Abstract
Immunological characterization of immune cells that reside in specific anatomic compartments often requires their isolation from the respective tissue on the basis of enzymatic tissue disintegration. Applying enzymatic digestion of primary splenocytes, we evaluated the impact of collagenase and dispase, two enzymes that are commonly used for the liberation of immune cells from tissues, on the detectability of 48 immunologically relevant surface molecules that are frequently used for flow cytometric identification, isolation, and characterization of immune cell subsets. Whereas collagenase treatment had only minor effects on surface expression of most molecules tested, dispase treatment considerably affected antibody-mediated detectability of the majority of surface markers in subsequent FACS analyses. This effect was long lasting and, in case of high-dose dispase treatment, evident for the majority of surface molecules even after 24 h of in vitro culture. Of note, high-dose dispase treatment not only affected surface expression of certain molecules but also impaired antigen-specific proliferation of CD4+ and CD8+ T cells. Together, our data indicate that enzymatic tissue disintegration can have profound effects on the expression of a variety of cell-surface molecules with direct consequences for phenotypic analysis, FACS- and MACS-based target cell isolation, and immune cell function in cell culture experiments.
Keywords: collagenase, dispase, enzymatic digestion, flow cytometry, surface molecules, T cell proliferation
Introduction
The immune system is highly compartmentalized, and the specific milieu existing in a given tissue greatly influences the phenotype and function of immune cells in the respective anatomic compartment. For instance, immune cells residing in the intestinal immune system may largely differ from those located in the peripheral blood, the skin, or the respiratory tract [1]. Therefore, the functional and molecular characterization of cellular subsets often necessitates investigation of immune cells obtained directly from tissues of particular interest. Whereas mechanical treatments are generally sufficient for isolation of cells from classical immunological tissues such as the blood, spleen, lymph nodes or thymus, high yield purification of vital cells from mucosal or other tissues often deserve the combined use of mechanical dissection and enzymatic tissue disintegration [2–7]. More recently, the immunological function of non-classical immune cells such as epithelial and endothelial cells has become increasingly appreciated. Of note, protocols to isolate these resident cells from tissues do often require even harsher conditions with respect to the nature or concentration of the enzyme to be used for efficient liberation of cells from tissues than those used for the isolation of lymphocytes or monocytes [8–10].
Enzymes typically used for tissue disintegration are collagenase and dispase I, often in combination with DNase, EDTA, and DTT to minimize cell clumping and to support cell dissociation. Although there is some evidence that these enzymes may affect the expression of surface antigens and immune cell function [6, 11, 12], many experimental approaches require cell numbers that cannot be obtained in sufficient quantity and quality by mechanical approaches alone. Enzyme-mediated reduced surface abundance of certain molecules may be a possible reason for the often observed discrepancy between data obtained by immunohistochemical staining of intact tissue, gene expression data, and data obtained by flow cytometry following surface staining of cells isolated on the basis of enzymatic tissue digestion. Moreover, since surface molecules mediate key immunological functions such as cell adhesion, pathogen recognition, and outside-in-signaling, lack of or reduced surface expression of certain molecules due to enzymatic cleavage may also impinge on immune cell functions giving rise to misleading results in subsequent in vitro cell culture assays.
Multicolor flow cytometry analysis with an increasing panel of commercially available antibodies and fluorochromes has developed a robust and reliable method for both the phenotypic characterization and the isolation of immune cell subsets from different tissues. Working with pre-clinical mouse models for intestinal and pulmonary inflammation, we routinely apply enzymatic digestion protocols for the isolation of cellular subsets from the gut and the lung [3, 7, 9, 13]. We observed that the experimental procedures applied for mucosal T cell isolation as well as for the isolation of alveolar type II epithelial cells from the murine lung, in particular dispase I treatment, may have a considerable impact on the expression level of certain surface molecules. In this study, we therefore systematically evaluated the impact of two commonly used enzymes, collagenase D and dispase I, on the detectability of as many as 48 different surface markers that are frequently used for the flow cytometric identification, isolation, and characterization of immune cell populations and/or exhibit important immunological functions. To further investigate the immunological consequences of enzyme-mediated reduction of surface marker expression, we performed in vitro T cell stimulation assays to directly test the impact of collagenase and dispase I treatment on T cell receptor-mediated antigen recognition and proliferation.
Materials and methods
Mice
BALB/c mice were obtained from Harlan (Borchen, Germany). Transgenic mice expressing a T cell receptor (TCR) specific for the MHC class II-restricted influenza hemagglutinin (HA) peptide 110–120 on CD4+ T cells (TCR/HA) [14] or the MHC class I-restricted HA-peptide 512–520 on CD8+ T cells (CL4) [15] were bred in our animal facility at the Helmholtz Centre for Infection Research, Braunschweig, and were kept under specific pathogen-free (SPF) conditions.
Cell preparation and enzymatic digestion
Spleens were rinsed with PBS to obtain single cell suspensions followed by erythrocyte lysis. Clumps and remaining tissue were removed by transfer through a 70-µm cell strainer. Splenocytes were digested with collagenase D from Clostridium histolyticum or dispase I from Bacillus polymyxa as specified in Table 1.
Table 1.
