Abstract
The movement of potexviruses through the cytoplasm to plasmodesmata (PD) and through PD to adjacent cells depends on the viral and host cellular proteins. Potexviruses encode three movement proteins [referred to as the triple gene block (TGB1–3)]. TGB1 protein moves cell-to-cell through PD and requires TGB2 and TGB3, which are endoplasmic reticulum (ER)-located proteins. TGB3 protein directs the movement of the ER-derived vesicles induced by TGB2 protein from the perinuclear ER to the cortical ER. TGB2 protein physically interacts with TGB3 protein in a membrane-associated form and also interacts with either coat protein (CP) or TGB1 protein at the ER network. Recent studies indicate that potexvirus movement involves the interaction between TGB proteins and CP with host proteins including membrane rafts. A group of host cellular membrane raft proteins, remorins, can serve as a counteracting membrane platform for viral ribonucleoprotein (RNP) docking and can thereby inhibit viral movement. The CP, which is a component of the RNP movement complex, is also critical for viral cell-to-cell movement through the PD. Interactions between TGB1 protein and/or the CP subunit with the 5′-terminus of genomic RNA [viral RNA (vRNA)] form RNP movement complexes and direct the movement of vRNAs through the PD. Recent studies show that tobacco proteins such as NbMPB2C or NbDnaJ-like proteins interact with the stem-loop 1 RNA located at the 5′-terminus of Potato virus X vRNA and regulate intracellular as well as intercellular movement. Although several host proteins that interact with vRNAs or viral proteins and that are crucial for vRNA transport have been screened and characterized, additional host proteins and details of viral movement remain to be characterized. In this review, we describe recent progress in understanding potexvirus movement within and between cells and how such movement is affected by interactions between vRNA/proteins and host proteins.
Keywords: potexvirus, intracellular trafficking, cell-to-cell movement, movement complexes, plasmodesmata, host proteins, host cellular membranes
INTRODUCTION
The infection cycle of plant RNA viruses includes the invasion of the host plant, RNA replication, cell-to-cell and long-distance movement in the host, and release from the host. Because their genomes are small and encode only a few genes, plant RNA viruses utilize many host factors during the infection cycle. The replication of viral RNAs (vRNAs) in host plants has been frequently studied but virus movement and other aspects of the viral infection cycle have received less attention (Mackenzie, 2005; Nagy and Pogany, 2010).
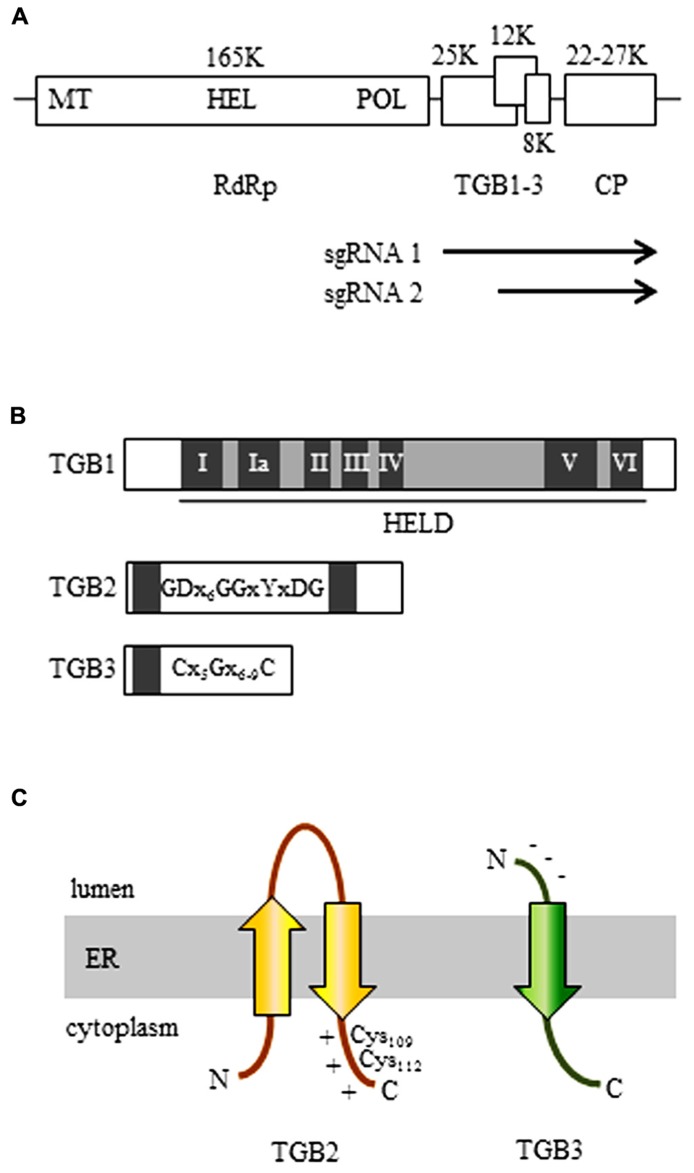
Potexviruses have been extensively studied and belong to the Alphaflexiviridae, a new family of plant RNA viruses, among which the genomes of the genus Potexvirus contain five open reading frames (ORFs) encoding an RNA-dependent RNA polymerase (RdRp; replicase), three overlapping movement proteins (MPs) [called the triple gene block (TGB1–3)], and the coat protein (CP; Adams et al., 2004; King et al., 2009; Figure 1A). All five of the virus-encoded proteins are used either in viral replication or in movement in infected host plants (Verchot-Lubicz, 2005; Verchot-Lubicz et al., 2010; Solovyev et al., 2012; Park et al., 2013). At the initial stage of infection, potexviruses, which have a plus (+)-stranded RNA genome, release vRNA from the virion and produce the virus-encoded replicase using host translation machinery. Replicase then forms a viral replication complex (VRC) along with several host factors and subsequently synthesizes (i) minus (-)-stranded vRNA from (+) vRNA and (ii) (+) vRNA or (+) subgenomic (sg) RNA from synthesized (-) vRNA. CP and TGB1–3 proteins are translated from the synthesized (+) sgRNAs and are used for encapsidation and movement of their progeny (+) vRNAs, which were produced from (-) vRNA as template, into neighboring uninfected cells through the plasmodesmata (PD). The movement of plant viruses is by definition essential if the progeny (+) vRNAs or virions are to spread into neighboring uninfected cells. In moving the progeny (+) vRNAs or virions via PD into adjacent cells, most plant viruses use their own MP(s). For potexviruses, substantial research has determined that viral cell-to-cell movement requires TGB proteins and CP (Chapman et al., 1992; Forster et al., 1992; Baulcombe et al., 1995; Morozov and Solovyev, 2003; Verchot-Lubicz et al., 2010; Niehl and Heinlein, 2011; Schoelz et al., 2011; Solovyev et al., 2012).
FIGURE 1.
The organization of the potexvirus genome. (A) The RNA-dependent RNA polymerase (RdRp, replicase) gene contains a methyltransferase domain (MET), a helicase domain (HEL), and an RNA polymerase domain (POL). The three genes of the triple gene block (TGB) are partially overlapped. Arrows indicate subgenomic (sg) RNAs for expression of TGBs. (B) The organization of the three TGB genes. TGB1: The first TGB ORF encodes the TGB1 protein and has a helicase-like domain (HELD), which contains seven typical motifs of a general helicase (I, Ia, II, III, IV, V, and VI; dark boxes). TGB2: the TGB2 protein is encoded in the second TGB ORF and has two transmembrane domains (dark boxes). The GDx6GGxYxDG sequence is conserved in TGB2-encoding viruses. TGB3: The TGB3 protein is encoded by the third TGB ORF and contains a transmembrane domain (dark box). Among the TGB3-encoding potexviruses, the TGB3 gene has a conserved C(x5)G(x6-9)C sequence. (C) 3-D structural models of TGB2 and TGB3 protein in the endoplasmic reticulum (ER). Potexvirus TGB2 protein integrates into ER membranes to form a U-like structure (with the central loop exposed to the ER lumen and with both the N- and C-terminus located in the ER cytoplasm) and has two conserved cysteine residues (Cys109 and Cys112) in the C-terminal region, which has a positive net charge (TGB2). Potexvirus TGB3 has an N-terminus that has a negative net charge and is exposed to the ER lumen and a C-terminus that is exposed to the ER cytoplasm (Morozov and Solovyev, 2003).
Most TGB-encoding plant RNA viruses belong to either the Virgaviridae or the Flexiviridae of the Alphaflexiviridae and Betaflexiviridae (Verchot-Lubicz et al., 2010). Recently, Solovyev et al. (2012) summarized information concerning TGB proteins and TGB-mediated plant viruses. The TGB proteins have been classified into two major groups, i.e., potex- and hordei-like TGBs, based on phylogeny and on differences in the viral movement mechanism (Morozov and Solovyev, 2003; Verchot-Lubicz et al., 2010). The potex-like viruses form filamentous virions containing a monopartite RNA genome and require the CP for cell-to-cell movement, whereas hordei-like viruses are rod-shaped, have multipartite RNA genomes, and do not require the CP for cell-to-cell movement (Morozov and Solovyev, 2003; Adams et al., 2004; Martelli et al., 2007; Verchot-Lubicz et al., 2010). Verchot-Lubicz et al. (2010) summarized and compared the movement strategies employed by TGB proteins in potex-like viruses and hordei-like viruses. In the current review, we supplement these earlier reviews by considering more recent findings on cell-to-cell movement of potexvirus vRNA and/or virions through the PD including the intracellular trafficking and intercellular transport of vRNA. We also describe the unique function of potexvirus RNA elements during intracellular trafficking.
