Abstract
While trade-offs between flight capability and reproduction is a common phenomenon in wing dimorphic insects, the molecular basis is largely unknown. In this study, we examined the transcriptomic differences between winged and wingless morphs of cotton aphids, Aphis gossypii, using a tag-based digital gene expression (DGE) approach. Ultra high-throughput Illumina sequencing generated 5.30 and 5.39 million raw tags, respectively, from winged and wingless A. gossypii DGE libraries. We identified 1,663 differentially expressed transcripts, among which 58 were highly expressed in the winged A. gossypii, whereas 1,605 expressed significantly higher in the wingless morphs. Bioinformatics tools, including Gene Ontology, Cluster of Orthologous Groups, euKaryotic Orthologous Groups and Kyoto Encyclopedia of Genes and Genomes pathways, were used to functionally annotate these transcripts. In addition, 20 differentially expressed transcripts detected by DGE were validated by the quantitative real-time PCR. Comparative transcriptomic analysis of sedentary (wingless) and migratory (winged) A. gossyii not only advances our understanding of the trade-offs in wing dimorphic insects, but also provides a candidate molecular target for the genetic control of this agricultural insect pest.
Keywords: Aphis gossypii, trade-off, migration, digital gene expression, wing polyphenism.
Introduction
Trade-offs between life-history traits have been studied in a variety of organisms 1-3. Individuals capable of fly benefit from the ability, but it is also costly on energy for flight mechanism and flight behavior. This energy, alternatively, can be used for other aspects, such as body size and reproduction 4. In the past forty years, trade-offs between flight and reproduction has been widely studied 5-9. Wing dimorphic insects have been studied extensively and have emerged as a great model system for the study of trade-offs in the past several decades 8-10. Wing polyphenism has been observed in a wide range of insect species, including Hemiptera, Orthoptera, and Coleoptera, to facilitate their range expansion 8, 10-12. Wing dimorphic insects display commonly dispersing and non-dispersing morphs 13, 14. Flight morphs, with long wings, flight muscles and flight capability, need additional energy and time for the migratory flight. Flightless morphs, with reduced or absent wings and underdeveloped flight muscles, show an earlier onset of oviposition and enhanced reproductive output 8, 15-17. Trade-offs have been studied at behavioral, morphological, physiological and biochemical levels in wing dimorphic insects, such as crickets, migratory locusts, and aphids 16, 18, 19. For instance, biochemical aspects and intermediary metabolisms are investigated in a wing dimorphic cricket, Gryllus firmus 20-22. However, the genetic basis of biochemistry and metabolism related to trade-offs remains unclear.
There are trade-offs related to dispersal and reproduction in aphids between two wing morphs. The wing polyphenism in aphids is well-studied 23, 24. Winged and wingless aphids exist in both sexual and parthenogenetic stages of its life cycle. The migrants in spring, summer and early autumn are parthenogenetic generations, and the migration normally occurs in spring and summer 25, 26. Wing polyphenism occurs primarily among parthenogenetic females, whereas wing polymorphism, which is genetically determined, has been found only in males 14, 24. Parthenogenetic females with different wing morphs are descended from the same ancestry. A single genotype can produce various phenotypes as a result of exposure to various environmental conditions 27. Such phenotypic plasticity is one of the main reasons contributing to the ecological success of aphids and their increasing pest status 26, 28. Plasticity is not only an adaptive behavioral strategy that has evolved in most insect orders for coping with complex and uncertain environments, but also a key factor that leads to population expansion and infestation. Wing-polyphenic aphids exhibited distinct morphological differences between two wing types of aphids, and showed differences in their life history, particularly in their behavioral and physiological traits 14, 29, 30. The biochemical basis of wing polyphenism related to energy resources, including glycogen, trehalose, and lipids, was investigated in cotton aphid, Aphis gossypii 30 and grain aphid, Sitobion avenae 31. Total lipid and triglyceride were significantly higher in winged morphs, whereas glycogen, soluble protein and water were higher in wingless morphs in both species. Recent genetic studies 32-34, especially the release of a draft genome of pea aphid Acyrthosiphon pisum 35, lays the foundation for a better understanding of trade-offs between flight and reproduction at the molecular level. Genome-wide transcriptome analyses provide a new avenue for the future functional characterization study.
Digital gene expression (DGE) is a tag-based RNA sequencing approach that allows millions of RNA reads, including differentially expressed transcripts, to be identified in a sample with or without any annotation information 36. For aphid wing polyphenism study, DGE has several advantages - a greater sequencing depth, detection of unknown transcripts, and sensitivity 37. Although external factors such as environmental cues, population density and nutrition status have been studied for years, the genetic basis of trade-offs in the cotton aphids, A. gossypii, a major cotton pest worldwide, has yet been examined in details. The objectives of this study are 1) to reveal differential gene expression profiles between winged and wingless A. gossypii adults, and 2) to establish a link between transcriptomic profiles and biochemical differences observed in previous studies. Our studies provide molecular basis that underpins trade-offs between flight capability and reproduction in A. gossypii and will facilitate the sustainable cotton aphid management through the RNA interference-mediated genetic disruption of its migratory behavior.
Materials and Methods
Aphis gossypii maintenance and sample preparation
A single A. gossypii female adult was collected from a cotton field (40°01′N, 116°17′E) at the China Agricultural University, Beijing, China, in July of 2009. Aphid colonies generated from this individual A. gossypii had been established in the laboratory for more than one year before being sacrificed in the subsequent experiments. Specifically, parthenogenetic adults were provisioned with cotton seedlings (three-leaf stage, cultivar: Zhong 35) and maintained in a growth chamber at 60% relative humidity, and 16: 8 L: D photoperiod with 24±1˚C in the light and 22±1˚C in the dark 30, 34, 38. Winged forms were induced by crowding in cotton leaves two weeks after A. gossypii was transferred to new host plants. The fourth instars were placed into individual dishes, and observed daily until molting. A total of 50 winged or wingless A. gossypii adults were collected separately within two days after molting. Aphids were frozen in liquid nitrogen immediately after the collection. RNA was extracted using the RNeasy Mini kit (Qiagen). Genomic DNA was digested by RNase-Free DNase Set (Qiagen). The quality and quantities of RNAs were determined using a Nanodrop2000 and Agilent bioanalyzer (Agilent).
