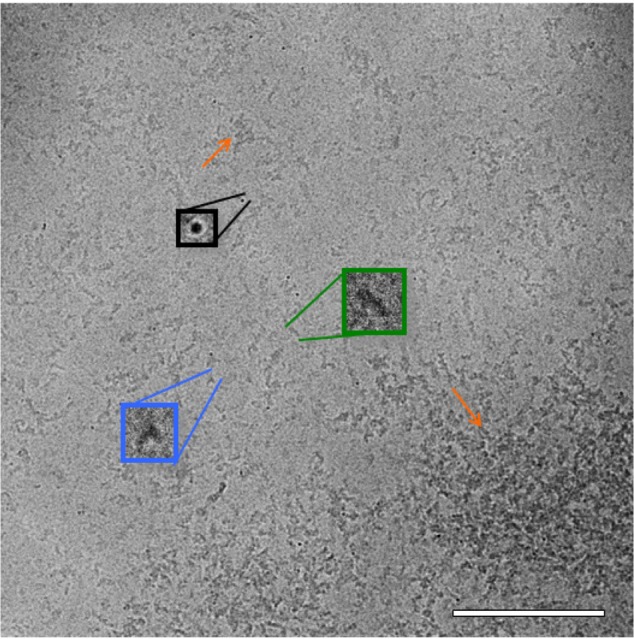
Cryo-TEM of ID93. Cryo-TEM image of ID93 (0.05 mg/mL) in 20 mM Tris buffer indicates the presence of very small particles appearing as black dots (black), small and medium length linear particles (green), tripod-shaped particles (blue), and various sizes of particle clusters (orange arrows). The sample was imaged at 52,000× magnification. Scale bar =200 nm.
Abbreviation: Cryo-TEM, cryogenic transmission electron microscopy.
Cryo-TEM of liposomes
Liposomes primarily appeared as unilamellar spherical vesicles consisting of a lipid bilayer (higher density bands) surrounding an aqueous core (same density as exterior aqueous phase; Figure 4). Liposome diameter was typically in the range of 45–100 nm, which is consistent with the Z-ave diameter detected by DLS (Figure 1), although some liposomes were found with diameters of 150–250 nm. Some multilamellar liposomes were also present, as well as rod-like particles and very small particles. Rod-like particles are lipid disk-shaped structures common in liposomal preparations, viewed edge-on.13
Figure 4.
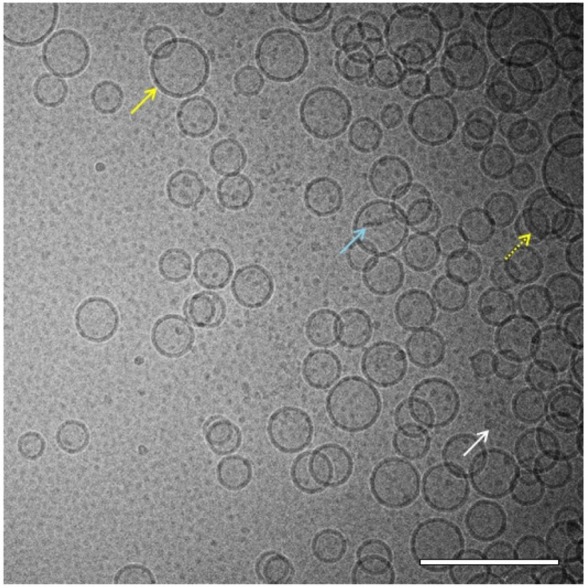
Cryo-TEM of liposomes. Cryo-TEM image of liposomes (8 mg/mL phospholipid) in PBS reveals mostly spherical unilamellar vesicles in the size range of ~45–100 nm (solid yellow arrow) although some multilamellar vesicles (dashed yellow arrow), rod-like particles (light blue arrow), and very small micelle particles (white arrow) were also present. The sample was imaged at 52,000× magnification. Scale bar =200 nm.
Abbreviations: Cryo-TEM, cryogenic transmission electron microscopy; PBS, phosphate-buffered saline.
