Abstract
Regulation of transcription in eukaryotes is considered in the light of recent findings demonstrating the presence of negative and positive superhelical tension in chromatin. This tension induces conformational transitions in DNA duplex. Particularly, the transition into A-form renders DNA accessible and waylaying for initiation of transcription producing RNA molecules long known to belong to the A-conformation. Competition between conformational transitions in various DNA sequences for the energy of elastic spring opens a possibility for understanding of fine tuning of transcription at a distance.
Keywords: DNA conformation, nucleosomes, transcription, RNA-polymerase, topoisomerase II
Background
Until recently, eukaryotic genome in its major part was considered to be devoid of torsional tension because of its association with nucleosomes. However, recent research revealed that considerable portion of human and yeast genomes possesses either positive or negative torsional tension.1,2 Moreover, those loops (topological domains) posed for transcription may change their sign of elastic superhelicity from negative to positive and vice versa in response to RNA-polymerase inhibition1 or shift to non-permissive temperature in topoisomerase II mutants.2
One nick per DNA loop is sufficient to abolish transcription in various species and various sized loops as evidenced from X-ray inhibition of transcription.3 The dose–response curves of transcription inactivation were found to correspond to an estimated X-ray target size of DNA loops. The transcription abrogated at the stage of initiation as was determined in run-on experiments performed on nuclei isolated from irradiated cells and tested for transcription in the presence of ammonium sulfate or sarkosyl. So the transcription of all the genes within transcriptionally active loop is affected by torsional tension.
This inference is supported by the fact that the majority of DNA sequences influencing transcription from any given transcription start site (TSS) are located within about 100 kb of neighboring DNA and rarely exceed this figure.4
Trying to understand the fact of interaction between DNA sequences competing for energy of elastic spring of DNA within any given loop, we first consider in the beginning of this article the situation with naked DNA rings and loops and then proceed to look at chromatin.
Notion on DNA and RNA conformations
Conformations of DNA duplex (double helix) are known to belong to several forms namely A, B, C, Z, and some others. A, C, and Z forms are known as crystalline forms with rather regular parameters of their double helices if only sufficient amount of positively charged counterions are present while B-form is known as semicrystalline or paracrystalline, that is why it exhibits high level of variation in the configuration of sugar-phosphate backbone. The arising of one or the other conformation depends on DNA sequence, counterions, and the level of hydration around DNA molecule. For example, certain amino acids and histone-like peptides provoke formation of C-form.5 Also some nearest neighboring nucleotides provoke the formation of A-form in certain sequences.6,7
Conformation of DNA duplex is also strongly dependent on torsion stress either negative or positive imposed on various DNA sequences (see below).
DNA–RNA hybrids as well as DNA–RNA triplexes always possess the A-conformation. The notable difference between A and B forms is their twist or the number of double helical turns per a given number of base pairs. Another most important feature of A-DNA is its “minor” groove width that becomes extremely wide (~8 Å) in comparison to B and C forms (~ 4 Å). This trait of A-form DNA divides all right-handed DNA conformations into two distinct alternative groups (see below).
The twist of DNA in the A-form is almost invariable and equals to pitch of 11 base pairs per turn or about 33° per base pair. The twist of DNA in B-conformation varies significantly depending on particular sequence of bases between 33 and 39° per base pair and a pitch of 9.0–11.0 base pairs per turn in water solutions. Now this variation in twist is explained by coexistence of B, C, and A conformations in one DNA molecule containing various nucleotide sequences.8–10
The “minor” groove width of A-form DNA allows to place into it the third polynucleotide chain such as RNA, thus allowing the formation of transient polynucleotide triplexes in the course of transcription.
Evidence
Conformational transitions in DNA rings
The first observations of conformational transitions in torsionally strained DNA were published by Wang et al.11,12 who employed novel method of two-dimensional electrophoresis of DNA topoisomers. They found that Z-form in (GC)16 sequence and cruciforms in palindrome sequences may arise in DNA in the result of negative torsional stress. The analysis of two-dimensional gels also revealed low-angular conformational transition recognized as A-form DNA in tensioned molecules.13 Further evidence reviewed below supported and substantiated this early analysis and inference. The arising of A-form in negatively tensioned DNA is depicted schematically in Figure 1 and in the legend of this figure.
Figure 1.