THREE TGB PROTEINS (TGB1, TGB2, AND TGB3)
First, we summarize the general information concerning potexvirus TGB proteins, which are required for vRNA trafficking through PD in host cells. Most potex-like viruses belonging to the genera Potexvirus, Allexivirus, and Mandarivirus of the family Alphaflexiviridae and the genera Carlavirus and Foveavirus of the family Betaflexiviridae encode three TGB proteins (Adams et al., 2004; King et al., 2009). The other TGB protein-containing viruses belong to the genera Hordeivirus, Pomovirus, Pecluvirus, and Benyvirus (Morozov and Solovyev, 2003; Adams et al., 2004). Among these nine genera, three partially overlapping ORFs encode TGB1–3 proteins and are usually expressed from two sgRNAs (Zhou and Jackson, 1996; Verchot et al., 1998; Figure 1A). In the TGB-containing potexviruses, TGB1 protein is translated from sgRNA1, whereas TGB2 and TGB3 proteins are co-translated from sgRNA2 (Verchot et al., 1998). Barley stripe mosaic virus (BSMV), in the genus Hordeivirus, temporally controls the level of TGB proteins accumulation so that the estimated ratio is 100:10:1 (TGB1:TGB2:TGB3) during replication in plant cells (Johnson et al., 2003; Jackson et al., 2009). Although the ratio of TGB proteins for potexviruses has not been reported, according to the evidence from BSMV, it seems that TGB-encoding plant viruses control the expression level of TGB proteins by controlling the synthesis of sgRNAs.
Potexvirus TGB1 protein is encoded by the first TGB ORF and contains a helicase-like domain (HELD) that has seven conserved typical motifs (I, Ia, II, III, IV, V, and VI) in superfamily 1 (SF1) among three SFs (SF1, SF2, and SF3) of RNA helicases (Gorbalenya and Koonin, 1993; Liou et al., 2000; Kalinina et al., 2002; Lin et al., 2004; Leshchiner et al., 2006; Han et al., 2007). Motif I has conserved GKS/T tripeptides in potexviruses, and motif II is responsible for binding ATP and Mg2+ to corresponding sites (“Walker A” and “Walker B”) found in numerous ATP-binding proteins (Gorbalenya and Koonin, 1993; Kadare and Haenni, 1997). Potexvirus TGB1 protein also functions as a suppressor of RNA silencing (Wung et al., 1999; Voinnet et al., 2000; Bayne et al., 2005; Lim et al., 2010b) and as a translational activator (Atabekov et al., 2000; Figure 1B; TGB1). Especially, a previous study suggested that TGB1 of Bamboo mosaic virus (BaMV) has RNA-binding activity, which might be associated with RNA silencing (Wung et al., 1999). In Potato virus X (PVX), it was shown that TGB1 mutants that lack ability to suppress RNA silencing are not able to contribute to viral movement, whereas some PVX TGB1 mutants that are movement defective still function as RNA silencing suppressor, indicating that these mechanisms of potexvirus TGB1 are integrated (Bayne et al., 2005). Interestingly, Senshu et al. (2009) showed different levels of the RNA silencing suppression by TGB1 in potexviruses including PVX, Plantago asiatica mosaic virus, Asparagus virus 3, White clover mosaic virus (WClMV), and Tulip virus X. These results indicate that potexvirus TGB1s contribute various levels in suppressing RNA silencing. In contrast, another study has shown that a single mutation in TGB1 from Alternanthera mosaic virus (AltMV) that showed a dramatic reduction of RNA silencing suppression activity still supports full cell-to-cell movement, indicating that potexvirus TGB1 protein appears to be uncoupled in silencing suppression and movement functions (Lim et al., 2010c). By using green fluorescent protein (GFP) fusion TGB1 of PVX, researchers have shown that, in addition to having RNA-binding and helicase activities, TGB1 protein increases the PD size-exclusion limits (SELs) for viral cell-to-cell movement (Angell et al., 1996; Tamai and Meshi, 2001).
Triple gene block 1-mediated X-body reorganization contributes to the compartmentalization of the viral gene products during viral infection (Tilsner et al., 2012; Yan et al., 2012). A TGB1 mutant that lost the movement function could not form rod-like structures, whereas those mutants that still supported cell-to-cell movement formed rod-like structures, indicating that TGB1’s movement function is closely associated with its function in the formation of rod-like structures (Yan et al., 2012). In addition, Tilsner et al. (2012) showed that the PVX TGB1 protein reorganizes actin and endomembranes (the endoplasmic reticulum [ER] and golgi) into the X-body as a VRC.
Potexvirus TGB2 protein is encoded by the second ORF of the TGB gene cluster and is an integral membrane protein that has two predicted transmembrane domains that interact with ER membranes (Mitra et al., 2003; Morozov and Solovyev, 2003). A topological study with BaMV showed that potexvirus TGB2 protein integrates into the ER membranes in a U-like structure with the central loop exposed to the ER lumen and with both the N- and C-terminus located on the cytoplasmic side of the ER (Hsu et al., 2008; Figure 1C; TGB2). Cysteine-to-alanine substitution analysis indicated that two conserved cysteine residues (Cys109 and Cys112) in the C-terminal region of potexvirus TGB2 protein are critical for both cell-to-cell and systemic movement of BaMV (Hsu et al., 2008; Tseng et al., 2009). In addition, potexvirus TGB2 protein has sequence-independent RNA-binding activity (Cowan et al., 2002; Hsu et al., 2009).
Like TGB2, potexvirus TGB3 protein, which is encoded by the third TGB ORF, is also an integral protein in ER membranes (Krishnamurthy et al., 2003; Figure 1C; TGB3). TGB3 protein has a conserved C(x5)G(x6-9)C sequence and a predicted transmembrane domain (Figure 1B; TGB3). The N-terminus of the TGB3 protein has a negative net charge and is exposed to the ER lumen, while the C-terminus is exposed to the cytoplasmic side of the ER (Krishnamurthy et al., 2003; Morozov and Solovyev, 2003; Figure 1C; TGB3). The cytoplasmic tail (C-terminus) of TGB3 protein contains a sorting signal that is necessary for TGB3 oligomerization and for the targeting of integral membrane proteins to cortical ER tubules (Wu et al., 2011). The TGB3 sorting signal is highly conserved among potexviruses, and a TGB3 mutant defective in the sorting signal showed impaired cell-to-cell viral movement (Wu et al., 2011).
Mutational analyses have shown that localization of TGB2 and TGB3 proteins into the ER is critical for viral cell-to-cell movement (Krishnamurthy et al., 2003; Mitra et al., 2003). Both TGB2 and TGB3 proteins enhanced cell-to-cell diffusion of free GFP or GFP–sporamin fusion proteins through the PD (Tamai and Meshi, 2001; Haupt et al., 2005), suggesting that TGB2 and TGB3 proteins are capable of gating the PD.
INTRACELLULAR TRAFFICKING OF POTEXVIRUS vRNA FROM THE CYTOPLASM TO PD
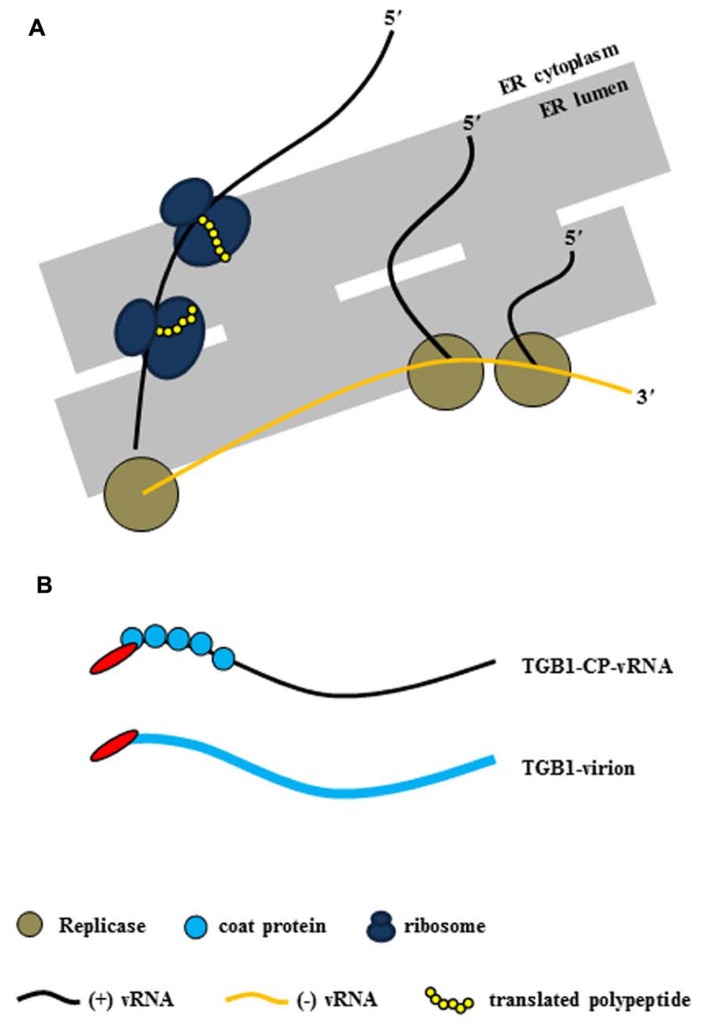
As mentioned earlier, potexvirus TGB1 protein has RNA-binding and helicase activities (Kalinina et al., 2002). Potexviruses form VRC containing (-) vRNA as templates, viral replicase, and host protein(s) at the host cellular membranes (Figure 2A). Following the replication of (+) vRNA by replicase, (+) vRNA is converted to the PD-transportable potexvirus vRNA form by TGB1 protein for the cell-to-cell movement through the PD. Two models have been proposed for the formation of PD-transportable potexvirus vRNA (intact virion or a ribonucleoprotein [RNP] movement complex containing vRNA, TGB1 protein, and CP) during the cell-to-cell movement of vRNA through the PD. Lough et al. (2000) provided experimental evidence that vRNAs of potexviruses including PVX and WClMV were transported by the formation of RNP movement complex involving vRNA, TGB1 protein, and CP rather than intact virion alone. Recently, however, Verchot-Lubicz et al. (2010) and Tilsner et al. (2013) have provided evidence that the PD-transportable potexvirus vRNA form is partially or fully encapsidated by the CP subunit and that the TGB1 protein is associated with the 5′ end of the CP-coated vRNA. Therefore, the formation of the RNP movement complex involves the association of the TGB1 protein with the 5′ end of the CP-coated vRNA (Verchot-Lubicz et al., 2010; Tilsner et al., 2013; Figure 2B).