Conditions for enzymatic digestion of splenocytes
| Enzyme | Manufacturer | Experimental conditions |
|---|---|---|
| Dispase I (high dose) | BD Bioscience | 200 U (50 U/ml, pre-warmed) for 45′ at room temperature |
| Dispase I (low dose) | Roche Diagnostics | 4 U (0.8 U/ml) for 45′ at 37 °C in IMDM + 5% FCS |
| Collagenase D | Roche Diagnostics | 0.221 U (0.044 U/ml) for 45′ at 37 °C in IMDM + 5% FCS |
Following enzymatic digestion, cells were collected by centrifugation, enzyme containing supernatant was discarded, and cells were resuspended in IMDM supplemented with 10% FCS, 1% Pen/Strep, and 0.25 mM β-mercaptoethanol (IMDM complete).
FACS analysis
Spleen cells (5×105) were stained either immediately after enzymatic digestion or after a 24 h recovery phase in IMDM complete at 37°C and 5% CO2. Splenocytes were stained for 10 min at 4 °C either directly with fluorescent-labeled antibodies or indirectly with biotin-conjugated antibodies followed by a secondary staining step with streptavidin-PE. Subsequently, cells were washed twice with PBS, fixed with 2% paraformaldehyde for 20 min at room temperature, and finally analyzed for cell surface expression of selected markers indicated in Table 2. Positively stained cells were quantified using a BD FACS Canto and analyzed with FlowJo software. Gates were set on single, living cells, according to their forward and sideward scatter characteristics. Percent marker positive enzyme-treated samples were normalized to marker positive untreated control samples, referred to as % expression. Unless otherwise stated, experiments were repeated three times, and one representative result out of three independent measurements is shown.
Table 2.
Antibodies and 2nd step reagents used for flow cytometry
| Antibody | Clone | Fluorochrome | Manufacturer |
|---|---|---|---|
| Anti-B7H4 | clone 9 | PE | BioLegend |
| Anti-CD3ε | 145-2C11 | PE | eBioscience |
| Anti-CD4 | GK1.5 | PE | eBioscience |
| Anti-CD8a | 53-6.7 | PE | BD Pharmingen |
| Anti-CD11b | M1/70 | PE | eBioscience |
| Anti-CD11c | HL3 | FITC | BD Pharmingen |
| Anti-CD14 | Sa2-8 | PE | eBioscience |
| Anti-CD19 | 1D3 | PE | eBioscience |
| Anti-CD25 | PC61 | PE | BD Pharmingen |
| Anti-CD27 | LG.3A10 | PE | BD Pharmingen |
| Anti-CD28 | 37.51 | PE | BD Pharmingen |
| Anti-CD34 | RAM34 | PE | eBioscience |
| Anti-CD38 | 90 | PE | BD Pharmingen |
| Anti-CD40 | Mar-23 | Biotin | BD Pharmingen |
| Anti-CD45 | 30-F11 | APC | BD Pharmingen |
| Anti-CD45R (B220) | RA3-6B2 | APC | eBioscience |
| Anti-CD49b (Pan-NK) | DX5 | FITC | BD Pharmingen |
| Anti-CD54 (ICAM-1) | YN1/1-7 | Bio | Kindly provided by S. Weiss |
| Anti-CD62L | MEL-14 | APC | BD Pharmingen |
| Anti-CD69 | H1.2F3 | PE | BD Pharmingen |
| Anti-CD70 | FR70 | PE | eBioscience |
| Anti-CD80 (B7.1) | 16-10A1 | Bio | eBioscience |
| Anti-CD86 (B7.2) | GL1 | Bio | BD Pharmingen |
| Anti-CD103 | 2E7 | FITC | BD Pharmingen |
| Anti-CD134 (OX40) | OX-86 | PE | eBioscience |
| Anti-CD137 (4-1BB) | 17B5 | PE | eBioscience |
| Anti-CD137L (4-1BBL) | TKS-1 | Bio | eBioscience |
| Anti-CD154 (CD40L) | MR1 | PE | BD Pharmingen |
| Anti-CD162 (PSGL-1) | 2PH1 | PE | BD Pharmingen |
| Anti-CD178 (FasL) | MLF3 | PE | BD Pharmingen |
| Anti-CD210 (IL-10R) | 1B1.3a | PE | BD Pharmingen |
| Anti-CD252 (OX40L) | RM134L | PE | BioLegend |
| Anti-CD272 (BTLA) | 6F7 | PE | eBioscience |
| Anti-CD273 (PD-2L) | TY25 | PE | BD Pharmingen |
| Anti-CD274 (PD-1L) | MIH5 | PE | eBioscience |
| Anti-CD275 (ICOSL) | HK5.3 | PE | BioLegend |
| Anti-CD278 (ICOS) | 7E.17G9 | PE | eBioscience |
| Anti-CD279 (PD-1) | J43 | PE | BD Pharmingen |
| Anti-CD282 (TLR4) | 6C2 | PE | eBioscience |
