Summary
Exomer is a cargo adaptor mediating the sorting of specific plasma membrane proteins into vesicles at the trans-Golgi network. Cargo adaptors must bind to multiple partners, including their cargo, regulatory proteins, and the membrane surface. During biogenesis of a vesicle, the membrane makes a transition from a relatively flat surface to one of high curvature, requiring cargo adaptors to somehow maintain protein-protein and protein-membrane interactions on a changing membrane environment. Here we present the crystal structure of a tetrameric Chs5/Bch1 exomer complex and use small angle x-ray scattering to demonstrate its flexibility in solution. The structural data suggest that the complex flexes primarily about the dimeric N-terminal domain of the Chs5 subunits, which adopts a non-canonical β-sandwich fold. We propose that this flexible hinge domain enables exomer to maintain interactions in the context of a dynamic membrane environment.
Introduction
The localization of membrane proteins is regulated spatiotemporally in eukaryotic cells by the action of soluble protein complexes, which serve as intermediary adaptors between the protein cargo and the machinery mediating the physical process of vesicle formation. By directly binding both cytoplasmic domains of transmembrane proteins and vesicle coats, these cargo adaptors package their cargo into specific vesicles, and indeed are core components of the vesicle coats themselves (Matsuoka et al., 1998; Robinson and Bonifacino, 2001; Takatsu et al., 2001). Structurally well-characterized examples of cargo adaptors include the GGA and AP adaptors for the clathrin coat (Owen et al., 2004), the core components βδ/γζ-COP and Sec23/24 of the unrelated COPI and COPII complexes respectively (Bi et al., 2002; Yu et al., 2012), and Vps26/29/35 of the more divergent retromer complex (Hierro et al., 2007).
Exomer is a recently discovered cargo adaptor (Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006a) possessing several unusual properties. Exomer acts in the poorly characterized direct trans-Golgi network (TGN) to plasma membrane (PM) transport pathway (De Matteis and Luini, 2008) to transport several PM proteins, including the chitin synthase Chs3 and the pheromone response mediator Fus1, in a temporally regulated manner (Santos et al., 1997; Santos and Snyder, 2003). While its Arf1-dependent recruitment to membranes parallels the Arf1 or Sar1 dependence of other cargo adaptors (Serafini et al., 1981; Oka et al., 1991; Donaldson et al., 1992; Yoshihisa et al., 1993), exomer neither exhibits the capacity to deform membranes (Wang et al., 2006a) nor is known to associate with canonical cage-like coats.
Exomer is composed of a Chs5 core protein and a suite of four homologous Chs5/Arf1 binding proteins, or ChAPs: Chs6, Bud7, Bch1, and Bch2 (Sanchatjate and Schekman, 2006; Trautwein et al., 2006; Wang et al., 2006a). While the stoichiometry of the complex in solution consists of two copies of Chs5 and two copies of a copurified ChAP (Paczkowski et al., 2012), following immunoprecipitation of the complex from yeast cells via one ChAP all three remaining ChAPs are detected (Sanchatjate and Schekman, 2006; Trautwein et al., 2006), indicative of a structural organization in which the four ChAPs interchangeably bind Chs5 dimeric cores. As the ChAPs produce different phenotypes when deleted from yeast cells (Trautwein et al., 2006) and are proposed to bind cargo sorting signals (Barfield et al., 2009; Starr et al., 2012), the modular architecture of exomer provides a molecular basis for the separate regulation of the transport of multiple proteins via the same complex.
Recently, we determined the crystal structure of a Chs5/Chs6 exomer complex, revealing a molecular organization of Chs5 in which exomer tetramerization, Chs5 binding of ChAPs, and Chs5 binding of Arf1 are mediated by distinct domains (Paczkowski et al., 2012). In contrast, Chs6 exhibits a complex fold topology in which an extended tetratricopeptide repeat (TPR) region, thought to be largely invariant between ChAPs (Rockenbauch et al., 2012), forms the binding surface for Chs5. However, extensive crystal packing interactions strained the N-terminal Chs5 dimerization interface to such a degree that little could be concluded regarding the interactions mediating tetramerization (Paczkowski et al., 2012).
Here we present the crystal structure of a tetrameric exomer complex, composed of the Chs5 N-terminal domains and a different ChAP, Bch1. The structure of the Chs5 dimerization interface is revealed to be a non-canonical trans β-sandwich. By comparing small-angle X-ray scattering (SAXS) and X-ray crystallographic data, we find that this motif exhibits an unexpected flexibility in solution with implications for exomer function as a cargo adaptor in a heterogenous membrane milieu.
