Abstract
Purpose
To screen the microarray expression of CDH1, ECM1, EIF1B, FXR1, HTR2B, ID2, LMCD1, LTA4H, MTUS1, RAB31, ROBO1, and SATB1 genes which are predictive of primary uveal melanoma metastasis, and NFKB2, PTPN18, MTSS1, GADD45B, SNCG, HHIP, IL12B, CDK4, RPLP0, RPS17, RPS12 genes that are differentially expressed in metastatic uveal melanoma in normal whole human blood and tissues prone to metastatic involvement by uveal melanoma.
Methods
We screened the GeneNote and GNF BioGPS databases for microarray analysis of genes predictive of primary uveal melanoma metastasis and those differentially expressed in metastatic uveal melanoma in normal whole blood, liver, lung and skin.
Results
Microarray analysis showed expression of all 22 genes in normal whole blood, liver, lung and skin, which are the most common sites of metastases. In the GNF BioGPS database, data for expression of the HHIP gene in normal whole blood and skin was not complete.
Conclusions
Microarray analysis of genes predicting systemic metastasis of uveal melanoma and genes differentially expressed in metastatic uveal melanoma may not be used as a biomarker for metastasis in whole blood, liver, lung, and skin. Their expression in tissues prone to metastasis may suggest that they play a role in tropism of uveal melanoma metastasis to these tissues.
Keywords: Eye, Uvea, Melanoma, Cancer, Metastasis, Microarray, Gene Expression Profiling
INTRODUCTION
The most common site for uveal melanoma metastasis is the liver which is involved in more than 90% of patients with metastatic disease, followed by the lung (31%), bone (23%), and skin (17%).1,2 Micrometastasis usually occurs before the diagnosis or treatment of uveal melanoma. Tumor doubling time indicates that the majority of tumors metastasize 5 years before the primary choroidal melanoma is diagnosed.3 Considering the lack of lymphatics in the eye and the fact that the uvea is a highly vascular organ, uveal melanoma cells should have hematogenous metastasis.
Detection of circulating tumor cells in the bloodstream could have an important role in identifying patients at higher risk of developing systemic metastasis and monitoring the response to adjuvant or systemic therapy. Polymerase chain reaction (PCR) and immunomagnetic cell isolation techniques showed the presence of circulating uveal melanoma cells in uveal melanoma patients.4-15 However, controversial results have been reported regarding the association between the presence of circulating uveal melanoma cells and the development of systemic metastasis.6,8,15
Harbour and coworkers16-19 described a gene expression profile predictive of systemic metastasis in uveal melanoma including CDH1, ECM1, EIF1B, FXR1, HTR2B, ID2, LMCD1, LTA4H, MTUS1, RAB31, ROBO1 and SATB1.6 Based on the gene expression profile, uveal melanomas are classified as class 1A, class 1B or class 2. Molecular classification was found to be correlated with tumor cytology, patient survival, and other prognostic cytogenetic features such as monosomy of chromosome 3 and duplication of chromosome 8q.16-18 Meir et al20 found that liver metastases from uveal melanomas have distant gene expression profile comparable to the primary uveal melanoma. These genes are NFKB2, PTPN18, MTSS1, GADD45B, SNCG, HHIP, IL12B, CDK4, RPLP0, RPS17 and RPS12.20 The gene expression profile of liver metastases was similar to that of the normal liver tissue. It is not known whether this similar gene expression profile contributes to tropism of uveal melanoma metastasis to the liver. The reported correlation between growth factors produced by the liver such as hepatocyte growth factor, epidermal growth factor, insulin-like growth factor-1 and their receptors, and the development of liver metastasis might also implicate homing and survival of metastatic melanoma cells to the liver.21-23
In the current study, we evaluated the microarray expression of genes predictive for metastasis of uveal melanoma and genes differentially expressed in uveal metastases in normal whole blood and in tissues prone to metastatic involvement by uveal melanoma.
