Abstract
The objective of this paper was to investigate whether retrospective pulsed-field gel electrophoresis (PFGE) of methicillin-resistant Staphylococcus aureus (MRSA) isolates at two-year intervals is suitable and sufficient to demonstrate changes in the clonal composition of MRSA isolates and to identify previously undetected local outbreaks. PFGE patterns of 400 MRSA isolates were collected between 2004 and 2008 at the University of Rostock Hospital in Germany, and were used to assess the prevalence of MRSA clones at different time points. Only minor changes were detected. The combined analysis of all isolates that were collected per year reduced the time needed to perform this laborious procedure. The retrospective identification of outbreaks may require shorter intervals. Improved infection prevention and control measures prevented further outbreaks in previously affected hospital departments. In conclusion, PGFE at two-year intervals is sufficient to detect changes in the clonal composition of local MRSA isolates. If time for identification is important during outbreak investigations, more rapid methods with a similarly high discriminatory power such as spa typing should be used.
Keywords: epidemic clone, methicillin-resistant Staphylococcus aureus, MRSA, outbreak, PFGE, pulsed-field gel electrophoresis, typing
Introduction
Pulsed-field gel electrophoresis (PFGE) is a reliable tool for comparing methicillin-resistant Staphylococcus aureus (MRSA) isolates to known epidemic clones. This technique, however, requires specialised equipment and a high level of technical expertise, and has the additional disadvantage of poor inter-laboratory comparability. As a result, sequence-based typing methods such as spa typing or multilocus sequence typing (MLST) are often preferred. The objective of this study was therefore to investigate whether PFGE is still a useful MRSA typing method for the purpose of infection prevention and control in a hospital setting. For this reason, the prevalence of endemic MRSA clones at the University of Rostock Hospital in Germany was assessed using MRSA strains isolated between 2004 and 2008. Six MRSA clones are considered endemic in Rostock, i.e. the ‘Northern German’ clone (spa type t051; sequence type ST247; clonal complex CC8), the ‘Southern German’ clone (t001; ST228; CC5), the ‘Hannover’ clone (t009; ST254; CC8), the ‘Berlin’ clone (t004, t038, t065; ST45; CC45), the ‘Barnim’ clone (t032, t022, t005; ST22; CC22), and the ‘Rhine-Hesse’ clone (t002, t045; ST5; CC5) as defined by the Robert Koch Institute (RKI, Wernigerode, Germany) (Table 1), which is the National Reference Centre for Staphylococci in Germany. A further objective of this study was to assess the usefulness of PFGE data for retrospectively identifying local outbreaks and modifying the infection prevention and control strategies of the hospital departments affected.
Table 1.
Phylogenetic classification of MRSA clones with known endemicity in Rostock (Germany) according to the National Reference Centre for Staphylococci Robert Koch Institute, Wernigerode (Germany)
| Clones | ʻNorthern Germanʼ | ʻSouthern Germanʼ | ʻHannoverʼ | ʻBerlinʼ | ʻBarnimʼ | ʻRhine-Hesseʼ |
|---|---|---|---|---|---|---|
| Corresponding spa types | t051 | t001 | t009 | t004, t038, t065 | t005, t022, t032 | t002, t045 |
| Corresponding sequence types | ST247 | ST228 | ST254 | ST45 | ST22 | ST5 |
| Corresponding clonal complexes | CC8 | CC5 | CC8 | CC45 | CC22 | CC5 |
Materials and methods
Strain collection
Methicillin-resistant S. aureus (MRSA) isolates are routinely collected and stored for 5 years at the Institute of Medical Microbiology of the University of Rostock Hospital. Only the first isolates that were obtained from new MRSA cases in 2004, 2006, and 2008 were included in the study. Copy strains were excluded. Screening was performed during the entire study period in accordance with the recommendations of the National Reference Centre for Infectious Diseases (Robert Koch Institute) [1].