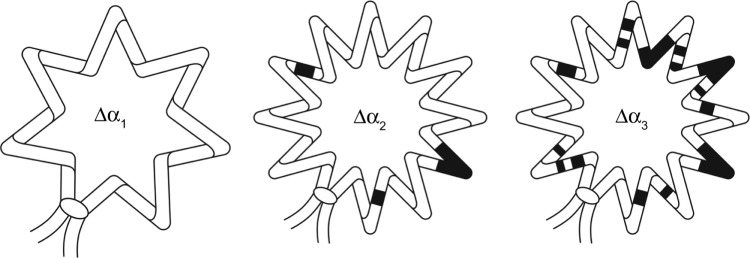
Schematic representation of B → A conformational transition in a closed superhelical DNA loop or ring with an increasing value of topological linking number difference (Δα1 < Δα2 < Δα3). DNA topological turns (topological linking number difference), which is the same as “titratable superhelical turns,” are partitioned between turns of “elastic spring” (shown as zigzag line) and turns spent for B → A (or any other) conformational transition. This is why the number of “zigzags” are the same (identical) in molecules where Δα2 < Δα3 – because all the supernumerary turns are spent to conformational transitions in DNA secondary structure (structure of DNA duplex or “double helix”). Black segments represent DNA segments having undergone conformational transition in DNA duplex structure into an underwound conformation (here an A-form, discussed in the present article). For the definition of Δα, see Ref. 36.
The coexistence of A, Z, and cruciform conformations and their competition for elastic turns and energy was analyzed at the quantitative level using original statistical–mechanical model of conformational transitions in closed DNA molecules.14 It was found that rather short GC or palindrome sequences may prevent the formation of A-form among thousands of base pairs but may not destroy all the A-form tracts.13
The analysis of electrophoretic behavior of superhelical DNA was extended into studies of sedimentation behavior of superhelical DNA rings and nucleoids containing bare superhelical DNA loops described in the next section.
Conformational transitions in superhelical DNA loops (nucleoids)
The density of DNA topological turns can be measured using nucleoids—structures containing superhelical loops of nuclear DNA held in place by some nonhistone proteins covalently linking the DNA to chromosome skeleton structures.
The method is based on gradual unwinding of negatively coiled superhelical DNA loops with increasing concentration of the intercalating drug ethidium bromide (EtdBr), which after reaching a certain critical concentration introduces positive superhelical turns into the DNA loops. These changes in the tertiary configuration of DNA affect the sedimentation rate of nucleoids. The concentration of EtdBr at which the sedimentation rate is minimal corresponds to complete DNA relaxation and may be used for calculation of the original density of DNA topological turns (“titratable superhelical turns”).
It is critically important in this connection to mention that the most “obstinate” DNA sequences go apart and abandon histones only at plateau levels of NaCl concentrations above 1.6–1.9 M NaCl.15 That is why measurements were performed in 15–30% of sucrose gradients containing 1.95 M NaCl and various concentrations of EtdBr.16
Nucleoids from various types of vertebrate cells always exhibited one and the same property of sedimentation behavior giving direct evidence for conformational transitions of DNA into A-form in negatively supercoiled loops and into C-form in positively supercoiled loops if only the sedimentation was performed in high salt conditions replacing electrostatic force of DNA–histone interaction in nucleosomes.
Studies with differentiating cells
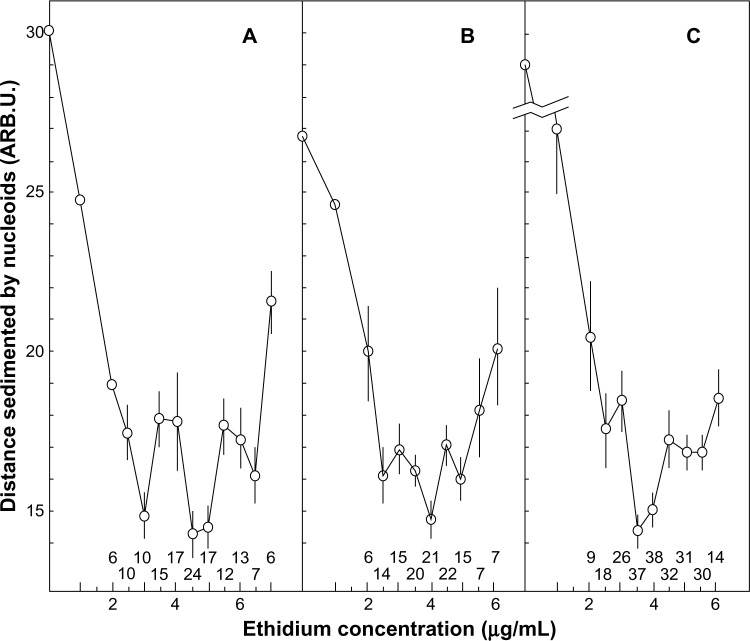
Primary cultures of vertebrate cells grown in vitro undergo gradual decrease in the doubling rate during serial passage. This phenomenon often referred to as in vitro cell aging correlates with the life span of species whose cells are being cultured and thus is considered to be the manifestation of organism senescence at the cellular level. Accumulated evidence suggests that in vitro cell aging is associated with progressive terminal differentiation of cells during serial passage. It was found that this phenomenon was accompanied by the decrease in the density of topological turns in nuclear superhelical DNA loops from Syrian hamster primary fibroblasts cells.16 The summarized results are presented in Figure 2.
