Abstract
Birth weight is an economically important trait in pig production because it directly impacts piglet growth and survival rate. In the present study, we performed a genome wide survey of candidate genes and pathways associated with individual birth weight (IBW) using the Illumina PorcineSNP60 BeadChip on 24 high (HEBV) and 24 low estimated breeding value (LEBV) animals. These animals were selected from a reference population of 522 individuals produced by three sires and six dam lines, which were crossbreds with multiple breeds. After quality-control, 43,257 SNPs (single nucleotide polymorphisms), including 42,243 autosomal SNPs and 1,014 SNPs on chromosome X, were used in the data analysis. A total of 27 differentially selected regions (DSRs), including 1 on Sus scrofa chromosome 1 (SSC1), 1 on SSC4, 2 on SSC5, 4 on SSC6, 2 on SSC7, 5 on SSC8, 3 on SSC9, 1 on SSC14, 3 on SSC18, and 5 on SSCX, were identified to show the genome wide separations between the HEBV and LEBV groups for IBW in piglets. A DSR with the most number of significant SNPs (including 7 top 0.1% and 31 top 5% SNPs) was located on SSC6, while another DSR with the largest genetic differences in FST was found on SSC18. These regions harbor known functionally important genes involved in growth and development, such as TNFRSF9 (tumor necrosis factor receptor superfamily member 9), CA6 (carbonic anhydrase VI) and MDFIC (MyoD family inhibitor domain containing). A DSR rich in imprinting genes appeared on SSC9, which included PEG10 (paternally expressed 10), SGCE (sarcoglycan, epsilon), PPP1R9A (protein phosphatase 1, regulatory subunit 9A) and ASB4 (ankyrin repeat and SOCS box containing 4). More importantly, our present study provided evidence to support six quantitative trait loci (QTL) regions for pig birth weight, six QTL regions for average birth weight (ABW) and three QTL regions for litter birth weight (LBW) reported previously by other groups. Furthermore, gene ontology analysis with 183 genes harbored in these 27 DSRs suggested that protein, metal, ion and ATP binding, viral process and innate immune response present important pathways for deciphering their roles in fetal growth or development. Overall, our study provides useful information on candidate genes and pathways for regulating birth weight in piglets, thus improving our understanding of the genetic mechanisms involved in porcine embryonic or fetal development.
Keywords: PorcineSNP60 BeadChip, Differentially selected regions, Pathways, Birth weight, Piglets.
Introduction
Birth weight is an economically important trait closely associated with piglet survival and growth after birth. For example, low birth weight in piglets often results in high mortality rates, slow growth and poor carcass quality 1-5. Generally speaking, the birth weight of a newborn piglet reflects the progress of its embryonic and fetal development, which mainly depends on the nutritional supply to the embryo/fetus and its ability to use the available substrates 6. On the other hand, individual birth weight (IBW) has a substantial genetic component. For example, the direct heritability for IBW ranges from 0.13 to 0.36 5, 7. Therefore, identification of IBW related candidate genes or their pathways are imperative for developing marker assisted selection to rapidly improve this phenotype in pigs.
In the past, several research groups have focused on candidate genes associated with piglet IBW, such as myogenin (MYOG) 8, myostatin (MSTN) 9, dopamine beta-hydroxylase (DBH) 10, POU class 1 homeobox 1 (POU1F1) 11, solute carrier family 27 (fatty acid transporter), member 4 (SLC27A4) 12, solute carrier family 6 (amino acid transporter), member 14 (SLC6A14) 13, proliferation associated nuclear element 1 (PANE1) 14, and secreted phosphoprotein 1 (SPP1) 15. Recent completion of the pig draft genome assembly and discovery of more than twenty million single nucleotide polymorphisms (SNPs) in the genome (http://www.ncbi.nlm.nih.gov/SNP/) have allowed the community to further advance genome wide association studies for the identification of QTLs related to many economically important traits in the species.
For instance, the Illumina PorcineSNP60 BeadChip is a powerful tool for identifying SNPs or genes associated with economically important quantitative traits, such as growth, body composition and structural soundness 16-17, reproduction and fertility 18-19, disease resistance 20-21, farrowing phenotypes 22, meat quality and fat deposition traits 23-25. Due to commercial services available to genotype these chips, such genome wide association studies can be done in a short period of time, but the cost can be high, particularly when a large number of samples are involved.
To date, only one group has focused on the applications of the Illumina PorcineSNP60 BeadChip in investigating quantitative trait loci (QTL) for litter birth weight (LBW) and average birth weight (ABW) 22. Using a total of 1,152 first parity gilts, the team found 33 QTL for LBW and 65 QTL for ABW, respectively 22. Therefore, the objective of this study was to use both high estimated breeding value (HEBV) and low estimated breeding value (LEBV) piglets for IBW to perform genome-wide analysis using the PorcineSNP60 BeadChip for identification of candidate genes and pathways associated with this economically important trait. Our results provide novel insights in molecular mechanisms involved in genetic differences of fetal development in piglets.
Materials and Methods
Animals and DNA Isolation
A composite population was developed at the Washington State University Swine Center during 2003-2004. When piglets were born, tails were docked and IBW were recorded. Tail docking is an industry routine practice as removal of part of the pigs' tail can reduce the risk of tail biting in older pigs. Tail biting is painful and can lead to serious injury, which might result in symptoms of physical or mental stress. As such, these previously collected tails were used for DNA extraction in the present study. The tissue was lysed and DNA prepared with the GenEluteTM Mammalian Genomic DNA Miniprep kit (Sigma, St. Louis, MO, USA) according to the manufacturer's instructions. The concentration and purity of DNA samples were measured using the NANODROP® spectrophotometer ND-1000 (Thermo Scientific, Waltham, MA, USA). Overall, the DNA concentrations, A260/280 ratios and A260/230 ratios ranged from 40.6 to 76.3 ng/μl, from 1.85 to 1.94 and from 1.98 to 2.31, respectively. Based on the pedigree information, we selected a total of 522 piglets derived from three sires and six dam lines to form our reference population. Both sire and dam lines were crossbreds, representing six breeds: Yorkshire, Landrace, Chester White, Large White, Hampshire, and Duroc.