Pulsed-field gel electrophoresis
MRSA isolates from each patient were analysed using pulsed-field gel electrophoresis (PFGE) as described elsewhere [2, 3]. Visual inspection was performed on the basis of the criteria established by Tenover et al. [4]. The results were analysed with GelCompar II software (Applied Maths, Sint-Martens-Latem, Belgium; settings: Dice coefficient, UPGMA, 0.5% optimisation, 1.0% position tolerance).
Outbreak identification
MRSA distribution in the various hospital departments and outbreaks were assessed retrospectively. An outbreak was defined as three or more different cases of nosocomial infection or colonisation by one MRSA clone per month. All repeated occurrences of an MRSA clone in a department were tracked, however, if the interval between occurrences did not exceed 2 months.
Statistical analysis
Non-numerical data were analysed by Fisher’s exact test. Numerical data were analysed by Student’s t-test with different variances using GraphPad Instat 3 and Microsoft® Excel software. Significance was accepted at p<0.05. If the p-value was smaller than 0.01, the differences were considered highly significant. If the p-value was smaller than 0.001, the differences were considered very highly significant.
Results and discussion
Strain collection and distribution of epidemic clones
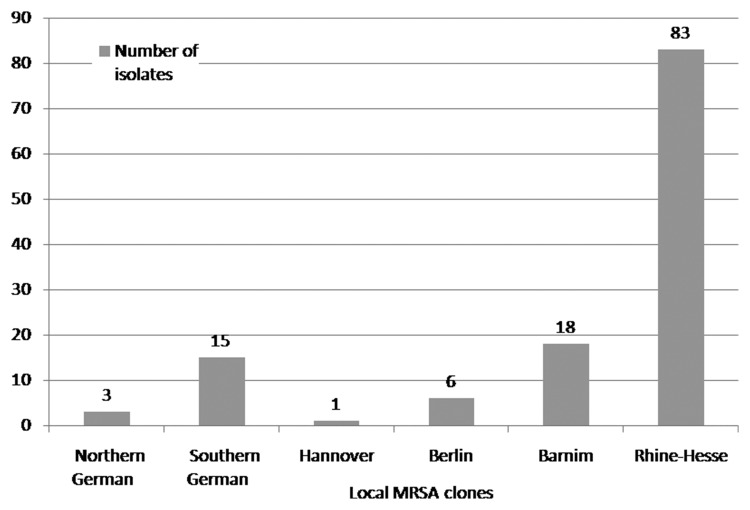
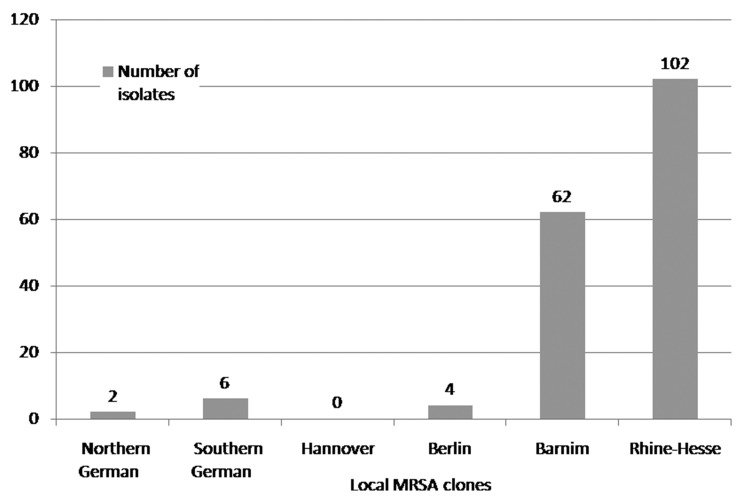
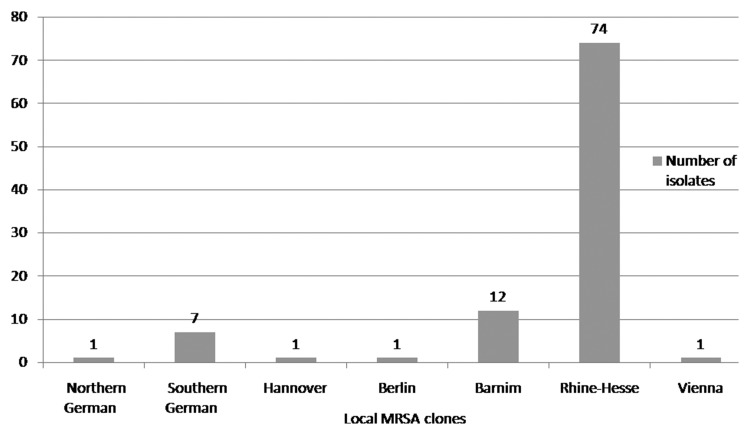
MRSA strains were collected inconsistently as a result of organisational problems in the routine laboratory. In total, 127 of 168 (76%) different isolates were collected and analysed in 2004, 97 of 165 (59%) in 2006, and 176 of 267 (66%) in 2008. The clinical isolates that were assigned to the ‘Northern German’ epidemic clone accounted for 2% (n=3) in 2004, 1% (n=1) in 2006, and 1% (n=2) in 2008. The isolates that clustered with the ‘Southern German’ epidemic clone accounted for 12% (n=15) in 2004, 7% (n=7) in 2006, and 3% (n=6) in 2008. The isolates that clustered with the ‘Hannover’ epidemic clone accounted for 1% (n=1) in 2004, 1% (n=1) in 2006, and 0% (n=0) in 2008. The clinical isolates that were assigned to the ‘Berlin’ epidemic clone accounted for 5% (n=6) in 2004, 1% (n=1) in 2006, and 