FIGURE 2.
Virus replication complex (VRC) and RNP movement complex in the ER tubule. (A) Potexvirus constructs the VRC, which consists of vRNA, replicase, and host proteins (i.e., ribosomes) for replication of vRNA. Replicase (brown circles) produces progeny (+) vRNA from (-) vRNA as a template in the ER structure, and host ribosomes (dark blue) translate viral proteins (yellow chain) from progeny (+) vRNA. (B) The two kinds of RNP movement complexes in potexviruses. In one case, the PD-transportable form of potexvirus vRNA is partially encapsidated by the CP subunit, and TGB1 protein is associated with the 5′ end of the partially CP-coated vRNA (TGB1-CP-vRNA). In the second case, TGB1 protein binds to the CP-encapsidated virion (TGB1-virion). The blue circles indicate CP in the first case, and the thick blue line indicates CP and virion in the second case.
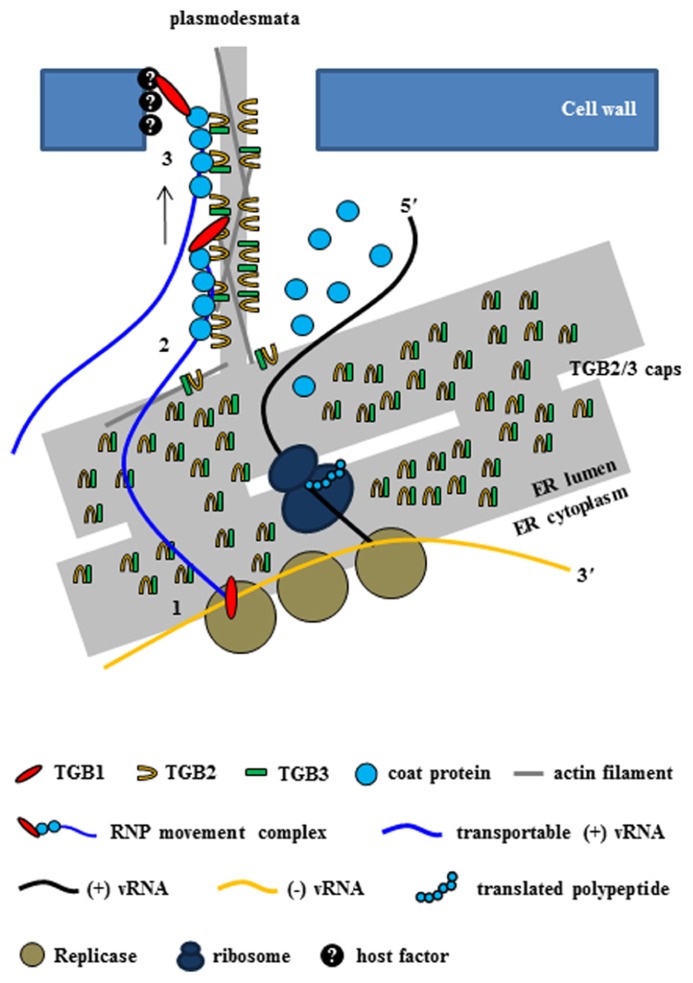
As mentioned earlier, cell-to-cell movement of potexvirus vRNA through the PD requires the three TGB proteins and the CP. Several host proteins might also be required for the formation of the RNP movement complex, but how host proteins cooperate with the RNP movement complex is unclear. The RNP movement complex containing TGB1 protein, CP, and vRNA subsequently binds to other viral proteins (TGB2 and TGB3 proteins) and possibly with several host proteins in the ER tubule (Figure 3).
FIGURE 3.
Intercellular transport of potexvirus vRNA to PD at the late stage of viral infection. The PD-transportable potexvirus vRNA (blue line) is released from the intermediate double-stranded RNA form by TGB1 protein acting as an RNA helicase (1). The PD-transportable potexvirus vRNA is partially or fully encapsidated by CP subunits, and then TGB1 protein associates with the 5′ end of the partially or fully CP-coated vRNA for the formation of the RNP movement complex at PD “cap” structures that consist of TGB2, TGB3 proteins, and VRC at the ER adjacent to the PD orifice (2). Then, the TGB1-driven RNP movement complex directs the nascent virions into the PD pore (3, Tilsner et al., 2013).
Potexvirus TGB1 protein is a component in the RNP movement complex (TGB1-CP-vRNA or TGB1-virion) and is able to transport vRNA into the PD as well as provide RNA helicase activity probably near the PD. However, plant expression experiments using GFP–PVX TGB1 to determine the movement of TGB1 protein within and between cells indicated that GFP–TGB1 was diffused throughout the cytoplasm, nucleoplasm, and rod-shape structure (Howard et al., 2004; Samuels et al., 2007). These findings indicate that potexvirus TGB1 protein requires viral CP in the RNP movement complex to move together with their vRNA into PD.
In addition, Rodionova et al. (2003) have shown that PVX TGB1 protein, which is translated from sgRNA at an early stage of viral infection, binds to polar CP subunits located at the end of PVX virion, resulting in the disassembling of the CP particles of the virion. This creates a translatable form of PVX vRNA in host plants. It seems that potexvirus TGB1 protein directly binds to CP but not to vRNA for disassembly of the PVX virion and thereby enables the release of a translatable form of vRNA. In the case of Plantago asiatica mosaic virus, the TGB1 protein directly interacts with CP and binds with the 5′ non-translated region (NTR) of vRNA for virion formation or for formation of the RNP movement complex (Ozeki et al., 2009).
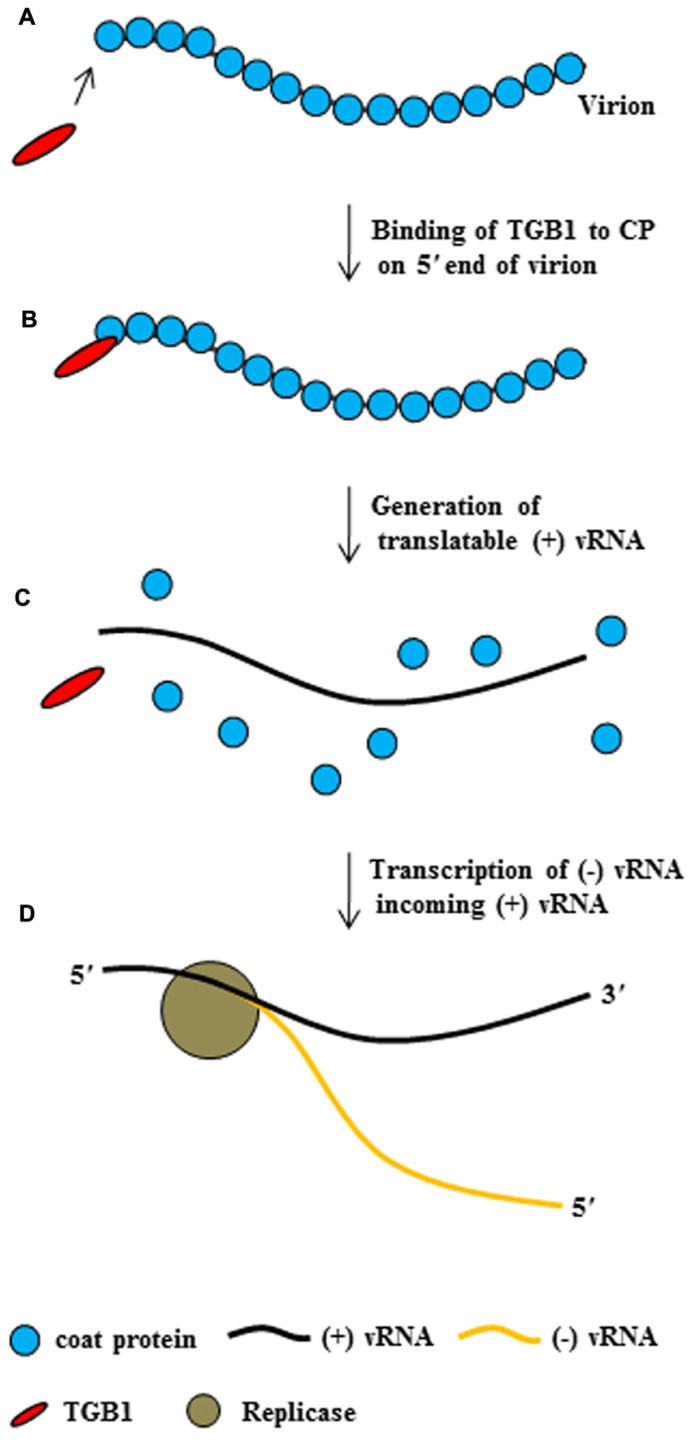
Taken together, these findings suggest that potexvirus TGB1 protein may have at least two functions during the virus infection cycle. First, TGB1 protein might convert non-translatable vRNA into a translatable form. To accomplish this first function, TBG1 protein early in infection in uninfected adjacent cells binds to the end of CP subunits on the virion (Figures 4A,B), leading to the disassembling of the CP from vRNA and the releasing of a translatable form of vRNA (Figure 4C). Finally, replicase transcribes (-) vRNA for incoming (+) RNA (Figure 4D). Second, TGB1 protein might facilitate the transport of vRNA to the PD. To accomplish this second function, TGB1 protein binds to the transportable form (virion form or partial CP-encapsidated vRNA) of progeny vRNA, which is generated after vRNA accumulates early in infection, and along with other viral proteins becomes part of the RNP movement complex (Figures 3 and 5).
FIGURE 4.