DGE library preparation and sequencing
High-throughput DGE was performed by Solexa/Illumina sequencing technology at the Beijing Genomics Institute, Shenzhen, China. DGE libraries (winged and wingless) were prepared using the Illumina Gene Expression Sample Prep Kit. Briefly, poly(A)+ RNA was enriched from 6 µg of total RNA using oligomagnetic beads. Double-stranded cDNAs were directly synthesized on the poly(A)+ RNA-bound beads and then digested with a 4 base recognition enzyme NlaIII. The fragmentized cDNAs were purified from the magnetic beads, containing 3' ends and a sticky end. Then the Illumina adaptor 1 (sense: 5'ACACTCTTTCCCTACACGACGCTCTTCCGATC3') was ligated to the 5' ends of these cDNA fragments. Digestion of MmeI and ligation of the Illumina adaptor 2 (sense: 5'GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG3') produced short DNA fragments, with two adaptors and 21bp tags started with the CATG sites. Later on, Primer GX1 and Primer GX2 were added for PCR. After 15 cycles of linear PCR amplification, 95bp fragments were purified using 6% TBE PAGE Gel electrophoresis. After denaturation, the single-stranded molecules were attached to the sequencing chip for sequencing via Illumina HiSeq™ 2000 System. During this process, adaptor 1 was used as sequencing primer. Each tunnel of chip (flow cell) generated millions of raw tags with sequencing length of 35 bp 39, 40.
Mapping of DGE tags
Clean tags were generated by removing 3' adaptor sequences, empty reads (with only 3' adaptor sequences), low quality tags (tags with unknown nucleotide ''N''), tags with erratic lengths, and tags with copy number of 1. A total of 88,851 A. gossypii ESTs were selected at NCBI on Oct, 28, 2010. After eliminating low quality sequences and Phrap assembly (minmatch: 42; minscore: 40, repeat_stringency:0.96), 23,630 unigenes were obtained and used as reference dataset (Supplementary Material: Table S1). Clean tags were aligned to the reference dataset with only one nucleotide mismatch tolerance. The number of unambiguous clean tag corresponding to each transcript was counted and normalized to TPM (number of transcripts per million clean tags) 36, 41.
Identification of differentially expressed transcripts
The differentially expressed transcripts between winged and wingless patterns were identified using an algorithm developed by Audic and Claverie 42. P value corresponds to differential gene expression test. FDR (False Discovery Rate) is a method of determining the threshold of P Value in multiple tests and analysis through manipulating FDR value 43. In this study, we considered a transcript as differentially expressed when there was at least a 2-fold difference in expression between the two samples with a FDR value≤ 0.001.
To functionally annotate these unigenes, distinct sequences were searched by BLASTx against the NCBI non-redundant protein database 44 with a cut-off E-value of 10-5, and assigned Cluster of Orthologous Groups (COG) and euKaryotic Orthologous Groups (KOG) protein classes with rpstblastn algorithm against COG and KOG databases in NCBI (E-value ≤10-5) 45, 46. The BLASTx result was imported into Blast2GO software (version 2.6.4) to map the Gene Ontology (GO) categories 47, and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways 48. The enriched GO terms were developed by Blast2Go using Fisher's Exact Test with Multiple Testing Correction (FDR<0.05)47. The enrichment analysis of pathway was performed to further understand genes biological functions.
qRT-PCR analysis
PCR reactions were performed in a 20 μl final volume containing 2× Power SYBR Green PCR Master Mix buffer (Applied Biosystems), 1 μM of each specific primer and 1 μl of diluted cDNA. PCR amplification was performed on an ABI Prism 7300 Sequence Detection System (Applied Biosystems) under the following conditions: 10 min at 95 °C for polymerase activation, and 40 cycles of 95°C for 15s, 60°C for 30s, and 72°C for 30s for amplification signal collection. A default melting curve was produced to confirm single gene-specific peaks without primer dimerization present. Housekeeping genes, β-actin and 18S rRNA were subjected to the reference gene selection experiment. Eventually, β-actin was used as a constitutive control to normalize the expressions of target genes using a -ΔΔCt method.
To validate DGE results, a total of 20 transcripts were chosen from the top 10 differentially expressed transcripts of winged and wingless libraries, respectively, and three significantly enriched pathways. The gene-specific primers designed for reference and target genes were listed in Supplementary Material: Table S2. Three independent replications were carried out for the qRT-PCR validation study, with approximately 30 winged or wingless females for each replication.
Results
Summary of digital gene expression sequencing
Reference sequences were obtained from 88,851 ESTs, of which 36 low quality (less than 100bp after remove vector) sequences were discarded. 23,630 unigenes, containing 12,507 contigs and 11,123 singletons, with a mean length of 715 bp, were assembled by Phrap. The generated unigenes were subjected to BLAST search and functional annotation using various databases.
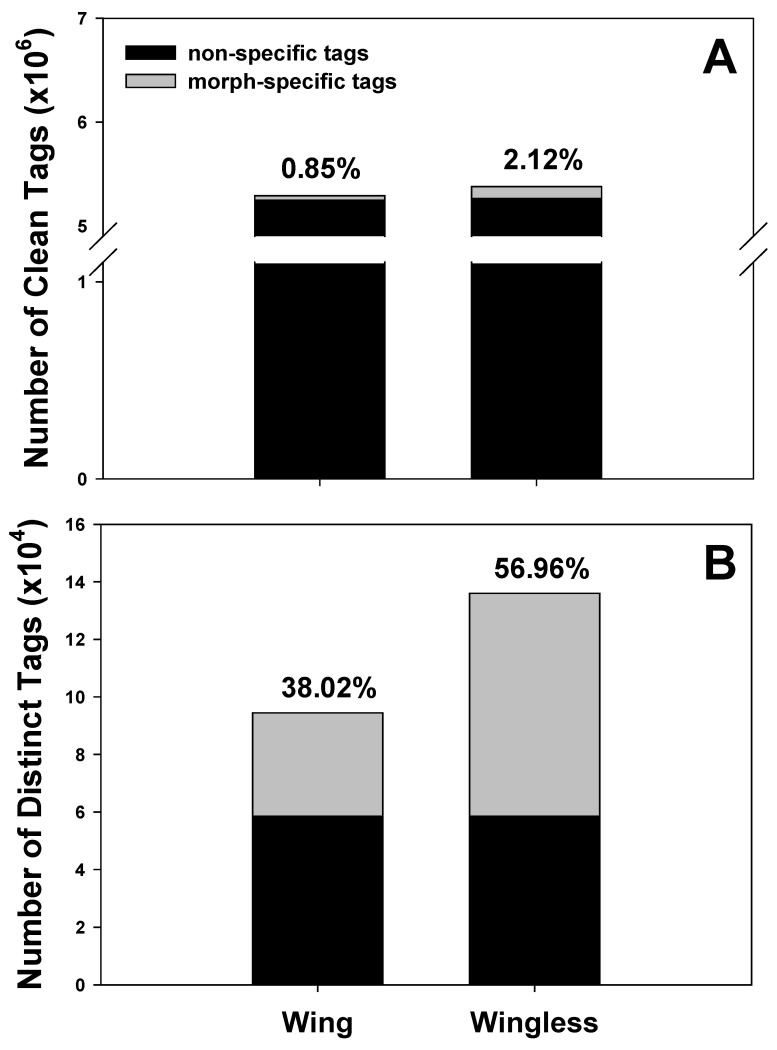
In total, 5.30 and 5.39 million raw tags were obtained from winged and wingless adults, respectively. Distribution details of tags were shown in Supplementary Material: Figure S1. Clean tags were mapped to 9,011 and 10161 unigenes, respectively (Table 1). Sequencing saturation analysis shows the number of detected genes almost ceases to increase when sequencing amount (total tag number) reaches 2 million (Supplementary Material: Figure S2), consequently, 5 million tags are sufficient for the expression profiling of cotton aphid. The total number of tags sequenced in the two samples was 10,479,972. Tags with the same sequence were identified as a distinct tag, and their distributions in different wing morphs were documented. Tags in the two wing forms were compared in total and in distinct (Figure 1). Surprisingly, morph-specific tags (tags detected in one morph only) accounted for a very small proportion (0.85% in winged morph, 2.12% in wingless morph) of the total clean tags, while the percentage of morph-specific tags increased obviously among the distinct tags.