Figure 2.
Conformational transitions in DNA accompanying changes in the level of positive or negative superhelical tension during EtdBr unwinding of DNA in 1.95 M NaCl (see text for the details). Transitions are obvious in “jumps” in sedimentation rates of superhelical DNA (“nucleoids”) from Syrian hamster primary fibroblasts. (A) Cells of 1st passage, (B) cells of 5th to 6th passage, and (C) cells of 11th to 12th passage (differentiated). Vertical bars represent standard errors. Figures below the graphs indicate the number of repeated measurements per point.
All curves exhibited three minima, the deepest one always being the central one. These three minima were not the result of heterogeneity of nucleoids but the intrinsic property of superhelical DNA found in homogeneous viral DNA preparations studied in high salt.17,18
This shape of curves could only be explained by conformational transitions in secondary structure in closed superhelical DNA molecules evident under high salt conditions in sedimentation experiments. The central minimum was always a point of transition from negative to positive DNA superhelix during EtdBr titration. The left minimum obviously demarcates the beginning of A- to B-form transition in DNA secondary structure during decrease of negative elastic torsional strain on increasing EtdBr concentration. The first maximum represents the end of A → B transition. The right minimum demarcates the end of B- to C-form transition (which begins in preceding maximum) in DNA secondary structure during increase of positive elastic torsional strain upon further increase in EtdBr concentration. The right and left minima were also evident in experiments with small circular DNAs homogeneous in size.
The main (central) minimum on the curve for non-differentiated cells of first passage always lies at 4.5–5.0 μg/mL of EtdBr, which corresponded to the density of −0.076 topological turns/10 bp of DNA. The main minimum on the curves for differentiated cells was shifted to the left, as compared with the curve for non-differentiated cells. It lies at 3.5 μg/mL of EtdBr, which corresponded to −0.062 topological turns/10 bp of DNA (Fig. 2). The DNA loops from cells of intermediate passage (panel B) exhibited rather shallow minima obviously connected with coexistence (mixture) of two types of cells within a population that had begun terminal differentiation.
The observed shift in overall DNA supercoiling during in vitro differentiation of Syrian hamster cells seems to be a universal phenomenon also found in other terminally differentiating cell types as it follows from experiments with Friend erythroleukemia (murine) cells.
The data on the densities of DNA topological turns in different cell types are summarized in Ref. 19. Taken together, these data may imply that the total change in the densities of topological turns in closed superhelical nuclear DNA loops in different cells during modulation of cellular proliferation and differentiation may be associated with the coordinate switch in transcription of many genes involved in genetic control of these processes (house-keeping genes).
Also the studies of nucleoids provide clear evidence for conformational transitions of DNA into A and C forms induced by either negative or positive torsional strain because no other low-angular cooperative conformational transitions are known in modern scientific knowledge. It is worth noting that the shape of curves was independent of individual loop size because the density of DNA topological turns is solely determined by nucleosome density and overall histone modifications in a given cell type (differentiating lineage).
The transition into A-form occurred at level of negative supercoiling far below the natural one. The transition into C-form also occurred at moderate levels of positive supercoiling.
Studies of transcriptionally active chromatin rings
Using SV40 model, it was found that a small part of SV40 chromatin extracted from infected cells unlike the bulk of extracted SV40 chromatin was transcriptionally active and also possessed an elastic torsional strain in DNA as was revealed after treatment of isolated minichromosomes with topoisomerase I. All minichromosomes extracted in 0.4 M NaCl contained full complement of core histones, but upon treatment with topoisomerase I the relaxing transcriptionally active fraction lost histones but retained subunits of RNA-polymerase capable to transcribe SV40 DNA in vitro in run-on experiments.20 This transcriptionally active fraction was also hypersensitive to DNase I in isolated nuclei—the feature intrinsic to transcriptionally active chromatin.21
Irradiation of nuclei from SV40-infected cells with X-rays to relax torsionally strained DNA because of a random nicking decreased DNase I hypersensitivity of SV40 minichromosomes. But some hypersensitivity still remained in intranuclear SV 40 minichromosomes.22 This fact got its explanation after extraction of minichromosomes in a low salt (0.1 M NaCl instead of 0.4 M NaCl)—the conditions not allowing the extraction of transcriptionally active fraction. These minichromosomes also exhibited DNase I hypersensitivity mapped in the same control region of SV40 minichromosome. It was concluded that DNase I hypersensitivity had been connected not only with torsional strain but also with nucleosome-free region found in SV40 minichromosomes by many authors. Thus, a fraction of torsionally strained minichromosomes was only a subfraction of minichromosomes with nucleosome-free region.
The results with SV40 minichromosomes were interpreted in terms of nucleosome unfolding so that DNA turns no longer were constrained by folded (“classical”) nucleosomes.