Estimated Breeding Value (EBV) Analysis and Selection of Piglets for Genotyping on the Illumina PorcineSNP60 BeadChip
The EBV analysis for IBW was performed using the following mixed linear model:
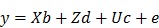 |
Where y is the observation of birth weight, b is a vector including the systematic effects of year-season (5), sex (2), parity (3), dam line (6) and sire (3). The vectors d, c, and e represent the direct additive genetic effects, common environmental litter effects (56), and the environmental residual effects, respectively. All computations were performed using AI-REML method with DMU v6.0 software 26. Based on the distribution of estimated breeding values for piglet IBW within 6 dam lines sired by 3 crossbred boars, 48 piglets with high (HEBV) and low estimated breeding values (LEBV) were selected to perform genome wide screening of candidate genes and pathways associated with piglet IBW. A student's t-test was used to determine the significant difference between HEBV and LEBV groups. Therefore, a total of 48 DNA samples were genotyped with standard procedures at GeneSeek (Lincoln, NE, USA) on the PorcineSNP60 BeadChip. The genotype quality control was done according to our previous report 27.
Detection of Differentially Selected Regions (DSRs) for IBW between HEBV and LEBV Piglets
As White et al (2012) 28 suggested, Hardy-Weinberg Equilibrium (HWE) of polymorphic SNPs was set to 0.000001 with PLINK v1.06 data analysis tools 29. The DSR algorithm was described previously 30-31, but with slight modifications: 1) raw values were ranked and used to identify regions; 2) Fisher's exact test was executed in R 2.14.0 to compare the allele frequencies between HEBV and LEBV, and SNPs with P values < 0.05 were considered statistically significant; 3) estimation of SNP FST were based on the model proposed by Nicholson et al (2002) 32 and Flori et al (2009) 33, and then the significant SNPs with 0.1% or 5% highest FST values were selected; and 4) by placing the top significant SNP(s) (0.1%) in the center of a DSR, adjacent SNP markers were collected to determine the region boundaries until more than three consecutive SNPs were not in the top significant 5% threshold. When DSRs overlapped, we combined them. In addition, when a region contained more than ten SNPs in the top significant 5%, but none were in the top significant 0.1%, we also considered it as a DSR region in our study.
SNP-specific FST values were smoothed over each chromosome with a local variable bandwidth kernel estimator 34. Genes in these DSRs were identified using the Sscrofa 10.2 assembly (http://www.animalgenome.org/cgi-bin/gbrowse/pig). Gene ontology information was collected from the NCBI database based on the summary of human gene functions.
Results and Discussion
Illumina PorcineSNP60 BeadChip Genotyping Basics
As described above, a total of 48 animals were selected for genotyping using the Illumina PorcineSNP60 BeadChip, including 24 HEBV and 24 LEBV piglets. The average IBW was 1.64 kg for all 522 piglets in our reference population. As shown in Table 1, the HEBV piglets were 0.65 kg heavier while LEBV animals were 0.63 kg lighter than the population average. As such, the 1.28 kg difference in birth weight between HEBV and LEBV was significant (t-test, P < 0.01). Genotyping revealed that among 61,565 SNPs on the Illumina PorcineSNP60 BeadChip, only 54,238 SNPs were exactly assigned to a unique chromosome position based on the Sus scrofa Build 10.2 assembly. Meanwhile, we observed that 3,725 SNPs failed to be scored on at least 95% of all the individuals, 7,235 SNPs had MAF ≤ 0.05 in the whole dataset, and 21 SNPs showed significant deviations from Hardy-Weinberg equilibrium (HWE) in any one of two groups. By excluding the SNPs described above, the remaining 43,257 SNPs, including 42,243 autosomal SNPs and 1,014 SNPs on chromosome X, were used for further analysis (Additional File 1: Figure S1). The HWE test was not performed on chromosome X because the SNPs on chromosome X in males carry only one copy.
Table 1.
EBVs and IBW in the reference population.
| Population | Number of piglets | EBVs | IBW (kg) |
|---|---|---|---|
| All pigs | 522 | -0.03 ± 0.40 | 1.64 ± 0.35 |
| HEBV | 24 | 0.73 ± 0.17a | 2.29 ± 0.14a |
| LEBV | 24 | -0.80 ± 0.12b | 1.01 ± 0.18b |
| P | < 0.01 | < 0.01 |
HEBV: high estimated breeding value population; LEBV: low estimated breeding value population; IBW: individual birth weight. Values are shown as mean ± standard deviation. A student's t-test was used to compare the significant difference between HEBV and LEBV in EBVs and IBW, respectively. P value < 0.05 or 0.01 is considered statistically significant or extremely significant level, respectively. The different lowercase letters between HEBV and LEBV indicate that the difference reached the significance level of P < 0.05.