2% (n=4) in 2008. The isolates that clustered with the ‘Barnim’ epidemic clone accounted for 15% (n=19) in 2004, 13% (n=12) in 2006, and 35% (n=62) in 2008. The isolates that were assigned to the ‘Rhine-Hesse’ epidemic clone accounted for 65% (n=83) in 2004, 76% (n=74) in 2006, and 58% (n=102) in 2008. One isolate that was found in 2006 could not be assigned to one of the six aforementioned epidemic clones and was identified by the National Reference Centre for Staphylococci (RKI) as belonging to the ‘Vienna’ epidemic clone (phylogenetic classification of the ‘Vienna’ epidemic clone: t037, t030; ST239; CC8) (Figs 1–3).
Fig. 1.
Distribution of endemic MRSA clones at the University of Rostock Hospital in 2004
Fig. 3.
Distribution of endemic MRSA clones at the University of Rostock Hospital in 2008
Fig. 2.
Distribution of endemic MRSA clones at the University of Rostock Hospital in 2006
The differences in the MRSA incidences in 2004, 2006, and 2008 were highly significant (p<0.01). Furthermore, there was a very highly significant increase (p<0.001) in the percentages of the ‘Barnim’ clone from 2004 to 2006 and from 2006 to 2008, and a highly significant decrease (p<0.01) in the percentages of the ‘Southern German’ clone from 2004 to 2008 and the ‘Rhine-Hesse’ clone from 2006 to 2008. No further significant changes were detected.
Identified outbreaks
Nine distinct outbreaks affecting three to six patients each were identified retrospectively. Of these, seven had already been known at the time of outbreak as a result of the investigations by the hospital department responsible for infection prevention and control. Four outbreaks occurred on surgical wards, three on medical wards, one on a nuclear medicine ward, and one on a neurological ward.
All four outbreaks that occurred on general surgical and surgical intensive care wards were detected in 2004 and were caused by the ‘Rhine-Hesse’ clone. In 2006, the ‘Rhine-Hesse’ clone was responsible for two outbreaks on general medical wards and one outbreak on a neurosurgical intensive care ward. In 2008, the ‘Rhine-Hesse’ clone was identified as the causative agent of two outbreaks on a general medical ward. The ‘Barnim’ clone was associated with an outbreak on a nuclear medicine ward.
