Abstract
Objective
In this paper, we focus on three aspects: (1) to annotate a set of standard corpus in Chinese discharge summaries; (2) to perform word segmentation and named entity recognition in the above corpus; (3) to build a joint model that performs word segmentation and named entity recognition.
Design
Two independent systems of word segmentation and named entity recognition were built based on conditional random field models. In the field of natural language processing, while most approaches use a single model to predict outputs, many works have proved that performance of many tasks can be improved by exploiting combined techniques. Therefore, in this paper, we proposed a joint model using dual decomposition to perform both the two tasks in order to exploit correlations between the two tasks. Three sets of features were designed to demonstrate the advantage of the joint model we proposed, compared with independent models, incremental models and a joint model trained on combined labels.
Measurements
Micro-averaged precision (P), recall (R), and F-measure (F) were used to evaluate results.
Results
The gold standard corpus is created using 336 Chinese discharge summaries of 71 355 words. The framework using dual decomposition achieved 0.2% improvement for segmentation and 1% improvement for recognition, compared with each of the two tasks alone.
Conclusions
The joint model is efficient and effective in both segmentation and recognition compared with the two individual tasks. The model achieved encouraging results, demonstrating the feasibility of the two tasks.
Keywords: Dual Decomposition, Segmentation, Named entity , Chinese Discharge Summary, Conditional Random Fields
Introduction
Electronic medical records have experienced a rapid growth in recent years.1 While sophisticated algorithms have been developed for English medical records,2–8 rare algorithms have been introduced to process Chinese medical records. On the other hand, China is the largest country in terms of the total number of population. Therefore, the large amount of information contained in Chinese medical texts, if processed properly, will certainly benefit the field of biomedical informatics.
In this paper, we mainly focus on two important tasks for Chinese medical text processing: word segmentation9 10 and named entity recognition.10 11 Due to the nature of the Chinese language, there is no space separating two different words. As a result, the first step for a Chinese language processor would be to separate Chinese characters into semantic words. For instance, the sentence ‘患者夜间无关节疼痛 (the patient does not have joint pain at night)’ should be segmented as ‘患者(patient)/夜间 (at night)/无 (no)/关节 (joint) 疼痛 (pain)’ (see figure 1). The second task (named entity recognition) involves detecting named entities from discharge summaries. In this paper, we consider the following four categories of named entities: (1) problems and symptoms like ‘咳嗽 (cough)’ and ‘肺炎 (pneumonia)’; (2) medical tests and assays like ‘血压 (blood pressure)’ and ‘CT’; (3) medications like ‘阿司匹林 (aspirin)’; and (4) treatments like ‘化疗 (chemotherapy)’. In the example sentence above, the phrase ‘关节疼痛 (joint pain)’ should be extracted as an entity of type ‘problem’ (figure 1).
Figure 1.

A sample for the two tasks of segmentation and recognition; white space: segmentation tag; blue color: name entity.
Noticeably, there exists a strong correlation between the two tasks. As words represent a group of characters that convey a certain semantic meaning, no named entities should be allowed to contain a fraction of a word, otherwise such a named entity will not have a clear meaning. On the other hand, a named entity is allowed to contain an arbitrary number of words (except for named entities of type ‘medication’, which will be explained in the Methods section). For instance, the named entity ‘失血性休克 (hemorrhagic shock)’ can be separated into two words: ‘失血性 (hemorrhagic)’ and ‘休克 (shock)’.
Out of vocabulary is another big challenge for word segmentation. In the medical domain, there is an enormous number of medical terms that cannot be found in current Chinese word dictionaries, especially medication names like ‘阿乐欣 (azlocillin sodium)’. According to the statistics of our data corpus, out-of-vocabulary words account for nearly 80% in all medication names. This large amount of out-of-vocabulary words make the word segmentation task especially more difficult, compared to word segmentation in other domains. For example, the standard segmentation for ‘肝内胆管多发结石 (many stones in the intrahepatic bile duct)’ is ‘肝内胆管/多发/结石 (in the intrahepatic bile duct|many|stones)’, while the segmentation result from the state-of-art segmentation tool is ‘肝/内胆/管/多发/结石 (in the intrahepatic|bile|duct|many|stones)’.