DSRs Discovered for IBW between HEBV and LEBV Groups
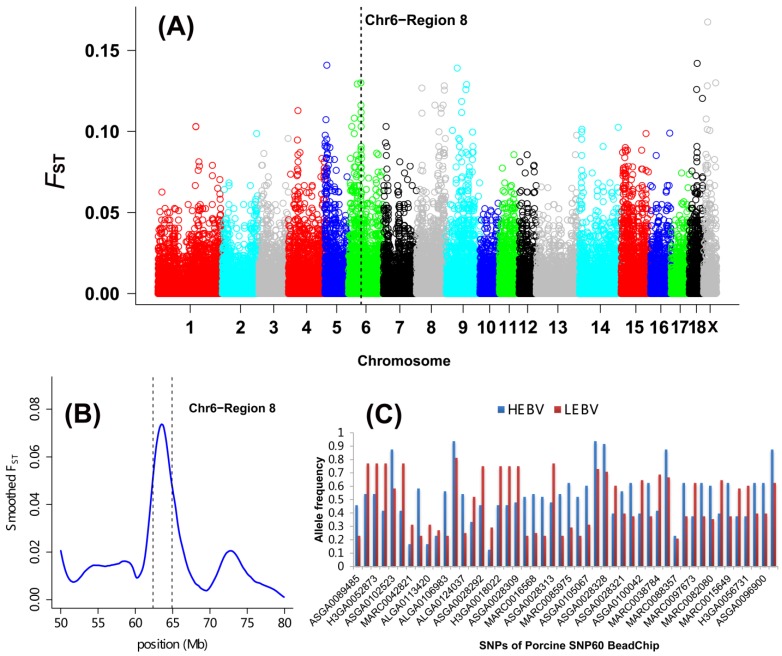
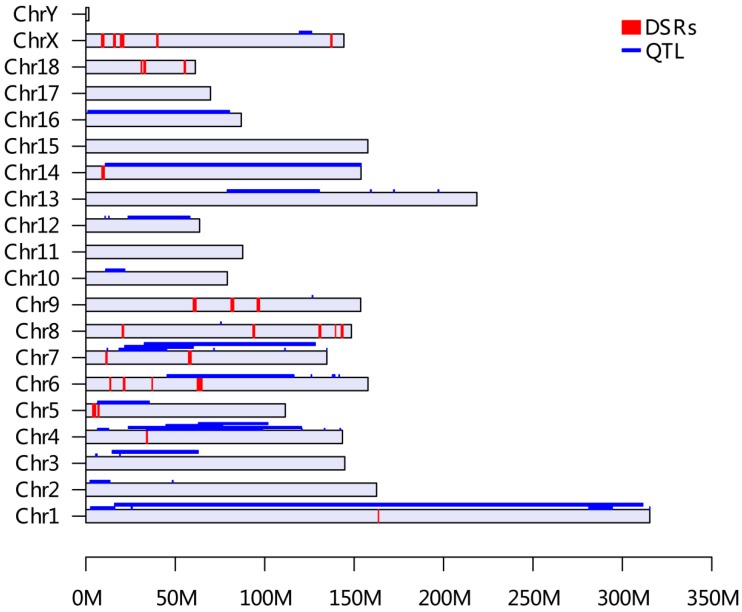
As shown in Table 2 and Figure 1A, a total of 27 DSRs across the entire pig genome were differentiated between the HEBV and LEBV groups for IBW, which included 43 significant top 0.1% SNPs (33 SNPs located on the autosomal chromosomes and 10 SNPs on chromosome X). Based on the current pig genome assembly (Sscrofa 10.2), these DSRs harbored a total of 183 unique genes: 168 of them are found within the autosomal DSRs and 15 genes were included in the chromosome X DSRs (Additional File 2: Table S1). Among these 27 DSRs, five supported the QTL regions previously reported for the birth weight in pigs: QTL#5232 on SSC1, QTL#369 on SSC4, QTL#3189 on SSC5, QTL#1012 on SSC6, QTL#187 and QTL#5197 on SSC 7 (Table 2 and Figure 2, http://cn.animalgenome.org/cgi-bin/QTLdb/index). More interestingly, six and three of these 27 DSRs for IBW were also located in relative close vicinity of QTLs identified for average birth weight (ABW) and litter body weight (LBW), respectively 22 (Additional File 2: Table S2). These results clearly indicated that some genome regions might be commonly responsible for IBW, ABW and LBW in piglets.
Table 2.
Differentially selected regions between high and low estimated breeding value populations.
| Region | PorcineSNP60 BeadChip Position (Mb) based on Sscrofa 10.2 assembly | Peak SNP (FST) | Top 0.1% | Top 5% | Candidate genes | Birth weight QTL number |
|---|---|---|---|---|---|---|
| 1 | chr.1:163614679..163648076 | ASGA0004841(0.1031) | 2 | 0 | NA | 5232 |
| 2 | chr.4:33947049..34338016 | ALGA0024390(0.1129) | 2 | 6 | NA | 369 |
| ASGA0019178(0.1129) | ||||||
| 3 | chr.5:3829983..5355995 | M1GA0007314(0.1073) | 1 | 10 | NA | NA |
| 4 | chr.5:6836955..7331522 | M1GA0007436(0.1409) | 1 | 3 | TMEM184B | 3189 |
| 5 | chr.6:13486031..13812780 | H3GA0017575(0.1031) | 1 | 4 | NA | NA |
| 6 | chr.6:21038055..21721506 | ALGA0034850(0.1082) | 1 | 7 | NA | NA |
| 7 | chr.6:37040339..37139826 | H3GA0054966(0.1293) | 1 | 2 | NA | NA |
| 8 | chr.6:62369260..64915744 | ASGA0095271(0.1300) | 7 | 31 | TNFRSF9, CA6 | 1012 |
| ALGA0117367(0.1300) | ||||||
| MARC0067004(0.1300) | ||||||
| 9 | chr.7:11223472..11802513 | ASGA0031182(0.1031) | 1 | 1 | NA | NA |
| 10 | chr.7:57441888..58806078 | ALGA0042164(0.0813) | 0 | 12 | CCDC37 | 187, 5197 |
| 11 | chr.8:20367972..21000928 | MARC0112253(0.1268) | 2 | 3 | TBC1D19 | NA |
| 12 | chr.8:93481336..94303744 | MARC0093552(0.1163) | 1 | 1 | NA | NA |
| 13 | chr.8:130415971..131295589 | ASGA0039827(0.1143) | 2 | 3 | ADH4 | NA |
| 14 | chr.8:139592943..139593441 | MARC0054584(0.1163) | 1 | 1 | NA | NA |
| 15 | chr.8:142867557..143732188 | ALGA0050003(0.1282) | 2 | 7 | WDFY3 | NA |
| 16 | chr.9:60085650..61556267 | ASGA0102114(0.1185) | 3 | 10 | NA | NA |
| 17 | chr.9:81214756..82621184 | ALGA0053850(0.1258) | 1 | 3 | LOC100738652 | NA |
| 18 | chr.9:95825746..97051426 | ALGA0054166(0.0857) | 0 | 16 | HDAC9 | NA |
| CASI0007446(0.0857) | ||||||
| ASGA0043971(0.0857) | ||||||
| 19 | chr.14:9010230..10254612 | ALGA0074932(0.1014) | 1 | 10 | NA | NA |
| 20 | chr.18:30963795..31174385 | ALGA0097763(0.1259) | 1 | 1 | NA | NA |
| 21 | chr.18:32516193..33286269 | INRA0055670(0.1420) | 1 | 2 | MDFIC | NA |
| 22 | chr.18:55026817..55614254 | ALGA0098742(0.1204) | 1 | 2 | NA | NA |
| 23 | chr.X:8818096..10005053 | MARC0040504(0.1077) | 1 | 9 | NA | NA |
| 24 | chr.X:15592452..16389800 | ASGA0080878(0.1282) | 2 | 2 | NA | NA |
| 25 | chr.X:19357189..21123895 | H3GA0051592(0.1675) | 1 | 10 | NA | NA |
| 26 | chr.X:39671416..40276593 | ASGA0081063(0.1007) | 5 | 3 | BCOR | NA |
| M1GA0023676(0.1007) | ||||||
| H3GA0051711(0.1007) | ||||||
| H3GA0051713(0.1007) | ||||||
| MARC0069431(0.1007) | ||||||
| 27 | chr.X:136986704..137562054 | MARC0046821(0.1300) | 1 | 1 | NA | NA |
Note: The candidate genes are given within the SNPs of top significant 0.1% or peak SNP for each region.