Results
Exomer assembles via a non-canonical trans β-sandwich assembly
The previously determined Chs5/Chs6 exomer structure contains the elongated FN3/BRCT of exomer (FBE) domain, connected to the main body of the complex by a 4-residue linker lacking regular secondary structure (Paczkowski et al., 2012). Removal of this flexible domain permitted us to generate crystals of Chs5(1–77) in complex with the ChAP Bch1. These crystals diffracted anisotropically to 2.9Å along the a* and c* axes and approximately 3.3Å along the b* axis as estimated by the Diffraction Anisotropy Server (Strong et al., 2006). Molecular replacement using the Chs5/Chs6 structure as a phase model located two Chs5/Bch1 dimers per asymmetric unit (Figure S1A), and the structure was further built and refined to yield appropriate statistics (Table 1).
Table 1.
Data collection and refinement statistics.
| Wavelength (Å) | 0.987 |
| Resolution range (Å) | 50 - 2.9 (2.95 - 2.9) |
| Space group | C 2 |
| Unit cell | a=141.39Å b=155.69Å c=99.05Å α=90° β=95.34° γ=90° |
| Total reflections | 152442 |
| Unique reflections | 43750 |
| Multiplicity | 3.5 (3.0) |
| Completeness (%) | 96.00 (68.70) |
| <I>/<σ(I)> | 8.59 (2.51) |
|
| |
| Wilson B-factor | 70.42 |
| Rsyma | 0.111 (0.567) |
| Rcrystb | 0.2365 (0.3253) |
| Rfreeb | 0.2882 (0.3929) |
| Number of atoms | 11082 |
| protein (1376 residues) | 11064 |
| Water | 18 |
| RMS(bonds) (Å) | 0.004 |
| RMS(angles) (°) | 0.90 |
| Ramachandran favored (%) | 94 |
| Ramachandran outliers (%) | 0.75 |
| Clash scorec | 21.50 |
| Average B-factor | 96.20 |
| Protein | 96.30 |
| Water | 60.80 |
Statistics for the highest-resolution shell are shown in parentheses.
Rfree was calculated with a random 5% of the reflections.
Calculated with MolProbity (Chen et al., 2010)
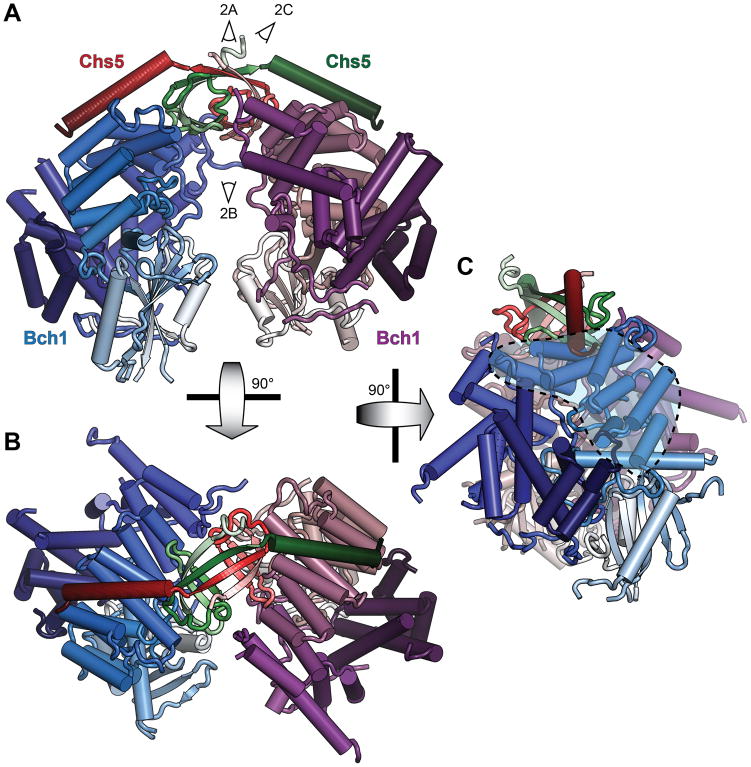
The Chs5/Bch1 asymmetric unit comprises a dimer of Chs5/Bch1 heterodimers as previously predicted for the native complex (Paczkowski et al., 2012), with the relatively flat faces of the Bch1 subunits apposed and separated by approximately 15–20Å (Figure 1; Movie S1); remarkably, despite their close proximity along their entire length, at no point do the two subunits contact each other. The primary Chs5/Bch1 interface is largely unchanged from that of Chs6, with the C-terminal half of the Chs5 helix packing against a conserved hydrophobic pocket formed by Bch1 helices 13–15.