Cryo-TEM of ID93 mixed with liposomes
As with the liposome sample, spherical unilamellar and some multilamellar liposomes were present in the ID93–liposome mixture (Figure 5). Likewise, various types of protein particles as seen in the ID93 sample were also evident. The liposomes did not appear to be uniformly coated with the particles found in the ID93 sample, and much of the protein remained unassociated with liposomes. However, some of the spherical liposomes show the appearance of black dots similar to those seen in the ID93 sample, indicating that the protein is possibly embedded into the lipid bilayer (Figure 6). Other structures observed in the mixture, but not seen in either of the individual samples include pairs of rod-like particles not contained in liposomes and faint round particles, both of which may have been decorated or surrounded by proteinaceous material. The rod-like particles often occurred in pairs in very close proximity, thus differing from the single rod-like particles noted in the liposome control sample. These rod-like paired structures also appear to be associated with black dots similar to those observed in the ID93 sample, possibly indicating protein embedded into their bilayers. Image analysis of 701 particles revealed 631 regular spherical liposomes and 70 rod-like pairs, representing 10% of the population.
Figure 5.
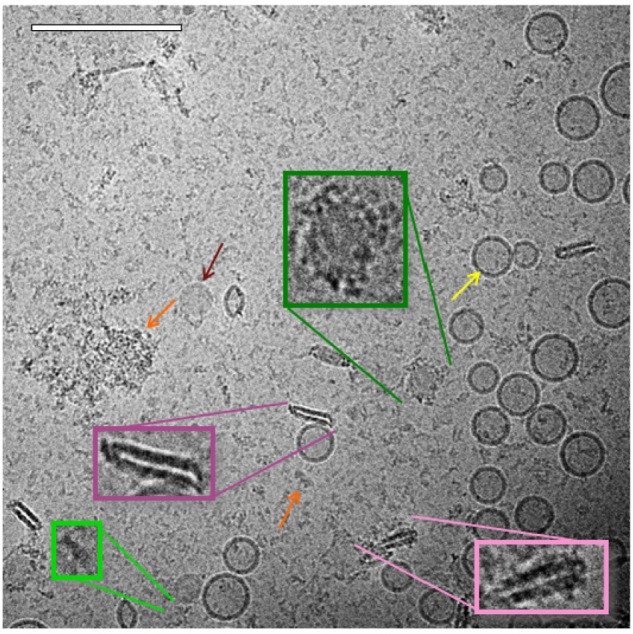
Cryo-TEM of ID93–liposomes. Cryo-TEM image of ID93–liposome mixture (0.25 mg/mL ID93, 4 mg/mL phospholipid) reveals many of the same liposome and protein particles present in the individual samples such as spherical unilamellar liposomes (yellow arrow), linear protein particles (light green), and clustered protein particles (orange arrow). Some of the spherical liposome bilayers contained black dots, possibly indicating embedded protein. Other novel structures included pairs of rod-like particles (pink), some of which were decorated by black dots or other proteinaceous structures (light pink), and faint round particles (maroon arrow) which also sometimes appeared decorated with protein (dark green). The sample was imaged at 52,000× magnification. Scale bar =200 nm.
Abbreviation: Cryo-TEM, cryogenic transmission electron microscopy.
Figure 6.