Methods
GeneNote is a full-genome database of gene expression in healthy human issues. GNF BioGPS is a gene annotation portal that provides access to hundreds of online gene annotation databases. We screened the GeneNote and GNF BioGPS databases at the website of GeneCards.org (Weizmann Institute of Science, Rehovot, Israel) for microarray expression of CDH1, ECM1, EIF1B, FXR1, HTR2B, ID2, LMCD1, LTA4H, MTUS1, RAB31, ROBO1, SATB1, NFKB2, PTPN18, MTSS1, GADD45B, SNCG, HHIP, IL12B, CDK4, RPLP0, RPS17 and RPS12 in normal whole blood, liver, lung, and skin.24
In screening the GeneNote and GNF BioGPS databases, the custom array, previously used to profile different human tissue, was designed to interrogate the expression of various human genes. Measurements were obtained for normal human tissues hybridized against Affymetrix GeneChips HG-U95A-E (GeneNote) and HG-U133A (GNF BioGPS). The Affymetrix MAS5 algorithm was used for array processing. The intensity values for each gene were averaged between duplicates, followed by averaging probeset values for each gene, and then global median-normalizing and scaling the median values to the same median, half-way between the GeneNote and GNF BioGPS medians. Normalized intensities were drawn on a root scale, which is an intermediate between log and linear scales.
Results
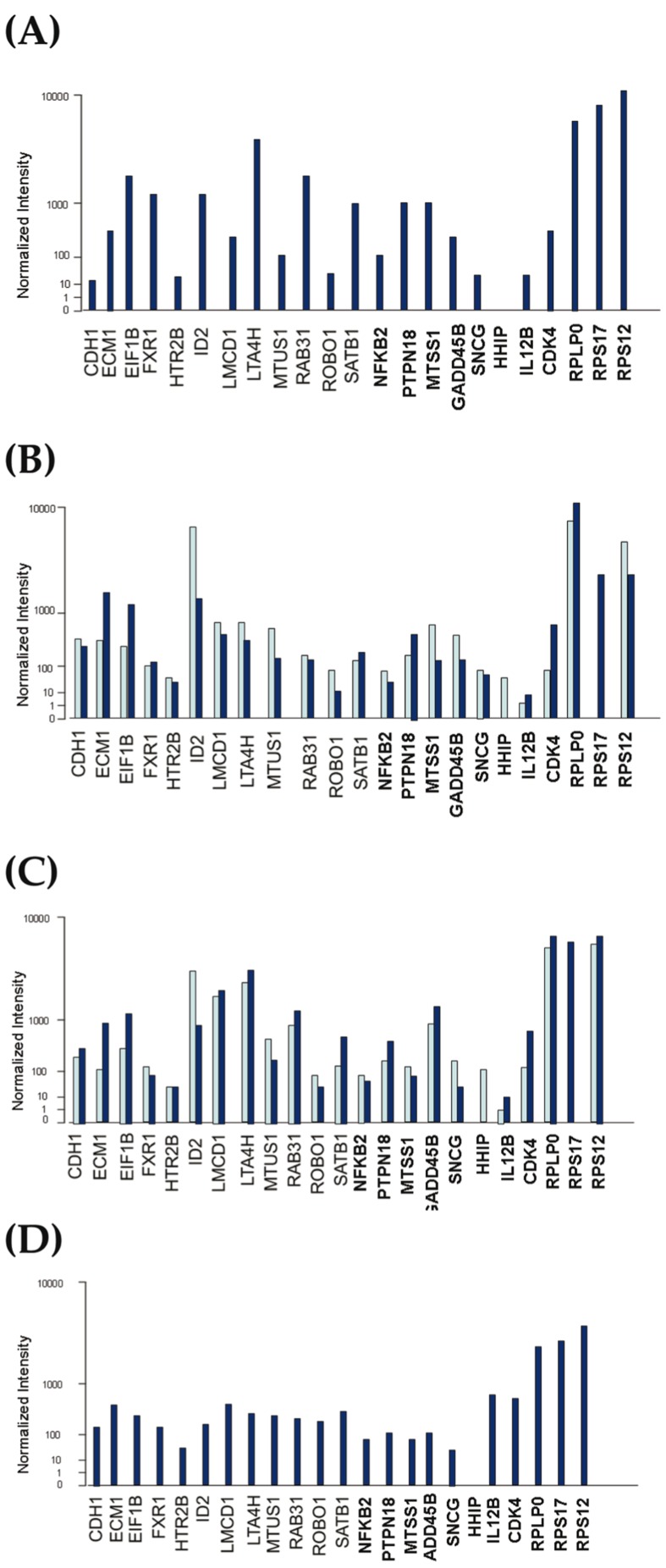
Results of microarray screening analysis for each gene in normal whole blood, liver, lung and skin are presented in Figure 1. All predictive genes and differentially expressed genes were expressed 10-fold greater than normalized intensity in all of the above-mentioned tissues except for IL12B expression in the liver and HHIP expression in whole blood and skin. IL12B was expressed around twice the normal values in the liver. In the GNF BioGPS database, no meaningful expression data for the HHIP gene was found in normal whole blood and skin after thresholding and normalization.
Figure 1.
Microarray expression of 12 predictive and 11 differently expressed genes in GNF BioGPS (dark blue bars), and Gene Note (light blue bars) databases for whole normal blood (A), liver (B), lung (C) and skin (D).
discussion