Resolving ambiguities is also a difficult problem for our tasks. For example, the standard results of segmentation for the phrase ‘无畏寒 (no chills)’ is ‘无|畏寒 (no | chills)’ for segmentation and ‘畏寒 (chills)’ should be identified as a named entity of type ‘problem’. However, the group of characters ‘无畏’ also stands for ‘fearless’ in Chinese. The segmentation system should be able to resolve these types of ambiguity in order to achieve a good performance.
Due to the complex challenges discussed in the previous paragraphs, existing segmentation tools had an undesirable performance on our data corpus. Our experiments showed that one of the state-of-art Chinese word segmentation tools, MSRSeg,9 obtained an F-measure of only 82.58%.
In our work, we first used two independently trained conditional random field (CRF) models to perform the two tasks separately. However, as we have mentioned, this method does not take the inherent correlation between segmentation and named entity recognition into account. To tackle these challenges, we designed joint models for both word segmentation and named entity recognition. We also attempted to exploit the relationship between the two tasks in order to boost the performance of our system. Our new framework uses the dual decomposition algorithm for inference, which solves a linear programing relaxation of the global inference problem. Experiments showed that the new framework achieved a better performance than baseline methods using only independent or incremental models. We also demonstrated that compared to other joint models with similar performance, our joint model using dual decomposition has a much smaller inference complexity and thus results in a much shorter running time.
The data corpus we used is Chinese discharge summaries provided by a Chinese hospital. It includes 336 labeled Chinese discharge summaries. After manual annotation, we labeled a total of 8881 medical problems, 1188 treatments, 782 medications, 1299 tests, and 71 355 words in the data corpus.
Our contributions in this paper are fourfold. First, we created a high-quality corpus with named entity and segmentation annotation in Chinese discharge summaries. Second, two independent models trained on combined labels were introduced to perform the segmentation and entity recognition tasks. Third, we furthermore proposed a new method using dual decomposition to improve the performance of the two tasks based on two independently trained CRF models. Finally, to the best of our knowledge, our work is the first to focus on Chinese discharge summaries.
Related work
Word segmentation12 and named entity recognition13 14 are traditional topics in natural language processing. For word segmentation, there are three major difficulties in the Chinese language: the construction of language resources, segmentation ambiguity, and out of vocabulary. For named entity recognition, most methods used the information of word segmentation and part-of-speech (POS) tags as features of CRF models. Almost all of these methods solved the two tasks separately or used a pipeline process. In addition, to the best of our knowledge, there is no related work for segmentation and named entity recognition in Chinese discharge summaries.
In natural language processing, some tasks are correlated such as segmentation and named entity recognition, POS tagging and dependency parsing, etc. Kruengkrai et al15 proposed an error-driven word-character hybrid joint model for Chinese word segmentation and POS tagging. The method achieved a superior performance. Hatori et al16 proposed a joint model for POS tagging and dependency parsing in Chinese. Their performance is considered as a new state-of-the-art performance on this joint task. Srikumar and Roth17 proposed a joint model that captured the interdependencies between verb semantic role labeling and relations expressed using prepositions, for extended semantic role labeling. In particular, some joint models were related to the dual decomposition algorithm, which was introduced by Dantzig and Wolfe in 1960.18 Dual decomposition was applied in natural language processing by Rush et al19 and Koo et al20 in 2010. They mainly applied the method for two types of problems: combining results from two lexicalized parsing models, and combination of results of a lexicalized parsing model and a trigram POS tagger. The dual decomposition approach has a major contribution by a 0.5% improvement. They also applied the decoding algorithm to phrase-based translation models.21 22 In addition, McClosky et al2,3 applied the dual decomposition algorithm in biomedical event extraction. Three models of joint trigger and argument extraction, capturing correlations between events, and consistency between arguments of the same event were presented. The dual decomposition was used as inference. Their group achieved the first place on the BioNLP 2009 shared task, the BioNLP 2011 Genia task and the BioNLP 2011 infectious diseases task.23–26 Luo et al27 proposed a dual decomposition method for Chinese predicate-argument structure analysis. Compared with state-of-the-art methods, the F-measure of the proposed method (85.97%) is better than that of state-of-the-art methods (85.3%). Hanamoto et al28 proposed a dual decomposition method for coordination structure. They combined head-driven phrase structure grammar (HPSG) parsing and coordinate structure analysis with alignment-based local features. The experiment shows that the joint model is better compared with each of the two algorithms alone. In this work, our joint models focus on segmentation and name entity in Chinese discharge summaries. Up to now, we cannot find a similar study with the two tasks in Chinese discharge summaries. Based on this, we proposed a joint model using dual decomposition for the two tasks in the corpus.