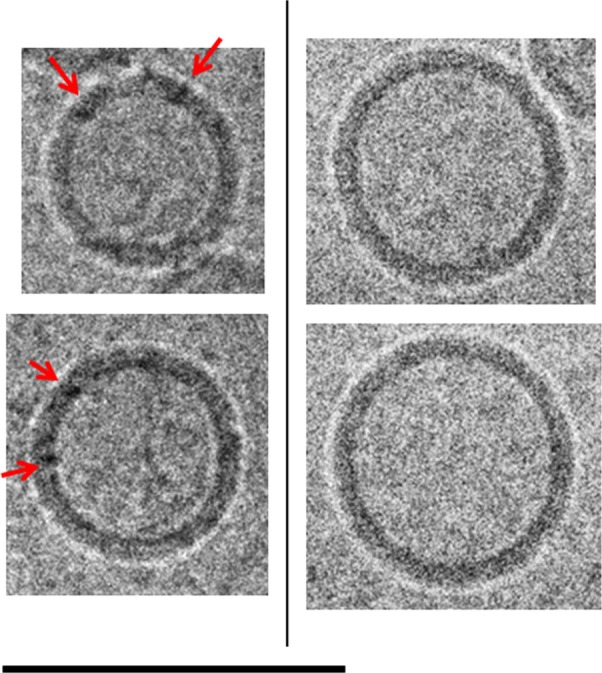
Cryo-TEM comparison of ID93–liposomes and liposomes. Cryo-TEM images of selected liposomes of ID93–liposome mixture (0.25 mg/mL ID93, 4 mg/mL phospholipid) on the left and liposomes (8 mg/mL phospholipid) in PBS on the right. Spherical unilamellar liposomes in the mixture appear to show features (black dots) in the bilayer that are consistent with the appearance expected for the embedded protein (red arrows). These features are not seen in liposomes in PBS. The sample was imaged at 52,000× magnification. Scale bar =100 nm.
Abbreviations: Cryo-TEM, cryogenic transmission electron microscopy; PBS, phosphate-buffered saline.
Tomography
In order to gain a better understanding of the morphology of the novel structures evident in the ID93–liposome mixture, we used electron tomography to obtain a 3D reconstruction of the sample. In an electron microscope the image that is formed of the specimen on the grid is a two-dimensional projection of the 3D volume. The 3D volume can be reconstructed from these projections using tomographic methods. In these methods, a series of projection images of the same sample area are acquired at incremental tilt angles relative to the electron beam. The tilted set of images can then be mathematically combined to reconstruct a 3D volume of the sample. A 3D tomographic reconstruction of the combined ID93 and liposomes sample appears to show that both the faint round particles and the pairs of rod-like particles are the same type of particle viewed from different angles and are flat disk-shaped liposomes (pairs of lipid disks or flattened liposomes) decorated by proteinaceous material that appears consistent with the material in the ID93 sample (Figure 7 and Video S1).
Figure 7.
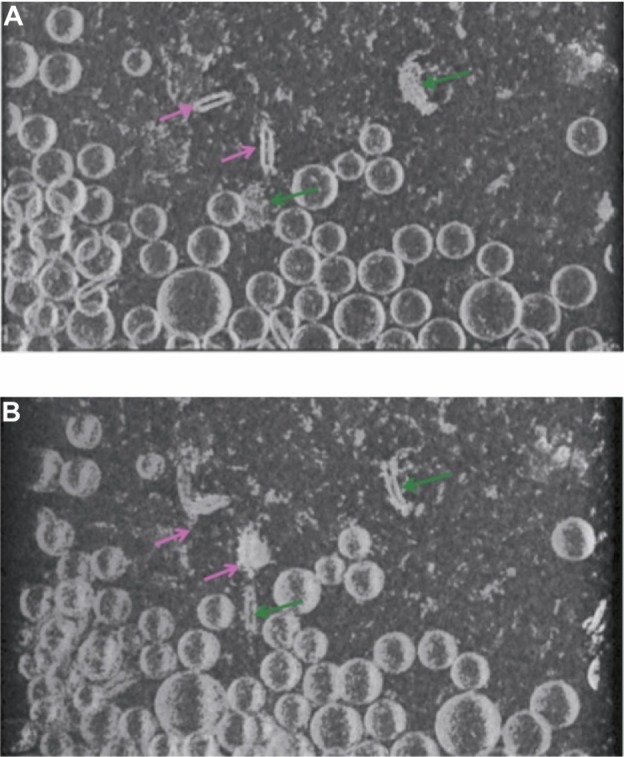
Cryo-TEM tomography. Views at different angles of the same rendered volume reveals pairs of rod-like particles in close proximity decorated with proteinaceous material (light pink arrow) and faint round particles decorated with proteinaceous material (dark green arrow). The rod-like pairs and the faint round particles in (A) appear as faint round particles and rod-like particles, respectively, in (B) due to the different viewing angle, but both are actually the same type of flattened liposome particle.