Figure 1. Crystal structure of the Chs5/Bch1 tetrameric exomer complex.
(A–C) The built Chs5(1–77)/Bch1 structure is shown in cartoon representation. Subunits are colored in light-to-dark N-to-C gradients as indicated, with additional texture added to the red Chs5 subunit. The TPR region is indicated by shading. View angles of the panels in Figure 2 are indicated. See also Figure S1 and Movie S1.
The N-terminal dimerization domain of Chs5, poorly resolved in the Chs5/Chs6 structure, here is well resolved and forms a symmetric β-sandwich with the dimer partner (Figures 2A, 2B). Anchored by strands 1 and 4, strands 2 and 3 of each copy of Chs5 reach across to pack against the opposing Bch1 protein in accordance with the prior domain swap model (Paczkowski et al., 2012); not predicted by the prior model, however, is an additional interaction between interstrand loops of the swapped domain with a highly conserved surface of the proximal Bch1 (Figure 2C). Strands 1 and 4 of both copies combine in a single four-stranded β-sheet, stabilizing the dimer interface together with a hydrophobic core. Notably, the opposing strands, 2 and 3 of each copy, form two parallel pairs of hairpins only, failing to assemble into the canonical 4-strand β-sheet expected of the sandwich (Figure 2B; Figure S1B); strand 3 of each copy are separated by nearly 7 Å relative to the more standard 5 Å within each hairpin. The entire Chs5 N-terminal domain is the most highly conserved region of any exomer domain (Figure 2A), indicative of a critical function in the complex.
Figure 2. The N-terminal Chs5/Chs5 dimerization domain is a non-canonical β-sandwich.
All view angles are indicated in Figure 1. (A) The Chs5/Chs5 β-sandwich shown from outside the complex. Left: Colored by light-to-dark N-to-C gradient as per Figure 1, including texture on one Chs5 subunit; strands are labeled by number, N-terminal to C-terminal. Right: Colored by conservation; red/green color begins to appear at the average BLOSUM62 value for residues in Chs5. β-strands are pleated to indicate direction of the side chain. (B) As per (A), but viewed from within the Bch1/Bch1 cleft; Bch1 subunits are cut away for visibility. (C) View of Chs5 trans packing against Bch1. Strands 1 and 4 are removed to improve visibility; strand 4 of chain c (red) is retained as a dotted line to indicate connectivity. Left: As per panel A. Right: Bch1 is drawn in surface representation and colored by conservation as per Chs5. Side chains are shown for all residues of Chs5 within 5Å of Bch1. See also Figure S2.
The Chs5 N-terminus forms a divergent oligosaccharide binding (OB) type fold, as the common RNA polymerase subunit Rpb8 forms a close monomeric match to the N-terminal dimer by Dali comparison (2E2H chain h, Z=9.9) (Figure S1C) (Krapp et al., 1998; Wang et al., 2006b; Holm and Rosenstrom, 2010). While proteins sharing this fold classification also include several domains responsible for binding a variety of metals and small molecules (Arcus, 2002), the dimeric composition of the Chs5 domain is highly atypical. No specific small molecule binding function has been ascribed to either exomer or Rpb8.
Comparison of divergent ChAP structures
S. cerevisiae possesses two homologous pairs of more closely related ChAP proteins: Bch1 and Bud7, and Bch2 and Chs6 (Trautwein et al., 2006). Other yeast species including the Candida and Pichia genera possess two ChAPs each corresponding to one S. cerevisiae pair. The structure presented here and our previously reported structure of a Chs5/Chs6 complex (Paczkowski et al., 2012) thus represent one example each of the two families. The TPR region forming the backbone of the protein (Figure 1C) is essentially identical between the two ChAPs. While similar elements (N-terminal β-sheet, overall protein topology) are present in the N- and C-terminal domains packing against the TPR repeat, their length and orientation varies somewhat (Figure S2A). Residue conservation within the respective ChAP families (Figure S2B) suggests that the face of the ChAPs formed by the N- and C-termini is likely to be responsible for mediating primary ChAP functions such as cargo binding. Outside of the highly conserved Chs5 binding interface, the TPR repeat region is largely unconserved on the solvent-exposed surface, suggesting a primarily structural role as a scaffold for the remainder of the protein.