Table 1.
Sequencing summary.
| Sequence | Winged Morph | Wingless Morph | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | % | Distinct | % | Total | % | Distinct | % | |
| Clean Tag | 5221975 | - | 44414 | - | 5257997 | - | 61655 | - |
| Mapped to gene | 4781916 | 91.57 | 21675 | 48.80 | 4345336 | 82.64 | 27998 | 45.41 |
| Unambiguous tag | 1329628 | 25.4 | 20748 | 46.71 | 2322979 | 44.18 | 26749 | 43.38 |
| Unknown tag | 440059 | 8.4 | 22739 | 51.20 | 912661 | 17.36 | 33657 | 54.59 |
Figure 1.
Morph-specific sequencing tags in the winged and wingless A. gossypii. Number and percentage of morph-specific tags were documented in total clean tags (A) and distinct clean tags (B), respectively.
Differentially expressed transcripts in winged and wingless aphids
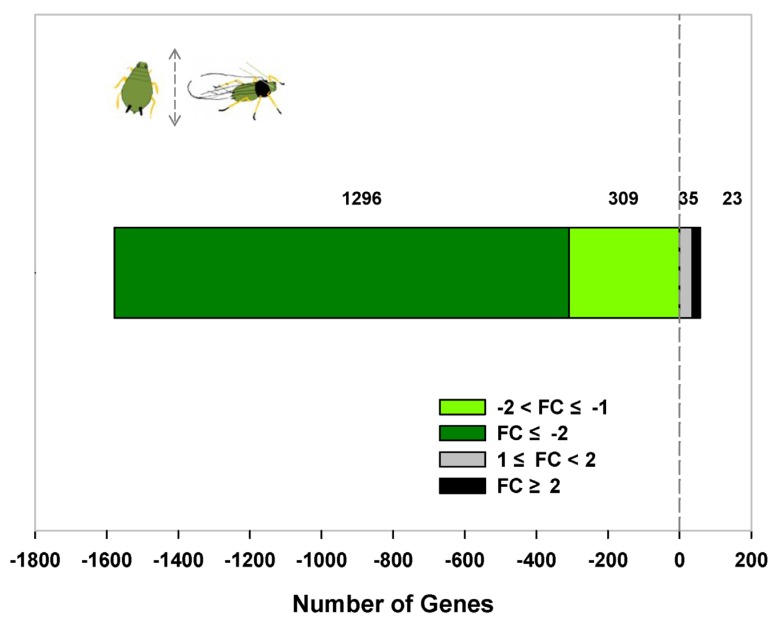
We obtained 1,663 differentially expressed transcripts from a total of 8,308 tag-mapped unigenes. Among them, 58 transcripts were expressed significantly higher in winged aphids, while 1,605 transcripts had a higher expression in wingless morphs (|log2 ratio|≥1; FDR value≤ 0.001). A total of 334 (20.69%) differentially expressed transcripts had a differential ratio higher than 2 (|log2 ratio|≥2; Figure 2), in which 3 transcripts were detected in winged forms only and 34 transcripts were expressed in wingless forms exclusively (Supplementary Material: Table S3). Top ten differentially expressed transcripts and their ratios are summarized in Table 2.
Figure 2.
Expression dynamics of differentially expressed transcripts between winged and wingless A. gossypii. Fold Change represents the log2 of the ratio of gene expression level in winged in relation to wingless adult females. Transcript expressed at a constant level (ratio of 1) has a log2 (ratio) of zero, which can be interpreted as "equally expressed among winged and wingless aphids". Positive number represents highly expressed transcripts in winged adult females. Negative number represents highly expressed transcripts in wingless adult females.
Table 2.
Top ten differentially expressed transcripts between winged and wingless A. gossypii.
| Gene ID | log2FC* | Description |
|---|---|---|
| AG10278 | 12.5 | Flightin-like |
| gi|289097293 | 11 | 3-ketoacyl-CoA thiolase, mitochondrial-like |
| AG4984 | 10.1 | MPA13 allergen-like |
| gi|289151497 | 8.3 | N/A |
| AG12533 | 8.1 | Phosphate carrier protein, mitochondrial-like |
| AG13078 | 5.7 | Adenine nucleotide translocator-like |
| AG6703 | 5.6 | PXMP2/4 family protein 4-like |
| AG11738 | 5.3 | Tectonin beta-propeller repeat-containing protein-like |
| AG12565 | 3.8 | Hypothetical protein LOC100165045 isoform 1 |
| AG11798 | 3.5 | Very long-chain specific acyl-CoA dehydrogenase mitochondrial-like isoform 1 |
| AG10938 | -9.7 | Seryl-tRNA synthetase, cytoplasmic-like |
| AG2471 | -9.3 | Hypothetical protein LOC100571294 |
| gi|289129407 | -9.2 | N/A |
| AG9276 | -9.1 | RNA-binding protein PNO1-like |
| AG11819 | -9 | T-complex protein 1 subunit alpha-like |
| gi|289076469 | -9 | N/A |
| AG7395 | -8.9 | Exosome complex exonuclease MTR3-like |
| gi|289138983 | -8.7 | N/A |
| AG4684 | -8.6 | N/A |
| AG3568 | -8.6 | N/A |
“*”: FC is the fold change of gene expression level between winged and wingless morphs. Positive and negative values, respectively, denote highly and lowly expressed gene expression in winged adult females relative to wingless individuals. Assembly details are provided in Supplementary Material: Table S3.