Direct evidence that nucleosome unfolding could be maintained because of superhelical strain was also presented by Garner et al.23 who used artificial minichromosomes and bacterial DNA gyrase to induce negative supercoiling in their experiments. Superhelical turns were found not to be constrained by unfolded nucleosomes.
Another piece of evidence for nucleosome unfolding came from studies of yeast 2 μm minichromosomes.24,25 These authors constructed artificial system of in vivo DNA relaxation in yeast using transfected plasmids bearing expressed bacterial topA gene. When yeast Δtop1 top2ts genes were blocked at non-permissive temperature in mutant cells, then bacterial topoisomerase slowly converted intracellular 2 μm minichromosomes into a form containing positively supercoiled DNA as was revealed by two-dimensional electrophoresis of DNA topoisomers. Relaxation of negative supercoils by bacterial topoisomerase led to slow interconversion of negatively supercoiled minichromosomes into positively supercoiled ones. Positively supercoiled minichromosomes accumulated at the expense of negatively supercoiled minichromosomes (negatively strained or “unconstrained” by histones). These in turn appeared at the expense of a pool of minichromosomes whose negative supercoils were fully constrained by nucleosomes as usually observed for the bulk of yeast 2 μm minichromosomes. Obviously, these constrained minichromosomes transformed into unconstrained (negatively strained) minichromosomes to replenish their pool depleted by bacterial topoisomerase converting them into positively supercoiled ones as argued by the authors.
The final level of positive supercoiling was at least two times lower than the native level of negative supercoiling. Also the nucleosome and chromatin structure in positively tensioned minichromosomes substantially differed from usual chromatin structure of the bulk 2 μm minichromosomes. Positively strained minichromosomes contained full complement of nucleosomes and exhibited features of transcriptionally active chromatin in respect to enhanced overall DNaseI and MNase sensitivities.25 The authors interpreted their results in terms of nucleosome unfolding in chromatin activated for transcription.
These inferences are in a good accordance with recent results on interconversion of negatively and positively stressed chromatin domains (loops) in human cells in response to inhibition of RNA-polymerase with α-amanitin or its withdrawal, respectively.1 Also these authors demonstrated disappearance of both negative and positive tensions in response to DNA nicking in vivo using bleomycin.
In this connection, the above-mentioned results on arising of A-form and C-form DNA at moderate levels of negative or positive supercoiling in DNA loops, respectively, look as an interesting possibility for arising of alternative DNA structures both in positively or negatively stressed chromatin loops.
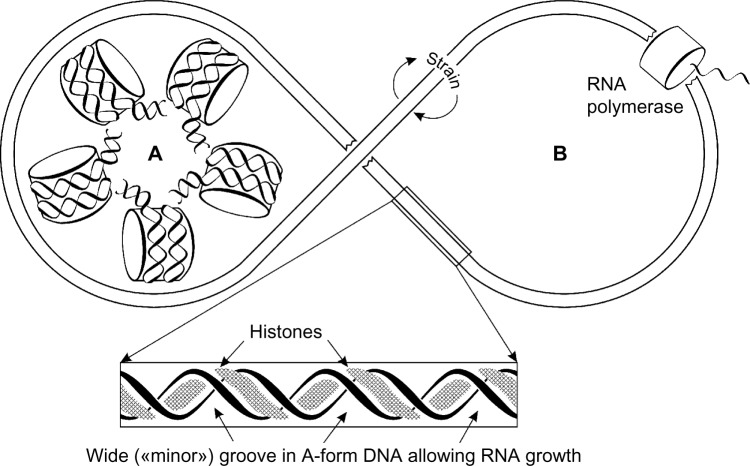
The conclusions from studies with SV40 minichromosomes and yeast 2 μm minichromosomes are summarized in Figure 3. Inactive minichromosome is shown as a classical beads-on-a-string structure, which lacks DNA torsional tension because DNA superhelix is firmly fixed and immobilized on a surface of nucleosome disk. The activation of minichromosomes for transcription occurs as a result of simultaneous and cooperative conformational transition both in DNA and histone core particles so that DNA assumes A-conformation with an expanded minor groove allowing formation of DNA–RNA triplexes, and histone core particles unfolded but retaining salt bonds with DNA phosphate groups. This transition seems to occur in all-or-none manner, which can be understood as cooperative transition of nucleoprotein fiber. The majority of nonelastic turns of DNA wound around the nucleosome core are transmitted into a crystal pitch of DNA intrinsic to the A-form (11 bp/turn), but the rest (minor part) of them gives rise to elastic strain necessary to maintain the unfolded structure under conditions of dynamic equilibrium.
Figure 3.
Scheme of conformational rearrangement of chromatin ring or loop from transcriptionally inactive state (A) to transcriptionally active one (B). Cooperative nucleosome unfolding is accompanied by conformational transition of DNA into transcriptionally active A-form. (This is a summary from described experiments with SV40 and yeast minichromosomes also tangible for giant chromatin loops in eukaryotic chromosomes.).