Figure 1.
Genome-wide distribution of SNPs between high and low estimated breeding value populations in piglets. (A) A global view of FST value distributions on pig autosomes 1 - 18 and sex chromosome X. A representative DSR (region 8 of SSC6) with 7 top significant 0.1% and 31 top significant 5% SNPs was indicated. (B) Smoothed FST estimates in region 8 of SSC6 showing the strong selection signals between the HEBV and LEBV groups. (C) Gene allele frequencies of each locus in region 8 of SSC6 between the HEBV and LEBV groups. HEBV: high estimated breeding value; LEBV: low estimated breeding value.
Figure 2.
The distribution of selected genomic regions (DSRs) and QTL for birth weight across the pig genome (based on Sus scrofa 10.2 Assembly). All QTL information for birth weight was retrieved from the Animal QTLdb release 21 (http://cn.animalgenome.org/cgi-bin/QTLdb/index). A total of 44 QTL from Animal QTLdb database and 27 DSRs of this study for IBW were placed on each pig chromosome based on their locations. The QTLs were painted in blue along each chromosome and DSRs were presented in red bars, respectively.
DSR with the Richest Top SNP for IBW Located on SSC6
A DSR region with 7 top significant 0.1% and 31 top significant 5% SNPs was found on SSC6 (named region 8, 62.37 to 64.92 Mb) (Table 2 and Figure 1B). The allele frequency differences among SNPs along the region are illustrated in Figure 1C. A major QTL for ABW was also discovered in this region using microsatellite markers 35 (Figure 2) as well as using the genome-wide association study 22 as described above. Two important candidates, namely tumor necrosis factor receptor superfamily member 9 (TNFRSF9) and carbonic anhydrase VI (CA6), are located in this region. TNFRSF9 is a member of the tumor necrosis factor receptor superfamily and it functions as an inducible co-stimulatory molecule for T cells 36. Recently, TNFRSF9 was found to have an important function during the implantation period of pregnancy in mice, which is a necessary process for the mother to adequately provide nutrients for fetal development 37. Moreover, TNFRSF9 plays an important role in reversing the hypo-responsiveness of pathogen-reactive maternal CD8+ T cells in the placenta 38. CA6 serves an important physiological role in the homeostasis of oral tissue by catalyzing the hydration of carbon hydroxide in body fluid and is the only isoform secreted in saliva, tears and milk of mammals. CA6 polymorphisms are known to be associated with the concentrations of secreted CA6 in young people, as well as salivary buffer capacity and dental plaque pH in children 39-40. There is currently limited information about the roles of TNFRSF9 and CA6 in livestock, and thus confirming the roles of these two genes in porcine fetal development warrants further investigation.
DSR with the Highest FST Signal for IBW Located on SSC18
Among autosomal DSRs, the highest FST signal (INRA0055670, FST = 0.1420, region 21; Table 2) was located at 33.29 Mb on SSC18 (Sscrofa 10.2 assembly) where the MyoD family inhibitor domain containing (MDFIC) gene resides. As a transcriptional regulator, this gene is a member of a family of proteins that include a specific cysteine-rich C-terminal domain for transcriptional regulation of viral genome expression 41 and have a modulatory role in immune cells 42, which, taken together, suggest that the function of this gene might be related to immune system capabilities. However, little is known about the role of MDFIC in livestock to date. Our results suggest that MDFIC might affect IBW, but additional studies are needed to confirm this speculation.
DSR with Four Imprinting Genes for IBW Located on SSC9
As shown in Additional File 2: Table S1, a DSR region of 81.21 - 82.62 Mb on SSC9 (Region 17; Table 2) harbors 4 putative candidate genes: PEG10 (paternally expressed 10), SGCE (sarcoglycan, epsilon), PPP1R9A (protein phosphatase 1, regulatory subunit 9A) and ASB4 (ankyrin repeat and SOCS box containing 4). These are imprinting genes that are associated with placental or embryonic growth in mouse 43. Therefore, we present here for the first time, to our knowledge, a potential imprinting region for IBW in pigs on SSC9. PEG10 and SGCE are candidate genes for embryonic lethality 44-45, while PPP1R9A and ASB4 play important roles in early development of multiple tissue types, which affect embryo growth and development in mouse and cattle 46-48. For pig, PEG10 may regulate the expression status in diploid parthenogenetic embryos and alter methylation at an imprinting control region 49. Furthermore, imprinting has been previously demonstrated for PEG10 and SGCE in porcine placenta at day 30 50.