Uveal melanoma spreads via hematogenous metastasis. Detection of melanoma cells in the bloodstream might be helpful for classifying the risk of metastasis and monitoring the development of metastasis and also the response to treatment. In uveal melanoma patients, melanoma cells have been detected in the whole blood by using biomicroscopy, immunomagmetic cell sorting and real-time PCR.4-15 Reverse transcription PCR has been used to detect tyrosinase mRNA as a surrogate marker for melanoma cells because it is involved in the synthesis of melanin. Pilot studies showed that patients who were PCR-positive for tyrosinase mRNA in the blood already had or subsequently developed clinically detectable liver metastasis after a mean follow-up of 9 months.5 However, follow-up studies with larger sample size did not confirm these findings.15 Later, PCR detection of Melan-A/MART1 and gp100 were found to be inconsistent in detecting uveal melanoma cells in the blood.9,15
Recently, Harbour et al identified 12 predictive genes that categorize uveal melanomas into 3 main classes: 1A, 1B and 2.16-19 Uveal melanoma patients with a tumor class 2 gene expression profile have a 70% risk of developing clinical metastasis over 5 years as compared to a 5% risk in patients with tumor class 1 profile.5 As seen in our screening of GeneNote and GNF BioGPS databases, these 12 genes are expressed in microarrays of normal whole blood, liver, lung, and skin. Therefore, they cannot be used for microarray detection of circulating melanoma cells or micrometastases in organs susceptible to uveal melanoma metastasis.
Meir et al20 compared gene expression in liver metastases of uveal melanoma with that of primary uveal melanoma and reported that metastatic melanoma has a distinct gene expression profile. None of the 11 genes differentially expressed in metastatic melanoma was among the 12 predictive genes identified by Harbour and associates.16-20 As seen in our screening of GeneNote and GNF BioGPS databases, all 11 genes except the HHIP gene were expressed in microarray of normal whole blood, liver, lung and skin. Data for the HHIP gene was not complete in the GNF BioGPS database for normal whole blood and skin.