Functional analysis of differentially expressed transcripts
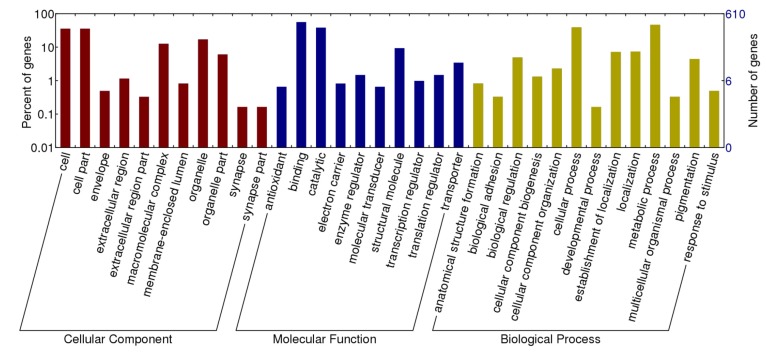
Following assignment of BLASTx top hits, 1,306 differentially expressed transcripts were functionally annotated with four additional databases. Among 8,308 sequenced transcripts, 3,184 transcripts were annotated with GO terms, including 2,318 involved in biological process, 2,547 in molecular function and 1,954 in cellular component. As to 1,663 differentially expressed transcripts, 763 transcripts were assigned with 3,168 GOs and can be classified into 42 secondary level categories (Figure 3). The category of biological process consists of 573 transcripts at 22 secondary levels. Molecular function category has 619 transcripts at 9 secondary levels. Cellular component includes 491transcripts in 11 secondary categories.
Figure 3.
Gene ontology classification of transcripts differentially expressed between winged and wingless A. gossypii. A total of 763 genes (45.88% of the differentially expressed transcripts) were categorized into three Gene Ontology classes, cellular components, molecular functions, and biological processes, respectively.
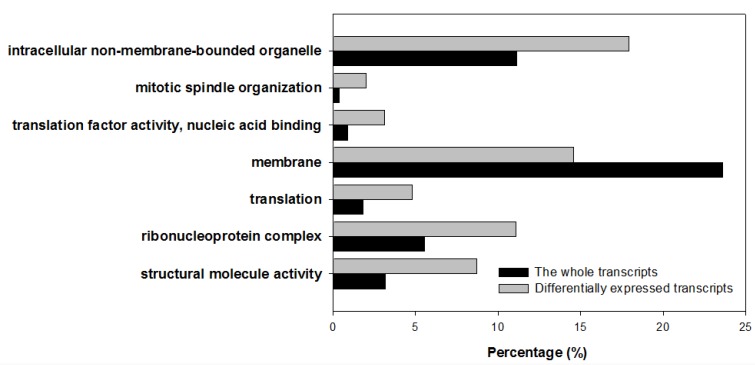
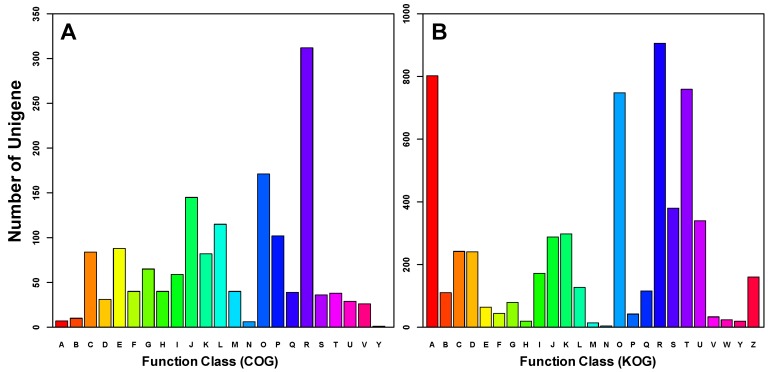
All differentially expressed transcripts were enriched in 16 GO group when compared to all the A. gossypii unigenes (Supplementary Material: Table S4). 7 of them were very deep level (specific) GO terms (Figure 4). Only one term (membrane) was underrepresented. In addition, transcripts were annotated by COG and KOG databases, by which 676 transcripts were categorized into 23 COG functional groups (Figure 5) and 1,144 transcripts into 25 KOG classes (Figure 5). With KOG classification, in addition to the category of general function prediction only, the number of transcripts in category A (RNA processing and modification), O (Posttranslational modification, protein turnover and chaperones), and U (Intracellular trafficking, secretion, and vesicular transport) were 802, 748 and 759, respectively, higher than the number in the other categories.
Figure 4.
Differentially expressed transcripts enriched specific Gene Ontology terms. All 763 differentially expressed transcripts are enriched in 16 GO groups. 7 deep level (specific) GO terms were gained. 6 terms were overrepresented, and one term was underrepresented.
Figure 5.
Functional classification of differentially expressed transcripts between winged and wingless A. gossypii. Based on Clusters of Orthologous Groups (COG, Figure_5A), 676 genes (40.65% of the Differentially expressed transcripts ) were annotated and categorized into 23 functional classes. In comparison, 1144 genes (68.79% of the Differentially expressed transcripts ) were annotated and categorized into 25 specific classes within the euKaryotic Orthologous Groups (KOG, Figure_5B). A: RNA processing and modification, B: Chromatin structure and dynamics, C: Energy production and conversion, D: Cell cycle control, cell division, chromosome partitioning, E: Amino acid transport and metabolism, F: Nucleotide transport and metabolism, G: Carbohydrate transport and metabolism, H: Coenzyme transport and metabolism, I: Lipid transport and metabolism, J: Translation, ribosomal structure and biogenesis, K: Transcription, L: Replication, recombination and repair, M: Cell wall/membrane/envelope biogenesis, N: Cell motility, O: Posttranslational modification, protein turnover, chaperones, P: Inorganic ion transport and metabolism, Q: Secondary metabolites biosynthesis, transport and catabolism, R: General function prediction only, S: Function unknown, T: Signal transduction mechanisms, U: Intracellular trafficking, secretion, and vesicular transport, V: Defence mechanisms, W: Extracellular structures, Y: Nuclear structure, Z: Cytoskeleton.
Pathway analysis of differentially expressed transcripts was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Among 1,663 differentially expressed transcripts, 878 transcripts could be annotated in KO based on sequence homologies and mapped to 211 KEGG pathways (Table 3). Pathway-oriented analysis showed that the top three significantly enriched metabolic pathways in DGEs were ribosome, pyruvate metabolism and proteasome. Differentially expressed transcripts were also enriched in pathways relating to protein synthesis and degradation, lipid metabolism, immunity, RNA transport, and some signaling pathways.
Table 3.