In addition, several clones that did not meet the aforementioned outbreak criteria were repeatedly detected during the study period and occurred particularly in intensive care units, and, to a lesser degree, in general surgical and medical departments. Only the Barnim and ‘Rhine-Hesse’ clones were associated with cumulative increases in the number of isolates.
The cumulative increases that did not meet the criteria for an outbreak and two of nine outbreaks were identified only retrospectively. These findings show that the real-time or near-real-time detection of outbreaks by molecular typing requires more rapid procedures.
Past and present methods of DNA-based MRSA typing
The literature describes a wide variety of methods for typing S. aureus and MRSA including a number of DNA-based techniques. Many methods are affected by limitations regarding their discriminatory power, reproducibility, stability, or immediate availability in the event of an outbreak [5, 6].
Plasmid profile analysis was the first molecular method of studying the epidemiology of MRSA. Its discriminatory power is, however, limited by the influences of the complex three-dimensional structure of native plasmids on the mobility of plasmids in gel and the ability of bacteria to spontaneously loose or acquire plasmids [5]. Despite its disadvantages, plasmid profile analysis is still in use today [7].
More recent methods are based on the analysis of chromosomal DNA such as the interpretation of DNA restriction patterns and Southern blotting followed by hybridisation with DNA probes [5]. These developments culminated in the first successful PFGE analysis that was described for yeast DNA in 1984 [8]. The main disadvantage of PFGE is that it is a time-consuming method and the results are not available for several days [9].
The retrospective typing of the MRSA-isolates at the University of Rostock Hospital between 2004 and 2008 was exclusively performed by PFGE. The assignments to spa types or sequence types and resulting clonal complexes were based upon the phylogenetic classification by German National Reference Centre for Staphylococci (Table 1).
Polymerase chain reaction (PCR)-based methods of typing MRSA include PCR-restriction fragment length polymorphism (PCR-RFLP), PCR ribotyping, arbitrarily primed-PCR (AP-PCR) also known as typing by random amplified polymorphic DNA (RAPD), which is performed with short primers (up to ten bases) that have no known homology to genome sequences of the target organism, and the repetitive palindromic extragenic elements PCR (rep-PCR). The discriminatory power of PCR-based approaches, however, is poorer than that of PFGE, especially when it comes to investigating an endemic circulation of clones [5, 9]. Commercially available rep-PCR-based typing systems are available as well, such as the DiversiLab microbial typing system (bioMérieux, Nuertingen, Germany) [10]. The advantage of commercial systems is that they provide rapid results and are easy to use [11]. Another PCR-based typing method is multiple-locus variable-number tandem-repeat (VNTR) analysis (MLVA). Repeat-containing DNA fragments are amplified by multiplex PCR, and the resulting band patterns in agarose gels are compared [12, 13]. High concordance between MLVA results and multilocus sequence typing (MLST) results can be achieved if appropriate cutoffs are used to distinguish between identical and different band patterns [14].
Sequence-based typing, especially MLST, correlates well with PFGE and facilitates supra-regional comparisons on the basis of numerical codes. The required sequencing of several genes is, however, laborious and expensive [5, 15]. For this reason, simpler techniques have been developed such as spa typing. This is a rapid and reproducible method based on analysing the polymorphic X region of the staphylococcal protein A (spa) gene, which consists of a variable number of 24 base pair repeats flanked by well-conserved regions [7, 16]. Sequence-based spa typing shows good correlation with PFGE and possesses a high discriminatory power [17, 18]. Another simple typing technique is double-locus sequence typing (DLST), which uses the spa and clfB genes, and shows an 88% concordance with PFGE [19].
The comparison of single-nucleotide polymorphisms (SNP) is also a sequence-based typing method [20] and provides the basis for real-time PCR or low-density array-based methods [21]. SNP analysis of three Staphylococcus enterotoxin genes (set2, set5, and set7) was reported to have even higher discriminatory power than MLST [22]. The use of sets of single-nucleotide polymorphisms in combination with binary markers, i.e. the presence or absence of certain pathogenicity factors, has been described for MRSA typing as well [23].