| Anti-CD284 (TLR2) | UT41 | PE | eBioscience |
| Anti-F4/80 | BM8 | APC | eBioscience |
| Anti-GITR | DTA-1 | PE | eBioscience |
| Anti-GITRL | YGL 386 | PE | BioLegend |
| Anti-Ly6G (Gr-1) | RB6-8C5 | PE-Cy7 | eBioscience |
| Anti-MHCI (H-2Kb) | AF6-88.5 | FITC | BD Pharmingen |
| Anti-MHCII | M5/114.15.2 | APC | eBioscience |
| Anti-NK1.1 | PK136 | PE | BD Pharmingen |
| Anti-Vβ8 | F23.1 | PE | BD Pharmingen |
| Streptavidin | PE | BD Pharmingen |
Proliferation assay
Collagenase- or dispase-treated splenocytes (5×105) from TCR/HA transgenic mice were plated in 96-well, round-bottom plates in a total volume of 200 µl IMDM supplemented with 10% FCS, 1% penicillin/streptomycin and 0.25 mM β-mercaptoethanol. CD4+ T cells were stimulated with HA peptide 110–120 and cultured for 24 h at 37 °C. Proliferation was detected following the addition of 3[H]-thymidine and culture for another 16 h at 37 °C. Incorporation of 3[H]-thymidine was measured by scintillation counting. Cell proliferation is presented as mean counts per minute (cpm) of triplicate wells, and one representative result out of three independent experiments with similar results is shown. In case CD8+ T cell proliferation was analyzed, 1×105 collagenase- or dispase-treated splenocytes obtained from CL4 transgenic mice were stimulated with the respective peptide HA 512–520 as described above and cultured for 24 h at 37 °C, and 3[H]-thymidine was measured by scintillation counting 12 h after the addition of 3[H]-thymidine HA-specific CD8+ T cell proliferation was evaluated in triplicates; the experiment was repeated twice with similar results.
Statistical analysis
Statistical analysis was performed using GraphPadPrism Software using unpaired, two-tailed t-test. Statistical significance is shown as p value of <0.05 indicated as *, p<0.01 as ** and p<0.001 as ***.
Results and discussion
Enzymatic digestion affects expression level of surface molecules on immune cells
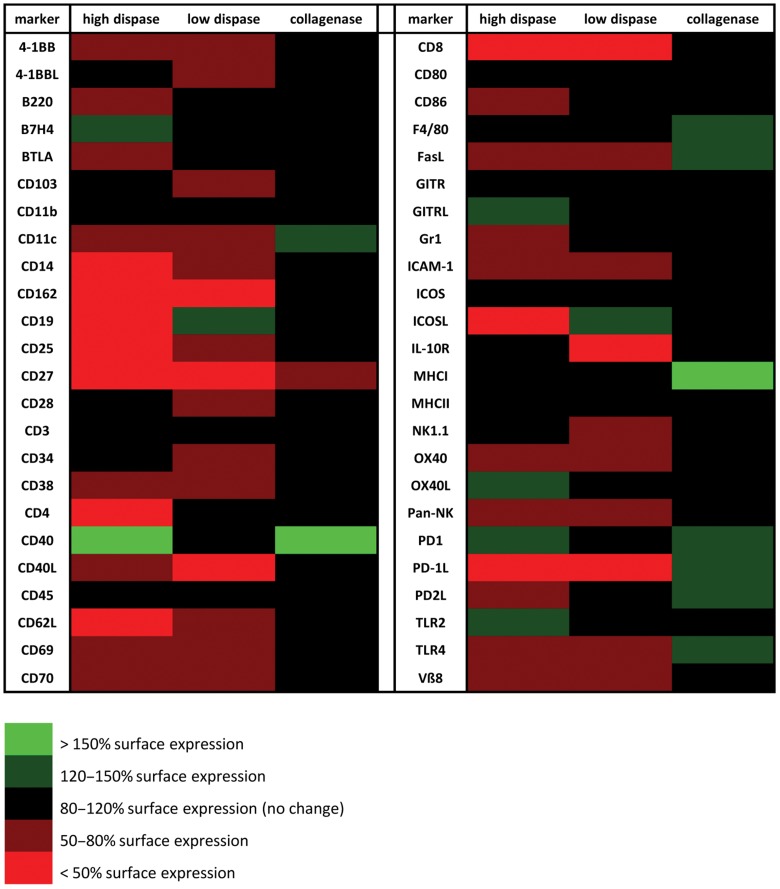
To get a focused starting point for deciphering the specific impact of different enzymes on surface molecule expression on immune cells, we performed broadly based digestion experiments. To this end, freshly isolated mouse splenocytes were incubated in vitro with collagenase D or dispase I as indicated in Table 1. Experimental conditions were adapted to established protocols routinely used for the liberation of lymphocytes and epithelial cells from tissues. Following enzymatic treatment, surface staining of selected molecular markers (refer to Table 2) was performed, and surface expression was determined by FACS analysis. Molecules analyzed included, upon others, typical T cell (CD3, CD4, CD8), B cell (B220, CD19), granulocyte (Gr-1), or NK cell (NK1.1, Pan-NK) markers, molecules commonly expressed on antigen presenting cells such as macrophages or dendritic cells (F4/80, CD11b, CD11c), molecules involved in antigen presentation and co-stimulation (MHC classes I and II, CD28, CD80, CD86, 4-1BB, PD1), cytokine receptors (IL-10R, CD25), integrins (CD103, CD11b, CD11c), and several other immunologically relevant surface molecules. As depicted in Figure 1 this initial screening of 48 different surface molecules revealed that dispase treatment markedly reduces the expression of numerous molecules on the surface of immune cells, including CD11c, ICAM-1, CD40L, CD162, CD8, CD25, CD69, and PD-1L both at high and low enzyme concentrations. Of note, collagenase treatment of splenocytes did not reduce surface expression of most tested molecules, but in contrast induced surface expression of certain markers such as CD11c, CD40, F4/80, and MHC class I (Fig. 1).