The active DNA molecule has histones smeared along and inside “major” groove and has a short piece of growing RNA within “minor” groove of the DNA. However, one has to keep in mind that in the A-form, “minor groove” becomes very wide and shallow with nucleotides looking and sticking out. But “major groove” becomes very deep and narrow according to crystallographic data of A and B forms of DNA. This indeed may cause some confusion in terminology oriented nowadays mainly in the B-form of DNA.
Conformational transitions in strained DNA in chromatin
The average pitch of DNA wrapped around nucleosomes core is about 10 bp per turn (see Ref. 26 and the references therein). (unlike free DNA in solution that relates to B-family and has a pitch varying between 9.0 and 11.0 bp/turn and an average pitch of 10.4–10.5 bp/turn).
Nucleosome unfolding according to the scheme presented in Figure 3 leads to formation of A-form of DNA (11 bp/turn). This means that after nucleosome unfolding, every base pair of DNA should consume 3.27° from negative supercoil (360°/10−360°/11 = 3.27°). For 144 bp of DNA in a core particle, this would give 470.1°. However, DNA wound around nucleosome core makes about ~1.67 physical turns of left coil (601.2°). Thus, remaining 130.3° should be spent for transition into A-form of a base belonging to linker DNA (30–60 bp). But a fraction of turns should remain elastic to maintain the unfolded chromatin fiber in a stable state.
Without partitioning of topological turns in elastic spring and conformational transition to underwound DNA secondary structure (A-form), such an unfolded chromatin fiber would be absolutely unstable. The partitioning of some turns in elastic spring follows from the cooperative response of active minichromosomes to the treatment with topoisomerase I or X-ray nicking. This again proves that elastic portion of topological turns is absolutely required for stable maintenance of unfolded nucleosome structure.
In these terms, B-DNA in solution may be understood as a combination of A and C forms arising in it because of thermal fluctuations. This explains the fact of paracrystalline nature of B-DNA. Thermal motions of DNA in chromatin are frozen in comparison to free DNA, and this fact alone could explain coexistence of crystal DNA forms in nucleosomes figured out recently to explain DNA curvature in nucleosome cores.27
Insights and Perspectives
Topoisomerase II as a tool for placing the DNA inside the RNA-polymerase upon activation of transcription
For the initiation of transcription and RNA elongation, the DNA template in the A-form is most fitting as discussed above. So transcription can start without denaturation, melting, or untwining of DNA. To do that RNA-polymerase should embrace active chromatin fiber putting it inside the enzyme. The same was firmly established for DNA polymerase subunits that become active only after their assemblage into a ring embracing DNA in all living organisms (in vivo), described in all reviews and handbooks (except some run-on in vitro reconstructions).
Thus, huge RNA-polymerase multi-subunit enzyme should represent a bagel or donut sliding along the DNA mole cule and having it always inside the donut hole.
The question arises, “How does this bagel sliding along A-form DNA add new ribonucleosides to a nascent RNA chain?” “Does it have 11 active centers to insert new ribonucleosides around one DNA turn while moving along the DNA duplex, or probably it has only one active center of enzymatic activity but polymerase bagel rotates rapidly while moving along the DNA chain?” It seems likely that both possibilities could be realized in different occasions as follows from run-on experiments with RNA-polymerase subunits. Also 12 subunits (or less) of RNA-polymerase II may alternate as active centers when catalytic atom of Mg2+ travels inside RNA-polymerase channel while moving around DNA duplex.
In any event, topoisomerase II is necessary to insert the DNA into a donut hole to initiate the transcription.28
Non-coding DNA
The DNA sequences that do not code for functional products such as proteins or functional RNA (rRNA, tRNA, etc.) are termed “non-coding.” For example, the sequences of major parts of introns that are transcribed but then are rapidly cut out and destroyed are not considered as “coding” in these terms. Also non-coding DNA are numerous satellite DNA sequences surrounding coding DNA. They comprise more than 50% of human genome (see Ref. 29 for a review).
Looking at genome polynucleotide sequences, one have to explain why coding DNA sequences comprise no more than 2–4% of all human genomic sequences and what is a function of 96–98% of DNA comprising the rest of the genome. (In non-human organisms, “junk DNA” is usually presented at a lower level, being the lowest at the lowest evolutionary scale step.)
Recently, annotated class of long non-coding RNAs (termed “lncRNAs” in ENCODE project) represents more than 9277 “genes” producing at least 14,880 transcripts containing only two small exons usually not exceeding 200 bp and only one intron usually exceeding 2000 bp. Processed lncRNAs mostly remain bound to chromatin.30 This novel class of RNA however adds only about 25 Mb of transcribed DNA mostly made of introns and contains no more than 2 Mb (in total) of non-coding exons.