PFGE for the detection of changes in the clonal distribution of MRSA isolates and outbreaks
The long time required for results is one of the limitations of PFGE [9]. However, this disadvantage plays only a minor role if no real-time surveillance is intended, e.g. if isolates are not typed immediately after isolation but are collected and typed retrospectively at yearly intervals. If entire sample series are analysed at a later time, the time spent handling individual isolates can be reduced. Complex and expensive procedures such as sequencing are not required at all.
Our results suggest that if PFGE is performed at regular intervals, it is a suitable technique to detect local changes in the distribution of endemic clones in the hospital setting over the course of several years. During the study period, the ‘Berlin’, ‘Hannover’, and ‘Northern German’ clones were almost absent, and the ‘Rhine-Hesse’ clone was predominant at our hospital. In addition, we observed a relative decrease in the ‘Southern German’ clone and a marked increase in the ‘Barnim’ clone from 2004 to 2008. Since these changes took place gradually, typing intervals of 2 years appear to be sufficient to give a general picture of the situation.
The latter finding is in good concordance with the previously described rareness of geographical spreads of MRSA clones over long distances or across cultural borders [24]. The increasing proportion of the ‘Barnim’ clone in Rostock reflects the epidemiological situation in Northern Germany. Two out of three MRSA isolates from the university hospital of Hannover (Northern Germany) could be assigned to the ‘Barnim’ clone in 2005 [25]. As recently described in 2011, the ‘Barnim’ clone was identified in 70% of MRSA isolates from nursing home residents in Brunswick (Northern Germany) [26]. An emergence of this clone was also monitored in Magdeburg in the centre of Germany between 2002 and 2005 [27]. In the meantime, the ‘Barnim’ clone is considered to be typical for hospital-acquired MRSA in the north of Germany. As published in 2008, the predominant clones in Eastern Saxony in the east of Germany comprise the ‘Rhine-Hesse’ clone, the ‘Southern German’ clone, the ‘Barnim’ clone, and the ‘Berlin’ clone, while, e.g. the ‘Hannover’ was only infrequently isolated [28]. Thus the clonal composition in Rostock reflects both the emergence of the ‘Barnim’ clone in the north of Germany and the stabile persistence of the ‘Rhine-Hesse’ clone in the east of the country. As published in 2009, the ‘Rhine-Hesse’ clone and the ‘Berlin’ clone are common in the Western German parts of the Euregio Meuse-Rhin (EMR) region, while – in addition to these clones – a higher clonal diversity was detected on the Dutch side. The predominant clonal complexes in the EMR region are CC5, CC8, CC30, and CC45 [29]. Another study identified the ‘Barnim’ clone as one of the predominant clones in the Dutch–German border area [30]. The ‘Barnim’ clone also emerged in 2006 near the borders of Belgium and Germany [31]. A differing distribution can be found in the south. While the ‘Southern German’ clone and the ‘Rhine-Hesse’ clone were frequent in Austria between 1996 and 2006, the ‘Northern German’ clone, the ‘Berlin’ clone, and the ‘Barnim’ clone were only occasionally isolated [32]. A great genetic diversity of MRSA isolates was observed at the university hospital Basel (Switzerland) between 2000 and 2005 [33].
In addition to epidemiological monitoring and even more important for the hygiene management of a hospital, PFGE allows for the confirmation of outbreak situations. SmaI macro-restriction-based PFGE is particularly useful for this purpose, because SmaI macro-restriction provides a higher discriminatory power than spa typing or multilocus sequence typing [34]. Several MRSA outbreaks and local cumulative increases in the number of different MRSA isolates were confirmed or identified retrospectively in different hospital departments. These findings allowed the infection prevention and control team to take appropriate measures, and provide relevant infection prevention and control training for clinical staff in order to prevent further outbreaks. Despite these measures, MRSA clones were repeatedly isolated in high-risk departments such as intensive care units. No further outbreaks, however, occurred in previously affected departments so that the infection control measures that were taken after retrospective PFGE analysis have apparently been effective.