The conversion of potexvirus virions into translatable vRNA. PVX TGB1 protein binds to polar CP subunits located at the end of PVX virion (A,B). Then, binding of TGB1 protein causes the CP to separate from the PVX virion (C) early in infection after movement of virion into uninfected adjacent cells, such that the virion become translatable. Replicase then replicates (-) vRNA from the translatable form of PVX (+) vRNA (D), replicase transcribed (-) vRNA from (+) vRNA, and finally (not shown), ribosomes bind to the (+) vRNA.
FIGURE 5.
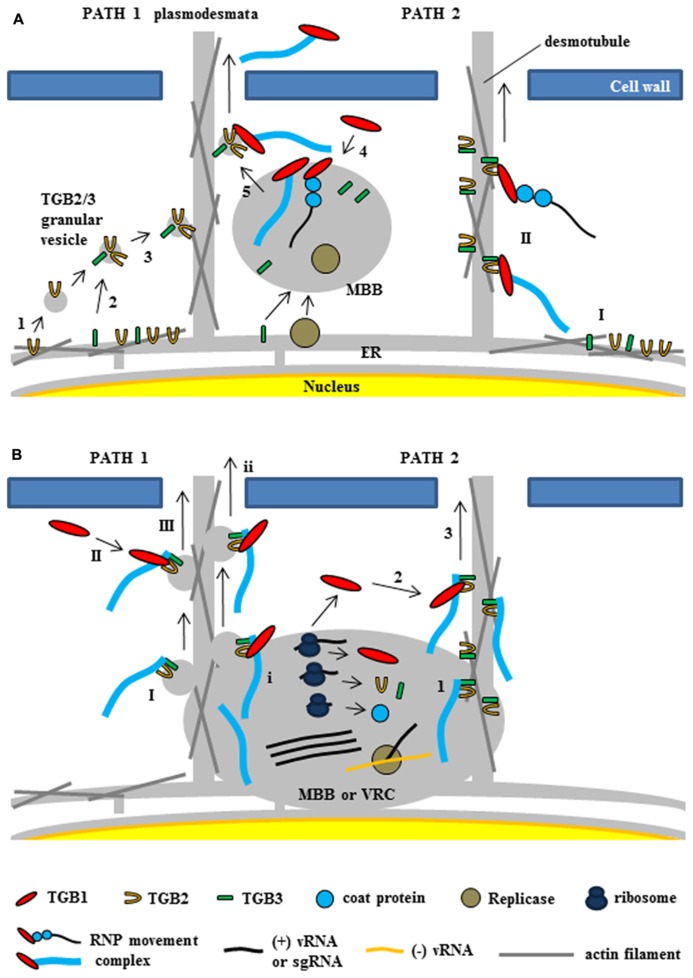
Two models for intracellular trafficking of potexviruses. These two models were partially adapted from Chou et al. (2013) and Verchot-Lubicz et al. (2010). (A) The first model proposed by Verchot-Lubicz et al. (2010). (PATH 1) Potexvirus RNP movement complex is trafficked by TGB2-induced granular vesicles with TGB3 protein (TGB2/3 granular vesicles) to the PD. TGB2 protein is able to induce the formation of granular vesicles derived from the ER membranes (1) and colocalizes with TGB3 protein (2). TGB3 protein directs movement of the TGB2-induced granular vesicles from the perinuclear ER to the cortical ER (3). vRNA or virion form of potexvirus mainly associates with membrane-bound bodies (MBB) derived the perinuclear ER that contains replicase and TGB3 protein. TGB1-bound vRNA or virion form (RNP movement complex released from MBB by TGB3 protein (4) and binds to TGB2/3 granular vesicles in the ER tubule for trafficking vRNA through the PD (5). (PATH 2) TGB2 protein localizes in the ER tubules with TGB3 protein, which can alone localize in membrane bodies at the cell periphery (I), and then RNP movement complex binds to TGB2 and TGB3 protein for vRNA trafficking to the PD (II). (B) The second model for TGB3-based intracellular trafficking of vRNA in BaMV (Chou et al., 2013). (PATH 1) Virion is associated by TGB2/3 granular vesicles (I) from ER-derived MBB or viral replication complex (VRC). Cytosolic TGB1 (II) or TGB1 protein (i) interacts with TGB2 protein into virion-TGB2/3 granular vesicles in cytosol and MBB or VCR, respectively and leads vRNA toward the PD (III and ii). (PATH 2) Virion interacts with TGB3 protein of TGB2/3 complex (1) and cytosolic TGB1 protein (2) interacts with TGB2 protein in virion-TGB2/3 complex. TBG1 protein drives virion-TGB1/2/3 complex toward the PD (3).
Potexvirus CP, which is another component in the RNP movement complex, is required for viral movement as well as for genome encapsidation in plants. Potexvirus CP mutants that were defective for interaction with TGB1 protein were able to form virus particles in vitro but were unable to move from cell-to-cell (Zayakina et al., 2008; Ozeki et al., 2009; Tilsner et al., 2012). This result suggests that the interaction between TGB1 protein and CP mediates the movement of the potexvirus RNP movement complex to the PD.
As mentioned earlier, research with GFP–TGB2 and GFP–TGB3 fusion proteins has demonstrated that both TGB2 and TGB3 proteins are integral membrane proteins in the ER or ER-associated vesicles located at actin filaments (Morozov and Solovyev, 2003; Hsu et al., 2008). In relation to the role of TGB2 and TGB3 proteins for potexvirus vRNA trafficking to PD, two models have been proposed (Verchot-Lubicz et al., 2010; Chou et al., 2013). Verchot-Lubicz et al. (2010) summarized the first model suggesting two pathways of potexvirus vRNA trafficking to PD based on the interactions between TGB2 and TGB3 proteins (Figure 5A, PATH 1 and PATH 2). One pathway suggests that the potexvirus RNP movement complex is transported by TGB2-induced granular vesicles as directed by TGB3 protein (TGB2/3 granular vesicles) to PD (Figure 5A, PATH 1). In support of this pathway, TGB2 protein is able to induce the formation of granular vesicles derived from the ER membranes that associate with actin filaments (Ju et al., 2005). Mutational analyses have shown that these TGB2-induced granular vesicles are necessary for cell-to-cell movement of PVX (Ju et al., 2007). Although subcellular localization assays have shown that PVX TGB3 protein is localized to the ER tubules when expressed alone in tobacco plant cells (Ju et al., 2005), other studies have shown that TGB3 protein is co-localized at the TGB2-induced granular vesicles in PVX-infected plant cells, suggesting that TGB2 protein is responsible for the localization of TGB3 protein at the ER membrane-associated granular vesicles (Schepetilnikov et al., 2005). TGB3 protein directs the movement of the TGB2-induced granular vesicles from the perinuclear ER to the cortical ER in both yeast and plant systems (Lee et al., 2010; Wu et al., 2011). These results indicate that potexvirus TGB3 protein serves as a driving factor for movement of vRNA to PD via TGB2-induced granular vesicles (Figure 5A, PATH 1).
PVX vRNA mainly associates with membrane-bound bodies (MBB) from the perinuclear ER that contain replicase and TGB1 protein (Tilsner et al., 2009), and TGB3 protein also associates with the cortical ER network and MBB (Krishnamurthy et al., 2003; Samuels et al., 2007; Bamunusinghe et al., 2009). Given these findings, the first pathway would indicate that the potexvirus RNP movement complex is released from MBB by TGB3 protein and that the released RNP movement complex then binds to the TGB2/3 granular vesicles in the ER tubule and moves to the PD (Verchot-Lubicz et al., 2010; Figure 5A, PATH 1).
The second pathway for the vRNA trafficking of potexvirus to PD by TGB3 protein is supported by interaction and localization assays between TGB2 and TGB3 proteins (Figure 5A, PATH 2). BaMV TGB2 protein localizes in the ER structures when expressed in the absence of TGB3 protein, whereas TGB3 protein expressed alone localizes in membrane bodies at the cell periphery (Lee et al., 2010). Bimolecular fluorescence complementation assay has indicated that BaMV TGB2 protein physically interacts with TGB3 protein in a membrane-associated form (Wu et al., 2011). The latter study also reported an interaction between TGB2 protein and either CP or TGB1 protein. The interactions of TGB1–TGB2 and TGB2–CP were associated with the ER network, which is consistent with the localization of TGB2 protein, and these interaction complexes were translocated to the TGB3-containing punctae within ER tubules by co-expression of TGB3 protein (Wu et al., 2011). In another study, which involved the mutation of the C-terminal domain of BaMV CP, the BaMV CP mutant did not interact with the HELD of BaMV replicase, and the mutation severely delayed the virus cell-to-cell movement independent of encapsidation (Lee et al., 2011). Thus, the authors suggested that movement of BaMV from an infected cell to a non-infected cell involves the RNP movement complex containing the replicase, TGB1 protein, CP, and vRNA. In PVX, the replicase is associated with ER membranes and co-localizes with the ER-derived TGB2/3 granular vesicles (Doronin and Hemenway, 1996; Bamunusinghe et al., 2009), indicating that the RNP movement complex containing replicase may be an intermediate form between the VRC and the final RNP movement complex.
Recently, the second model suggesting the stable association of the virion cargo with the TGB2- and TGB3-based membrane complex and recruitment of TGB1 protein to the PD by this complex for cell-to-cell movement of BaMV has been proposed (Chou et al., 2013; Figure 5B). The authors suggested that TGB3 protein assists the targeting of the TGB2-induced vesicles or the ER-localized TGB2 protein to the cell periphery (Ju et al., 2005, 2007; Lee et al., 2010) and the movement of virus across cells (Verchot-Lubicz et al., 2010; Wu et al., 2011). TGB3 protein associates with TGB2 protein as a complex and this TGB3-based complex containing TGB2 protein (TGB2/TGB3 complex) stably associates with CP-encapsidated vRNA (virion form) but not partially CP-encapsidated vRNA containing TGB1 protein (non-virion form of RNP movement complex; Chou et al., 2013). They also showed that the stable TGB2–TGB3–virion complex associates with TGB1 protein for targeting to the PD and proposed the refined model for potexvirus vRNA trafficking to PD (Chou et al., 2013). These authors also suggested two pathways in the refined model (Figure 5B, PATH 1 and PATH 2) and speculated that TGB2 protein in the membrane-associated TGB2–TGB3–virion complex of BaMV cooperatively interacts with TGB1 protein for efficient PD-localization, while TGB3 protein in the same complex drives the whole movement complex to the cortical ER of cell periphery (Chou et al., 2013).