The fact of evolutionary arising of non-coding DNA including satellite DNA is readily explained by the crucial role of conformational transitions in torsionally strained DNA competing for the energy of elastic spring, thus making it possible to transmit signals along huge stretches of DNA within topological domains. It is reminiscent of transmission of electric energy through a metal wire when electric tension (voltage) is applied to the ends of wire. One nick per DNA topological domain abolishes all the transcription within particular loop3 as well as one cut of wire in an electric circuit switches off all the electric power in a given circuit.
We proceed further arguing leaning against the theory of conformational transitions in closed circular DNA molecules14 substantiated by numerous experiments discussed above. Also we expand and detail biological implications of this theory.
Transmission of genetic signals at a distance
Unlike metal wires, which are homogeneous, the DNA “wire” has variable base pair composition along its length; hence, its conformational flexibility is variable. Nature had only limited possibilities to change nucleotides within coding triplets without destroying the structure and function of important enzymes and other proteins. So not only the base pair composition of genes is important but also the exact sequence (within a framework of the degeneracy (redundancy) of the genetic code). For this reason, the conformational flexibility of coding sequences is heavily narrowed by their obligation to code for proteins.
To compensate this obstacle, evolution created many more sequences scattered around genes and within introns to achieve necessary conformational flexibility in the net polynucleotide chains within every particular covalently closed DNA topological domain (loop) in the genome.
To create necessary level of torsional tension in coding sequences and their conformational transition into transcriptionally active A-form arising after nucleosome unfolding within a given loop, compensatory sequences are scattered around genes and within introns. These non-coding DNA sequences are able to act in two different ways. (1) They take up some redundant topological turns or vice versa give up additional topological turns necessary to activate coding sequences upon nucleosome unfolding and arising of torsional tension in closed DNA loops. (2) Some non-coding DNA sequences are known to be excised during differentiation of many cell lineages in the course of ontogenesis, thus creating novel topological possibilities for gene expression in particular loops. However, this second way is characterized in experiment very scarcely. So here we will consider the first possibility in more detail. The second one will be considered in another publication.
The crucial line of evidence comes from well-established intron-mediated enhancement of transcription (see Ref. 31 and the references therein for review). It has been shown that introns are necessary to enhance transcription of neighboring coding sequences sometimes as much as by several orders of magnitude. The genetic constructs or transgenes with genes artificially devoid of introns are poorly transcribed and sometimes fail to be transcribed at all.
Intron-mediated enhancement of transcription can be understood in two ways: (1) the enhancement of cooperativity of conformational transition into A-form in coding sequences linked to nearby intron sequences so to exclude the formation of patches of C-form capable to block the transcription through such patches and (2) the prevention of cooperativity in the formation of C-form in coding sequences by splitting the genes into several pieces.
DNA “conformational code”
To analyze DNA sequences in introns and exons, one has to pay attention to the so-called “Chargaff second parity rule.”32 According to this rule, the amount of adenine equals to amount of thymine (A = T) as well as G = C in any one of the two strands of DNA duplex in all living organisms (except some organelles) if only the length of separated strands exceeds 10 kb in eukaryotes (and much less in viruses). This rule is still unexplained by current theories but is often used for deeper investigation of conformational properties of DNA strands.
Also the general observation is that T and G are more abundant in introns than in exons in higher eukaryotes (considering “sense” (non-transcribed) ie corresponding to RNA transcript in its sequence DNA strand).32,33 This difference disappears in integral exon–intron stretches longer than 10 kb.
Moreover, the bias between exons and introns was evident at the level of oligonucleotides. Thus considering short oligonucleotide sequences by “moving window frame” of a certain nucleotide length along the DNA sequence shifting it one by one nucleotide and counting resulting in equal-sized oligonucleotides varying in sequence but not in length (using computer program), it was found that resulting frequencies of all possible hexanucleotides varied between exons and introns drastically (see datasets S1 and S2 in supplementary material of Ref. 33).
These variations can only be understood if we look at a DNA sequence not only as information message coding for proteins, functional RNAs, or regulatory protein binding sites. Rather, we should recognize in DNA molecule some “conformational code” with cooperative conformational properties existing in stretches of DNA longer than 3–10 nucleotides. Moreover, these stretches do not have certain boundaries because they overlap each other. And also the energies required to shift equilibrium between different conformational states vary with variations in nucleotide neighboring.
This inference follows from meticulous analysis of nearest-neighbor effects on a base pair conformation, which is heavily dependent on nucleotides surrounding it, even if remote by several nucleotides.7 For example, CG steps were heavily prone to adopt A-conformation if only they were not flanked by A (adenine) from the 5′ end, or flanked by C or T nucleotides from the 3′ end. Other numerous nucleotide flankings favored A-conformation arising within a CG step.7 For some other dinucleotide steps, the influence of 5′ and 3′ neighbors was also dramatic (see supplementary material of Ref. 7). For example, some dinucleotide steps strongly prefer to adopt C-form but certain neighbors enhanced their capacity to adopt A-form. The most reliable parameters dividing all DNA conformations into two alternative states were twist and “minor” groove width, always giving bimodal distributions under various neighborhoods.