Top ten enriched metabolic pathways among differentially expressed transcripts.
| Pathway ID | Description | DETa | Referenceb | P-valuec | Q-value |
|---|---|---|---|---|---|
| ko03010 | Ribosome | 49 | 144 | 1.35E-14 | 2.85E-12 |
| ko00620 | Pyruvate metabolism | 21 | 83 | 8.96E-05 | 9.45E-03 |
| ko03050 | Proteasome | 15 | 53 | 0.000239 | 1.68E-02 |
| ko03040 | Spliceosome | 44 | 268 | 0.001539 | 7.15E-02 |
| ko04141 | Protein processing in endoplasmic reticulum | 42 | 254 | 0.001695 | 7.15E-02 |
| ko00280 | Valine, leucine and isoleucine degradation | 15 | 65 | 0.002402 | 8.45E-02 |
| ko00062 | Fatty acid elongation in mitochondria | 6 | 16 | 0.004098 | 1.24E-01 |
| ko04612 | Antigen processing and presentation | 15 | 75 | 0.009839 | 2.60E-01 |
| ko04146 | Peroxisome | 23 | 134 | 0.011384 | 2.67E-01 |
| ko01040 | Biosynthesis of unsaturated fatty acids | 9 | 38 | 0.014408 | 2.67E-01 |
“a”: Differentially expressed transcripts
“b”: Reference sequences were extracted from NCBI (see Materials and Methods for details).
“c”: P-value is calculated based on Hypergeometric Test. Pathways with Q value≤0.05 are significantly enriched in differentially expressed transcripts. Pathway ID can be found in KEGG database.
qRT-PCR validation study
qRT-PCR analysis was carried out to validate differentially expressed transcripts identified by DGE. All 20 genes showed the similar expression pattern as indicated by the DGE analysis (Table 4), suggesting that DGE approach can be a powerful tool for the discovery of candidate genes in a high-throughput manner.
Table 4.
Differentially expressed transcripts validated by qRT-PCR analysis.
| Gene ID | Description | Fold Change* | SE | |
|---|---|---|---|---|
| DGE | qRT-PCR | |||
| AG10278 | Flightin | 5961.13 | 1631.77 | 159.36 |
| gi|289097293 | Kat | 2068.00 | 11.06 | 1.93 |
| AG4984 | Fabp | 1092.00 | 12.25 | 2.18 |
| AG12533 | phosphate carrier protein, mitochondrial-like | 270.69 | 129.31 | 12.41 |
| AG13078 | Ant | 52.62 | 55.20 | 0.78 |
| AG11798 | very long-chain specific acyl-CoA dehydrogenase, mitochondrial-like | 11.63 | 12.14 | 2.33 |
| AG10938 | SerRS | -837.00 | -3.32 | 0.21 |
| AG13124 | ribosomal protein S28e-like | 11.45 | 26.07 | 5.96 |
| AG13172 | ribosomal protein LP0 | -4.53 | -2.03 | 0.54 |
| AG12143 | ribosomal protein S27A | -4.37 | -3.42 | 1.07 |
| AG13161 | 40S ribosomal protein S4-like | -4.15 | -1.18 | 0.17 |
| AG11336 | 40S ribosomal protein S21-like | -4.03 | -2.46 | 0.18 |
| AG13187 | ribosomal protein L15 | -4.03 | -3.76 | 1.16 |
| AG8620 | acylphosphatase | 11.18 | 26.64 | 6.96 |
| gi|289143076 | acylphosphatase-2-like | -9.64 | -2.06 | 0.46 |
| AG8620 | pyruvate kinase-like | -7.07 | -1.87 | 0.69 |
| AG10855 | malate dehydrogenase, cytoplasmic-like | -6.52 | -2.70 | 0.99 |
| AG12539 | pyruvate dehydrogenase E1 component subunit alpha, mitochondrial-like | -4.16 | -2.51 | 1.21 |
| AG9726 | proteasome activator complex subunit 3-like | -4.88 | -1.69 | 0.18 |
| AG12585 | proteasome inhibitor-like | -4.32 | -1.10 | 0.05 |
“*”: Fold change of gene expression level between winged and wingless morphs. SE is the standard error of qRT-PCT. Positive and negative values, respectively, denote highly and lowly expressed gene expression in winged adult females relative to wingless individuals. Morph-specific genes are highlighted in shade. Assembly details are provided in Supplementary Material: Table S3.
Discussion
DGE analysis
We sequenced wing-polyphenic aphids using the DGE approach and obtained 10.69 million raw base sequences, 10.48 million of which were clean tags. Clean tags were compared in different libraries. Interestingly, compared to the clean tags in non-specific forms, clean tags of morph-specific forms account for a small fraction of the total clean tags (Figure 1A), however, they comprise a higher level of percentage in distinct tags (Figure 1B), indicating that morph-specific tags have a high diversity. This result may suggest that expression level of common genes which maintain the basic growth and development is higher relative to specific genes that play crucial and multiple roles in trait variation. Typically, the reference sequences used by DGE analysis are selected from genome sequence information of evolutionarily related species to the target species, especially when there is no genome information available for the target species. In current DGE analysis, we chose EST sequence of A. gossypii as reference sequences, 46,766 tags were mapped to 18,141 transcripts (Table 1).
Differentially expressed transcripts
Through DGE analysis, we obtained a total of 1,663 differentially expressed transcripts, in which 58 transcripts were expressed significantly higher in winged aphids and 1,605 transcripts had a higher expression in wingless A. gossypii (Figure 2). This discrepancy is a reflection of the flight-reproduction trade-offs, in which genes associated with reproduction is likely over-numbered the flight capability. Among these differentially expressed transcripts, Flightin and seryl-tRNA synthetase (SerRS) was highly expressed in winged and wingless A. gossypii, respectively. Transcripts related to lipid metabolism and energy production were found at higher expression levels in migratory morphs (Table 2, Supplementary Material: Table S2). Differential energy allocation is an important part of trade-offs between flight capability and reproduction. This result is consistent with the previous observation of the significantly higher triglyceride content in winged aphid versus the wingless aphid 30, 31.
Associated with lipid synthesis and degradation, transcripts of 3-ketoacyl-CoA thiolase (KAT), very long-chain specific acyl-CoA dehydrogenase, medium-chain specific acyl-CoA dehydrogenase and long-chain-fatty-acid-CoA ligase 1 were shown significantly higher expression levels in winged morphs. Furthermore, 15 highly expressed “mitochondrial like” transcripts in winged morphs may contribute to energy production and thought to be involved in aspects of the citric acid cycle, respiratory chain, ATP synthesis and transport. These results suggest that lipid is the fuel for aphid migration. Similarly, transcripts involved in energy production showed elevated transcriptions in winged morphs of A. pisum 34 and the brown planthopper, Nilaparvata lugens 40.
Differentially expressed transcripts were significantly enriched in 7 GO terms, and 5 of them, structural molecule activity, ribonucleoprotein complex, translation, translation factor activity/nucleic acid binding, mitotic spindle organization, were directly or indirectly associated with protein translation and assembly. This is consistent with the pathway enrichment analysis. The remaining 2 term, membrane and intracellular non-membrane-bounded organelle, were associated with organelle, in which a series of biological process occurs, including partial protein synthesis processes in endoplasmic reticulum, fatty acid elongate in mitochondria, peroxisome break down very long chain fatty acids.
qRT-PCR validation study confirmed the results from DGE analysis (Table 4). Fully replicated qRT-PCR analysis, to certain extent, complements the lack of DGE replications. All 20 genes subjected to the qRT-PCR analysis showed the similar trends with the DGE results. In comparison to DGE analysis, however, the fold changes in gene expression of Flightin, Fabp, Kat and SerRS were less dramatic in qRT-PCR analysis. Such discrepancy is not without precedent 40. Due to the computation needs (the lowest expression level of any gene in the DGE analysis, by default, is 0.01 even if it is not expressed), the differential gene expression level is typically overestimated in the DGE analysis. In this study, the expression of Fabp, Kat and SerRS were morph-specific, and consequently, the result from the DGE analysis is likely overestimated.