Abbreviation: Cryo-TEM, cryogenic transmission electron microscopy.
The association of ID93 with anionic liposomes in a buffer with pH well above the pI of the protein is not expected based on overall electrostatic considerations alone. However, association (detected by ultracentrifugation) between anionic liposomes and anionic protein has been reported previously.14 ID93 is a large fusion protein composed of 62% molar non-polar residues and 5% molar cationic residues.15 Moreover, spectroscopic data on ID93 indicate that it has little tertiary structure (data not shown), which could result in exposed hydrophobic residues. Indeed, ID93 has low solubility below pH 7. Therefore, association between liposomes and protein could be explained by hydrophobic interactions between lipid acyl chains and exposed hydrophobic subdomains on the protein. Moreover, electrostatic screening by buffer salts could help increase the relevance of hydrophobic interactions. Consistent with this reasoning, Figures 5 and 6 indicate that the protein localizes with the hydrophobic core of the lipid bilayer.
Light-scattering-based particle sizing
We analyzed ID93 liposome mixtures by light-scattering-based methods to determine if they could complement or confirm the results obtained by cryo-TEM. Specifically, we employed two distinct methods: DLS and NTA. A comparison of these methods has been published,16 indicating that NTA is more applicable to characterizing heterogeneous size populations, whereas DLS has greater sensitivity for particle sizes <30 nm.
DLS characterization of ID93 diluted in 20 mM Tris buffer indicated a somewhat heterogeneous distribution of size (larger PDI compared to the liposomes), with Z-ave of 37.8 nm (Figure 8), which is consistent with cryo-TEM analysis. It is important to note, however, that Z-ave values obtained by DLS are weighted to light intensity and therefore the signal from larger particles (which scatter significantly more light due to the 106 proportionality factor of scattering intensity to particle diameter) will be more heavily represented in the Z-ave value than smaller particles. Therefore smaller protein particle signal may be dominated by signal from larger aggregate particles. DLS analysis of liposomes diluted in PBS revealed a mostly homogeneous distribution of 78.8 nm particles consistent with the cryo-TEM analysis (Figure 8). DLS characterization of the ID93–liposome mixture did not differ significantly from that of the liposomes alone (Z-ave 77.6 nm), likely due to the much larger number of liposome particles compared to protein particles. Moreover, no indication of a separate population of particles such as disks was evident, which is not surprising since it is unlikely that DLS could detect a small population of disks of similar diameter as a larger population of spheres since the estimated hydrodynamic diameter of a disk with linear (length-wise) diameter of 75 nm is Dh=62 nm,17 assuming bilayer thickness of 5 nm18 and disk thickness of 15 nm.
Figure 8.
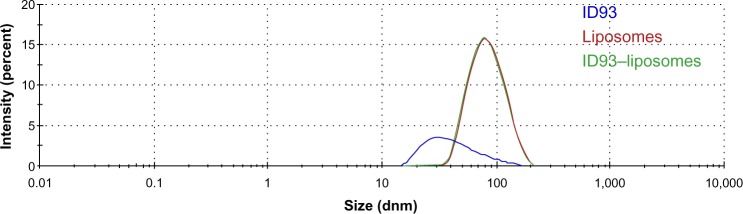
Dynamic light scattering analysis. Representative intensity-based size distributions of ID93 (0.05 mg/mL) in 20 nM Tris buffer, liposomes (4 mg/mL phospholipid in PBS), and ID93–liposome mixture (0.25 mg/mL protein, 4 mg/mL phospholipid). These are the same batches of material employed in the electron microscopy studies.
Abbreviation: PBS, phosphate-buffered saline.