Materials and annotation
Dataset
The data, 336 discharge summaries, were collected by random sampling from diverse departments in a hospital in China. We list some statistics, including the average number of characters, sentences, entities, etc, in supplementary material A, tables S1 and S2 (available online only). The annotation guidelines and the annotated corpus are available online at http://research.microsoft.com/en-us/projects/ehuatuo/.
Annotation guidelines
The annotation for named entities is more involved and we list the detailed annotation guideline for named entity annotation in the appendix file (see supplementary material B, available online only).
Our segmentation annotation is similar to that of Gao et al.9 However, in the medical domain, we experienced several issues that were not included in Gao et al.9 Therefore, aside from rules in Gao et al,9 we also employ a general rule in order to guarantee a consistent segmentation labeling: if the English translation of a group of Chinese characters is only one word, then the corresponding group of characters is labeled as a single word. For instance, ‘癌转移 (cancerometastasis)’ and ‘球结膜水肿 (chemosis)’ are labeled as single words, although according to Gao et al9 they should be labeled as separated words. We also employed several additional rules to make the segmentation annotation more consistent, which we list in the appendix (see supplementary material C, available online only).
Annotation flow
To find a gold standard, annotations were made by annotators with relevant domain backgrounds. Three doctors (A1, A2, and A3) were asked to make annotations. Each record was annotated by two doctors (A1 and A2) independently, and a third doctor (A3) would judge the results as correct or incorrect. The results from the doctors were merged as follows. If the two doctors hold the same opinion, it is decided. If the opinions of the two are different, then the annotation will be determined by the third doctor. However, if the third doctor still cannot decide on the annotation, further discussion must be conducted until an agreement on the annotation is reached. Doctors proficient in medical terminologies are able to find the content that should be annotated accurately; however, they are inconsistent in identifying the boundaries of records due to the diversity of language.
Most of the inconsistency comes from the doctors’ dealing with word boundaries. For example, in the phrase ‘偶发结石 (stones occasionally found)’, the doctors were inconsistent on whether to include ‘偶发 (occasionally found)’ in the named entity. Similarly, when annotating drug names like ‘阿莫西林胶囊 (amoxicillin capsules)’, the doctors were inconsistent with whether ‘胶囊 (capsules)’ was part of the medication entity. Occasionally, doctors may annotate two different entities as a single entity because the two entities often occur simultaneously, such as ‘咳嗽咳痰 (cough and expectoration)’.
Inconsistency in annotation may affect the learning model and subsequently lower the performance of our system. Therefore, to obtain a more consistently labeled corpus, a second round of annotation is required. In the second round, three annotators with backgrounds in computer linguistics (B1, B2, and B3) were asked to annotate the record following the same procedures as in the first round. They were given the annotation produced by doctors and were asked to annotate on top of them so that no entities were mislabeled due to a lack of medical expertise. Refined results and a final gold standard were obtained by combining the results from the above two rounds of annotation.
Inter-annotator agreement
Disagreements exist between annotators with different domain backgrounds, but ultimately benefit us in building a more completely annotated corpus, showing the importance of a mixture of both medical background and computer linguistic background in achieving an optimized result. The Kappa statistics were used in this paper to assess the agreements of different annotators.
As shown in supplementary material D, table S3 (available online only), there is a great difference in inter-annotator agreements (IAA) between annotators and the gold standard. This difference substantially demonstrates the complementary nature of the annotation from different domains. Therefore, it is reasonable to state that IAA between annotators and the final gold standard is of great importance as it denotes the accuracy and the completeness of the annotations.
With high values of k in B1 and B2, the annotation result is very satisfying. Shown in table S3, the k for doctor annotators and the gold standard is rather rough (k=71.94% and 71.87%), and the k for computer linguistic annotators and the gold standard is higher (k=91.59% and 90.98%). The experiment shows the substantial agreement between them, which represents the completeness of the annotations.