Fig. 1.
Enzymatic digestion broadly affects surface marker expression. BALB/c splenocytes were digested with collagenase D (0.044 U/ml) or high (50 U/ml) or low (0.8 U/ml) concentrations of dispase I, respectively, followed by antibody staining of 48 selected surface markers and FACS analysis. Percent positive cells obtained after enzymatic digestion was normalized to surface marker expression on untreated splenocytes which was arbitrarily set to 100%. Green color indicates elevated surface expression following enzymatic digestion, red color indicates reduced surface expression; black indicates no change in surface expression
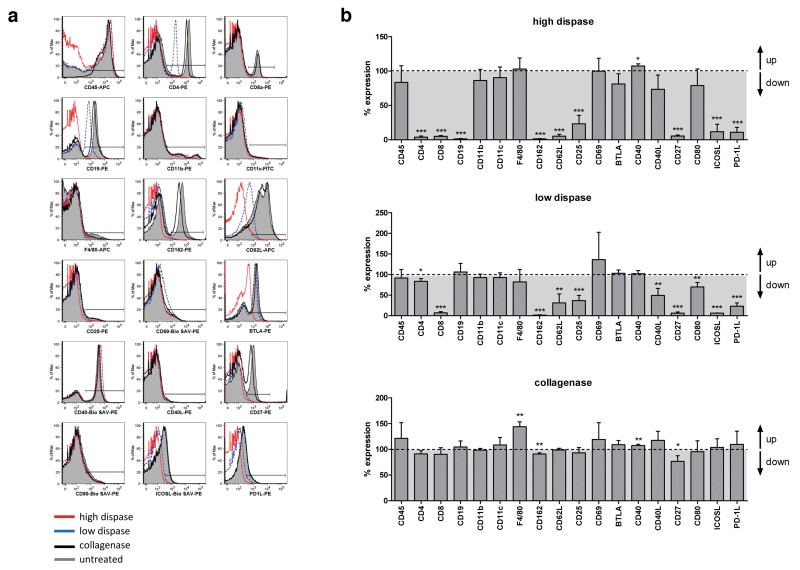
Of the 48 pre-screened surface molecules, 18 markers exhibiting pronounced sensitivity to enzymatic digestion were selected for further analysis. Following enzymatic digestion, splenocytes were surface stained and analyzed by flow cytometry as described before. As depicted in Figure 2, immune cell digestion with high dispase concentrations resulted in dramatic decrease in surface expression of CD4, CD8, CD19, CD162, CD62L, CD25, CD27, ICOS-L, and PD-1L. Diminished surface expression of most of these markers on splenocytes was also evident following low-dose dispase digestion although to a lower extend (Fig. 2). As observed before, only a minor impact on surface marker expression was found in case of collagenase digestion. Whereas Figure 2a shows representative histograms obtained in one selected experiment, Figure 2b summarizes data obtained in three independent digestion experiments, scrutinizing significant reduction of immunologically relevant molecules from the surface of immune cells especially following dispase treatment.
Fig. 2.