NTA is more suitable to characterize heterogeneous size populations and particles of different shape, as well as to provide particle concentrations. Furthermore, in the case of monodisperse particle size populations, NTA data corresponds closely to DLS data.16,19 Since NTA collects images of individual diffusing particles, it is possible to detect non-spherical shapes due to asymmetric flashing (Video S2). However, since the solution of ID93 contained non-spherical protein aggregates, it was not possible to distinguish these from the presence of flattened liposomes once ID93 was mixed with liposomes. NTA in this case was not able to specifically reveal the presence of the disk-shaped lipid-protein particles, which were possibly masked by the presence of non-spherical particles in both the protein and liposome control samples (Figure 9 and Video S2). It is interesting to note that the liposome size mean and especially the ID93 size mean detected by NTA were larger than that detected by DLS, indicating that both the liposomes and the ID93 solution contained small particles (<~30 nm) that were not detectable by NTA, although one cannot be certain of this conclusion since NTA measurements were conducted at more dilute sample concentrations than the DLS measurements. Another interesting result is found in the particle concentrations detected by NTA, a feature not available in DLS systems: the particle concentration of the ID93–liposome mixture was approximately the sum of the particle concentrations of the protein alone and the liposomes alone, indicating that most of the protein particles may not associate with the liposomes.
Figure 9.
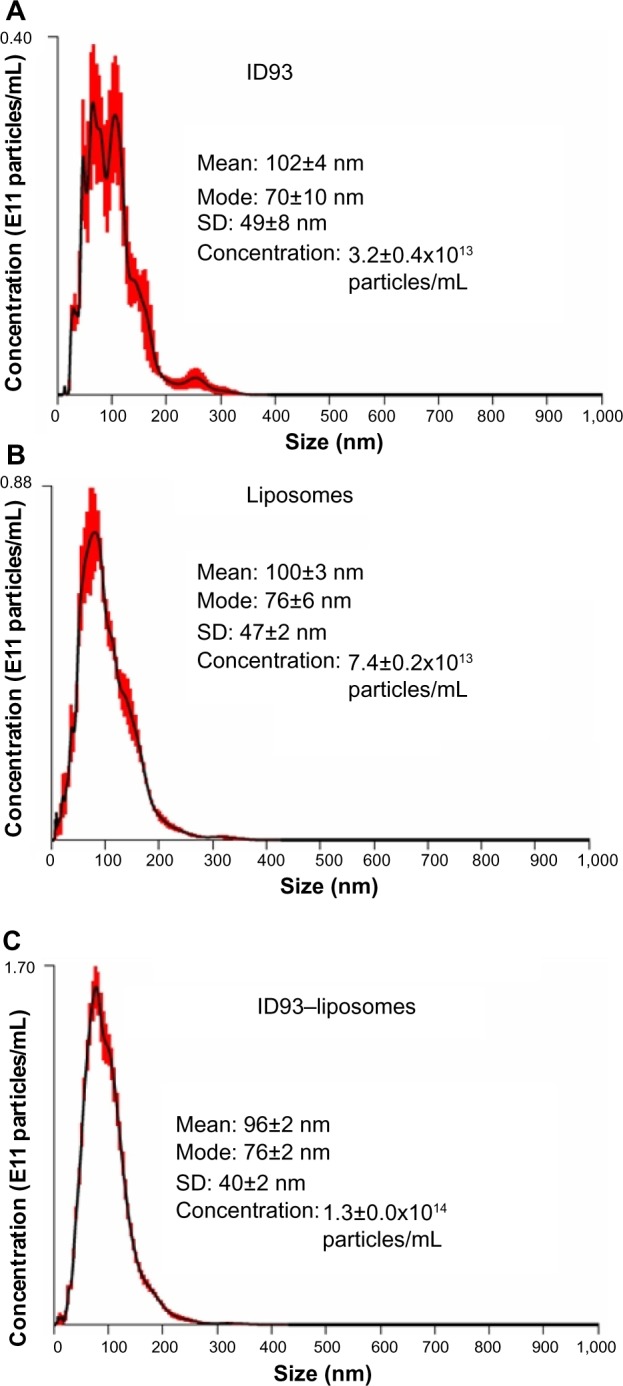
Nanoparticle tracking analysis. Nanoparticle tracking analysis of (A) ID93 in 20 mM Tris buffer, (B) liposomes in PBS, and (C) ID93–liposome mixture. These are the same batches of material employed in the electron microscopy studies.