We also computed the IAA for segmentation annotation, which is shown in supplementary material D, table S4 (available online only).
Methods
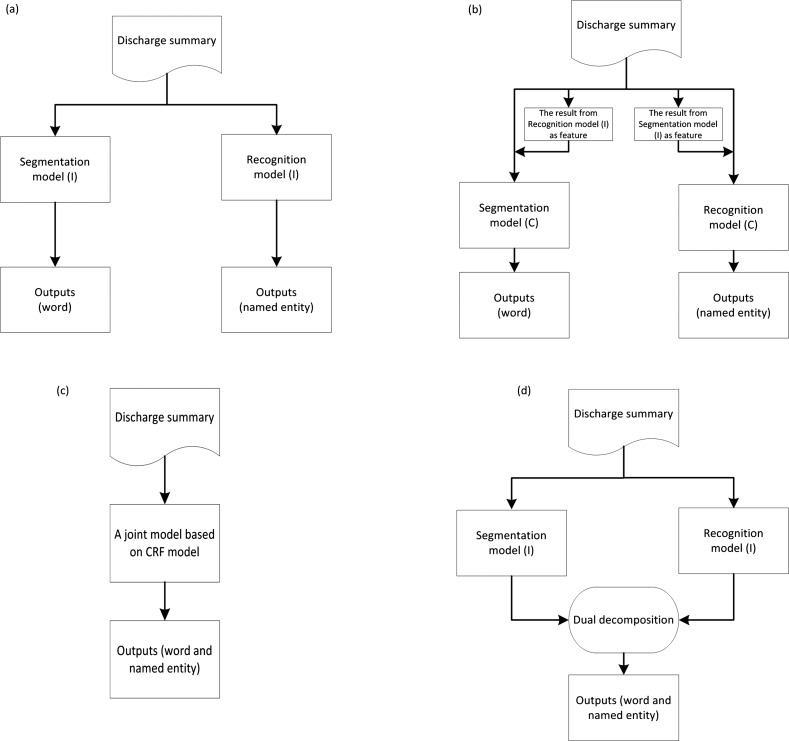
In this section, we first briefly describe the CRF,29 which are widely used to solve sequential labeling problems in the literature of natural language processing.30 We then introduce baseline methods, including independent models and incremental models based on two CRF models. We also introduce a joint model trained on combined labels. Next, we describe the concept of dual decomposition.19 Finally, we present the joint model using a dual decomposition algorithm for the two tasks. Figure 2 shows the flow charts of the four methods (independent model, incremental model, joint model trained on combined labels (joint_CRF) and joint model using dual decomposition (joint_DD)).
Figure 2.
The flow charts of the four methods (independent model, incremental model, joint_CRF and joint_DD). CRF, conditional random field; DD, dual decomposition.
Conditional random fields
We give a brief description of CRF models29 used for sequential labeling. In natural language processing, we consider CRF whose dependency graph is a chain. Let 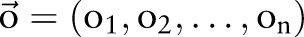 be an observation sequence. Our goal is to assign sequential labels
be an observation sequence. Our goal is to assign sequential labels 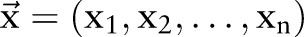 where
where 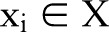 is the label assigned for the ith instance. For instance, in our segmentation task
is the label assigned for the ith instance. For instance, in our segmentation task 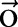 is the sequence of all Chinese characters of a sentence and
is the sequence of all Chinese characters of a sentence and 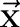 can be used to represent a segmentation solution of the sentence
can be used to represent a segmentation solution of the sentence 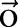 . When using CRF models to solve this problem, we first define classes of features
. When using CRF models to solve this problem, we first define classes of features 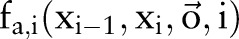 , where
, where 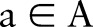 is the type of feature (eg, punctuation, capital letters, etc.) and
is the type of feature (eg, punctuation, capital letters, etc.) and 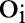 is the instance to be labeled. A set of parameters
is the instance to be labeled. A set of parameters 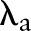 are then trained on the training data and the loss function for a label sequence
are then trained on the training data and the loss function for a label sequence 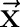 on an observation sequence
on an observation sequence 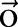 can then be expressed as
can then be expressed as
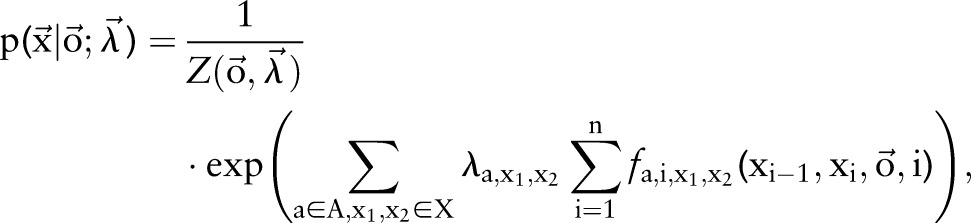 |
1 |
where 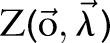 is a normalization factor and can be omitted when we only care about the best label sequence. Equation (2) is then used to find the optimal label sequence
is a normalization factor and can be omitted when we only care about the best label sequence. Equation (2) is then used to find the optimal label sequence 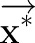 .