The microenvironment of the end organ and crosstalk between melanoma cells and their microenvironment are important for colonization, survival and growth of the metastatic uveal melanoma cells. This concept is supported by recent studies that have shown circulating melanoma cells in patients with uveal melanoma independent of tumor size, type of treatment, or length of follow-up.10 Therefore, circulating melanoma cells are thought to colonize in distant organs, form micrometastases, and remain dormant while sporadically seeding tumor cells into the circulating blood. Although these two sets of genes showed expression of two different tumor microenvironments which were not comparable, microarray analysis for 12 predictive genes described by Harbour and coworkers and 11 differentially expressed genes described by Meir and coworkers16-20 are not useful for detecting circulating uveal melanoma cells or tissues which are commonly colonized by them. It remains unclear whether similarity in the gene expression profile of the primary tumor to tissues prone to development of metastasis contributes to the tropism of uveal melanoma for metastasis to the liver.
Acknowledgments
The authors would like to thank Mrs. and Mr. Witham for their generous support.
Footnotes
Conflicts of Interest
None.
REFERENCES
- 1.Lorigan JG, Wallace S, Mavligit GM. The prevalence and location of metastases from ocular melanoma: imaging study in 110 patients. AJR Am J Roentgenol. 1991;157:1279–1281. doi: 10.2214/ajr.157.6.1950883. [DOI] [PubMed] [Google Scholar]
- 2.Borthwick NJ, Thombs J, Polak M, Gabriel FG, Hungerford JL, Damato B, et al. The biology of micrometastases from uveal melanoma. J Clin Pathol. 2011;64:666–671. doi: 10.1136/jcp.2010.087999. [DOI] [PubMed] [Google Scholar]
- 3.Eskelin S, Pyrhonen S, Summanen P, Hahka-Kemppinen M, Kivela T. Tumor doubling times in metastatic malignant melanoma of the uvea: tumor progression before and after treatment. Ophthalmology. 2000;107:1443–1449. doi: 10.1016/s0161-6420(00)00182-2. [DOI] [PubMed] [Google Scholar]
- 4.Horodenski J. Studies on the presence of free cells of malignant melanoma of the uvea in peripheral blood. Klin Oczna. 1969;39:407–412. [PubMed] [Google Scholar]
- 5.Fernandes BF, Belfort RN, Di Cesare S, Burnier MN. Circulating uveal melanoma cells: should we test for them? Can J Ophthalmol. 2008;43:155–158. doi: 10.3129/i08-011. [DOI] [PubMed] [Google Scholar]
- 6.Tobal K, Sherman LS, Foss AJ, Lightman SL. Detection of melanocytes from uveal melanoma in peripheral blood using the polymerase chain reaction. Invest Ophthalmol Vis Sci. 1993;34:2622–2625. [PubMed] [Google Scholar]
- 7.El-Shabrawi Y, Langmann G, Hutter H, Kenner L, Hoefler G. Comparison of current methods and PCR for the diagnosis of metastatic disease in uveal malignant melanoma. Ophthalmologica. 1998;212:80. [PubMed] [Google Scholar]
- 8.Boldin I, Langmann G, Richtig E, Schwantzer G, Ardjomand N, Wegscheider B, et al. Five-year results of prognostic value of tyrosinase in peripheral blood of uveal melanoma patients. Melanoma Res. 2005;15:503–507. doi: 10.1097/00008390-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Keilholz U, Goldin-Lang P, Bechrakis NE, Max N, Letsch A, Schmittel A, et al. Quantitative detection of circulating tumor cells in cutaneous and ocular melanoma and quality assessment by real-time reverse transcriptase-polymerase chain reaction. Clin Cancer Res. 2004;10:1605–1612. doi: 10.1158/1078-0432.ccr-0610-3. [DOI] [PubMed] [Google Scholar]