Abbreviation: SD, standard deviation.
In this work, we employed cryo-TEM to identify morphological interactions between a recombinant vaccine antigen (ID93) and an adjuvant formulation platform (anionic liposomes). In an earlier related study, we found that the anionic liposomes did not strongly associate with the protein as detected by SEC and SDS-PAGE.7 Nevertheless, anionic liposomes proved to be a potent adjuvant formulation platform when combined with a TLR4 agonist and the vaccine antigen. While some reports conclude that electrostatic or covalent antigen association with liposomes enhances vaccine potency,20–22 other researchers have reported that simple mixtures of anionic liposomes with anionic vaccine antigens result in potent antibody and cellular immune responses, where significant antigen–liposome association was evident even though both components were anionic.14 It is possible that more subtle interactions between liposomes and proteins of the same charge are occurring but not detectable by traditional methods involving SEC or ultracentrifugation.
Through cryo-TEM analysis, we have identified interactions between anionic protein and anionic liposomes resulting in protein–lipid disk formation. Protein–lipid disks or PEGylated lipid mixtures have been reported in the literature, with disk formation likely attributable to multiple factors including lipid–protein packing behavior, surface curvature effects, lipid phase structure, and charge.17,23,24 However, such disks consist of a single bilayer and so differ from those detected in the present work, which may more appropriately be termed flattened liposomes since they apparently consist of pairs of opposing bilayer disks. These flattened liposome structures were distinguishable by employing cryo-TEM analysis but not DLS or NTA. Flattened liposomes have been reported earlier including for photo-oxidized liposomes25 or liposomes adsorbed to a surface.26 In these studies, the formation of flattened liposomes was related to lipid raft formation25 or electrostatic surface adsorption interactions.26 However, the present work is the first report to our knowledge of flattened liposomes involving a lipid-based vaccine formulation. Since the liposomes included multiple components it is possible that upon interaction with protein new domains are formed, with disk edges comprised of components more favorable for high curvature conditions, analogous to the system proposed by Yuan et al.25 Indeed, many of the flattened liposomes appear to have disconnected lipid bilayers at one or both of the edges, with higher protein content than lipid content in the edge regions. Lipid domain formation could plausibly help reduce repulsive electrostatic interactions between like-charged DPPG and protein components. It is possible that there are also effects related to the different buffer systems employed for the liposomes and the protein.
An important remaining question to be investigated is whether the protein–lipid disks are more biologically active compared to the spherical liposomes containing relatively less associated protein. It is quite plausible that the shape of the disks as well as the density of associated protein could alter their interactions with and uptake by antigen-presenting cells and corresponding adjuvant activity. Indeed, particle shape has been shown to influence uptake of microparticles by antigen-presenting cells.27,28 For instance, Sharma et al found that phagocytosis of polymeric microparticles (with volumes in the range of 0.075 to 0.69 μm3) by macrophages was enhanced for oblate ellipsoids compared to prolate ellipsoids or spheres.28 Moreover, increased protein densities, which are apparent on the flattened liposomes, could alter cell uptake and immunogenicity due to multimeric antigen presentation.29 Also it remains to be determined whether the presence of TLR agonists alters the formation of the flattened liposomes. Finally, the effects of different buffers and other common vaccine excipients on lipid structures should be evaluated. Such studies will help elucidate further the relevance of the findings reported here. The unique capabilities of cryo-TEM, compared to light scattering methods, in the analysis of lipid-vesicle-based vaccine formulations have been demonstrated in this work. Cryo-TEM is a unique, informative analytical method that should be considered for vaccine formulation systems in order to elucidate interactions between vaccine antigens and formulation components.
Supplementary materials
Zeta potential measurement at different pH values. Two separate autotitrations were performed to measure zeta potential versus pH of ID93. Three zeta potential measurements were obtained at each pH value. Isoelectric point of ID93 determined by a third-degree polynomial fit was determined to be 4.7 (A) or 4.8 (B).
Cryo-TEM tomography of ID93–liposomes.
Abbreviation: Cryo-TEM, cryogenic transmission electron microscopy.