Important candidate genes involved in DSRs on SSC9 and SSCX
Two DSRs, one located in region 95.83 - 97.05 on SSC9 and another in region 39.67 - 40.28 on SSCX, appear to be important in early development. The former DSR contains HDAC9 (histone deacetylase), while the latter contains BCOR (BCL6 corepressor). HDCA9 is a member of the HDAC family of proteins that function in deacetylation of histones, which change chromatin structure and gene transcription, and thus regulate different development and differentiation processes 51-52. This gene is sensitized to hypertrophic signals and exhibits stress-dependent cardiomegaly 53 and can suppress the transcriptional activity of myocyte enhancer factor (MEF2), which is a key regulator of muscle development in embryonic mice that forms a negative-feedback loop in the transcriptional circuitry of muscle differentiation 54. Moreover, the gene is also strongly expressed in skeletal muscle and heart during murine embryogenesis 54. Taken together, these data suggest that HDAC9 might play a role in cardiogenesis and/or myogenesis in pigs as well. The BCOR gene is a key transcriptional regulator of embryonic stem cell differentiation and early embryonic development, in particular in the differentiation of multiple tissue lineages or embryonic stem cells into ectoderm, mesoderm and hematopoietic lineages 55-56. Furthermore, in situ analysis showed that BCOR expression is significantly increased throughout the embryo after embryonic turning in mouse 55, suggesting a significant role in embryonic development. In addition, BCOR is differentially expressed in diverse tissue lineages such as eye, brain, neural tube and branchial arches during organogenesis and fetal period, which correlate with tissues affected in oculofaciocardiodental (OFCD) patients 55. Most recently, a mutation in BCOR was found to be associated with OFCD syndrome in human 57. Interestingly, our results suggested HDCA9 and BCOR genes might be associated with IBW, but how the tissue-specific expression patterns of these two genes specifically affect IBW during piglet embryogenesis requires further investigation.
A View of Gene Ontology from Candidate Genes
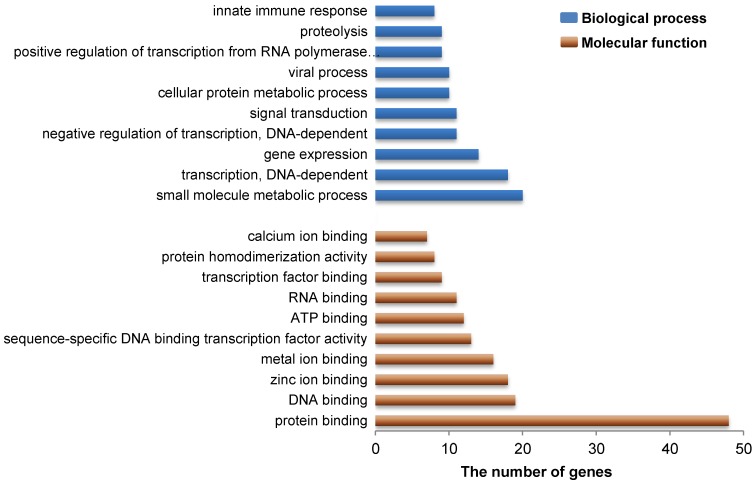
As indicated above, our present study revealed a total of 183 genes that reside in 27 DSRs for IBW in piglets (Additional File 2: Table S1). Gene ontology analysis revealed that these genes are mainly involved in functional or biological processes related to protein/metal/calcium/zinc ion binding, ATP binding, viral process and innate immune response (Figure 3 and Additional File 2: Table S3 and S4). This is not surprising because these processes are necessary to meet the requirements of embryonic or fetal development. For example, zinc is an essential element during early murine morphogenesis 58, and its deficiency causes congenital malformations in rat fetuses 59. Zinc deficiency can affect birth weight of offspring of rats or ewes when the degree of deficiency is severe 60-61. Moreover, zinc supplementation had significant effects on birth weight in pregnant adolescents who were underweight multiparas or from poor urban communities 62-64. Adequate maternal calcium intake is significantly correlated with appropriate birth weight in newborns 65 and a suitable concentration of calcium may meet fetal bone requirements in human 66. Our results provide further evidence that calcium/zinc are important factors associated with IBW in pigs. However, the influence of maternal calcium level on human infant growth remains unclear because other studies did not show this positive correlation 67-68. Therefore, additional studies need to be conducted to confirm the exact role of calcium ions in pig fetal development. On the other hand, viral process and innate immune response are also important to developing fetuses. Actually, low birth weight is often related to intrauterine growth restriction (IUGR), which is defined as impaired growth and development of the mammalian embryo or other organs during pregnancy and is found in multiple mammalian species 69. Up to now, several studies have discovered that IUGR decreased the levels of proteins related to immune function and reduced the immune response in pigs 70-71. In human, many immune response related genes are down-regulated in low birth weight newborns compared to normal birth weight newborns 72. Interestingly, some genes in our study, including RPL34, RPS17 and RPL9 (Additional File 2: Table S1), were the same down-regulated genes previously reported in human 72, suggesting that these genes might be important in antiviral innate immune response affected by a maternal viral infection in fetal development. Given their known developmental role in Rab GTPase activator activity, metal ion binding, and zinc ion binding (Additional File 2: Table S3), additional candidate genes (TBC1D19, WDFY3, and ADH4) were also identified in this study and suggest that these genes might be involved in embryonic or fetal development in pigs, respectively. These data provide fodder for future functional research that may aid in the discovery of genetic mechanisms of embryonic or fetal development in pigs.
Figure 3.
Gene ontology analysis related to the candidate genes from DSRs between high and low estimated breeding value populations. These 183 functional genes harbored in 27 DSRs for IBW in piglets were identified to represent a total of 264 molecular functions or 706 biological processes. Here the top 10 molecular functions and the top 10 biological processes are shown by brown and blue bars, respectively. Vertical axis represents gene ontology categories, while horizontal axis indicates the number of genes in each ontology category.
In summary, we selected two groups of animals with high and low estimated breeding values (HEBV and LEBV) for IBW from a reference population of 522 piglets and revealed a total of 27 DSRs between them using Illumina PorcineSNP60 BeadChips. Twelve of these DSRs were consistent with QTL regions identified for piglet birth weight, average birth weight and litter birth weight reported previously 22. Moreover, several putative candidate genes and pathways in these regions based on Sscrofa 10.2 assembly were identified. The innovative feature of this study is that it provides an economic and effective way to confidently determine important genes and pathways related to economically important traits in pigs. More importantly, this study provides a novel genome-wide view of the genetic foundation for IBW in pigs.
Supplementary Material
Figure S1.
Table S1-S4.
Acknowledgments
This work was supported by the Agricultural Research Center, Washington State University, USA to ZJ and the National Natural Science Foundation of China (No: 31000997) to LZ.