Quantification of the impact of collagenase and dispase treatment on surface expression of selected markers. (a) Splenocytes were digested with either collagenase D or different dispase I concentrations as described in Materials and methods, followed by antibody staining of selected surface molecules and FACS analysis. Untreated splenocytes served as internal control. Histograms show representative results obtained in one out of three independent experiments. (b) Data obtained from three independent digestion experiments were combined to quantify the impact of enzymatic treatment on surface expression levels of all 18 markers tested. For every marker analyzed, percent expression after enzymatic treatment was normalized to marker expression on untreated splenocytes which was defined as 100%. p values were calculated using t-test, and p<0.05 is indicated as *, p<0.01 is indicated as **, and p<0.001 is indicated as ***
Recovery of surface molecule expression on immune cells after in vitro culture
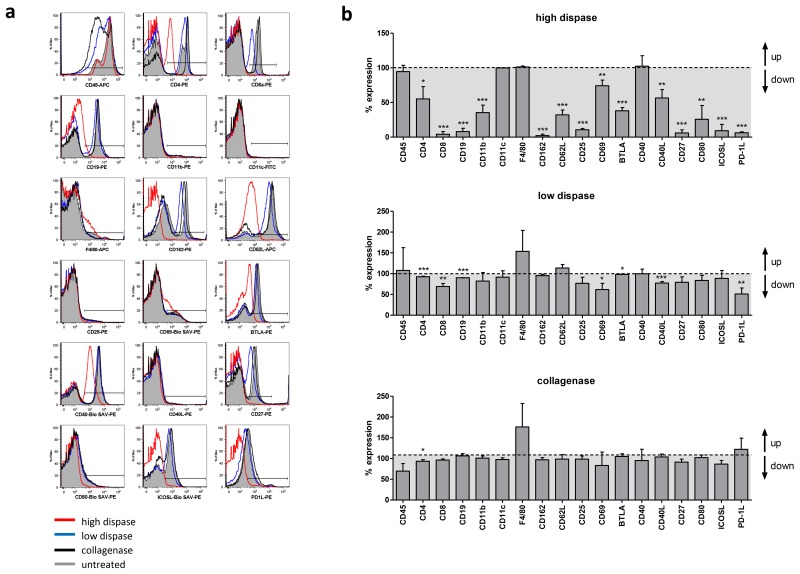
In order to test whether surface molecule expression may be restored after short-term in vitro culture, splenocytes that underwent enzymatic digestion were cultured for 24 h before reassessment of surface marker expression by flow cytometry. As depicted in Figure 3, surface expression to baseline level was found for CD162, CD62L, CD25, CD27, CD80, and ICOS-L after in vitro culture following low dispase treatment. Expression of other markers such as CD4, CD8, CD40L, and PD-1L was only partially restored and remained below the surface expression level of untreated cells. However, 1 day of culture was sufficient to restore >50% surface expression for all markers analyzed. In contrast to this, high-dose dispase treatment rendered the cells almost completely negative for surface expression of CD8, CD19, CD162, CD25, CD27, ICOSL, and PD-1L even 24 h after the initial treatment. Other markers such as CD4, CD11b, CD62L, and CD80, which were fully restored 24 h after low dispase digestion, still did not reach surface expression level found in untreated control cells. Of note, surface molecule expression pattern of splenocytes treated with collagenase largely resembled that of undigested immune cells following 24 h in vitro culture.
Fig. 3.
Partial recovery of surface molecule expression on immune cells following 24 h in vitro culture after enzymatic digestion. (a) Splenocytes were digested with either collagenase D or different dispase I concentrations as described in Materials and methods, followed by 24 h in vitro culture in IMDM compl. at 37 °C and 5% CO2. Cells were harvested and stained for subsequent FACS analysis with the antibodies indicated on the x axis of the histograms. Untreated splenocytes served as internal control. Histograms show representative results obtained in one out of three independent experiments. (b) Data obtained from three independent digestion experiments were combined to quantify the impact of enzymatic treatment on surface expression levels of all 18 markers tested. For every marker analyzed, percent expression after enzymatic treatment was normalized to marker expression on untreated splenocytes which was defined as 100%. p values were calculated using t-test; p<0.05 is indicated as *, p<0.01 is indicated as ** and p<0.001 is indicated as ***
High dose dispase treatment impairs antigen specific CD4+ and CD8+ T cell proliferation
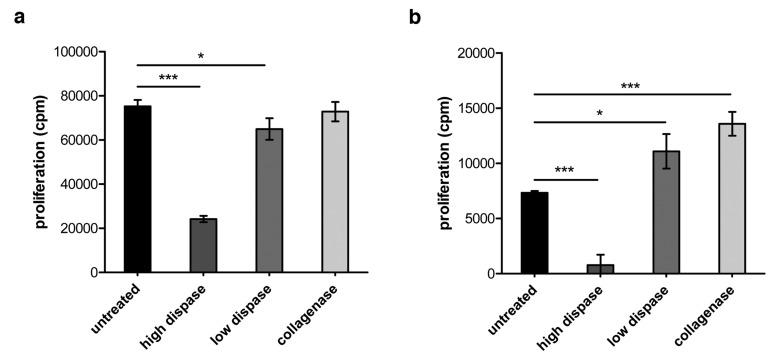
Since surface molecules play a fundamental role in cell–cell interaction and as part of the immunological synapse are crucially involved in the transmission of positive or negative signals from one immune cell to another, we studied the impact of enzymatic digestion on the outcome of antigen-specific T cell proliferation. To this end, splenocytes were isolated from TCR-HA and CL4 transgenic mice, respectively, that harbor a certain population of hemagglutinin (HA)-specific CD4+ (TCR-HA) or CD8+ (CL4) T cells within their T cell repertoire. Splenocytes were subjected to collagenase, and high- and low-dose dispase treatment followed by in vitro culture in the presence of the corresponding MHC class I or MHC class II HA-peptide. As depicted in Figure 4, high dispase treatment of splenocytes drastically reduces the proliferative capacity of CD4+ as well as of CD8+ T cells. In contrast and consistent with the far less pronounced impact of collagenase treatment on surface molecule expression, collagenase digestion of immune cells does not impair antigen-specific proliferation of CD4+ T cells (Fig. 4a) but even elevates proliferation in case of antigenic stimulation of HA-specific CD8+ T cells (Fig. 4b). Together, these data indicate that isolation of cellular subsets by means of enzymatic tissue disintegration may have sustained effects on surface expression of a broad range of immunologically relevant molecules with direct consequences for functionality of immune cell subsets in subsequent in vitro test assays.
Fig. 4.