Also the energies separating conformations were extremely low in terms of number of milliseconds spent by a given sequence in an alternative conformation under conditions of constant temperature. This means a certain dynamic equilibrium between conformations (for naked DNA). In torsionally strained DNA, equilibrium is easily shifted as discussed in the beginning of this article.
In transcriptionally active chromatin (strained DNA bound to unfolded nucleosomes), the situation is more complex and interesting as follows from nucleosome “crystal-like” behavior in vivo34 and the fact that the majority of exons have a mean size close to 147 bp (nucleosome core DNA length).35 These intriguing aspects await their further developments.
Conclusions
Experimental evidence for conformational transitions of DNA duplexes into A and C forms both in naked DNA and in chromatin loops and rings upon arising of torsional tension is considered. This evidence is reevaluated in the light of recent findings.
Transcription of DNA without its melting and rotation is understood and explained by the transition of DNA template into A-conformation. The “minor” groove width of A-form DNA allows to place into it a third polynucleotide chain (RNA), thus allowing the formation of transient polynucleotide triplexes in the course of transcription. Topoisomerase II is obviously required to insert DNA into RNA-polymerase channel to dispose the enzyme ready for transcription of DNA as suggested by recent findings.
The existence and indispensability of non-coding “junk” DNA sequences is explained and understood through the occurrence of conformational transitions in these sequences, competing at large distances for turns and energy of torsionally stressed DNA at the sites of transcription, thus performing fine tuning of transcription.
Acknowledgments
I thank Yury Safonov for help in preparation of figures.
Footnotes
ACADEMIC EDITOR: James Willey, Editor in Chief
FUNDING: Author discloses no funding sources.
COMPETING INTERESTS: Author discloses no potential conflicts of interest.
Author Contributions
Conceived the concepts: ANL. Analyzed the data: ANL. Wrote the first draft of the manuscript: ANL. Made critical revisions: ANL. The author reviewed and approved of the final manuscript.
DISCLOSURES AND ETHICS
As a requirement of publication the author has provided signed confirmation of compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
REFERENCES
- 1.Naughton C, Avlonitis N, Corless S, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–95. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermudez I, Garcia-Martinez J, Perez-Ortin JE, Roca J. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 2010;38:e182. doi: 10.1093/nar/gkq687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luchnik AN, Hisamutdinov TA, Georgiev GP. Inhibition of transcription in eukaryotic cells by X-irradiation: relation to the loss of topological constraint in closed DNA loops. Nucleic Acids Res. 1988;16:5175–90. doi: 10.1093/nar/16.11.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos JL, Portugal J, Subirana JA, Mayer R. Influence of amino acid and peptide counterions on the conformation of DNA. J Mol Biol. 1986;187:441–7. doi: 10.1016/0022-2836(86)90444-4. [DOI] [PubMed] [Google Scholar]
- 6.Svosil D, Kalina J, Omelka M, Schneider B. DNA conformations and their sequence preferences. Nucleic Acids Res. 2008;36:3690–706. doi: 10.1093/nar/gkn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavery R, Zakrzewska K, Beveridge D, et al. A systematic molecular dynamics study of nearest-neighbor effects on base pair and base pair step conformations and fluctuations in B-DNA. Nucleic Acids Res. 2010;38:299–313. doi: 10.1093/nar/gkp834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dam L, Levitt MH. B II nucleotides in the B and C forms of natural-sequence polymeric DNA: a new model for the C forms of DNA. J Mol Biol. 2000;304:541–61. doi: 10.1006/jmbi.2000.4194. [DOI] [PubMed] [Google Scholar]
- 9.Heddi B, Oguey C, Lavelle C, Foloppe N, Hartmann B. Intrinsic flexibility of B-DNA: the experimental TRX scale. Nucleic Acids Res. 2010;38:1034–47. doi: 10.1093/nar/gkp962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng HL, Kopka ML, Dickerson RE. The structure of a stable intermediate in the A <--> B DNA helix transition. Proc Natl Acad Sci U S A. 2000;97:2035–9. doi: 10.1073/pnas.040571197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JC, Peck LJ, Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harbor Symp Quant Biol. 1983;47:85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Courey AJ, Wang JC. Cruciform formation in negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983;33:817–29. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- 13.Luchnik AN. Evidence for a novel conformational transition in superhelical DNA. J Biol Phys. 1985;13:48–54. [Google Scholar]