Pathway enrichment analysis
Through the Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation system, differentially expressed transcripts were significantly enriched (Q<0.05) in three pathways, ribosome, pyruvate metabolism and proteasome (Table 3). Of the differentially expressed transcripts 5.58% were in the ribosome pathway. While in the whole reference background, only 1.72% of transcripts were annotated in this pathway. The ribosome is required for protein synthesis, which is critical for all aspects of cell growth and division 49. They are usually recognized as stable expression genes. However, in the current study, 48 highly expressed transcripts in wingless morph and 1 in winged morph were coded for the ribosomal proteins. Ribosomal genes are typically highly expressed genes. Previously reported research results showed gene products that are components of ribosomes were over-representation in A. pisum 34. One 40S ribosomal protein S20 displayed a higher expression level in brachypterous morph of N. lugens than in macropterous adult females 40. This evidence convinces us of difference in ribosomal proteins between two morphs of A. gossypii. Our data indicated that more protein synthesis was detected in the non-dispersing morphs, which results in faster developmental rates and higher fecundity 14, 50.
It is expected that pyruvate metabolism was a differentially expressed transcripts enriched pathway. 21 transcripts in this pathway were screened as differentially expressed transcripts. Pyruvate is a key intersection in several metabolic pathways, and known as the “hub” of carbohydrate, fats and proteins 16, 46. Pyruvate can be generated from glucose through glycolysis 16. In recent study, wingless morphs showed significantly higher soluble sugar and glycogen content than winged morphs during adulthood in A. gossypii 30 and in S. avenae 31. The result of gene expression was consistent with our previous research in biochemical composition. Most transcripts in this pathway were highly expressed in wingless morphs. Our results suggest that sugar or glycogen metabolism is faster in wingless morphs and those carbohydrates were used for development and production. This is consistent with the contention that sugar and glycogen is not the fuel of aphids for migration 51.
26S proteasome, working with ubiquitin, operates the energy-dependent regulated proteolysis process in eukaryotic cells. The immunotype proteasomes, whose catalytic subunits are replaced by the related counterparts, were discovered recently in vertebrates 52. The protein degradation process including immunity may be diverse in different morphs. In previous study, soluble protein content were higher in dispersal morphs than in reproduction morphs in A. gossypii 30 and S. avenae 31. As such, proteasome for protein degradation and protein turnover increased.
Genes of interests
Genes which we are interested in include Flightin, fatty acid binding protein gene (Fabp), adenine nucleotide translocase gene (Ant), abnormal wing discs1 (Awd1) and abnormal wing discs2 (Awd2) (Supplementary Material: Table S2).
The primary difference between winged and wingless adults is the existence of flight apparatus, including wing and indirect flight muscle. For winged adults, genes expressed in muscle were identified in this study, Flightin and Fabp. Flightin is a multiply phosphorylated, myofibrillar protein in Drosophila indirect flight muscles (IFM), and an essential part of the flight muscle contractile mechanism for thick filament assembly and sarcomere stability in Drosophila 53-56. Particularly, Flightin exhibits the greatest differential transcript accumulation in winged morphs of A. pisum 34 and N. lugens 40. Another gene coding FABP in muscle was one of the top ten most differentially expressed genes in winged morphs. FABP will increase the solubility of fatty acids and thus lead to a rapid transport through the cytosol, since the lipid is the preferred fuel for insect migrating 57 In recent research, free fatty acid content was significantly higher in winged than wingless adults before flight behavior in A. gossypii 30 and A. pisum 31.
ANT belongs to the mitochondria carrier protein which is embedded in the inner membrane of mitochondria and required for importing ADP into the mitochondrial matrix throughout the ATP synthesis 33. The gene expression of Ant gene is significantly higher in winged morphs than wingless morphs in A. gossypii (Table 2). The expression pattern of Ag Ant is similar as that of Mp Ant 33, suggesting the need of energy for winged morphs during the radiation to diverse food resources and habitats 58.
The awd1 and awd2 showed a significantly higher expression level in wingless morphs than winged morphs in A. gossypii. The awd of Drosophila encodes a nucleoside diphosphate kinase (NDPK). Complete loss of function of the awd gene causes lethality in Drosophila after the larval stage. Mutations in awd gene cause abnormal tissue morphology and aberrant differentiation. 59, 60. The awd plays an important role in development and differentiation of insects. They are not involved in the presumptive wing-patterning gene network 61. Little attention was given to awd genes in the previous studies related to wing polyphenism.
Perspectives and future research
This study sheds lights on the genetic basis of trade-offs between flight capability and reproduction. Genes involved in lipid metabolism show higher expression levels in winged A. gossypii than wingless morphs. In contrast, genes involved in sugar metabolism are elevated in wingless morphs. Our combined results suggest aphids using lipid rather than sugar or glycogen as fuel during migration. Flight ability of alate aphids depends on the energy supply of lipids. KAT, FABP, and very long-chain specific acyl-CoA dehydrogenase seems to be more important for winged morphs. Silencing of these genes using RNA interference (RNAi) may disrupt their energy supply, and compromise their ability to fly. RNAi through injection 62, 63 and feeding 64 methods have been well established in A. pisum. It is germane to investigate the pest control potential of these genes, including Flightin, through genetically interrupting the dispersal of A. gossypii.
Supplementary Material
Table S1-S4, and Fig.S1 - S2.
Acknowledgments
Special thanks go to Timothy Moural (University of Washington State) and Drs. Tracy Lu and John Obrycki (University of Kentucky) for their comments on an earlier version and Wenbo Chen for his assistance in the data analysis. This research was supported by the National Science & Technology Pillar Program (2012BAD19B05). Xiaowei Yang was supported by a start-up fund to Zhou and College of Agriculture, University of Kentucky. The granting agencies have no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Deposition
The accession numbers for DGE libraries derived from sedentary and migratory morphs of cotton aphids, Aphis gossypii, are SRX246107 and SRX242773, respectively.
Abbreviations
- DGE
Digital gene expression
- EST
Expressed Sequence Tag
- TPM
Number of transcripts per million clean tags
- FDR
False Discovery Rate
- GO
Gene Ontology
- COG
Cluster of Orthologous Groups
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG ontology
- KOG
euKaryotic Orthologous Groups
- qRT-PCR
Quantitative real-time PCR, IFM: indirect flight muscles
- SerRS
seryl-tRNA synthetase
- KAT
3-ketoacyl-CoA thiolase
- FABP
fatty acid binding protein
- NDPK
nucleoside diphosphate kinase
- ANT
adenine nucleotide translocase.