.
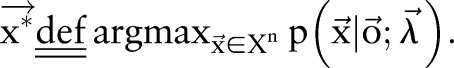 |
2 |
Baseline methods: independent models and incremental models
Baseline methods are to view the two tasks as completely independent tasks. Both word segmentation and named entity recognition can be viewed as sequential labeling tasks, which are often solved using CRF models in the literature of natural language processing. To perform the two tasks, two sets of different training data (one is for word segmentation and the other is for named entity recognition) were used in two different CRF models to perform them.
Needless to say, independent CRF models do not give satisfactory performance on the two tasks because this method does not take the inherent correlation between the two tasks into account. For example, almost all medication named entities serve as an isolated single word in word segmentation (such as ‘阿司匹林 (aspirin)’) and no named entities will contain a fraction of a segmented word. Therefore, knowing the result of one task can significantly benefit the inference of another task. To this end, we developed incremental models, in which the labeled results of word segmentation of named entity recognition were used as features to train new (and better performed) models incrementally. Note that the results used as features of segmentation and entity recognition were produced by two independent models. Please see figure 2 for a graphical depiction of independent and incremental models.
Although the incremental model can capture the correlation between word segmentation and named entity recognition to some extent, it has two major fallacies: (1) Segmentation and entity recognition results produced by independent CRF models may not be accurate. As a result, using these inaccurate results as features may lead to the problem of error propagation and lower the performance of the final incremental models. (2) Due to the nature of the CRF algorithm, the incremental models are unable to capture complex correlations between segmentation and entity recognition. For example, the models cannot capture the fact that all medication entities will be segmented as a single word, because these entities may contain many words and such inference is beyond the capability of chain CRF models.
A joint model using a CRF model
Aside from independent and incremental models, we proposed a joint model that was trained on combined labels. That is, instead of training two separate CRF models for word segmentation and named entity recognition, we combined the sequential labels of the two tasks and trained a joint CRF model that performs the two tasks simultaneously. Mathematically, we construct a new label set 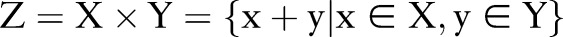 , where ‘+’ means the concatenation of two strings. Subsequently, the loss function of two sequential labels (ie, segmentation results and named entity recognition results) can then be expressed as
, where ‘+’ means the concatenation of two strings. Subsequently, the loss function of two sequential labels (ie, segmentation results and named entity recognition results) can then be expressed as
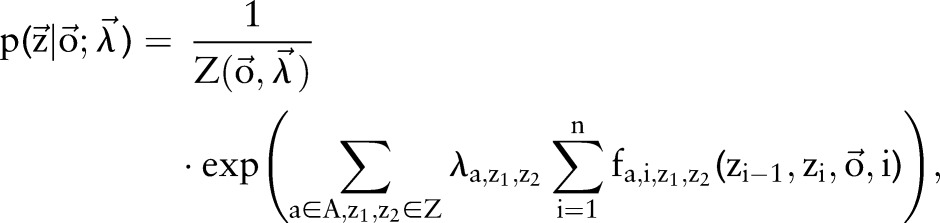 |
3 |
which can also be written as
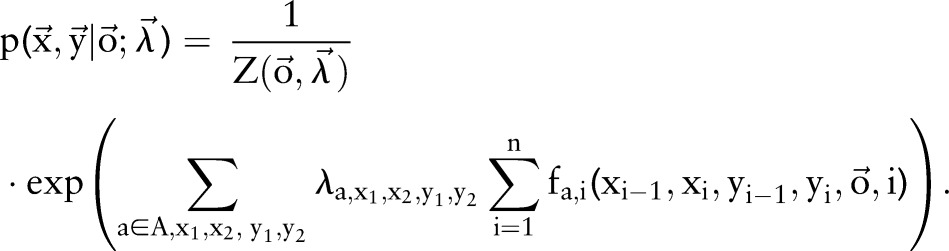 |
4 |
Clearly, this method not only utilizes the correlation between segmentation and entity recognition tasks, but it also avoids the error propagation problem in the incremental model because the joint model performs the two tasks at the same time. Our experiments also show that the joint model trained on combined labels outperforms both independent and incremental models in various settings.