References
- 1.Pomeroy RW. Infertility and neonatal mortality in the sow III. Neonatal mortality and foetal development. J Agric Sci. 1960;54:31–56. [Google Scholar]
- 2.Pond WG, Mersmann HJ, Yen JT. Severe feed restriction of pregnant swine and rats: effects on postweaning growth and body-composition of progeny. J Nutr. 1985;115:179–189. doi: 10.1093/jn/115.2.179. [DOI] [PubMed] [Google Scholar]
- 3.Milligan BN, Fraser D, Kramer DL. Within-litter birth weight variation in the domestic pig and its relation to pre-weaning survival, weight gain, and variation in weaning weights. Livest Prod Sci. 2002;76:181–191. [Google Scholar]
- 4.Quiniou N, Dagorn J, Gaudre D. Variation of piglets' birth weight and consequences on subsequent performance. Livest Prod Sci. 2002;78:63–70. [Google Scholar]
- 5.Kapell DNRG, Ashworth CJ, Knap PW. et al. Genetic parameters for piglet survival, litter size and birth weight or its variation within litter in sire and dam lines using Bayesian analysis. Livest Sci. 2011;135:215–224. [Google Scholar]
- 6.Rehfeldt C, Kuhn G. Consequences of birth weight for postnatal growth performance and carcass quality in pigs as related to myogenesis. J Anim Sci. 2006;84(Suppl):E113–E123. doi: 10.2527/2006.8413_supple113x. [DOI] [PubMed] [Google Scholar]
- 7.Roehe R, Shrestha NP, Mekkawy W. et al. Genetic parameters of piglet survival and birth weight from a two-generation crossbreeding experiment under outdoor conditions designed to disentangle direct and maternal effects. J Anim Sci. 2010;88:1276–1285. doi: 10.2527/jas.2009-2287. [DOI] [PubMed] [Google Scholar]
- 8.te Pas MF, Soumillion A, Harders FL. et al. Influences of myogenin genotypes on birth weight, growth rate, carcass weight, backfat thickness, and lean weight of pigs. J Anim Sci. 1999;77:2352–2356. doi: 10.2527/1999.7792352x. [DOI] [PubMed] [Google Scholar]
- 9.Jiang YL, Li N, Fan XZ. et al. Associations of T→A mutation in the promoter region of myostatin gene with birth weight in Yorkshire pigs. Asian-Aust J Anim Sci. 2002;15:1543–1545. [Google Scholar]
- 10.Tomás A, Casellas J, Ramírez O. et al. Polymorphisms of the porcine dopamine beta-hydroxylase gene and their relation to reproduction and piglet survivability in an Iberian x Meishan F2 intercross. Anim Genet. 2006;37:279–282. doi: 10.1111/j.1365-2052.2006.01457.x. [DOI] [PubMed] [Google Scholar]
- 11.Song CY, Gao B, Teng SH. et al. Polymorphisms in intron 1 of the porcine POU1F1 gene. J Appl Genet. 2007;48:371–374. doi: 10.1007/BF03195234. [DOI] [PubMed] [Google Scholar]
- 12.Xu ZY, Xiong YZ, Lei MG. et al. Genetic polymorphisms and preliminary association analysis with production traits of the porcine SLC27A4 gene. Mol Biol Rep. 2009;36:1427–1432. doi: 10.1007/s11033-008-9332-2. [DOI] [PubMed] [Google Scholar]
- 13.Yang GL, Ren J, Zhang SH. et al. Isolation and molecular characterization of the porcine SLC6A14 gene excludes it as a candidate gene for fat deposition and growth. J Appl Genet. 2010;51:299–308. doi: 10.1007/BF03208859. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Deng H, Yang Y. et al. Molecular characterization and association analysis of porcine PANE1 gene. Mol Biol Rep. 2010;37:2571–2577. doi: 10.1007/s11033-009-9775-0. [DOI] [PubMed] [Google Scholar]
- 15.Han SH, Shin KY, Lee SS. et al. Porcine SPP1 gene polymorphism association with phenotypic traits in the Landrace × Jeju (Korea) Black pig F2 population. Mol Biol Rep. 2012;39:7705–7709. doi: 10.1007/s11033-012-1606-z. [DOI] [PubMed] [Google Scholar]
- 16.Fan B, Onteru SK, Du ZQ. et al. Genome-wide association study identifies Loci for body composition and structural soundness traits in pigs. PLoS One. 2011;6:e14726. doi: 10.1371/journal.pone.0014726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández AI, Pérez-Montarelo D, Barragán C. et al. Genome-wide linkage analysis of QTL for growth and body composition employing the PorcineSNP60 BeadChip. BMC Genet. 2012;13:41. doi: 10.1186/1471-2156-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uimari P, Sironen A, Sevón-Aimonen ML. Whole-genome SNP association analysis of reproduction traits in the Finnish Landrace pig breed. Genet Sel Evol. 2011;43:42. doi: 10.1186/1297-9686-43-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Onteru SK, Fan B, Du ZQ. et al. A whole-genome association study for pig reproductive traits. Anim Genet. 2012;43:18–26. doi: 10.1111/j.1365-2052.2011.02213.x. [DOI] [PubMed] [Google Scholar]
- 20.Nonneman DJ, Brown-Brandl T, Jones SA. et al. A defect in dystrophin causes a novel porcine stress syndrome. BMC Genomics. 2012;13:233. doi: 10.1186/1471-2164-13-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preissler R, Tetens J, Reiners K. et al. A genome-wide association study to detect genetic variation for postpartum dysgalactia syndrome in five commercial pig breeding lines. Anim Genet. 2013;44:502–508. doi: 10.1111/age.12047. [DOI] [PubMed] [Google Scholar]
- 22.Schneider JF, Rempel LA, Snelling WM. et al. Genome-wide association study of swine farrowing traits. Part II: Bayesian analysis of marker data. J Anim Sci. 2012;90:3360–3367. doi: 10.2527/jas.2011-4759. [DOI] [PubMed] [Google Scholar]
- 23.Luo W, Cheng D, Chen S. et al. Genome-wide association analysis of meat quality traits in a porcine Large White × Minzhu intercross population. Int J Biol Sci. 2012;8:580–595. doi: 10.7150/ijbs.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontanesi L, Schiavo G, Galimberti G. et al. A genome wide association study for backfat thickness in Italian Large White pigs highlights new regions affecting fat deposition including neuronal genes. BMC Genomics. 2012;13:583. doi: 10.1186/1471-2164-13-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uimari P, Sironen A, Sevón-Aimonen ML. Evidence for three highly significant QTL for meat quality traits in the Finnish Yorkshire pig breed. J Anim Sci. 2013;91:2001–2011. doi: 10.2527/jas.2012-5811. [DOI] [PubMed] [Google Scholar]
- 26.Madsen P, Jensen J. A User's Guide to DMU. A package for analysing multivariate mixed models. 2007. Version 6, release 4.7, University of Aarhus, Faculty of Agricultural Sciences, Tjele, Denmark.