References
- 1.Stearns SC. Trade-offs in life-history evolution. Functional ecology. 1989;3:259–68. [Google Scholar]
- 2.Roff DA. Evolution of life histories: theory and analysis. New York, USA: Chapman & Hall; 1992. [Google Scholar]
- 3.Stearns SC. The evolution of life histories. New York, USA: Oxford University Press; 1992. [Google Scholar]
- 4.Brackenbury J. Insects in flight. London, UK: Blandford; 1992. [Google Scholar]
- 5.Moller Andersen N. Seasonal polymorphism and developmental changes in organs of flight and reproduction in bivoltine pondskaters (Hem. Gerridae) Insect Systematics & Evolution. 1973;4:1–20. [Google Scholar]
- 6.Gunn A, Gatehouse A, Woodrow K. Trade-off between flight and reproduction in the African armyworm moth, Spodoptera exempta. Physiological Entomology. 1989;14:419–27. [Google Scholar]
- 7.Kullberg C, Houston DC, Metcalfe NB. Impaired flight ability—a cost of reproduction in female blue tits. Behavioral Ecology. 2002;13:575–9. [Google Scholar]
- 8.Zera AJ, Denno RF. Physiology and ecology of dispersal polymorphism in insects. Annu Rev Entomol. 1997;42:207–30. doi: 10.1146/annurev.ento.42.1.207. [DOI] [PubMed] [Google Scholar]
- 9.Guerra PA. Evaluating the life-history trade-off between dispersal capability and reproduction in wing dimorphic insects: a meta-analysis. Biological reviews of the Cambridge Philosophical Society. 2011;86:813–35. doi: 10.1111/j.1469-185X.2010.00172.x. [DOI] [PubMed] [Google Scholar]
- 10.Roff DA. The Evolution of Wing Dimorphism in Insects. Evolution. 1986;40:1009–20. doi: 10.1111/j.1558-5646.1986.tb00568.x. [DOI] [PubMed] [Google Scholar]
- 11.Harrison RG. Dispersal Polymorphisms in Insects. Annual Review of Ecology and Systematics. 1980;11:95–118. [Google Scholar]
- 12.Wang ZL, Liu TT, Huang ZY, Wu XB, Yan WY, Zeng ZJ. Transcriptome Analysis of the Asian Honey Bee Apis cerana cerana. PLoS One. 2012;7:e47954. doi: 10.1371/journal.pone.0047954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roff DA. The Evolution of Wing Dimorphism in Insects. Evolution. 1986;40:1009. doi: 10.1111/j.1558-5646.1986.tb00568.x. [DOI] [PubMed] [Google Scholar]
- 14.Braendle C, Davis GK, Brisson JA, Stern DL. Wing dimorphism in aphids. Heredity (Edinb) 2006;97:192–9. doi: 10.1038/sj.hdy.6800863. [DOI] [PubMed] [Google Scholar]
- 15.Castañeda LE, Figueroa CC, Bacigalupe LD, Nespolo RF. Effects of wing polyphenism, aphid genotype and host plant chemistry on energy metabolism of the grain aphid, Sitobion avenae. Journal of Insect Physiology. 2010;56:1920–4. doi: 10.1016/j.jinsphys.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Simpson SJ, Sword GA, Lo N. Polyphenism in insects. Current Biology. 2011;21:R738–R49. doi: 10.1016/j.cub.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Xu X, Liu X, Zhang Q, Wu J. Morph-specific differences in life history traits between the winged and wingless morphs of the aphid, Sitobion avenae (Fabricius) (Hemiptera: Aphididae) Entomol Fennica. 2011;22:56–64. [Google Scholar]
- 18.Zera AJ. Wing polymorphism in Gryllus (Orthoptera: Gryllidae): Proximate endocrine, energetic and biochemical mechanisms underlying morph specialization for flight vs. reproduction. Phenotypic Plasticity of Insects: Mechanisms and Consequences; 2009. pp. 609–53. [Google Scholar]
- 19.Tanaka S, Nishide Y. First Record of the Occurrence and Genetics of a Short-Winged Morph in the Migratory Locust,Locusta migratoria(Orthoptera: Acrididae) Journal of Orthoptera Research. 2012;21:169–74. [Google Scholar]
- 20.Zera AJ, Sall J, Otto K. Biochemical aspects of flight and flightlessness in Gryllus: flight fuels, enzyme activities and electrophoretic profiles of flight muscles from flight-capable and flightless morphs. J Insect Physiol. 1999;45:275–85. doi: 10.1016/s0022-1910(98)00123-1. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Z, Zera AJ. Differential lipid biosynthesis underlies a tradeoff between reproduction and flight capability in a wing-polymorphic cricket. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16829–34. doi: 10.1073/pnas.262533999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zera AJ, Zhao Z. Intermediary metabolism and life-history trade-offs: differential metabolism of amino acids underlies the dispersal-reproduction trade-off in a wing-polymorphic cricket. Am Nat. 2006;167:889–900. doi: 10.1086/503578. [DOI] [PubMed] [Google Scholar]
- 23.Müller CB, Williams IS, Hardie J. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecological Entomology. 2001;26:330–40. [Google Scholar]
- 24.Braendle C, Friebe I, Caillaud MC, Stern DL. Genetic variation for an aphid wing polyphenism is genetically linked to a naturally occurring wing polymorphism. Proceedings Biological sciences / The Royal Society. 2005;272:657–64. doi: 10.1098/rspb.2004.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simon JC, Rispe C, Sunnucks P. Ecology and evolution of sex in aphids. Trends in Ecology & Evolution. 2002;17:34–9. [Google Scholar]
- 26.Moran NA. The Evolution of Aphid Life Cycles. Annual Review of Entomology. 1992;37:321–48. [Google Scholar]
- 27.Simpson SJ, Sword GA, Lo N. Polyphenism in insects. Current biology: CB. 2011;21:R738–49. doi: 10.1016/j.cub.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Bhatia V, Uniyal PL, Bhattacharya R. Aphid resistance in Brassica crops: challenges, biotechnological progress and emerging possibilities. Biotechnol Adv. 2011;29:879–88. doi: 10.1016/j.biotechadv.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Ishikawa A, Hongo S, Miura T. Morphological and histological examination of polyphenic wing formation in the pea aphid Acyrthosiphon pisum (Hemiptera, Hexapoda) Zoomorphology. 2008;127:121–33. [Google Scholar]
- 30.Shi SL, Liu XX, Zhang QW, Zhao ZW. Morph-specific differences in metabolism related to flight in the wing-dimorphic Aphis gossypii. Insect Science. 2010;17:527–34. [Google Scholar]
- 31.Xu XL, Liu XX, Zhang QW, Wu JX. Morph-specific differences in biochemical composition related to flight capability in the wing-polyphenic Sitobion avenae. Entomologia Experimentalis Et Applicata. 2011;138:128–36. [Google Scholar]