Interestingly, AltMV TGB3 protein was localized at the chloroplast membrane which may be the main site of AltMV replication and accumulated preferentially in mesophyll cells, whereas PVX TGB3 is localized to the ER and accumulates primarily in the epidermis (Lim et al., 2010a). The AltMV TGB3 interacts with Photosystem II (PSII) oxygen-evolving complex (OEC) protein (PsbO), a nuclear-encoded major component of the chloroplast-localized OEC of PS II, surrounding chloroplast in mesophyll cells. The finding raised the possibility that the interaction possibly draws the chloroplasts together and induces symptom development (Jang et al., 2013). Comparative sequence analyses revealed that AtlMV TGB3 has limited similarity to the TGB3 protein homologs of other potexviruses. Mutational analyses showed that AltMV TGB3 protein contains a novel signal for chloroplast membrane localization. The mutations of the chloroplast-targeting signal in AltMV TGB3 protein resulted in very limited cell-to-cell movement, suggesting that this signal is required for the systemic movement of AltMV (Lim et al., 2010a). Together, these data indicate that the mechanisms for viral movement may differ among potexviruses (Lin et al., 2006).
INTERCELLULAR TRANSPORT OF POTEXVIRUS vRNA THROUGH THE PD
For transport of vRNA to the adjacent cell, most plant viruses must increase the PD SEL and exit through the PD. As mentioned earlier, potexvirus and plant viruses in general move their vRNA through the PD as its RNP movement complex or virion form. Lough et al. (1998, 2000) showed that TGB1 protein is essential for plasmodesmal gating rather than CP which is involved in RNP movement complex.
In a new model for intercellular transport of PVX vRNA at the entrances of PD at the late stage of infection that was recently proposed by Tilsner et al. (2013), vRNA processing and trafficking are highly compartmentalized at PD, i.e., replication occurs at the PD so that vRNA is rapidly moved through PD and to adjacent cells soon after replication. The authors, who termed the model coreplicational insertion, reported that fluorescent protein–TGB1 and –CP fusions (FP–TGB1 and FP–CP) co-localize in the PD channel with the intercellular wall space, but that FP–TGB2 and FP–TGB3 fusion proteins localize in punctate “caps” at the cytoplasmic orifices of the PD; these punctate caps harbor the replicase and vRNA, such that the CP aligns with the vRNA inside the pores, whereas CP without the vRNA localizes inside the PD (Tilsner et al., 2013). The caps at the PD orifices during PVX infection have been reported to be replication sites containing non-encapsidated vRNA whorls like those observed in VRC (Tilsner et al., 2009, 2013; Tilsner and Oparka, 2012). At 2–3 days post infiltration (dpi), TGB2 protein localizes in the ER tubule or ER-derived granular vesicles (Ju et al., 2005). However, at 1–2 dpi, when expression levels are still low, TGB2 protein is concentrated in punctae along the lateral walls where PD are present rather than in the ER (Tilsner et al., 2013). This finding suggests that TGB2 protein first targets PD and localizes at the ER or ER-associated granules. TGB3 protein also localizes at PD caps with TGB2 protein (Schepetilnikov et al., 2005; Tilsner et al., 2013), and TGB2 protein increases PD-located TGB3 protein (Tilsner et al., 2013). These results support the inference that TGB2 and TGB3 proteins act as a complex (Lee et al., 2010). Thus, Tilsner et al. (2013) have provided convincing evidence that although the three TGB proteins are localized to PD during PVX infection, TGB1 protein is localized inside the pores together with CP while TGB2/3 complexes are localized in caps at the PD orifices (Figure 3). In contrast to earlier models, the new model indicates that virus replication and movement are not spatially separated within the cell. However, some questions concerning the interactions between TBG proteins have yet to be experimentally confirmed, i.e., how three TGB proteins cooperate to facilitate vRNA transport (Tilsner et al., 2013) and whether other factors including host protein(s) are required for these interactions and for vRNA transport.
With respect to vRNA transport through the PD at the late stage of infection, the new model indicates that the TGB1 protein at the PD caps first functions as a RNA helicase and diverts progeny vRNA away from replication/translation and toward the assembly of the RNP movement complex (Tilsner et al., 2012, 2013; Figure 3). Next, in the presence of locally translated CP, the nascent progeny RNA is encapsidated in cis, and TGB1 protein binds specifically to the 5′ terminal CP subunits of partially or fully encapsidated virions (Karpova et al., 2006; Zayakina et al., 2008). Finally, TGB1 protein directs the CP-encapsidated nascent virions as a RNP movement complex into the gated PD pore (Figure 3).
HOST CELLULAR FACTORS INVOLVED IN CELL-TO-CELL MOVEMENT OF POTEXVIRUS
The viral RNP movement complex interacts with host cellular factors at the orifice of the PD, which allows cell-to-cell movement (Morozov and Solovyev, 2003; Verchot-Lubicz, 2005; Lucas, 2006). However, the interactions between the viral RNP movement complex and host cellular factor(s) remain unclear.
In addition to documenting that TGB proteins and CP have crucial roles in potexvirus cell-to-cell movement, recent research has identified and characterized cellular host proteins that might be involved in intercellular movement of potexvirus. The casein kinase 2 family (CK2) phosphorylates PVX TGB1 protein as well as the Tomato mosaic virus (ToMV) P30 MP (Matsushita et al., 2003; Modena et al., 2008). Modena et al. (2008) showed that recombinant and native PVX TGB1 protein is phosphorylated by N. tabacum extracts from PVX-infected leaves and that PVX TGB1 protein is efficiently phosphorylated by the recombinant tobacco CK2 subunit. Using a phosphopeptide mass mapping approach, they also found that TGB1 protein is phosphorylated at Ser-165 within a CK2 consensus sequence (Modena et al., 2008). Few studies concerning the phosphorylation of plant viral MPs have been reported. Most studies have been performed with the tobamoviruses Tobacco mosaic virus (TMV) and ToMV (Citovsky et al., 1993; Kawakami et al., 1999; Waigmann et al., 2000). TMV MP is phosphorylated by a kinase in cell wall-enriched extracts (Waigmann et al., 2000), and the kinase is associated with PD and belongs to the CK1 family (CK1; Lee et al., 2005). Experiments with phosphorylation mutants have shown that phosphorylation of TMV MP negatively regulates viral transport through PD in a host-dependent manner (Waigmann et al., 2000). A recent study showed that tobacco Serine/threonine kinase-like protein NbSTKL, which is a membrane-associated protein, is involved in the cell-to-cell movement of the potexvirus BaMV (Cheng et al., 2013); that finding suggests the possibility that membrane-associated host proteins interact with the TGB1-containing RNP movement complex and aid cell-to-cell trafficking of potexviruses. However, how host kinase proteins directly or indirectly affect the assembly of the RNP movement complex and/or PD gating has not been studied.
A yeast two-hybrid screening assay has also identified three host proteins (TIP1, TIP2, and TIP3) that interact with PVX TGB2 protein (Fridborg et al., 2003). These proteins interact with β-1,3-glucanase to regulate the degradation of PD-accumulated callose and to increase the PD SEL (Fridborg et al., 2003). PVX TGB1 protein interacts with remorin (REM), a protein located in the cytosolic leaflet of plasma membrane microdomains (lipid rafts) and in clusters at PD. REM can be phosphorylated in the presence of oligogalacturonides and can serve as a counteracting membrane platform for docking of the viral RNP movement complex (Raffaele et al., 2009). In addition, when REM was overexpressed in plants, it inhibited the movement of PVX through PD. This finding suggests that membrane rafts might have a critical role in virus intercellular trafficking.
A recent study suggested that several host cellular heat shock proteins (Hsps), which function as chaperones and folding enzymes, are also related to potexvirus movement (Verchot, 2012). Hsps are conserved molecular polypeptide chaperones that primarily ensure protein quality. Potexviruses rely on chaperone systems in the ER to support cell-to-cell movement. TGB3 protein induces the expression of ER-resident chaperones via the bZIP60 transcription factor (Verchot, 2012). The CP of another potexvirus, Pepino mosaic virus, interacts with Hsp cognate 70 (Hsc70), which has APTase activity, and it seems to take the translocation ability of CP-Hsc70 interaction to facilitate viral cell-to-cell movement through the PD (Mathioudakis et al., 2012). In addition, another host cellular protein, named NbPCIP1, was characterized from a Nicotiana benthamiana cDNA library as a PVX CP-interacting protein based on the yeast two-hybrid system. NbPCIP1 enhanced PVX replication and movement in N. benthamiana (Park et al., 2009).
Overall, it is quite clear that the identified host cellular proteins play important roles in potexvirus replication and cell-to-cell movement. However, many of them still remain unclear and need to be elucidated in regard to identifying and functionally characterizing host cellular proteins associated with the infection cycles of potexviruses.
HOST CELLULAR FACTORS INTERACTING WITH VIRAL RNA ELEMENTS IN POTEXVIRUS MOVEMENT
The (+) SL1 RNA or (-) SL1 RNA of PVX behaves as a cis-acting element and can affect cell-to-cell movement of PVX (Lough et al., 2006). As noted earlier, many host proteins that interact with viral proteins and that might affect intra-/inter-cellular trafficking of potexviruses have been identified. Little is known, however, about how host proteins interact with vRNAs because functional studies of plant host proteins usually concern interactions between viral protein and host protein rather than between vRNA and host protein. In this regard, PVX SL1 RNAs can efficiently bind to proteins extracted from tobacco protoplasts, as confirmed by a systematic evolution of ligands by exponential enrichment in vitro (Kwon and Kim, 2006). Northwestern blot analysis and matrix-assisted laser desorption ionization time-of-flight mass spectrometry have been recently used to identify tobacco proteins that bind to PVX SL1 RNAs (Cho et al., 2012a).