The Chs5 β-sandwich is a flexible molecular hinge
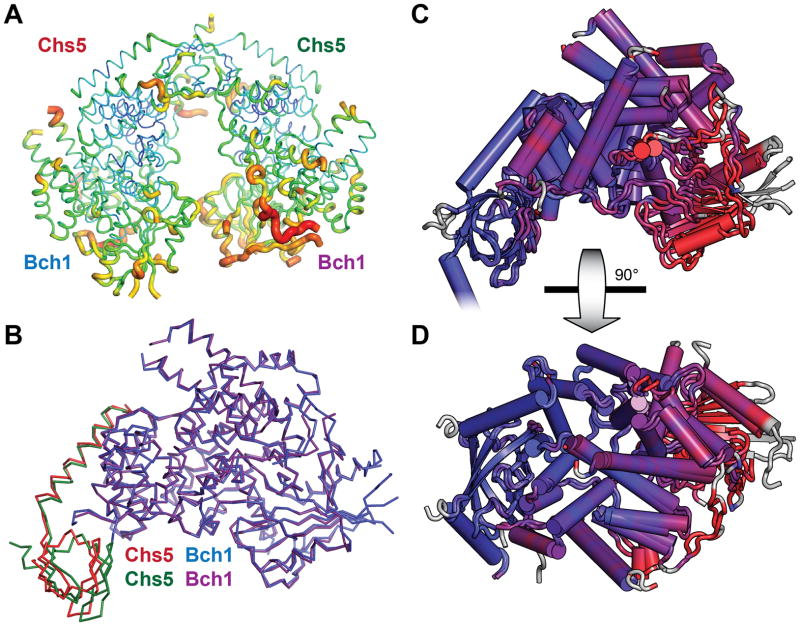
The atomic B-factors of the structure display a striking gradient along the body of the Bch1 subunits, ranging from low proximal to Chs5 to extremely high at the distal tips (Figure 3A). Given the position of the Chs5 dimerization domain to one side of the complex, this led us to hypothesize that the structure may be more flexible than the static crystal structure suggests, and that the two Bch1 subunits may act as lever arms on a Chs5 hinge. Alignment of the two Chs5/Bch1 heterodimers in the asymmetric unit identifies a significant difference in the positioning of the two Chs5 N-terminal domains relative to Bch1 (Figure 3B; Figure S3). This indicates an inherent flexibility of the Bch1 subunit positions relative to the dimeric Chs5 N-terminal domains and to each other. Accordingly, aligning the two halves of the asymmetric unit by superimposing the two Chs5 N-terminal domains results in displacement of the Bch1 subunits relative to each other, with the greatest displacement found at the distal ends (Figures 3C and 3D). Interestingly, the TPR region does not display any significant deviation, suggestive of some additional degree of flexibility in the packing of the N- and C-terminal domains of Chs6 against the TPR backbone.
Figure 3. Crystallographic evidence of exomer flexibility.
(A) The Chs5/Bch1 structure is shown in putty format, with residues possessing higher B-factors shown in warmer colors and as a wider backbone ribbon. (B) The two Chs5/Bch1 heterodimers in the asymmetric unit are superimposed. The Chs5 N-terminus adopts a different conformation relative to each Bch1 molecule. (C) The two Chs5/Bch1 heterodimers are aligned via the Chs5 N-terminal domain and colored by RMSD variation between residues, from blue at RMSD ≤ 0.5 Å to red at RMSD ≥ 3.0 Å; grey residues have no counterpart in the other chain. Both copies of Chs5 are shown in the diagram for clarity. (D) As per C, rotated 90° as indicated. The TPR repeat region is at the upper left. See also Figure S3.