Conclusions
A time interval of 2 years is sufficient for PFGE to detect changes in the clonal composition of local MRSA isolates. If time for identification is important during outbreak investigations, more rapid methods with a similarly high discriminatory power such as spa typing should be used. These methods can contribute to the early identification of outbreaks and the immediate institution of appropriate infection control measures.
aParts of this paper were presented at the 61st Annual Meeting of the German Society for Hygiene and Microbiology (Deutsche Gesellschaft für Hygiene und Mikrobiologie – DGHM) in 2009 and in the German-language journal Der Mikrobiologe in 2011 but have not yet been published in an international journal.
Contributor Information
H. Frickmann, 1Department of Tropical Medicine at the Bernhard Nocht Institute, German Armed Forces Hospital of Hamburg, Hamburg, Germany; 2Institute of Medical Microbiology, Virology and Hygiene, University of Rostock Hospital, Rostock, Germany.
P. P. Gawlik, 3Department of Internal Medicine, German Armed Forces Hospital of Ulm, Ulm, Germany.
S. Crusius, 2Institute of Medical Microbiology, Virology and Hygiene, University of Rostock Hospital, Rostock, Germany.
A. Podbielski, 2Institute of Medical Microbiology, Virology and Hygiene, University of Rostock Hospital, Rostock, Germany.
References
- 1.Gastmeier P, Witte W, Kola A, Mattner F, Vonberg RP, Ziesing S, Suerbaum S, Reischl U, Weist K, Wendt C. Management of MRSA Screening. Epi Bull. 2005;42:385–389. [Article in German] [Google Scholar]
- 2.Struelens MJ, Deplano A, Godard C, Maes N, Serruys E. Epidemiologic typing and delineation of genetic relatedness of methicillin-resistant Staphylococcus aureus by macrorestriction analysis of genomic DNA by using pulsed-field gel electrophoresis. J Clin Microbiol. 1992 Oct;30(10):2599–2605. doi: 10.1128/jcm.30.10.2599-2605.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winters MA, Goering RV, Boon SE, Morin R, Sorensen M, Snyder L. Epidemiological analysis of methicillin-resistant Staphylococcus aureus comparing plasmid typing with chromosomal analysis by field inversion gel electrophoresis. Med Microbiol Lett. 1993;2:33–41. [Google Scholar]
- 4.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995 Sep;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trindade PA, McCulloch JA, Oliveira GA, Mamizuka EM. Molecular techniques for MRSA typing: current issues and perspectives. Braz J Infect Dis. 2003 Feb;7(1):32–43. doi: 10.1590/s1413-86702003000100005. [DOI] [PubMed] [Google Scholar]
- 6.Weller TM. Methicillin-resistant Staphylococcus aureus typing methods: which should be the international standard? J Hosp Infect. 2000 Mar;44(3):160–172. doi: 10.1053/jhin.1999.0701. [DOI] [PubMed] [Google Scholar]
- 7.Wildemauwe C, De Brouwer D, Godard C, Buyssens P, Dewit J, Joseph R, Vanhoof R. The use of spa and phage typing for characterization of a MRSA population in a Belgian hospital: comparison between 2002 and 2007. Pathol Biol (Paris) 2010 Feb;58(1):70–72. doi: 10.1016/j.patbio.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz DC, Cantor CR. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 9.Strandén A, Frei R, Widmer AF. Molecular typing of methicillin-resistant Staphylococcus aureus: can PCR replace pulsed-field gel electrophoresis? J Clin Microbiol. 2003 Jul;41(7):3181–3186. doi: 10.1128/JCM.41.7.3181-3186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ross TL, Merz WG, Farkosh M, Carroll KC. Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005 Nov;43(11):5642–5647. doi: 10.1128/JCM.43.11.5642-5647.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shutt CK, Pounder JI, Page SR, Schaecher BJ, Woods GL. Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains. J Clin Microbiol. 2005 Mar;43(3):1187–1192. doi: 10.1128/JCM.43.3.1187-1192.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malachowa N, Sabat A, Gniadkowski M, Krzyszton-Russjan J, Empel J, Miedzobrodzki J, Kosowska-Shick K, Appelbaum PC, Hryniewicz W. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J Clin Microbiol. 2005 Jul;43(7):3095–3100. doi: 10.1128/JCM.43.7.3095-3100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tenover FC, Vaughn RR, McDougal LK, Fosheim GE, McGowan JE., Jr. Multiple-locus variable-number tandem-repeat assay analysis of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2007 Jul;45(7):2215–2219. doi: 10.1128/JCM.02451-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pourcel C, Hormigos K, Onteniente L, Sakwinska O, Deurenberg RH, Vergnaud G. Improved multiple-locus variable-number tandem-repeat assay for Staphylococcus aureus genotyping, providing a highly informative technique together with strong phylogenetic value. J Clin Microbiol. 2009 Oct;47(10):3121–3128. doi: 10.1128/JCM.00267-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000 Mar;38(3):1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallin M, Friedrich AW, Struelens MJ. spa typing for epidemiological surveillance of Staphylococcus aureus. Methods Mol Biol. 2009;551:189–202. doi: 10.1007/978-1-60327-999-4_15. [DOI] [PubMed] [Google Scholar]
- 17.Narukawa M, Yasuoka A, Note R, Funada H. Sequence-based spa typing as a rapid screening method for the areal and nosocomial outbreaks of MRSA. Tohoku J Exp Med. 2009 Jul;218(3):207–213. doi: 10.1620/tjem.218.207. [DOI] [PubMed] [Google Scholar]
- 18.Petersson AC, Olsson-Liljequist B, Miörner H, Haeggman S. Evaluating the usefulness of spa typing, in comparison with pulsed-field gel electrophoresis, for epidemiological typing of methicillin-resistant Staphylococcus aureus in a low-prevalence region in Sweden 2000-2004. Clin Microbiol Infect. 2010 May;16(5):456–462. doi: 10.1111/j.1469-0691.2009.02881.x. [DOI] [PubMed] [Google Scholar]
- 19.Basset P, Senn L, Prod'hom G, Bille J, Francioli P, Zanetti G, Blanc DS. Usefulness of double locus sequence typing (DLST) for regional and international epidemiological surveillance of methicilin-resistant Staphylococcus aureus. Clin Microbiol Infect. 2010 Aug;16(8):1289–1296. doi: 10.1111/j.1469-0691.2009.03070.x. [DOI] [PubMed] [Google Scholar]
- 20.McDonald M, Dougall A, Holt D, Huygens F, Oppedisano F, Giffard PM, Inman-Bamber J, Stephens AJ, Towers R, Carapetis JR, Currie BJ. Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J Clin Microbiol. 2006 Oct;44(10):3720–3727. doi: 10.1128/JCM.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens AJ, Huygens F, Inman-Bamber J, Price EP, Nimmo GR, Schooneveldt J, Munckhof W, Giffard PM. Methicillin-resistant Staphylococcus aureus genotyping using a small set of polymorphisms. J Med Microbiol. 2006 Jan;55(Pt 1):43–51. doi: 10.1099/jmm.0.46157-0. [DOI] [PubMed] [Google Scholar]