The host protein NbDnaJ (or Hsp40) binds to SL1 (-) RNA of PVX and also interacts with PVX CP. As indicated by deletion assay, the C-terminal region of NbDnaJ is essential for the interaction with PVX CP (Cho et al., 2012c). According to the latter study, when NbDnaJ is overexpressed in transient assay, PVX movement is suppressed. This study showed that NbDnaJ plays a role in the early stages of viral infection by suppressing PVX replication and movement. A previous study had identified another host protein, NbMPB2Cb, that binds with both SL1 (-) and SL1 (+) RNAs of PVX (Cho et al., 2012b). NbMPB2Cb localizes at microtubules in the ER and suppresses PVX movement (Cho et al., 2012b). Although it is not immediately relevant to the intra-/intercellular movement of potexviruses, SL1 (+) RNA binds to the N. tabacum WRKY1 transcription factor (NtWRKY1; Kim et al., 2002; Park and Kim, 2012). By increasing PVX accumulation, silencing of NtWRKY1 in N. benthamiana caused plants to exhibit lethal apical necrosis, suggesting that NtWRKY1 regulates multiple defense response genes (Park and Kim, 2012). Together, these results suggest that a plant vRNA element greatly affect viral movement and replication, possibly by interacting with several host factors. The latter study might be the first to report that the interaction between a plant vRNA element and a host cellular protein is required for viral movement.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Substantial information has been obtained concerning the cellular mechanisms underlying intra- and intercellular movement of plant RNA viruses. Here, we have summarized the intracellular and intercellular movement of vRNA in potexviruses based on recent studies. As indicated, intracellular and intercellular vRNA movement is mediated by viral MPs and host proteins, indicating that the plasmodesmal transport of vRNA is a highly regulated process. Here, we have tried to summarize the cell-to-cell movement of potexvirus vRNA with intracellular trafficking and intercellular transport of vRNA in plant cells. Although we have provided two general models that describe how potexvirus vRNA is trafficked to PD (Figure 5; Verchot-Lubicz et al., 2010; Chou et al., 2013), TGB protein interactions and many other aspects of intra- and intercellular potexvirus movement remain unclear. To further investigate the interactions between TGBs in relation to potexvirus movement, additional experimental evidence clarifying whether each TGB directly binds to the other TGBs or requires other factors at each step during the movement process(es) is necessary. As for the possible involvement of the SL1 of PVX and other cis-acting elements for potexvirus movement, we should determine whether viral and host cellular proteins compete for binding to the viral element and how viral elements regulate the viral movement.
The viral RNP movement complex interacts with host cellular factors at the orifice of the PD, which enables cell-to-cell movement (Morozov and Solovyev, 2003; Verchot-Lubicz, 2005; Lucas, 2006). Although it is well-known that host proteins interacting with PVX TGB2 protein (TIP1, TIP2, and TIP3) interact with β-1,3-glucanase to regulate the degradation of PD callose and to increase the PD SEL (Fridborg et al., 2003), the interactions between the viral RNP movement complex and host cellular factor(s) remain unclear. Little is also known about how potexviruses interact with host cellular proteins to mediate virus movement. Identifying new host cellular proteins required for cell-to-cell movement should be facilitated by the use of microarrays and/or RNA-Seq with high-throughput gene silencing and other new methods.
It also appears that potexviruses do not always have the same movement mechanisms even if all have TGB proteins (Lim et al., 2010a). Thus, research is needed on different viruses in the same genus. Research is also needed regarding the functions of PD that mediate vRNA movement and how PD-associated proteins regulate vRNA movement through PD. In addition to increasing our understanding of the virus infection cycle, understanding the details of virus movement is valuable because it provides insight into fundamental aspects of the cellular machinery. We suspect that potexviruses will continue to be useful models for studying intra- and inter-cellular vRNA movement in plants.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported in part by grants from the National Research Foundation (grant no. 20110012328) funded by the Ministry of Education, Science, and Technology (MEST); the Vegetable Breeding Research Center (no. 710001-03) through the Agriculture Research Center program from the Ministry for Food, Agriculture, Forestry and Fisheries; and the Next-Generation BioGreen 21 Program of Rural Development Administration (no. PJ00819801).
REFERENCES
- Adams M. J., Antoniw J. F., Bar-Joseph M., Brunt A. A., Candresse T., Foster G. D., et al. (2004). The new plant virus family Flexiviridae and assessment of molecular criteria for species demarcation. Arch. Virol. 149 1045–1060 10.1007/s00705-004-0304-0 [DOI] [PubMed] [Google Scholar]
- Angell S. M., Davies C., Baulcombe D. C. (1996). Cell-to-cell movement of potato virus X is associated with a change in the size-exclusion limit of plasmodesmata in trichome cells of Nicotiana clevelandii. Virology 216 197–201 10.1006/viro.1996.0046 [DOI] [PubMed] [Google Scholar]
- Atabekov J. G., Rodionova N. P., Karpova O. V., Kozlovsky S. V., Poljakov V. Y. (2000). The movement protein-triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology 271 259–263 10.1006/viro.2000.0319 [DOI] [PubMed] [Google Scholar]
- Bamunusinghe D., Hemenway C. L., Nelson R. S., Sanderfoot A. A., Ye C. M., Silva M. A., et al. (2009). Analysis of Potato virus X replicase and TGBp3 subcellular locations. Virology 393 272–285 10.1016/j.virol.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Baulcombe D. C., Chapman S, Santa Crus S. (1995). Jellyfish green fluorescent protein as a as a reporter for virus infections. Plant J. 7 1045–1053 10.1046/j.1365-313X.1995.07061045.x [DOI] [PubMed] [Google Scholar]
- Bayne E. H., Rakitina D. V., Morozov S. Y., Baulcombe D. C. (2005). Cell-to-cell movement of potato virus X is dependent on suppression of RNA silencing. Plant J. 44 471–482 10.1111/j.1365-313X.2005.02539.x [DOI] [PubMed] [Google Scholar]
- Chapman S., Hills G., Watts J. E., Baulcombe D. C. (1992). Mutational analysis of the coat protein gene of potato virus X: effects of virion morphology and viral pathogenicity. Virology 191 223–230 10.1016/0042-6822(92)90183-P [DOI] [PubMed] [Google Scholar]
- Cheng S. F., Tsai M. S., Huang C. L., Huang Y. P., Chen I. H., Lin N. S., et al. (2013). Ser/Thr kinase-like protein of Nicotiana benthamiana is involved in the cell-to-cell movement of Bamboo mosaic virus. PLoS ONE 8:e62907 10.1371/journal.pone.0062907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.-Y., Cho W. K., Kim K.-H. (2012a). Identification of tobacco proteins associated with the stem-loop 1 RNAs of Potato virus X. Mol. Cells 33 379–384 10.1007/s10059-012-2298-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.-Y., Cho W. K., Choi H. S., Kim K.-H. (2012b). Cis-acting element (SL1) of Potato virus X controls viral movement by interacting with the NbMPB2Cb and viral proteins. Virology 427 166–176 10.1016/j.virol.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Cho S.-Y., Cho W. K., Sohn S. H., Kim K.-H. (2012c). Interaction of the host protein NbDnaJ with Potato virus X minus-strand stem-loop 1 RNA and capsid protein affects viral replication and movement. Biochem. Biophys. Res. Commun. 417 451–456 10.1016/j.bbrc.2011.11.137 [DOI] [PubMed] [Google Scholar]
- Chou Y. L., Hung Y. J., Tseng Y. H., Hsu H. T., Yang J. Y., Wung C. H., et al. (2013). The stable association of virion with the triple-gene-block protein 3-based complex of Bamboo mosaic virus. PLoS Pathog. 9:e1003405 10.1371/journal.ppat.1003405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., McLean B. G., Zupan J. R., Zambryski P. (1993). Phosphorylation of tobacco mosaic virus cell-to-cell movement protein by a developmentally regulated plant cell wall-associated protein kinase. Genes Dev. 7 904–910 10.1101/gad.7.5.904 [DOI] [PubMed] [Google Scholar]
- Cowan G. H., Lioliopoulou F., Ziegler A., Torrance L. (2002). Subcellular localisation, protein interactions, and RNA binding of Potato mop-top virus triple gene block proteins. Virology 298 106–115 10.1006/viro.2002.1435 [DOI] [PubMed] [Google Scholar]
- Doronin S. V., Hemenway C. (1996). Synthesis of potato virus X RNAs by membrane-containing extracts. J. Virol. 70 4795–4799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster R. L. S., Beck D. L., Guilford P. J., Voot D. M., van Dolleweerd C. J., Andersen M. T. (1992). The coat protein of white clover mosaic potexvirus has a role in facilitating cell-to-cell transport in plants. Virology 191 480–484 10.1016/0042-6822(92)90215-B [DOI] [PubMed] [Google Scholar]
- Fridborg I., Grainger J., Page A., Coleman M., Findlay K., Angell S. (2003). TIP, a novel host factor linking callose degradation with the cell-to-cell movement of Potato virus X. Mol. Plant Microbe Interact. 16 132–140 10.1094/MPMI.2003.16.2.132 [DOI] [PubMed] [Google Scholar]