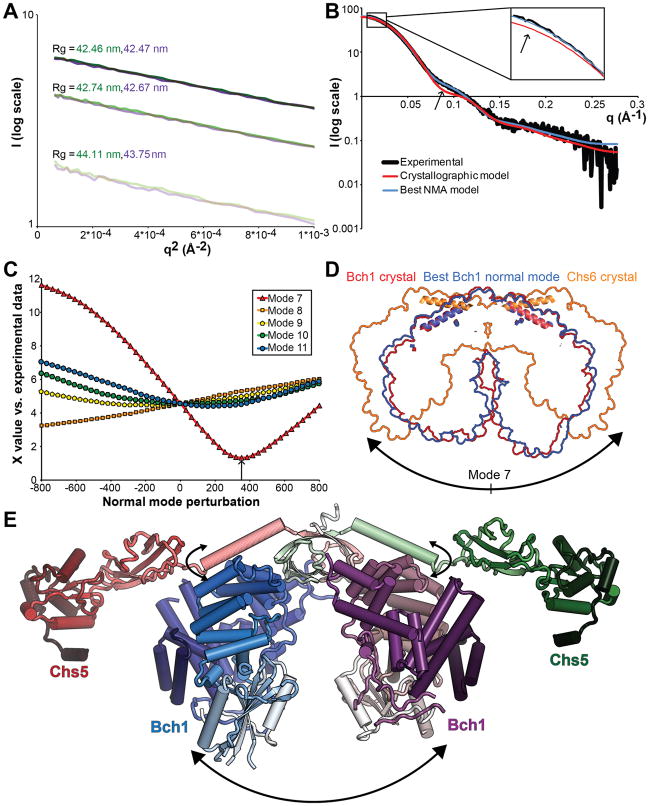
To address the likelihood that crystal packing may exogenously stabilize a more flexible structure, we performed SAXS analysis to gain insight into the conformation of exomer in solution. The Chs5(1–77)/Bch1 complex proved ideal for SAXS analysis, displaying no detectable native, radiation-dependent, or concentration dependent aggregation as assessed by Rg and linearity in the Guinier region (Figure 4A). The theoretical scattering curve calculated from the crystal structure, while displaying features qualitatively similar to the experimental data, deviated significantly (Figure 4B), indicating a significant discrepancy between the crystal and solution conformations of exomer.
Figure 4. SAXS analysis of exomer solution structure indicates flexibility.
(A) Guinier plots of Chs5(1–77)/Bch1 scattering curves at 9 mg/ml (dark), 6 mg/ml, and 3 mg/ml (light). Green and purple represent the first and second sequential exposures of the same sample. Radius of gyration calculated from the Guinier slope is indicated for each. (B) Fit of theoretical scattering curves calculated from the indicated structures to experimental scattering data at 9 mg/ml. Inset: zoom on low-angle region. Arrows: Primary regions of divergence between the crystal structure and the experimental SAXS data. (C) Quality of fit to experimental data of theoretical scattering curves of models evenly spaced along the indicated normal mode vibrations. Arrow: best-fit NMA model. (D) Overlaid space-filling outlines of the indicated structures aligned with the approximate Chs5 hinge axis perpendicular to the page. Chs5 helices of each structure are added for reference. (E) Assembled Chs5/Bch1 structure based on the best-fit NMA model, with FBE domains modeled from the Chs5/Chs6 structure via alignment of the shared Chs5 helix. Flexible motions are indicated with arrows. See also Figure S4 and Movie S2.
To determine the nature of this structural discrepancy, we employed normal mode analysis (NMA). NMA predicts likely flexible movements of proteins and protein complexes on the basis of their backbone conformation (Tirion, 1996). Empirically, the lowest-frequency vibrational modes incorporating movements of the majority of the molecule often accurately model transitions between pairs of crystallographic conformations, especially when calculated from open conformations and without significant energetic contribution from binding ligands (Tama and Sanejouand, 2001; Petrone and Pande, 2006) as is the case for exomer. Calculation of the normal modes of the Chs5/Bch1 complex with elNémo (Suhre and Sanejouand, 2004) yielded five normal modes with low frequency and broad residue coverage, numbered 7–12 after excluding the six trivial rotations and translations (Movie S2).
Comparison of the theoretical scattering of the resulting structural models to the experimental SAXS curve identified a model produced by perturbation along the lowest-frequency mode, mode 7, fitting the experimental data with a X-value of 1.3, comparable to model data used in testing CRYSOL (Svergun et al., 1995) (Figures 4B and 4C). This normal mode represents a simple opening and closing of the complex along an axis running through the Chs5 β-sandwich (Figures 4D and 4E, Figure S4A). Extending this opening motion further leads to the orientation observed in the previous Chs5/Chs6 crystal structure (Figure 4D) (Paczkowski et al., 2012); while the dimerization domain is largely disrupted at this extreme, enough flexibility must exist in exomer to permit formation of elements of the crystal packing without complete disruption of the trans Chs5-Chs6 interaction.
While a single conformational model suffices to describe the experimental scattering curve, this may represent either a true dominant conformation or the average of an ensemble spanning a subset of the conformational space described by normal mode 7. Either interpretation indicates a degree of flexibility of exomer in solution in the absence of external forces such as those exerted by crystal packing or binding to partner proteins. Attempts to use de novo modeling to calculate a molecular envelope were unsuccessful, likely because the enforcement of a roughly globular formation is a critical element of such modeling (Franke and Svergun, 2009), and exomer exhibits a long crevice between the Bch1 subunits running nearly the entire length of the complex.