- 22.Aguiar-Alves F, Medeiros F, Fernandes O, Gudziki Pereira RM, Perdreau-Remington F, Riley LW. New Staphylococcus aureus genotyping method based on exotoxin (set) genes. J Clin Microbiol. 2006 Aug;44(8):2728–2732. doi: 10.1128/JCM.00376-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huygens F, Inman-Bamber J, Nimmo GR, Munckhof W, Schooneveldt J, Harrison B, McMahon JA, Giffard PM. Staphylococcus aureus genotyping using novel real-time PCR formats. J Clin Microbiol. 2006 Oct;44(10):3712–3719. doi: 10.1128/JCM.00843-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2008 Sep 16;105(37):14130–14135. doi: 10.1073/pnas.0804178105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaberny IF, Bindseil A, Sohr D, Gastmeier P. A point-prevalence study for MRSA in a German university hospital to identify patients at risk and to evaluate an established admission screening procedure. Infection. 2008 Dec;36(6):526–532. doi: 10.1007/s15010-008-7436-1. [DOI] [PubMed] [Google Scholar]
- 26.Pfingsten-Würzburg S, Pieper DH, Bautsch W, Probst-Kepper M. Prevalence and molecular epidemiology of meticillin-resistant Staphylococcus aureus in nursing home residents in northern Germany. J Hosp Infect. 2011 Jun;78(2):108–112. doi: 10.1016/j.jhin.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 27.Ghebremedhin B, König W, Witte W, Hardy KJ, Hawkey PM, König B. Subtyping of ST22-MRSA-IV (Barnim epidemic MRSA strain) at a university clinic in Germany from 2002 to 2005. J Med Microbiol. 2007 Mar;56(Pt 3):365–375. doi: 10.1099/jmm.0.46883-0. [DOI] [PubMed] [Google Scholar]
- 28.Monecke S, Jatzwauk L, Weber S, Slickers P, Ehricht R. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from Eastern Saxony. Clin Microbiol Infect. 2008 Jun;14(6):534–545. doi: 10.1111/j.1469-0691.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 29.Deurenberg RH, Nulens E, Valvatne H, Sebastian S, Driessen C, Craeghs J, De Brauwer E, Heising B, Kraat YJ, Riebe J, Stals FS, Trienekens TA, Scheres J, Friedrich AW, van Tiel FH, Beisser PS, Stobberingh EE. Cross-border dissemination of methicillin-resistant Staphylococcus aureus, Euregio Meuse-Rhin region. Emerg Infect Dis. 2009 May;15(5):727–734. doi: 10.3201/eid1505.071618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Köck R, Brakensiek L, Mellmann A, Kipp F, Henderikx M, Harmsen D, Daniels-Haardt I, von Eiff C, Becker K, Hendrix MG, Friedrich AW. Cross-border comparison of the admission prevalence and clonal structure of meticillin-resistant Staphylococcus aureus. J Hosp Infect. 2009 Apr;71(4):320–326. doi: 10.1016/j.jhin.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 31.Nulens E, Stobberingh EE, Smeets E, van Dessel H, Welling MA, Sebastian S, van Tiel FH, Beisser PS, Deurenberg RH. Genetic diversity of methicillin-resistant Staphylococcus aureus in a tertiary hospital in The Netherlands between 2002 and 2006. Eur J Clin Microbiol Infect Dis. 2009 Jun;28(6):631–639. doi: 10.1007/s10096-008-0686-0. [DOI] [PubMed] [Google Scholar]
- 32.Krziwanek K, Luger C, Sammer B, Stumvoll S, Stammler M, Sagel U, Witte W, Mittermayer H. MRSA in Austria--an overview. Clin Microbiol Infect. 2008 Mar;14(3):250–259. doi: 10.1111/j.1469-0691.2007.01896.x. [DOI] [PubMed] [Google Scholar]
- 33.Fenner L, Widmer AF, Dangel M, Frei R. Distribution of spa types among meticillin-resistant Staphylococcus aureus isolates during a 6 year period at a low-prevalence University Hospital. J Med Microbiol. 2008 May;57(Pt 5):612–616. doi: 10.1099/jmm.0.47757-0. [DOI] [PubMed] [Google Scholar]
- 34.Strommenger B, Kettlitz C, Weniger T, Harmsen D, Friedrich AW, Witte W. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol. 2006 Jul;44(7):2533–2540. doi: 10.1128/JCM.00420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]