Impact of enzymatic digestion on proliferative capacity of CD4+ and CD8+ T cells. (a) Splenocytes were isolated from TCR-HA mice and treated with collagenase or dispase as described in the section Materials and methods. Subsequently, 5 × 105 enzyme-treated spleen cells and untreated control cells were seeded per well of a 96-well microtiter plate and stimulated in vitro with 0.01 µg/ml of the corresponding HA peptide 110–120 for 24 h. 3[H]-thymidine was added for another 16 h of culture before harvesting the cells and scintillation counting to determine antigen-specific CD4+ T cell proliferation. (b) 1 × 105 splenocytes isolated from CL4 mice and treated with the respective enzymes were stimulated in vitro with 0.001 µg/ml of the corresponding HA peptide 512–520 for 24 h before the addition of 3[H]-thymidine and further culture for 12 h. Radioactively labeled cells were harvested, and antigen-specific CD8+ T cell proliferation was determined by scintillation counting. p values were calculated using t-test; p<0.05 is indicated as *, and p<0.001 is indicated as ***
The observation that the use of enzymes to isolate cellular subsets from complex tissues may have side effects with respect to surface marker expression and cell function is not entirely new. Consistent with our data, dispase treatment has been shown before to dramatically reduce CD4 and CD25 surface expression on murine T cells [11], whereas collagenase/DNase digestion of human peripheral blood leukocytes had only marginal impact on expression level of surface molecules such as MHC class II, CD3, CD8, and interleukin-2 receptor on human peripheral blood leukocytes [16]. Whereas we found sustained effect of dispase treatment on CD4 and CD8 surface expression even 24 h after treatment, White and colleagues demonstrated recovery of CD4 and CD8 expression after overnight culture following treatment of human female reproductive tract cells with an enzyme cocktail composed of pancreatin, hyaluromidase, and collagenase [12]. Since many different protocols exist with, in part, great variations regarding enzyme concentrations, combination of enzymes used for the isolation of cells, target cells/tissue, incubation times, nature of markers analyzed, etc., existing data are difficult to compare or to generalize. We were facing the problem that enzymatic digestion may have profound effects on surface marker expression first when we applied the standard protocol routinely used in our laboratory for the isolation of lung CD4+ T cells [3] to pulmonary CD8+ T cells. Consistent with Figure 2, we found that the CD8 molecule appears to be extremely vulnerable to the enzymatic activity of dispase I, even at low concentrations, whereas collagenase treatment does not affect CD8 expression and is at the same time sufficient for isolation of high lymphocyte numbers from the murine lung (data not shown). It would therefore be highly recommended to set up optimized experimental conditions with respect to the enzyme(s) used for liberation of cells dependent on the cell type of interest for subsequent magnetic or flow cytometric isolation or FACS analysis to avoid difficulties in the detection of selected markers applying flow cytometric analysis or to prevent low yield of cells when applying classical cell isolation protocols such as FACS or MACS that are based on positive selection of antibody-stained target cells.
Enzymatic removal of surface molecules may also account for the potential discordance between FACS data and data obtained by gene expression analysis. In order to isolate primary alveolar type II epithelial cells (AECII) from the murine lung, we are generally applying a protocol that is based on high dispase treatment [13]. Whereas gene expression profiling using Affymetrix GeneChips and quantitative real-time RT-PCR revealed in part dramatic differences in the expression level of several surface molecules between AECII isolated from the lung of healthy mice and mice suffering from autoimmune-mediated lung inflammation [13], we were not able to confirm differential expression for all of the markers tested on the protein level by applying antibody-mediated surface staining and FACS analysis (unpublished data). Thus, to circumvent this obvious technical limitation of enzyme-mediated tissue disintegration, which is on the one hand the prerequisite for the liberation of AECII from the lung tissue but at the same time destroys potential surface marker epitopes needed for subsequent antibody staining and FACS-based characterization, we are currently establishing protocols to apply iterative chip-based cytometry (iCBC) to whole lung tissue slices. Like multi-epitope-ligand cartography (MELC) described by Schubert et al., iCBC represents a microscopic robot technology for high-throughput protein co-localization studies that runs cycles of fluorescence tagging, imaging, and bleaching in situ and thereby allows for the analysis of virtually unlimited numbers of intracellular and surface markers even on living immune cells [17, 18]. Automated single-cell recognition software provides numerous options for marker combination analysis and strategic experimental conception, and thus MELC and iCBC may represent powerful alternatives for the characterization of cells within tissues in case their liberation would require experimental conditions being incompatible with subsequent flow cytometric analysis of surface markers of particular interest.