- 32.Brisson JA. Aphid wing dimorphisms: linking environmental and genetic control of trait variation. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2010;365:605–16. doi: 10.1098/rstb.2009.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghanim M, Dombrovsky A, Raccah B, Sherman A. A microarray approach identifies ANT, OS-D and takeout-like genes as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae (Sulzer) Insect Biochem Mol Biol. 2006;36:857–68. doi: 10.1016/j.ibmb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Brisson JA, Davis GK, Stern DL. Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum) Evol Dev. 2007;9:338–46. doi: 10.1111/j.1525-142X.2007.00170.x. [DOI] [PubMed] [Google Scholar]
- 35.International Aphid Genomics C. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao S, Mo D, Wang Q, Jia J, Qin L, Yu X. et al. Aberrant host immune response induced by highly virulent PRRSV identified by digital gene expression tag profiling. BMC Genomics. 2010;11:544. doi: 10.1186/1471-2164-11-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.t Hoen PA, Ariyurek Y, Thygesen HH, Vreugdenhil E, Vossen RH, de Menezes RX. et al. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundu R, Dixon AFG. Evolution of Complex Life-Cycles in Aphids. Journal of Animal Ecology. 1995;64:245–55. [Google Scholar]
- 39.Harhay GP, Smith TP, Alexander LJ, Haudenschild CD, Keele JW, Matukumalli LK. et al. An atlas of bovine gene expression reveals novel distinctive tissue characteristics and evidence for improving genome annotation. Genome Biol. 2010;11:R102. doi: 10.1186/gb-2010-11-10-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue J, Bao YY, Li BL, Cheng YB, Peng ZY, Liu H. et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS One. 2010;5:e14233. doi: 10.1371/journal.pone.0014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu F, Li W, Li Z, Zhang S, Chen S, Su S. High-abundance mRNAs in Apis mellifera: comparison between nurses and foragers. J Insect Physiol. 2011;57:274–9. doi: 10.1016/j.jinsphys.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–95. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 43.Yoav B, Daniel Y. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics. 2001;29:1165–88. [Google Scholar]
- 44.Rosengard AM, Krutzsch HC, Shearn A, Biggs JR, Barker E, Margulies IMK. et al. Reduced Nm23/Awd protein in tumour metastasis and aberrant Drosophila development. Nature. 1989;342:177–80. doi: 10.1038/342177a0. [DOI] [PubMed] [Google Scholar]
- 45.Tatusov RL. A Genomic Perspective on Protein Families. Science. 1997;278:631–7. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 46.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV. et al. The COG database: an updated version includes eukaryotes. BMC bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–6. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 48.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin JI, Mitchell NC, Kalcina M, Tchoubrieva E, Stewart MJ, Marygold SJ. et al. Drosophila ribosomal protein mutants control tissue growth non-autonomously via effects on the prothoracic gland and ecdysone. PLoS Genet. 2011;7:e1002408. doi: 10.1371/journal.pgen.1002408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartzberg EG, Kunert G, Westerlund SA, Hoffmann KH, Weisser WW. Juvenile hormone titres and winged offspring production do not correlate in the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 2008;54:1332–6. doi: 10.1016/j.jinsphys.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 51.Dixon AFG, Kindlmann P. Cost of Fligth Apparatus and Optimum Body Size of Aphid Migrants. Ecology. 1999;80:1678–90. [Google Scholar]
- 52.Tanaka K, Mizushima T, Saeki Y. The proteasome: molecular machinery and pathophysiological roles. Biological Chemistry. 2012;393(4):217–34. doi: 10.1515/hsz-2011-0285. [DOI] [PubMed] [Google Scholar]
- 53.Vigoreaux JO, Hernandez C, Moore J, Ayer G, Maughan D. A genetic deficiency that spans the flightin gene of Drosophila melanogaster affects the ultrastructure and function of the flight muscles. Journal of Experimental Biology. 1998;201:2033–44. doi: 10.1242/jeb.201.13.2033. [DOI] [PubMed] [Google Scholar]
- 54.Vigoreaux JO, Saide JD, Valgeirsdottir K, Pardue ML. Flightin, a novel myofibrillar protein of Drosophila stretch-activated muscles. The Journal of Cell Biology. 1993;121:587–98. doi: 10.1083/jcb.121.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reedy MC, Bullard B, Vigoreaux JO. Flightin Is Essential for Thick Filament Assembly and Sarcomere Stability in Drosophila Flight Muscles. The Journal of Cell Biology. 2000;151:1483–500. doi: 10.1083/jcb.151.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ayer G, Vigoreaux J. Flightin is a myosin rod binding protein. Cell Biochem Biophys. 2003;38:41–54. doi: 10.1385/CBB:38:1:41. [DOI] [PubMed] [Google Scholar]
- 57.Haunerland NH. Transport and utilization of lipids in insect flight muscles*. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 1997;117:475–82. [Google Scholar]
- 58.Santamaria M, Lanave C, Saccone C. The evolution of the adenine nucleotide translocase family. Gene. 2004;333:51–9. doi: 10.1016/j.gene.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 59.Timmons L, Shearn A. Role of AWD/nucleoside diphosphate kinase in Drosophila development. J Bioenerg Biomembr. 2000;32:293–300. doi: 10.1023/a:1005545214937. [DOI] [PubMed] [Google Scholar]
- 60.Dearolf CR, Hersperger E, Shearn A. Developmental consequences of awdb3, a cell-autonomous lethal mutation of Drosophila induced by hybrid dysgenesis. Developmental Biology. 1988;129:159–68. doi: 10.1016/0012-1606(88)90170-4. [DOI] [PubMed] [Google Scholar]
- 61.Brisson JA, Ishikawa A, Miura T. Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Mol Biol. 2010;19(Suppl 2):63–73. doi: 10.1111/j.1365-2583.2009.00935.x. [DOI] [PubMed] [Google Scholar]
- 62.Mutti NS, Park Y, Reese JC, Reeck GR. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum. Journal of insect science (Online) 2006;6:1–7. doi: 10.1673/031.006.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jaubert-Possamai S, Le Trionnaire G, Bonhomme J, Christophides GK, Rispe C, Tagu D. Gene knockdown by RNAi in the pea aphid Acyrthosiphon pisum. BMC biotechnology. 2007;7:63. doi: 10.1186/1472-6750-7-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. Silencing of aphid genes by dsRNA feeding from plants. PloS one. 2011;6:e25709. doi: 10.1371/journal.pone.0025709. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1-S4, and Fig.S1 - S2.