- Gorbalenya A. E., Koonin E. V. (1993). Helicases: amino acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3 419–429 10.1016/S0959-440X(05)80116-2 [DOI] [Google Scholar]
- Han Y. T., Tsai C. S., Chen Y. C., Lin M. K., Hsu Y. H., Meng M. (2007). Mutational analysis of a helicase motif-based RNA 5′-triphosphatase/NTPase from Bamboo mosaic virus. Virology 367 41–50 10.1016/j.virol.2007.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt S., Cowan G. H., Ziegler A., Roberts A. G., Oparka K. J., Torrance L. (2005). Two plant-viral movement proteins traffic in the endocytic recycling pathway. Plant Cell 17 164–181 10.1105/tpc.104.027821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A. R., Heppler M. L., Ju H. J., Krishnamurthy K., Payton M. E., Verchot-Lubicz J. (2004). Potato virus X TGBp1 induces plasmodesmata gating and moves between cells in several host species whereas CP moves only in N. benthamianaleaves. Virology 328 185–197 10.1016/j.virol.2004.06.039 [DOI] [PubMed] [Google Scholar]
- Hsu H. T., Chou Y. L., Tseng Y. H., Lin Y. H., Lin T. M., Lin N. S., et al. (2008). Topological properties of the triple gene block protein 2 of Bamboo mosaic virus. Virology 379 1–9 10.1016/j.virol.2008.06.019 [DOI] [PubMed] [Google Scholar]
- Hsu H. T., Tseng Y. H., Chou Y. L., Su S. H., Hsu Y. H., Chang B. Y. (2009). Characterization of the RNA-binding properties of the triple-gene-block protein 2 of Bamboo mosaic virus. Virol. J. 6 50 10.1186/1743-422X-6-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. O., Lim H. S., Bragg J., Ganesan U., Lee M. Y. (2009). Hordeivirus replication, movement, and pathogenesis. Annu. Rev. Phytopathol. 47 385–422 10.1146/annurev-phyto-080508-081733 [DOI] [PubMed] [Google Scholar]
- Jang C., Seo E. Y., Nam J., Bae H., Gim Y. G., Kim H. G., et al. (2013). Insights into Alternanthera mosaic virus TGB3 functions: interactions with Nicotiana benthamiana PsbO correlate with chloroplast vesiculation and veinal necrosis caused by TGB3 over-expression. Front. Plant Sci. 4:5 10.3389/fpls.2013.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. A., Bragg J. N., Lawrence D. M., Jackson A. O. (2003). Sequence elements controlling expression of Barley stripe mosaic virus subgenomic RNAs in vivo. Virology 313 66–80 10.1016/S0042-6822(03)00285-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H. J., Brown J. E., Ye C. M., Verchot-Lubicz J. (2007). Mutations in the central domain of Potato virus X TGBp2 eliminate granular vesicles and virus cell-to-cell trafficking. J. Virol. 81 1899–1911 10.1128/JVI.02009-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju H. J., Samuels T. D., Wang Y. S., Blancaflor E., Payton M., Mitra R., et al. (2005). The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol. 138 1877–1895 10.1104/pp.105.066019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadare G., Haenni A. L. (1997). Virus-encoded RNA helicases. J. Virol. 71 2583–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina N. O., Rakitina D. V., Solovyev A. G., Schiemann J., Morozov S. Y. (2002). RNA helicase activity of the plant virus movement proteins encoded by the first gene of the triple gene block. Virology 296 321–329 10.1006/viro.2001.1328 [DOI] [PubMed] [Google Scholar]
- Karpova O. V., Zayakina O. V., Arkhipenko M. V., Sheval E. V., Kiselyova O. I., Poljakov V. Y., et al. (2006). Potato virus X RNA-mediated assembly of single-tailed ternary ‘coat protein-RNA-movement protein’ complexes. J. Gen. Virol. 87 2731–2740 10.1099/vir.0.81993-0 [DOI] [PubMed] [Google Scholar]
- Kawakami S., Padgett H. S., Hosokawa D., Okada Y., Beachy R. N., Watanabe Y. (1999). Phosphorylation and/or presence of serine 37 in the movement protein of tomato mosaic tobamovirus is essential for intracellular localization and stability in vivo. J. Virol. 73 6831–6840 10.1101/gad.7.5.904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.-H., Kwon S.-J., Hemenway C. (2002). Cellular protein binds to sequences near the 5′ terminus of Potato virus X RNA that are important for virus replication. Virology 301 305–312 10.1006/viro.2002.1559 [DOI] [PubMed] [Google Scholar]
- King A. M. Q., Lefkowitz E., Adams M. J., Carstens E. B. (2009). Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. London: Academic Press [Google Scholar]
- Krishnamurthy K., Heppler M., Mitra R., Blancaflor E., Payton M., Nelson R. S., et al. (2003). The Potato virus X TGBp3 protein associates with the ER network for virus cell-to-cell movement. Virology 309 135–151 10.1016/S0042-6822(02)00102-2 [DOI] [PubMed] [Google Scholar]
- Kwon S.-J., Kim K.-H. (2006). The SL1 stem-loop structure at the 5′-end of Potato virus X RNA is required for efficient binding to host proteins and for viral infectivity. Mol. Cells 21 63–75 [PubMed] [Google Scholar]
- Lee C. C., Ho Y. N., Hu R. H., Yen Y. T., Wang Z. C., Lee Y. C., et al. (2011). The interaction between Bamboo mosaic virus replication protein and coat protein is critical for virus movement in plant hosts. J. Virol. 85 12022–12031 10.1128/JVI.05595-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. Y., Taoka K., Yoo B. C., Ben-Nissan G., Kim D. J., Lucas W. J. (2005). Plasmodesmal-associated protein kinase in tobacco and Arabidopsis recognizes a subset of non-cell-autonomous proteins. Plant Cell 17 2817–2831 10.1105/tpc.105.034330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. C., Wu C. H., Wang C. W. (2010). Traffic of a viral movement protein complex to the highly curved tubules of the cortical endoplasmic reticulum. Traffic 11 912–930 10.1111/j.1600-0854.2010.01064.x [DOI] [PubMed] [Google Scholar]
- Leshchiner A. D., Solovyev A. G., Morozov S. Y., Kalinina N. O. (2006). A minimal region in the NTPase/helicase domain of the TGBp1 plant virus movement protein is responsible for ATPase activity and cooperative RNA binding. J. Gen. Virol. 87 3087–3095 10.1099/vir.0.81971-0 [DOI] [PubMed] [Google Scholar]
- Lim H. S., Vaira A. M., Bae H., Bragg J. N., Ruzin S. E., Bauchan G. R., et al. (2010a). Mutation of a chloroplast-targeting signal in Alternanthera mosaic virus TGB3 impairs cell-to-cell movement and eliminates long-distance virus movement. J. Gen. Virol. 91 2102–2115 10.1099/vir.0.019448-0 [DOI] [PubMed] [Google Scholar]
- Lim H. S., Vaira A. M., Domier L. L., Lee S. C., Kim H. G., Hammond J. (2010b). Efficiency of VIGS and gene expression in a novel bipartite potexvirus vector delivery system as a function of strength of TGB1 silencing suppression. Virology 402 149–163 10.1016/j.virol.2010.03.022 [DOI] [PubMed] [Google Scholar]
- Lim H. S., Vaira A. M., Reinsel M. D., Bae H., Bailey B. A., Domier L. L., et al. (2010c). Pathogenicity of Alternanthera mosaic virus is affected by determinants in RNA-dependent RNA polymerase and by reduced efficacy of silencing suppression in a movement-competent TGB1. J. Gen. Virol. 91 277–287 10.1099/vir.0.014977-0 [DOI] [PubMed] [Google Scholar]
- Lin M. K., Chang B. Y., Liao J. T., Lin N. S., Hsu Y. H. (2004). Arg-16 and Arg-21 in the N-terminal region of the triple-gene-block protein 1 of Bamboo mosaic virus are essential for virus movement. J. Gen. Virol. 85 251–259 10.1099/vir.0.19442-0 [DOI] [PubMed] [Google Scholar]
- Lin M. K., Hu C. C., Lin N. S., Chang B. Y., Hsu Y. H. (2006). Movement of potexviruses requires species-specific interactions among the cognate triple gene block proteins, as revealed by a trans-complementation assay based on the Bamboo mosaic virus satellite RNA-mediated expression system. J. Gen. Virol. 87 1357–1367 10.1099/vir.0.81625-0 [DOI] [PubMed] [Google Scholar]
- Liou D. Y., Hsu Y. H., Wung C. H., Wang W. H., Lin N. S., Chang B. Y. (2000). Functional analyses and identification of two arginine residues essential to the ATP-utilizing activity of the triple gene block protein 1 of bamboo mosaic potexvirus. Virology 277 336–344 10.1006/viro.2000.0610 [DOI] [PubMed] [Google Scholar]
- Lough T. J., Lee R. H., Emerson S. J., Forster R. L., Lucas W. J. (2006). Functional analysis of the 5′ untranslated region of potexvirus RNA reveals a role in viral replication and cell-to-cell movement. Virology 351 455–465 10.1016/j.virol.2006.03.043 [DOI] [PubMed] [Google Scholar]
- Lough T. J., Netzler N. E., Emerson S. J., Sutherland P., Carr F., Beck D. L., et al. (2000). Cell-to-cell movement of potexviruses: evidence for a ribonucleoprotein complex involving the coat protein and first triple gene block protein. Mol. Plant Microbe Interact. 13 962–974 10.1094/MPMI.2000.13.9.962 [DOI] [PubMed] [Google Scholar]
- Lough T. J., Shash K., Xoconostle-Cázares B., Hofstra K. R., Beck D. L., Balmori E., et al. (1998). Molecular dissection of the mechanism by which potexvirus triple gene block proteins mediate cell-to-cell transport of infectious RNA. Mol. Plant Microbe Interact. 11 801–814 10.1094/MPMI.1998.11.8.801 [DOI] [Google Scholar]
- Lucas W. J. (2006). Plant viral movement proteins: agents for cell-to-cell trafficking of viral genomes. Virology 344 169–184 10.1016/j.virol.2005.09.026 [DOI] [PubMed] [Google Scholar]