Nevertheless, there is a major drawback with this joint method: the number of parameters in the joint model is much larger than the number in independent and incremental models. To be more specific, let 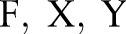 denote the feature set, label sets for word segmentation and named entity recognition, respectively. Assuming we are using a chain CRF, the joint model will require a total of
denote the feature set, label sets for word segmentation and named entity recognition, respectively. Assuming we are using a chain CRF, the joint model will require a total of 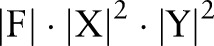 parameters (since
parameters (since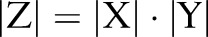 ), while the independent and incremental models only require
), while the independent and incremental models only require 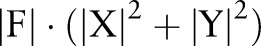 parameters. As a result, the training time required by the joint model is significantly longer than required by independent and incremental models. The large amount of parameters also makes the model perform worse on a smaller corpus, because there might not be enough data to infer the joint model's parameters accurately.
parameters. As a result, the training time required by the joint model is significantly longer than required by independent and incremental models. The large amount of parameters also makes the model perform worse on a smaller corpus, because there might not be enough data to infer the joint model's parameters accurately.
To overcome the shortcomings of this method, we proposed another joint model based on the technique of dual decomposition. Our proposed method provides a flexible way to represent the correlation between segmentation and named entity recognition tasks. Furthermore, the number of parameters in our new joint model is much smaller compared to the joint model trained on combined labels. To begin with, we first introduce the concept of dual decomposition.
Dual decomposition
Dual decomposition, proposed by Rush et al,19 is a classic method to solve optimization problems whose objective function can be divided into subproblems that can be solved easily.22 31 A typical optimization problem that can be solved using dual decomposition is presented as follows.
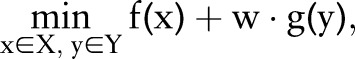 |
5 |
where w is the weight of the second task and 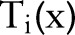 ,
,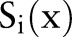 ,
,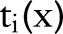 ,
,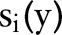 are usually predicates and the constraints require the two variables (ie,
are usually predicates and the constraints require the two variables (ie,  and
and  ) to satisfy properties
) to satisfy properties 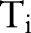 ,
, 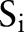 and
and 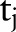 ,
, 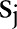 simultaneously for all
simultaneously for all 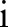 and
and  . While the primal optimization problem is often hard and cannot be solved efficiently, its Lagrange dual problem can be solved (or approximated) efficiently as the two subproblems
. While the primal optimization problem is often hard and cannot be solved efficiently, its Lagrange dual problem can be solved (or approximated) efficiently as the two subproblems 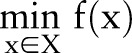 and
and 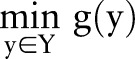 can be solved efficiently when considered separately as two independent optimization problems.
can be solved efficiently when considered separately as two independent optimization problems.
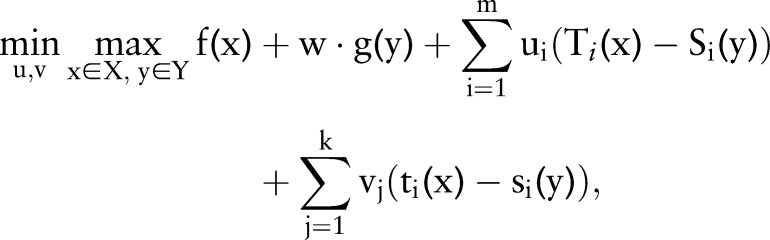 |
6 |
To find the optimal  and
and 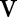 , we used a subgradient method,19
32 which involves iteratively solving the easy subproblems and updates weights
, we used a subgradient method,19
32 which involves iteratively solving the easy subproblems and updates weights 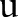 accordingly.
accordingly.