To address the possibility that flexibility may be specific to the Bch1 ChAP, we collected SAXS data on both Chs5(1–77)/Chs6 and Chs5(1–77)/Bud7. While the Chs6-containing complex displayed significant dependence of Rg on concentration, precluding meaningful analysis of the scattering data (Figure S4B), analysis of the Bud7-complex using the Chs5/Bch1 structure as a low-resolution approximation revealed a very similar dependence on normal mode perturbation (Figures S4C and S4D). This indicates that the observed flexibility occurs independently of ChAP identity.
By superimposing the ChAP-interacting helix of Chs5 common to the Chs5(1–299)/Chs6 and Chs5(1–77)/Bch1 structures, a composite model can be constructed of intact exomer incorporating our NMA/SAXS based solution results (Figure 4E). In this model, the two FBE domains extend in opposite directions, although their precise orientation appears to be flexible on the basis of the Chs5/Chs6 crystal structure.
Discussion
Cargo adaptors function in vesicle biogenesis to recruit a multitude of cargo proteins to the budding vesicle. Thus, they serve as intermediaries between the variable sizes and distribution of the cargo and the rapidly changing curvature of the budding vesicle on one hand, and the constrained geometries of the small GTPases and cage-like coat proteins on the other. One expectation of cargo adaptors, therefore, is that they must be structurally capable of mediating these multiple divergent interactions during dynamic membrane shape changes. Our results demonstrate flexibility of the exomer cargo adaptor, mediated by the N-terminus of Chs5 acting as a molecular hinge.
Structural comparison of the Chs5 β-sandwich to that of Rpb8 highlights an unusual absence of stabilizing β-sheet formation between the parallel and only marginally separated strand 2/strand 3 hairpins. As this feature of the protein backbone lies in a well-resolved region of electron density and was not present in the homology model used for molecular replacement, it is unlikely to represent an artifact of model building. As such, the “flaw” in the Chs5 β-sandwich between strand 3 of each copy of Chs5 represents a likely source of flexibility causing the hinge behavior. Mutations H25D/H26D and M47P previously have been made in this domain and shown to result in outright disassembly of the tetramer into Chs5/Chs6 dimers (Paczkowski et al., 2012). The other source of flexibility in the hinge domain appears to be the domain’s contact with the adjacent ChAP in the loop between strands 1 and 2.
With respect to membrane binding, one likely possibility is that the twofold symmetric axis of the tetramer lies perpendicular to the surface of the membrane. In light of the marked lack of conservation of the outer face of Chs5, we predict that the Chs5-distal tips of the ChAP subunits may lie proximal to the membrane following recruitment by the FBE domain-Arf1 interaction (Paczkowski et al., 2012). Flexibility of the Chs5 dimer interface would allow exomer to accommodate the curvature and proteins of the heterogenous membrane environment, as well as the binding of multiple cargos by a single complex.
The limited degree of flexibility seen in exomer stands in contrast to the dichotomy between rigid structures and complete flexibility seen in other cargo adaptors. The Sec23/24 COPII cargo adaptor forms a seemingly rigid “pre-budding complex” with its cognate GTPase Sar1 (Bi et al., 2002). The clathrin adaptor AP complexes, while exhibiting a significant conformational rearrangement upon membrane and cargo binding, appear to exist in a single, stable conformation when membrane bound (Collins et al., 2002; Heldwein et al., 2004; Kelly et al., 2008; Jackson et al., 2010). We note that both Sec23/Sec24 and the AP complexes bind to vesicle cage and accessory proteins via flexible linkers (Owen et al., 2004; Bi et al., 2007). Exomer, however, employs a hinge domain as a primary source of flexibility, implying that constrained flexibility, rather than the complete conformational freedom allowed by a flexible linker, is critical for its function at the TGN.
Experimental Procedures
Protein expression and purification
S. cerevisiae Chs5 residues 1–77 and C-terminally 6His-tagged Bch1 were cloned into the pETDuet-1 expression vector (Novagen) for recombinant expression in the Rosetta2 strain of E. coli (Novagen). Cultures were grown in TB to an OD600 of ~3.5, followed by reduction of temperature to 18°C and induction of expression by addition of 250 μM IPTG. After overnight expression, cultures were collected by centrifugation, resuspended in 450 mM KOAc, 10% glycerol, 50 mM HEPES pH 7.4, 20 mM imidazole pH 8, and 10 mM β-mercaptoethanol, and lysed by sonication. The protein was then purified via nickel affinity in batch (Ni-NTA, Qiagen), anion exchange (MonoQ, GE Healthcare), and gel filtration (Superdex 200, GE Healthcare) without removal of the 6His tag. The final protein was concentrated to 10 mg/ml in 20 mM HEPES pH 7.4, 150 mM NaCl, and 2 mM DTT. Complexes of Chs5 residues 1–77 with ChAPs Chs6 or Bud7 were purified for SAXS following the same protocol, with the exception that Bud7 was concentrated to a final concentration of 4 mg/ml due to low yield.