- 10.Callejo SA, Antecka E, Blanco PL, Edelstein C, Burnier MN. Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye (Lond) 2007;21:752–759. doi: 10.1038/sj.eye.6702322. [DOI] [PubMed] [Google Scholar]
- 11.Schuster R, Bechrakis NE, Stroux A, Busse A, Schmittel A, Scheibenbogen C, et al. Circulating tumor cells as prognostic factor for distant metastases and survival in patients with primary uveal melanoma. Clin Cancer Res. 2007;13:1171–1178. doi: 10.1158/1078-0432.CCR-06-2329. [DOI] [PubMed] [Google Scholar]
- 12.Ulmer A, Beutel J, Süsskind D, Hilgers RD, Ziemssen F, Lüke M, et al. Visualization of circulating melanoma cells in peripheral blood of patients with primary uveal melanoma. Clin Cancer Res. 2008;14:4469–4474. doi: 10.1158/1078-0432.CCR-08-0012. [DOI] [PubMed] [Google Scholar]
- 13.Suesskind D, Ulmer A, Schiebel U, Fierlbeck G, Spitzer B, Spitzer MS, et al. Circulating melanoma cells in peripheral blood of patients with uveal melanoma before and after different therapies and association with prognostic parameters: a pilot study. Acta Opthalmol. 2011;89:17–24. doi: 10.1111/j.1755-3768.2009.01617.x. [DOI] [PubMed] [Google Scholar]
- 14.Eide N, Faye RS, Høifødt HK, Øvergaard R, Jebsen P, Kvalheim G, et al. Immunomagnetic detection of micrometastatic cells in bone marrow in uveal melanoma patients. Acta Ophthalmol. 2009;87:830–836. doi: 10.1111/j.1755-3768.2008.01378.x. [DOI] [PubMed] [Google Scholar]
- 15.Foss AJ, Guille MJ, Occleston NL, Hykin PG, Hungerford JL, Lightman S. The detection of melanoma cells in peripheral blood by reverse transcription-polymerase chain reaction. Br J Cancer. 1995;72:155–159. doi: 10.1038/bjc.1995.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004;64:7205–7209. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010;12:461–468. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onken MD, Worley LA, Harbour JW. Association between gene expression profile, proliferation and metastasis in uveal melanoma. Curr Eye Res. 2010;35:857–863. doi: 10.3109/02713683.2010.493265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang SH, Worley LA, Onken MD, Harbour JW. Prognostic biomarkers in uveal maelanoma: evidence fro a stem cell-like phenotype associated with metastasis. Melanoma Res. 2008;18:191–200. doi: 10.1097/CMR.0b013e3283005270. [DOI] [PubMed] [Google Scholar]
- 20.Meir T, Dror R, Yu X, Qian J, Simon I, Pe’er J, et al. Molecular characteristics of liver metastases from uveal melanoma. Invest Ophthalmol Vis Sci. 2007;48:4890–4896. doi: 10.1167/iovs.07-0215. [DOI] [PubMed] [Google Scholar]
- 21.Topcu-Yilmaz P, Kiratli H, Saglam A, Soylemezoglu F, Hascelik G. Correlation of clinicopathological parameters with HGF, c-Met, EGFR, and IGF-1R expression in uveal melanoma. Melanoma Res. 2010;20:126–132. doi: 10.1097/CMR.0b013e328335a916. [DOI] [PubMed] [Google Scholar]
- 22.All-Ericsson C, Girnita L, Seregard S, Bartolazzi A, Jager MJ, Larsson O. Insulin-like growth factor-1 receptor in uveal melanoma: a predictor for metastatic disease and a potential therapeutic target. Invest Ophthalmol Vis Sci. 2002;43:1–8. [PubMed] [Google Scholar]
- 23.Mallikarjuna K, Pushparaj V, Biswas J, Krishnakumar S. Expression of insulin-like growth factor receptor (IGF-1R), c-Fos, and c-Jun in uveal melanoma: an immunohistochemical study. Curr Eye Res. 2006;31:875–883. doi: 10.1080/02713680600878790. [DOI] [PubMed] [Google Scholar]
- 24.Stelzer G, Dalah I, Stein TI, Satanower Y, Rosen N, Nativ N, et al. In-silico human genomics with GeneCards. Human Genomics. 2011;5:709–717. doi: 10.1186/1479-7364-5-6-709. [DOI] [PMC free article] [PubMed] [Google Scholar]