- 27.Michelizzi VN, Wu X, Dodson MV. et al. A global view of 54,001 single nucleotide polymorphisms (SNPs) on the Illumina BovineSNP50 BeadChip and their transferability to water buffalo. Int J Biol Sci. 2010;7:18–27. doi: 10.7150/ijbs.7.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White SN, Mousel MR, Herrmann-Hoesing LM. et al. Genome-wide association identifies multiple genomic regions associated with susceptibility to and control of ovine lentivirus. PLoS One. 2012;7:e47829. doi: 10.1371/journal.pone.0047829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, Neale B, Todd-Brown K. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kijas JW, Lenstra JA, Hayes B. et al. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection. PLoS Biology. 2012;10:e1001258. doi: 10.1371/journal.pbio.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Mousel MR, Wu X. et al. Genome-wide genetic diversity and differentially selected regions among Suffolk, Rambouillet, Columbia, Polypay, and Targhee sheep. PLoS One. 2013;8:e65942. doi: 10.1371/journal.pone.0065942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholson G, Smith AV, Jónsson F. et al. Assessing population differentiation and isolation from single-nucleotide polymorphism data. J R Stat Soc Ser B Stat Methodol. 2002;64:695–715. [Google Scholar]
- 33.Flori L, Fritz S, Jaffrézic F. et al. The genome response to artificial selection: a case study in dairy cattle. PLoS One. 2009;4:e6595. doi: 10.1371/journal.pone.0006595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrmann E. Local bandwidth choice in kernel regression estimation. J Comput Graph Statist. 1997;6:35–54. [Google Scholar]
- 35.Yue G, Stratil A, Kopecny M. et al. Linkage and QTL mapping for Sus scrofa chromosome 6. J Anim Breed Genet. 2003;120(Suppl 1):45–55. [Google Scholar]
- 36.Melero I, Shuford WW, Newby SA. et al. Monoclonal antibodies against the 4-1BB T-cell activation molecule eradicate established tumors. Nat Med. 1997;3:682–685. doi: 10.1038/nm0697-682. [DOI] [PubMed] [Google Scholar]
- 37.Eckstrum K, Bany BM. Tumor necrosis factor receptor subfamily 9 (Tnfrsf9) gene is expressed in distinct cell populations in mouse uterus and conceptus during implantation period of pregnancy. Cell Tissue Res. 2011;344:567–576. doi: 10.1007/s00441-011-1171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim KH, Choi BK, Kim JD. et al. 4-1BB Signaling breaks the tolerance of maternal CD8+ T cells that are reactive with alloantigens. PLoS One. 2012;7:e45481. doi: 10.1371/journal.pone.0045481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peres RC, Camargo G, Mofatto LS. et al. Association of polymorphisms in the carbonic anhydrase 6 gene with salivary buffer capacity, dental plaque pH, and caries index in children aged 7-9 years. Pharmacogenomics J. 2010;10:114–119. doi: 10.1038/tpj.2009.37. [DOI] [PubMed] [Google Scholar]
- 40.Aidar M, Marques R, Valjakka J. et al. Effect of genetic polymorphisms in CA6 Gene on the expression and catalytic activity of human salivary carbonic anhydrase VI. Caries Res. 2013;47:414–420. doi: 10.1159/000350414. [DOI] [PubMed] [Google Scholar]
- 41.Thébault S, Gachon F, Lemasson I. et al. Molecular cloning of a novel human I-mfa domain-containing protein that differently regulates human T-cell leukemia virus type I and HIV-1 expression. J Biol Chem. 2000;275:4848–4857. doi: 10.1074/jbc.275.7.4848. [DOI] [PubMed] [Google Scholar]
- 42.Gu L, Dean J, Oliveira AL. et al. Expression profile and differential regulation of the Human I-mfa domain-Containing protein (HIC) gene in immune cells. Immunol Lett. 2009;123:179–184. doi: 10.1016/j.imlet.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 43.Tunster SJ, Jensen AB, John RM. Imprinted genes in mouse placental development and the regulation of fetal energy stores. Reproduction. 2013;145:R117–R137. doi: 10.1530/REP-12-0511. [DOI] [PubMed] [Google Scholar]
- 44.Piras G, El Kharroubi A, Kozlov S. et al. Zac1 (Lot1), a potential tumor suppressor gene, and the gene for epsilon-sarcoglycan are maternally imprinted genes: identification by a subtractive screen of novel uniparental fibroblast lines. Mol Cell Biol. 2000;20:3308–3315. doi: 10.1128/mcb.20.9.3308-3315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono R, Nakamura K, Inoue K. et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat Genet. 2006;38:101–106. doi: 10.1038/ng1699. [DOI] [PubMed] [Google Scholar]
- 46.Nakabayashi K, Makino S, Minagawa S. et al. Genomic imprinting of PPP1R9A encoding neurabin I in skeletal muscle and extra-embryonic tissues. J Med Genet. 2004;41:601–608. doi: 10.1136/jmg.2003.014142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kile BT, Viney EM, Willson TA. et al. Cloning and characterization of the genes encoding the ankyrin repeat and SOCS box-containing proteins Asb-1, Asb-2, Asb-3 and Asb-4. Gene. 2000;258:31–41. doi: 10.1016/s0378-1119(00)00402-9. [DOI] [PubMed] [Google Scholar]
- 48.Zaitoun I, Khatib H. Comparative genomic imprinting and expression analysis of six cattle genes. J Anim Sci. 2008;86:25–32. doi: 10.2527/jas.2007-0150. [DOI] [PubMed] [Google Scholar]