Crystallization and structure determination
Purified Chs5/Bch1 complex was crystallized by hanging drop vapor diffusion using a crystallization solution of 15% PEG-8000, 19% glycerol, 0.1M Tris pH 8, and 0.2M NaCl in a 1:1 drop ratio. Cryoprotection was accomplished by a one-step transfer to a similar solution with glycerol increased to 25%. Diffraction data were collected at CHESS beamline A1 using a ADSC Quantum-210 CCD detector. The crystal was indexed to a space group of C2 with unit cell dimensions a=141 Å, b=156Å, c=99Å and angle β=95.34°, and diffraction data were processed with HKL-2000 (Otwinowski and Minor, 1997). Phases were determined in PHENIX (Adams et al., 2010) by molecular replacement using the 22.7% identical 4GNS, processed by SCULPTOR on the basis of a MUSCLE (Edgar, 2004)-generated alignment manually adjusted to match predicted secondary structure, as a search model. The core of the protein was automatically built in PHENIX, followed by many rounds of manual building in COOT (Emsley et al., 2010) alternating with refinement in PHENIX. To minimize model bias, early- and mid-stage refinements were conducted with NCS restraints and simulated annealing; TLS using domains calculated by TLSMD (Painter and Merritt, 2006) was employed to model the strongly anisotropic B-factors of the structure. Structural figures were generated with PyMol (Schrödinger, LLC).
Small-angle X-ray scattering data collection and analysis
SAXS data were collected at CHESS beamline F2 with equipment previously described (Adams et al., 2010). Protein samples were identical to those used for crystallization as described above; the buffer eluate from the final protein concentration step was used as a matched blank and for all dilutions. Scattering images were collected from two serial 180 second exposures of the stock protein solution and 2:1 and 1:2 dilutions in matched buffer to assess radiation- and concentration-dependent aggregation. Data were processed with RAW to generate scattering curves. Theoretical scattering curves of molecular models were generated and fit to the experimental scattering curve using CRYSOL (Svergun et al., 1995).
Accession number
Coordinates and structure factors have been deposited in the Protein Data Bank (4IN3).
Supplementary Material
Highlights.
We determine the Chs5/Bch1 exomer complex crystal structure to 2.9Å
The Chs5 dimerization domain is a non-canonical trans β-sandwich
Exomer is flexible about the Chs5 dimerization domain
Flexibility may facilitate function on dynamic membranes
Acknowledgments
We gratefully acknowledge the assistance and advice of CHESS A1 and F2 beamline staff, especially that of Richard Gillilan with respect to SAXS data collection and analysis. We thank Holger Sondermann for helpful discussion regarding normal mode analysis, Julia Kumpf for Chs5/Bud7 complex purification, Mike Henne for critical reading of the manuscript, and Jon Paczkowski for assistance with x-ray data collection, Chs5/Chs6 complex purification, and critical reading of the manuscript. This work is based upon research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation and the National Institutes of Health/National Institute of General Medical Sciences under NSF award DMR-0936384, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award GM103485 from the National Institutes of Health, through its National Institute of General Medical Sciences. This work was funded by Cornell University startup funds.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcus V. OB-fold domains: a snapshot of the evolution of sequence, structure and function. Current Opinion in Structural Biology. 2002;12:794–801. doi: 10.1016/s0959-440x(02)00392-5. [DOI] [PubMed] [Google Scholar]
- Barfield RM, Fromme JC, Schekman R. The Exomer Coat Complex Transports Fus1p to the Plasma Membrane Via a Novel Plasma Membrane Sorting Signal in Yeast. Mol Biol Cell. 2009;20:4985–4996. doi: 10.1091/mbc.E09-04-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. Insights into COPII Coat Nucleation from the Structure of Sec23•Sar1 Complexed with the Active Fragment of Sec31. Developmental Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular Architecture and Functional Model of the Endocytic AP2 Complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Cassel D, Kahn RA, Klausner RD. ADP-ribosylation factor, a small GTP-binding protein, is required for binding of the coatomer protein beta-COP to Golgi membranes. PNAS. 1992;89:6408–6412. doi: 10.1073/pnas.89.14.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. 2009 doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed]
- Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. PNAS. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, Steven AC, Bonifacino JS, Hurley JH. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Research. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Höning S, Evans PR, Owen DJ. A Large-Scale Conformational Change Couples Membrane Recruitment to Cargo Binding in the AP2 Clathrin Adaptor Complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BT, McCoy AJ, Späte K, Miller SE, Evans PR, Höning S, Owen DJ. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456:976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp S, Kelly G, Reischl J, Weinzierl ROJ, Matthews S. Eukaryotic RNA polymerase subunit RPB8 is a new relative of the OB family. Nature Structural & Molecular Biology. 1998;5:110–114. doi: 10.1038/nsb0298-110. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-Coated Vesicle Formation Reconstituted with Purified Coat Proteins and Chemically Defined Liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- Oka T, Nishikawa S, Nakano A. Reconstitution of GTP-binding Sar1 protein function in ER to Golgi transport. J Cell Biol. 1991;114:671–679. doi: 10.1083/jcb.114.4.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Charles J, Carter W, editors. Methods in Enzymology. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR. ADAPTORS FOR CLATHRIN COATS: Structure and Function. Annual Review of Cell and Developmental Biology. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Paczkowski JE, Richardson BC, Strassner AM, Fromme JC. The exomer cargo adaptor structure reveals a novel GTPase-binding domain. The EMBO Journal. 2012;31:4191–4203. doi: 10.1038/emboj.2012.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J Appl Crystallogr. 2006;39:109–111. [Google Scholar]
- Petrone P, Pande VS. Can Conformational Change Be Described by Only a Few Normal Modes? Biophys J. 2006;90:1583–1593. doi: 10.1529/biophysj.105.070045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS, Bonifacino JS. Adaptor-related proteins. Current Opinion in Cell Biology. 2001;13:444–453. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- Rockenbauch U, Ritz AM, Sacristan C, Roncero C, Spang A. The complex interactions of Chs5p, the ChAPs and the cargo Chs3p. Mol Biol Cell. 2012;23:4402–4415. doi: 10.1091/mbc.E11-12-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchatjate S, Schekman R. Chs5/6 Complex: A Multiprotein Complex That Interacts with and Conveys Chitin Synthase III from the Trans-Golgi Network to the Cell Surface. Mol Biol Cell. 2006;17:4157–4166. doi: 10.1091/mbc.E06-03-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Duran A, Valdivieso M. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2485–2496. doi: 10.1128/mcb.17.5.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B, Snyder M. Specific Protein Targeting during Cell Differentiation: Polarized Localization of Fus1p during Mating Depends on Chs5p in Saccharomyces cerevisiae. Eukaryotic Cell. 2003;2:821–825. doi: 10.1128/EC.2.4.821-825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothmant JE. ADP-Ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: A novel role for a GTP-binding protein. Cell. 1981;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Starr TL, Pagant S, Wang CW, Schekman R. Sorting Signals That Mediate Traffic of Chitin Synthase III between the TGN/Endosomes and to the Plasma Membrane in Yeast. PLoS ONE. 2012;7:e46386. doi: 10.1371/journal.pone.0046386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. PNAS. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhre K, Sanejouand YH. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Research. 2004;32:W610–W614. doi: 10.1093/nar/gkh368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Takatsu H, Futatsumori M, Yoshino K, Yoshida Y, Shin HW, Nakayama K. Similar Subunit Interactions Contribute to Assembly of Clathrin Adaptor Complexes and COPI Complex: Analysis Using Yeast Three-Hybrid System. Biochemical and Biophysical Research Communications. 2001;284:1083–1089. doi: 10.1006/bbrc.2001.5081. [DOI] [PubMed] [Google Scholar]
- Tama F, Sanejouand Y-H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- Tirion MM. Large Amplitude Elastic Motions in Proteins from a Single-Parameter, Atomic Analysis. Phys Rev Lett. 1996;77:1905–1908. doi: 10.1103/PhysRevLett.77.1905. [DOI] [PubMed] [Google Scholar]
- Trautwein M, Schindler C, Gauss R, Dengjel J, Hartmann E, Spang A. Arf1p, Chs5p and the ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 2006;25:943–954. doi: 10.1038/sj.emboj.7601007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-W, Hamamoto S, Orci L, Schekman R. Exomer: a coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J Cell Biol. 2006a;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural Basis of Transcription: Role of the Trigger Loop in Substrate Specificity and Catalysis. Cell. 2006b;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- Yu X, Breitman M, Goldberg J. A Structure-Based Mechanism for Arf1-Dependent Recruitment of Coatomer to Membranes. Cell. 2012;148:530–542. doi: 10.1016/j.cell.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.