- Mackenzie J. (2005). Wrapping things up about virus RNA replication. Traffic 6 967–977 10.1111/j.1600-0854.2005.00339.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martelli G. P., Adams M. J., Kreuze J. F., Dolja V. V. (2007). Family Flexiviridae: a case study in virion and genome plasticity. Annu. Rev. Phytopathol. 45 73–100 10.1146/annurev.phyto.45.062806.094401 [DOI] [PubMed] [Google Scholar]
- Mathioudakis M. M., Veiga R., Ghita M., Tsikou D., Medina V., Canto T., et al. (2012). Pepino mosaic virus capsid protein interacts with a tomato heat shock protein cognate 70. Virus Res. 163 28–39 10.1016/j.virusres.2011.08.007 [DOI] [PubMed] [Google Scholar]
- Matsushita Y., Ohshima M., Yoshioka K., Nishiguchi M., Nyunoya H. (2003). The catalytic subunit of protein kinase CK2 phosphorylates in vitro the movement protein of Tomato mosaic virus. J. Gen. Virol. 84 497–505 10.1099/vir.0.18839-0 [DOI] [PubMed] [Google Scholar]
- Mitra R., Krishnamurthy K., Blancaflor E., Payton M., Nelson R. S., Verchot-Lubicz J. (2003). The Potato virus X TGBp2 protein association with the endoplasmic reticulum plays a role in but is not sufficient for viral cell-to-cell movement. Virology 312 35–48 10.1016/S0042-6822(03)00180-6 [DOI] [PubMed] [Google Scholar]
- Modena N. A., Zelada A. M., Conte F., Mentaberry A. (2008). Phosphorylation of the TGBp1 movement protein of Potato virus X by a Nicotiana tabacum CK2-like activity. Virus Res. 137 16–23 10.1016/j.virusres.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Morozov S. Y., Solovyev A. G. (2003). Triple gene block: modular design of a multifunctional machine for plant virus movement. J. Gen. Virol. 84 1351–1366 10.1099/vir.0.18922-0 [DOI] [PubMed] [Google Scholar]
- Nagy P. D., Pogany J. (2010). Global genomics and proteomics approaches to identify host factors as targets to induce resistance against Tomato bushy stunt virus. Adv. Virus Res. 76 123–177 10.1016/S0065-3527(10)76004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehl A., Heinlein M. (2011). Cellular pathways for viral transport through plasmodesmata. Protoplasma 248 75–99 10.1007/s00709-010-0246-1 [DOI] [PubMed] [Google Scholar]
- Ozeki J., Hashimoto M., Komatsu K., Maejima K., Himeno M., Senshu H., et al. (2009). The N-terminal region of the Plantago asiatica mosaic virus coat protein is required for cell-to-cell movement but is dispensable for virion assembly. Mol. Plant Microbe Interact. 22 677–685 10.1094/MPMI-22-6-0677 [DOI] [PubMed] [Google Scholar]
- Park M.-R., Park S.-H., Cho S.-Y., Kim K.-H. (2009). Nicotiana benthamiana protein, NbPCIP1, interacting with Potato virus X coat protein plays a role as susceptible factor for viral infection. Virology 386 257–269 10.1016/j.virol.2008.12.044 [DOI] [PubMed] [Google Scholar]
- Park M.-R., Seo J.-K., Kim K.-H. (2013). Viral and nonviral elements in potexvirus replication and movement and in antiviral responses. Adv. Virus Res. 87 75–112 10.1016/B978-0-12-407698-3.00003-X [DOI] [PubMed] [Google Scholar]
- Park S.-H., Kim K.-H. (2012). Virus-induced silencing of the WRKY1 transcription factor that interacts with the SL1 structure of Potato virus X leads to higher viral RNA accumulation and severe necrotic symptoms. Plant Pathol. J. 28 40–48 10.5423/PPJ.OA.11.2011.0226 [DOI] [Google Scholar]
- Raffaele S., Bayer E., Lafarge D., Cluzet S., German Retana S., Boubekeur T., et al. (2009). Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs Potato virus X movement. Plant Cell 21 1541–1555 10.1105/tpc.108.064279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionova N. P., Karpova O. V., Kozlovsky S. V., Zayakina O. V., Arkhipenko M. V., Atabekov J. G. (2003). Linear remodeling of helical virus by movement protein binding. J. Mol. Biol. 333 565–572 10.1016/j.jmb.2003.08.058 [DOI] [PubMed] [Google Scholar]
- Samuels T. D., Ju H. J., Ye C. M., Motes C. M., Blancaflor E. B., Verchot-Lubicz J. (2007). Subcellular targeting and interactions among the Potato virus X TGB proteins. Virology 367 375–389 10.1016/j.virol.2007.05.022 [DOI] [PubMed] [Google Scholar]
- Schepetilnikov M. V., Manske U., Solovyev A. G., Zamyatnin A. A., Jr., Schiemann J., Morozov S., Yu., et al. (2005). The hydrophobic segment of Potato virus X TGBp3 is a major determinant of the protein intracellular trafficking. J. Gen. Virol. 86 2379–2391 10.1099/vir.0.80865-0 [DOI] [PubMed] [Google Scholar]
- Schoelz J. E., Harries P. A., Nelson R. S. (2011). Intracellular transport of plant viruses: finding the door out of the cell. Mol. Plant 4 813–831 10.1093/mp/ssr070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senshu H., Ozeki J., Komatsu K., Hashimoto M., Hatada K., Aoyama M., et al. (2009). Variability in the level of RNA silencing suppression caused by triple gene block protein 1 (TGBp1) from various potexviruses during infection. J. Gen. Virol. 90 1014–1024 10.1099/vir.0.008243-0 [DOI] [PubMed] [Google Scholar]
- Solovyev A. G., Kalinina N. O., Morozov S. Y. (2012). Recent advances in research of plant virus movement mediated by triple gene block. Front. Plant Sci. 3:276 10.3389/fpls.2012.00276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai A., Meshi T. (2001). Cell-to-cell movement of Potato virus X: the role of p12 and p8 encoded by the second and third open reading frames of the triple gene block. Mol. Plant Microbe Interact. 14 1158–1167 10.1094/MPMI.2001.14.10.1158 [DOI] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Christensen N. M., Bell K., Roberts I. M., Lacomme C., et al. (2009). Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J. 57 758–770 10.1111/j.1365-313X.2008.03720.x [DOI] [PubMed] [Google Scholar]
- Tilsner J., Oparka K. J. (2012). Missing links? – The connection between replication and movement of plant RNA viruses. Curr. Opin. Virol. 2 705–711 10.1016/j.coviro.2012.09.007 [DOI] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Louveaux M., Roberts I. M., Chapman S. N., Oparka K. J. (2013). Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J. Cell Biol. 201 981–995 10.1083/jcb.201304003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilsner J., Linnik O., Wright K. M., Bell K., Roberts A. G., Lacomme C., et al. (2012). The TGB1 movement protein of Potato virus X reorganizes actin and endomembranes into the X-body, a viral replication factory. Plant Physiol. 158 1359–1370 10.1104/pp.111.189605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y. H., Hsu H. T., Chou Y. L., Hu C. C., Lin N. S., Hsu Y. H., et al. (2009). The two conserved cysteine residues of the triple gene block protein 2 are critical for both cell-to-cell and systemic movement of Bamboo mosaic virus. Mol. Plant Microbe Interact. 22 1379–1388 10.1094/MPMI-22-11-1379 [DOI] [PubMed] [Google Scholar]
- Verchot J. (2012). Cellular chaperones and folding enzymes are vital contributors to membrane bound replication and movement complexes during plant RNA virus infection. Front. Plant Sci. 3:275 10.3389/fpls.2012.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot J., Angell S. M., Baulcombe D. C. (1998). In vivo translation of the triple gene block of potato virus X requires two subgenomic mRNAs. J. Virol. 72 8316–8320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verchot-Lubicz J. (2005). A new cell-to-cell transport model for potexviruses. Mol. Plant Microbe Interact. 18 283–290 10.1094/MPMI-18-0283 [DOI] [PubMed] [Google Scholar]
- Verchot-Lubicz J., Torrance L., Solovyev A. G., Morozov S. Y., Jackson A. O., Gilmer D. (2010). Varied movement strategies employed by triple gene block-encoding viruses. Mol. Plant Microbe Interact. 23 1231–1247 10.1094/MPMI-04-10-0086 [DOI] [PubMed] [Google Scholar]
- Voinnet O., Lederer C., Baulcombe D. C. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103 157–167 10.1016/S0092-8674(00)00095-7 [DOI] [PubMed] [Google Scholar]
- Waigmann E., Chen M. H., Bachmaier R., Ghoshroy S., Citovsky V. (2000). Regulation of plasmodesmal transport by phosphorylation of tobacco mosaic virus cell-to-cell movement protein. EMBO J. 19 4875–4884 10.1093/emboj/19.18.4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. H., Lee S. C., Wang C. W. (2011). Viral protein targeting to the cortical endoplasmic reticulum is required for cell-cell spreading in plants. J. Cell Biol. 193 521–535 10.1083/jcb.201006023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wung C. H., Hsu Y. H., Liou D. Y., Huang W. C., Lin N. S., Chang B. Y. (1999). Identification of the RNA-binding sites of the triple gene block protein 1 of bamboo mosaic potexvirus. J. Gen. Virol. 80 1119–1126 [DOI] [PubMed] [Google Scholar]
- Yan F., Lu Y., Lin L., Zheng H., Chen J. (2012). The ability of PVX p25 to form RL structures in plant cells is necessary for its function in movement, but not for its suppression of RNA silencing. PLoS ONE 7:e43242 10.1371/journal.pone.0043242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayakina O., Arkhipenko M., Kozlovsky S., Nikitin N., Smirnov A., Susi P., et al. (2008). Mutagenic analysis of Potato virus X movement protein (TGBp1) and the coat protein (CP): in vitro TGBp1-CP binding and viral RNA translation activation. Mol. Plant Pathol. 9 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Jackson A. O. (1996). Expression of the barley stripe mosaic virus RNA beta “triple gene block”. Virology 216 367–379 10.1006/viro.1996.0072 [DOI] [PubMed] [Google Scholar]