- 14.Luchnik AN. Conformational transitions in closed circular DNA molecules I. Topological and energetical considerations. Mol Biol Rep. 1980;6:3–9. doi: 10.1007/BF00775746. [DOI] [PubMed] [Google Scholar]
- 15.Chua EY, Vasudevan D, Davey GE, Wu B, Davey CA. The mechanics behind DNA sequence-dependent proportions of the nucleosome. Nucleic Acids Res. 2012;40:6338–52. doi: 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glaser VM, Luchnik AN. Decrease in the density of DNA topological turns during in vitro aging of Syrian hamster cells. Exp Cell Res. 1982;139:249–55. doi: 10.1016/0014-4827(82)90249-x. [DOI] [PubMed] [Google Scholar]
- 17.Upholt WB, Gray HB, Vinograd J. Sedimentation velocity behavior of closed circular SV40 DNA as a function of superhelix density, ionic strength, counterion and temperature. J Mol Biol. 1971;62:21–38. doi: 10.1016/0022-2836(71)90128-8. [DOI] [PubMed] [Google Scholar]
- 18.Hinton DM, Bode VC. Purification of closed circular lambda deoxiribonucleic acid and its sedimentation properties as a function of sodium chloride concentration and ethidium binding. J Biol Chem. 1975;250:1071–9. [PubMed] [Google Scholar]
- 19.Luchnik AN, Bakayev VV, Glaser VM. DNA supercoiling: Changes during cellular differentiation and activation of chromatin transcription. Cold Spring Harbor Symp Quant Biol. 1983;47:793–801. doi: 10.1101/sqb.1983.047.01.091. [DOI] [PubMed] [Google Scholar]
- 20.Luchnik AN, Bakayev VV, Zbarsky IB, Georgiev GP. Elastic torsional strain in DNA within a fraction of SV40 minichromosomes: relation to transcriptionally active chromatin. EMBO J. 1982;1:1353–8. doi: 10.1002/j.1460-2075.1982.tb01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luchnik AN, Bakayev VV, Yugai AA, Zbarsky IB, Georgiev GP. DNAasel-hypersensitive minichromosomes of SV40 possess an elastic torsional strain in DNA. Nucleic Acids Res. 1985;13:1135–49. doi: 10.1093/nar/13.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakayev VV, Yugai AA, Luchnik AN. Effect of X-ray induced DNA damage on DNAase I hypersensitivity of SV40 chromatin: relation to elastic torsional strain in DNA. Nucleic Acids Res. 1985;13:7079–93. doi: 10.1093/nar/13.19.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garner MM, Felsenfeld G, O’Dea MH, Gellert M. Effects of DNA supercoiling on the topological properties of nucleosomes. Proc Natl Acad Sci U S A. 1987;84:2620–3. doi: 10.1073/pnas.84.9.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eucaryotic cells. Cell. 1988;55:849–56. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 25.Lee M-S, Garrard WT. Positive DNA supercoiling generates a chromatin conformation characteristic of highly active genes. Proc Natl Acad Sci U S A. 1991;88:9675–9. doi: 10.1073/pnas.88.21.9675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaffney DJ, McVicker G, Pai AA, et al. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olson WK, Zhurkin VB. Working the kinks out of nucleosomal DNA. Curr Opin Struct Biol. 2011;21:348–57. doi: 10.1016/j.sbi.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperling AS, Jeong KS, Kitada T, Grunstein M. Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast. Proc Natl Acad Sci U S A. 2011;108:12693–8. doi: 10.1073/pnas.1106834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richard G-F, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev. 2008;72:686–727. doi: 10.1128/MMBR.00011-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derrien T, Johnson R, Bussotti, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose AB, Emami S, Bradnam K, Korf I. Evidence for a DNA-based mechanism of intron-mediated enhancement. Front Plant Sci. 2011;2:98. doi: 10.3389/fpls.2011.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albrecht-Buehler G. Asymptotically increasing compliance of genomes with Chargaff second parity rules through inversions and inverted transpositions. Proc Natl Acad Sci USA. 2006;103:17828–33. doi: 10.1073/pnas.0605553103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C, Li W-H, Krainer AR, Zhang MQ. RNA landscape of evolution for optimal exon and intron discrimination. Proc Natl Acad Sci U S A. 2008;105:5797–802. doi: 10.1073/pnas.0801692105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaillant C, Palmeira L, Chevereau G, et al. A novel strategy of transcription regulation by intragenic nucleosome ordering. Genome Res. 2010;20:59–67. doi: 10.1101/gr.096644.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bolisetty MT, Beemon KL. Splicing of internal large exons is defined by novel cis-acting sequence elements. Nucleic Acids Res. 2012;40:9244–54. doi: 10.1093/nar/gks652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer W, Vinograd J. The interaction of closed circular DNA with intercalative dyes. I. The superhelix density of SV40 DNA in the presence and absence of dye. J Mol Biol. 1968;33:141–71. doi: 10.1016/0022-2836(68)90286-6. [DOI] [PubMed] [Google Scholar]