In addition to the obvious obstacle that proteolytic enzymes diminish expression levels of certain key cell surface molecules, and thereby complicate identification of cellular subsets by FACS-based approaches and their isolation using magnetic beads or flow cytometry, these molecular alterations obviously can also affect immune cell functions. Holt and colleagues investigated the impact of collagenase/DNase digestion procedure of human lung parenchyma on polyclonal T cell proliferation, antigen presentation, IL-1 production, and NK cell activity [16]. Consistent with the relatively minor effect of the enzyme combination on the expression of T cell, B cell, monocyte, and macrophage markers, the cells behaved almost normally in all functional assays [16]. Interestingly, background proliferation of T cells and spontaneous IL-1 production tend to be elevated in enzyme-treated cells, which may be indicative for their transient activation during collagenase digestion. This is in line with our observation that of the 48 markers tested, nine were found to be induced rather than removed from the surface of immune cells (Fig. 1). Of note, not only enzymatic but also mechanical cell isolation techniques can affect immune cell function in subsequent in vitro studies. In this context, Bland and colleagues have shown that isolation of human large bowel lymphoid cells on the basis of mechanic processes stimulated the synthesis of prostaglandin E2, which directly suppressed cytotoxic activity in mechanically-liberated lymphocytes. This inhibitory effect was not observed when using a protocol based on collagenase digestion [19].
Using a well-defined experimental setup based on transgenic HA-specific T cells, we could show that treatment of immune cells with high dispase concentration drastically reduces antigen-specific proliferation of both CD4+ and CD8+ T cells (Fig. 4). This was not unexpected, since dispase-mediated cleavage of key surface molecules including the two co-receptors CD4 and CD8 on T cells may directly hamper optimal formation of the immunological synapse, thereby interfering with T cell receptor-mediated recognition of MHC/peptide complexes on the surface of antigen presenting cells [20]. In addition, molecules such as CD27 and the high-affinity IL-2 receptor CD25 (and possibly other important receptors not included in this survey) were found to be extremely sensitive to dispase treatment, which may have direct effects on the expansion and survival of activated T cells [21, 22]. Interestingly, whereas high-dose dispase digestion dampens in vitro T cell proliferation, collagenase treatment even seems to promote CD8+ T cell activation (Fig. 4). This is well in line with our observation that collagenase does not reduce surface expression of molecular markers on immune cells (Figs 1 and 2), and moreover, expression level of MHC class I molecules is even increased in collagenase-treated cells, suggesting improved MHC class I mediated antigen display in support of improved CD8+ T cell proliferation.
Together our data indicate that enzymatic tissue disintegration can have profound effects on the expression of a variety of cell-surface molecules with direct consequences for phenotypic analysis, FACS- and MACS-based target cell isolation, and immune cell function in cell culture experiments. This underscores the importance of designing adequate pre-experiments to decipher the specific impact of a particular enzyme cocktail used for the liberation of immune cells from a tissue on their phenotype and function, and thereby to prevent drawing invalid conclusions.
Acknowledgments
This work was supported by grants from the German Research Foundation (DFG) to GH (SFB587, TP B14) and DB (SFB587, TP B12), and a stipend from the Hanover Biomedical Research School to AA (DFG GSC 108).
Contributor Information
A. Autengruber, 1Immune Regulation Group, Helmholtz Centre for Infection Research, Braunschweig, Germany.
M. Gereke, 1Immune Regulation Group, Helmholtz Centre for Infection Research, Braunschweig, Germany; 2Infection Immunology Group, Institute of Medical Microbiology, Faculty of Medicine, Otto-von-Guericke-University, Magdeburg, Germany.
G. Hansen, 3Department of Pediatric Pneumology, Allergology and Neonatology, Hannover Medical School, Hannover, Germany.
C. Hennig, 3Department of Pediatric Pneumology, Allergology and Neonatology, Hannover Medical School, Hannover, Germany.
D. Bruder, 1Immune Regulation Group, Helmholtz Centre for Infection Research, Braunschweig, Germany; 2Infection Immunology Group, Institute of Medical Microbiology, Faculty of Medicine, Otto-von-Guericke-University, Magdeburg, Germany.
References
- 1.Kroemer G, Cuende E, Martínez C. Compartmentalization of the peripheral immune system. Adv Immunol. 1993;53:157–216. doi: 10.1016/s0065-2776(08)60500-3. [DOI] [PubMed] [Google Scholar]
- 2.Arstila T, Arstila TP, Calbo S, Selz F, Malassis-Seris M, Vassalli P, Kourilsky P, Guy-Grand D. Identical T cell clones are located within the mouse gut epithelium and lamina propia and circulate in the thoracic duct lymph. J Exp Med. 2000 Mar 6;191(5):823–834. doi: 10.1084/jem.191.5.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruder D, Westendorf AM, Geffers R, Gruber AD, Gereke M, Enelow RI, Buer J. CD4 T Lymphocyte-mediated lung disease: steady state between pathological and tolerogenic immune reactions. Am J Respir Crit Care Med. 2004 Dec 1;170(11):1145–1152. doi: 10.1164/rccm.200404-464OC. [DOI] [PubMed] [Google Scholar]