- 49.Park CH, Uh KJ, Mulligan BP. et al. Analysis of imprinted gene expression in normal fertilized and uniparental preimplantation porcine embryos. PLoS One. 2011;6:e22216. doi: 10.1371/journal.pone.0022216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bischoff SR, Tsai S, Hardison N. et al. Characterization of conserved and nonconserved imprinted genes in swine. Biol Reprod. 2009;81:906–920. doi: 10.1095/biolreprod.109.078139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chatterjee TK, Idelman G, Blanco V. et al. Histone deacetylase 9 is a negative regulator of adipogenic differentiation. J Biol Chem. 2011;286:27836–27847. doi: 10.1074/jbc.M111.262964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang CL, McKinsey TA, Chang S. et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haberland M, Arnold MA, McAnally J. et al. Regulation of HDAC9 gene expression by MEF2 establishes a negative-feedback loop in the transcriptional circuitry of muscle differentiation. Mol Cell Biol. 2007;27:518–525. doi: 10.1128/MCB.01415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wamstad JA, Bardwell VJ. Characterization of Bcor expression in mouse development. Gene Expr Patterns. 2007;7:550–557. doi: 10.1016/j.modgep.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wamstad JA, Corcoran CM, Keating AM. et al. Role of the transcriptional corepressor Bcor in embryonic stem cell differentiation and early embryonic development. PLoS One. 2008;3:e2814. doi: 10.1371/journal.pone.0002814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feberwee H, Feenstra I, Oberoi S. et al. Novel BCOR mutations in patients with oculofaciocardiodental (OFCD) syndrome. Clin Genet. 2013 doi: 10.1111/cge.12125. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 58.Kambe T, Weaver BP, Andrews GK. The genetics of essential metal homeostasis during development. Genesis. 2008;46:214–228. doi: 10.1002/dvg.20382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hurley LS, Swenerton H. Congenital malformations resulting from zinc deficiency in rats. Proc Soc Exp Biol Med. 1966;123:692–696. doi: 10.3181/00379727-123-31578. [DOI] [PubMed] [Google Scholar]
- 60.Apgar J. Effects of zinc deficiency deprivation from day 12, 15, or 18 of gestation on parturition in the rat. J Nutr. 1972;102:343–348. doi: 10.1093/jn/102.3.343. [DOI] [PubMed] [Google Scholar]
- 61.Apgar J, Fitzgerald JA. Effect on the ewe and lamb of low zinc intake throughout pregnancy. J Anim Sci. 1985;60:1530–1538. doi: 10.2527/jas1985.6061530x. [DOI] [PubMed] [Google Scholar]
- 62.Cherry FF, Sandstead HH, Rojas P. et al. Adolescent pregnancy: associations among body weight, zinc nutriture and pregnancy outcome. Am J Clin Nutr. 1989;50:945–954. doi: 10.1093/ajcn/50.5.945. [DOI] [PubMed] [Google Scholar]
- 63.Castillo- Durán C, Marín VB, Alcázar LS. et al. Controlled trial of zinc supplementation in Chilean pregnant adolescents. Nutr Res. 2001;21:715–724. [Google Scholar]
- 64.Castillo- Durán C. Weisstaub G. Zinc supplementation and growth of the fetus and low birth weight infant. J Nutr. 2003;133(5 Suppl 1):1494S–1497S. doi: 10.1093/jn/133.5.1494S. [DOI] [PubMed] [Google Scholar]
- 65.Sabour H, Hossein-Nezhad A, Maghbooli Z. et al. Relationship between pregnancy outcomes and maternal vitamin D and calcium intake: A cross-sectional study. Gynecol Endocrinol. 2006;22:585–589. doi: 10.1080/09513590601005409. [DOI] [PubMed] [Google Scholar]
- 66.Hacker AN, Fung EB, King JC. Role of calcium during pregnancy: maternal and fetal needs. Nutr Rev. 2012;70:397–409. doi: 10.1111/j.1753-4887.2012.00491.x. [DOI] [PubMed] [Google Scholar]
- 67.Jarjou LM, Prentice A, Sawo Y. et al. Randomized, placebo-controlled, calcium supplementation study in pregnant Gambian women: effects on breast-milk calcium concentrations and infant birth weight, growth, and bone mineral accretion in the first year of life. Am J Clin Nutr. 2006;83:657–666. doi: 10.1093/ajcn.83.3.657. [DOI] [PubMed] [Google Scholar]
- 68.Abdel-Aleem H, Merialdi M, Elsnosy ED. et al. The effect of calcium supplementation during pregnancy on fetal and infant growth: a nested randomized controlled trial within WHO calcium supplementation trial. J Matern Fetal Neonatal Med. 2009;22:94–100. doi: 10.1080/14767050802464569. [DOI] [PubMed] [Google Scholar]
- 69.Wu G, Bazer FW, Wallace JM. et al. Board-invited review: intrauterine growth retardation: implications for the animal sciences. J Anim Sci. 2006;84:2316–2337. doi: 10.2527/jas.2006-156. [DOI] [PubMed] [Google Scholar]
- 70.Wang J, Chen L, Li D. et al. Intrauterine growth restriction affects the proteomes of the small intestine, liver, and skeletal muscle in newborn pigs. J Nutr. 2008;138:60–66. doi: 10.1093/jn/138.1.60. [DOI] [PubMed] [Google Scholar]
- 71.Zhong X, Li W, Huang X. et al. Impairment of cellular immunity is associated with overexpression of heat shock protein 70 in neonatal pigs with intrauterine growth retardation. Cell Stress Chaperones. 2012;17:495–505. doi: 10.1007/s12192-012-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh VV, Chauhan SK, Rai R. et al. Decreased pattern recognition receptor signaling, interferon-signature, and bactericidal/permeability-increasing protein gene expression in cord blood of term low birth weight human newborns. PLoS One. 2013;8:e62845. doi: 10.1371/journal.pone.0062845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.
Table S1-S4.