Proposed method: a joint model using dual decomposition
Let 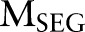 and
and 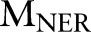 be two CRF models trained separately for word segmentation and named entity recognition. Let also
be two CRF models trained separately for word segmentation and named entity recognition. Let also 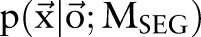 and
and 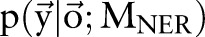 be conditional probabilities of segmentation
be conditional probabilities of segmentation 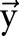 and named entity recognition
and named entity recognition 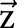 . Clearly, given the CRF models
. Clearly, given the CRF models 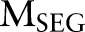 ,
, 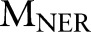 and the observation (ie, feature) sequence
and the observation (ie, feature) sequence 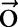 , the optimal
, the optimal 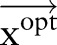 and
and 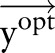 can be computed efficiently.
can be computed efficiently.
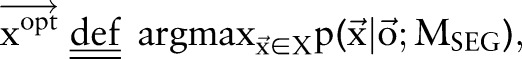 |
7 |
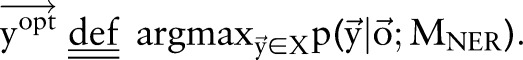 |
Constraints in the primal optimization problem are correlations between word segmentation and named entity recognition. In our system, we implement two classes of constraints:
A named entity shall never contain a fraction of a segmented word. In other words, no group of Chinese characters can be segmented as a single word if these characters contain the border of a named entity.
A medication named entity should always be segmented as a single word.
The above constraints are natural rules for word segmentation and named entity recognition and were also carried out when annotating the corpus. Please refer to our description of data corpus to find detailed rules guiding our annotation. Mathematically, for an observation sequence 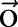 ,
, 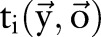 were used to represent whether the ith character is inside a named entity of type ‘medication’;
were used to represent whether the ith character is inside a named entity of type ‘medication’; 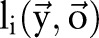 and
and 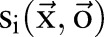 was used to represent whether the ith character and the
was used to represent whether the ith character and the 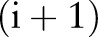 th character belong to different named entities or segmented words. A solution
th character belong to different named entities or segmented words. A solution 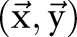 is considered feasible if
is considered feasible if 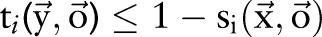 and
and  for all
for all 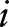 . We then used the algorithm of dual decomposition, which is detailed in Box 1 to find the optimal
. We then used the algorithm of dual decomposition, which is detailed in Box 1 to find the optimal 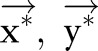 given the two CRF models
given the two CRF models 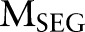 and
and 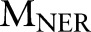 . In our experiment, the maximum number of iterations
. In our experiment, the maximum number of iterations 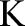 is set to 50.
is set to 50.
Box 1. Subgradient algorithm for dual decomposition.
Input Two CRF models 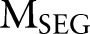 ,
, 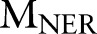 ; the observation sequence
; the observation sequence 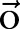 and its length
and its length 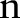 .
.
Output A pair of solution 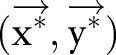 .
.
Parameters
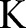 , the maximum number of iterations;
, the maximum number of iterations; 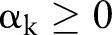 , the step sizes.
, the step sizes.
Initialize Weight of each constraint 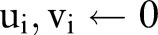 .
.
for
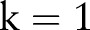 to
to 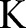 do
do
 |
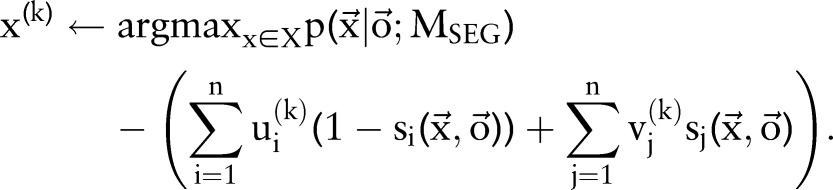 |
if
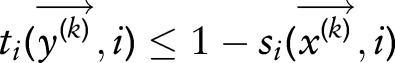 and
and 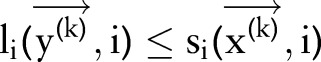 for all
for all  then
then
return
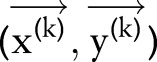 .