- 4.Derbinski J, Schulte A, Kyewski B, Klein L. Promiscuous gene expression in medullary thymic epithelial cells mirrors the peripheral self. Nat Immunol. 2001 Nov;2(11):1032–1039. doi: 10.1038/ni723. [DOI] [PubMed] [Google Scholar]
- 5.Sedgwick JD, Schwender S, Imrich H, Dörries R, Butcher GW, ter Meulen V. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):7438–7442. doi: 10.1073/pnas.88.16.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weigmann B, Tubbe I, Seidel D, Nicolaev A, Becker C, Neurath MF. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat Protoc. 2007;2(10):2307–2311. doi: 10.1038/nprot.2007.315. [DOI] [PubMed] [Google Scholar]
- 7.Westendorf AM, Templin M, Geffers R, Deppenmeier S, Gruber AD, Probst-Kepper M, Hansen W, Liblau RS, Gunzer F, Bruder D, Buer J. CD4+ T cell mediated intestinal immunity: chronic inflammation versus immune regulation. Gut. 2005 Jan;54(1):60–69. doi: 10.1136/gut.2003.037663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corti M, Brody AR, Harrison JH. Isolation and primary culture of murine alveolar type II cells. Am J Respir Cell Mol Biol. 1996 Apr;14(4):309–315. doi: 10.1165/ajrcmb.14.4.8600933. [DOI] [PubMed] [Google Scholar]
- 9.Gereke M, Jung S, Buer J, Bruder D. Alveolar type II epithelial cells present antigen to CD4(+) T cells and induce Foxp3(+) regulatory T cells. Am J Respir Crit Care Med. 2009 Mar 1;179(5):344–355. doi: 10.1164/rccm.200804-592OC. [DOI] [PubMed] [Google Scholar]
- 10.Lienenklaus S, Cornitescu M, Zietara N, Łyszkiewicz M, Gekara N, Jabłónska J, Edenhofer F, Rajewsky K, Bruder D, Hafner M, Staeheli P, Weiss S. Novel reporter mouse reveals constitutive and inflammatory expression of IFN-beta in vivo. J Immunol. 2009 Sep 1;183(5):3229–3236. doi: 10.4049/jimmunol.0804277. [DOI] [PubMed] [Google Scholar]
- 11.Ford AL, Foulcher E, Goodsall AL, Sedgwick JD. Tissue digestion with dispase substantially reduces lymphocyte and macrophage cell-surface antigen expression. J Immunol Methods. 1996 Jul 17;194(1):71–75. doi: 10.1016/0022-1759(96)00067-1. [DOI] [PubMed] [Google Scholar]
- 12.White HD, Prabhala RH, Humphrey SL, Crassi KM, Richardson JM, Wira CR. A method for the dispersal and characterization of leukocytes from the human female reproductive tract. Am J Reprod Immunol. 2000 Aug;44(2):96–103. doi: 10.1111/j.8755-8920.2000.440205.x. [DOI] [PubMed] [Google Scholar]
- 13.Gereke M, Gröbe L, Prettin S, Kasper M, Deppenmeier S, Gruber AD, Enelow RI, Buer J, Bruder D. Phenotypic alterations in type II alveolar epithelial cells in CD4+ T cell mediated lung inflammation. Respir Res. 2007 Jul 4;8:47. doi: 10.1186/1465-9921-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirberg J, Baron A, Jakob S, Rolink A, Karjalainen K, von Boehmer H. Thymic selection of CD8+ single positive cells with a class II major histocompatibility complex-restricted receptor. J Exp Med. 1994 Jul 1;180(1):25–34. doi: 10.1084/jem.180.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan DJ, Liblau R, Scott B, Fleck S, McDevitt HO, Sarvetnick N, Lo D, Sherman LA. CD8(+) T cell-mediated spontaneous diabetes in neonatal mice. J Immunol. 1996 Aug 1;157(3):978–983. [PubMed] [Google Scholar]
- 16.Holt PG, Robinson BW, Reid M, Kees UR, Warton A, Dawson VH, Rose A, Schon-Hegrad M, Papadimitriou JM. Extraction of immune and inflammatory cells from human lung parenchyma: evaluation of an enzymatic digestion procedure. Clin Exp Immunol. 1986 Oct;66(1):188–200. [PMC free article] [PubMed] [Google Scholar]
- 17.Hennig C, Adams N, Hansen G. A versatile platform for comprehensive chip-based explorative cytometry. Cytometry A. 2009 Apr;75(4):362–370. doi: 10.1002/cyto.a.20668. [DOI] [PubMed] [Google Scholar]
- 18.Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Böckelmann R, Malykh Y, Gollnick H, Friedenberger M, Bode M, Dress AW. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol. 2006 Oct;24(10):1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 19.Bland PW, Richens ER, Britton DC, Lloyd JV. Isolation and purification of human large bowel mucosal lymphoid cells: effect of separation technique on functional characteristics. Gut. 1979 Dec;20(12):1037–1046. doi: 10.1136/gut.20.12.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gascoigne NR, Zal T, Yachi PP, Hoerter JA. Co-receptors and recognition of self at the immunological synapse. Curr Top Microbiol Immunol. 2010;340:171–189. doi: 10.1007/978-3-642-03858-7_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendriks J, Xiao Y, Borst J. CD27 promotes survival of activated T cells and complements CD28 in generation and establishment of the effector T cell pool. J Exp Med. 2003 Nov 3;198(9):1369–1380. doi: 10.1084/jem.20030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Parijs L, Biuckians A, Ibragimov A, Alt FW, Willerford DM, Abbas AK. Functional responses and apoptosis of CD25 (IL-2R alpha)-deficient T cells expressing a transgenic antigen receptor. J Immunol. 1997 Apr 15;158(8):3738–3745. [PubMed] [Google Scholar]