NTA of ID93, liposomes, and ID93–liposome mixture. (A) NTA of ID93; presence of highly aggregated protein or contaminant particles. Asymmetric flashing suggests non-spherical shape. (B) NTA of liposomes; mostly small, low scattering intensity spherical particles with a few high intensity scattering particles. (C) NTA of ID93–liposomes; qualitatively more high intensity scattering particles. Asymmetric flashing suggests non-spherical particles. Flashing particles could be changes associated with protein–liposome interactions or could be attributed to protein aggregates.
Abbreviation: NTA, nanoparticle tracking analysis.
Acknowledgments
The authors thank Lucien Barnes V, Tony Phan, Mark Orr, Traci Mikasa, Susan Lin, Sandra Sivananthan, and Ryan Kramer for helpful discussions and excellent technical assistance. This project has been funded in part with funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No HHSN272200800045C.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bertholet S, Ireton GC, Ordway DJ, et al. A defined tuberculosis vaccine candidate boosts BCG and protects against multidrug-resistant Myco-bacterium tuberculosis. Sci Transl Med. 2010;2(53):53ra74. doi: 10.1126/scitranslmed.3001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coler RN, Bertholet S, Pine SO, et al. Therapeutic immunization against Mycobacterium tuberculosis is an effective adjunct to antibiotic treatment. J Infect Dis. 2013;207(8):1242–1252. doi: 10.1093/infdis/jis425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baldwin SL, Bertholet S, Reese VA, Ching LK, Reed SG, Coler RN. The importance of adjuvant formulation in the development of a tuberculosis vaccine. J Immunol. 2012;188(5):2189–2197. doi: 10.4049/jimmunol.1102696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Infectious Disease Research Institute Phase 1 ID93 + GLA-SE Vaccine Trial in BCG-Vaccinated Healthy Adult Volunteers. [Accessed December 9, 2013]. Available from: http://clinicaltrials.gov/ct2/show/NCT01927159?term=ID93+GLA-SE&rank=2. NLM identifier: NCT01927159.
- 5.Infectious Disease Research Institute Phase 1 ID93 + GLA-SE Vaccine Trial in Healthy Adult Volunteers. [Accessed December 9, 2013]. Available from: http://clinicaltrials.gov/ct2/show/NCT01599897?term=ID93+GLA-SE&rank=1. NLM identifier: NCT01599897.
- 6.Owusu-Agyei S, Ansong D, Asante K, et al. Randomized controlled trial of RTS, S/AS02D and RTS, S/AS01E malaria candidate vaccines given according to different schedules in Ghanaian children. PLoS ONE. 2009;4(10):e7302. doi: 10.1371/journal.pone.0007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orr MT, Fox CB, Baldwin SL, et al. Adjuvant formulation structure and composition is critical for the development of an effective vaccine against tuberculosis. J Control Rel. 2013;172(1):190–200. doi: 10.1016/j.jconrel.2013.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuntsche J, Horst JC, Bunjes H. Cryogenic transmission electron microscopy (cryo-TEM) for studying the morphology of colloidal drug delivery systems. Int J Pharm. 2011;417(1–2):120–137. doi: 10.1016/j.ijpharm.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Suloway C, Pulokas J, Fellmann D, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151(1):41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120(3):343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 11.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 12.Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38(50):16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 13.Almgren M, Edwards K, Karlsson G. Cryo transmission electron microscopy of liposomes and related structures. Colloids Surf A Physicochem Eng Asp. 2000;174(1–2):3–21. [Google Scholar]
- 14.Yanasarn N, Sloat BR, Cui Z. Negatively charged liposomes show potent adjuvant activity when simply admixed with protein antigens. Mol Pharm. 2011;8(4):1174–1185. doi: 10.1021/mp200016d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice P, Longden I, Bleasby A. EMBOSS: The European molecular biology open software suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 16.Filipe V, Hawe A, Jiskoot W. Critical evaluation of nanoparticle tracking analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnsson M, Edwards K. Liposomes, disks, and spherical micelles: aggregate structure in mixtures of gel phase phosphatidylcholines and poly(ethylene glycol)-phospholipids. Biophys J. 2003;85(6):3839–3847. doi: 10.1016/S0006-3495(03)74798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonenko ZV, Finot E, Ma H, Dahms TES, Cramb DT. Investigation of temperature-induced phase transitions in DOPC and DPPC phospholipid bilayers using temperature-controlled scanning force microscopy. Biophys J. 2004;86(6):3783–3793. doi: 10.1529/biophysj.103.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misquith A, Fung M, Dowling QM, Guderian JA, Vedvick TS, Fox CB. In vitro evaluation of TLR4 agonist activity: formulation effects. Coll Surf B Biointerfaces. 2014;113:312–319. doi: 10.1016/j.colsurfb.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henriksen-Lacey M, Christensen D, Bramwell VW, et al. Liposomal cationic charge and antigen adsorption are important properties for the efficient deposition of antigen at the injection site and ability of the vaccine to induce CMI response. J Control Rel. 2010;145(2):102–108. doi: 10.1016/j.jconrel.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 21.Shahum E, Thérien HM. Immunopotentiation of the humoral response by liposomes: encapsulation versus covalent linkage. Immunology. 1988;65(2):315–317. [PMC free article] [PubMed] [Google Scholar]
- 22.Watson DS, Endsley AN, Huang L. Design considerations for liposomal vaccines: influence of formulation parameters on antibody and cell-mediated immune responses to liposome associated antigens. Vaccine. 2012;30(13):2256–2272. doi: 10.1016/j.vaccine.2012.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584(9):1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wessman P, Edwards K, Mahlin D. Structural effects caused by spray- and freeze-drying of liposomes and bilayer disks. J Pharm Sci. 2010;99(4):2032–2048. doi: 10.1002/jps.21972. [DOI] [PubMed] [Google Scholar]
- 25.Yuan J, Hira SM, Strouse GF, Hirst LS. Lipid bilayer discs and banded tubules: photoinduced lipid sorting in ternary mixtures. J Am Chem Soc. 2008;130(6):2067–2072. doi: 10.1021/ja710305c. [DOI] [PubMed] [Google Scholar]
- 26.Goksu EI, Vanegas JM, Blanchette CD, Lin WC, Longo ML. AFM for structure and dynamics of biomembranes. Biochim Biophys Acta. 2009;1788(1):254–266. doi: 10.1016/j.bbamem.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 27.Champion JA, Mitragotri S. Role of target geometry in phagocytosis. Proc Natl Acad Sci U S A. 2006;103(13):4930–4934. doi: 10.1073/pnas.0600997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma G, Valenta DT, Altman Y, et al. Polymer particle shape independently influences binding and internalization by macrophages. J Control Release. 2010;147(3):408–412. doi: 10.1016/j.jconrel.2010.07.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox CB, Kramer RM, Barnes VL, Dowling QM, Vedvick TS. Working together: interactions between vaccine antigens and adjuvants. Ther Adv Vaccines. 2013;1(1):7–20. doi: 10.1177/2051013613480144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Zeta potential measurement at different pH values. Two separate autotitrations were performed to measure zeta potential versus pH of ID93. Three zeta potential measurements were obtained at each pH value. Isoelectric point of ID93 determined by a third-degree polynomial fit was determined to be 4.7 (A) or 4.8 (B).
Cryo-TEM tomography of ID93–liposomes.
Abbreviation: Cryo-TEM, cryogenic transmission electron microscopy.
NTA of ID93, liposomes, and ID93–liposome mixture. (A) NTA of ID93; presence of highly aggregated protein or contaminant particles. Asymmetric flashing suggests non-spherical shape. (B) NTA of liposomes; mostly small, low scattering intensity spherical particles with a few high intensity scattering particles. (C) NTA of ID93–liposomes; qualitatively more high intensity scattering particles. Asymmetric flashing suggests non-spherical particles. Flashing particles could be changes associated with protein–liposome interactions or could be attributed to protein aggregates.
Abbreviation: NTA, nanoparticle tracking analysis.