.
for all
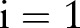 to
to 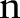 do
do
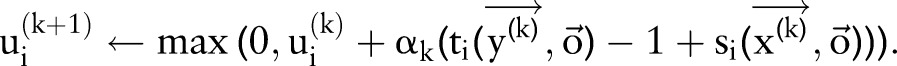 |
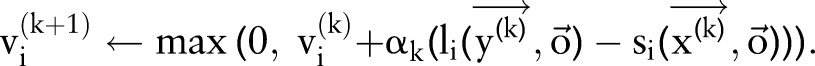 |
return

CRF, conditional random field.
Experiment
Experiment settings
We now describe the settings of our experiments. The CRF toolkit we used is CRF++.33 Four methods (independent models, incremental models, a joint model trained on combined labels and a joint model using dual decomposition) were compared for both word segmentation and named entity recognition under three different sets of features. For feature set 1 (Ftr. Set 1), only trigrams of Chinese characters were used as features; for feature set 2 (Ftr. Set 2), we added some other basic features, such as punctuations, digits, English characters, conjunctions (like ‘和 (and)’, ‘伴 (with)’), positions (like ‘左 (left)’, ‘右 (right)’), etc.; for feature set 3 (Ftr. Set 3), apart from features from the two previous feature sets, we added special dictionaries for problem, test and medication entities as features. Please refer to Xu et al34 for a detailed description of the method used to construct these special dictionaries. We also gave more detailed descriptions of the features we used in supplementary material E (available online only).
Note that we did not integrate any POS features into our recognition systems because mainstream Chinese POS tagging systems perform considerably poorer on Chinese texts in the clinical domain. There are at least two reasons for the poor performance: first, most drug names consist of several Chinese characters and most Chinese POS tagging systems tend to mislabel these characters completely. Take the word ‘氯硝西泮’ (clonazepam, literally ‘chlorine saltpeter west pam’) as an example. The Stanford Chinese POS tagger35 tags this word as ‘氯/CD 硝/M 西/NN 泮/NN’, which is completely wrong. Second, most disease names have a long list of modifying words before the actual disease, which is a syntactical pattern rarely found in ordinary texts like newspapers. For example, the word ‘慢性阻塞性肺炎 (chronic obstructive pulmonary disease)’ has two modifiers ‘慢性 (chronic)’ and ‘阻塞性 (obstructive)’ that modifes the central word ‘肺炎 (pneumonia)’. The second modifier ‘阻塞性 (obstructive)’ is tagged by the Stanford Chinese POS tagger as a verb and is clearly wrong. In summary, as mainstream Chinese POS taggers performs considerably worse on our corpus, we did not use POS tags as features for fear that these features will lower the performance of our system.
Evaluation metric
The three standard performance metrics, pecision (P), recall (R) and F-measure (F), are used as evaluation metrics in our task.
In the experiment, we followed the standard leave-one-out method and the cross-validation method. In this paper, fivefold cross-validation and averaged metrics were used.
Results
The experimental results are shown in table 1. In general, joint models achieved better results than independent and incremental models on both tasks. The joint model using dual decomposition achieved an increase of 0.30%, 0.58% and 0.17% on the named entity recognition task under three different sets of features, compared to the joint model using combined task labels. The two joint models also achieved comparable performance on the word segmentation task. Furthermore, as we have mentioned in the previous section, the joint model using dual decomposition inference has a much smaller number of parameters than the joint model using combined task labels, which means the training time required by the former model is much less than required by the latter one. In our experiment, the joint model using dual decomposition used nearly 2 h to finish the fivefold cross-validation while the joint model trained on combined labels used more than 10 h to finish the same task (both models were trained using the third set of features). The experimental results clearly demonstrate the efficiency and effectiveness of our proposed joint model using dual decomposition inference.
Table 1.
Performances of four models for two tasks
| Named entity recognition | Word segmentation | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | 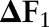
|
P | R | F1 | 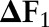
|
||
| MSRSeg | – | – | – | – | 79.42 | 86.00 | 82.58 | ||
| Ftr. Set 1 | Independent | 89.61 | 84.19 | 86.82 | 95.20 | 94.75 | 94.97 | ||
| Incremental | 89.84 | 85.69 | 87.72 | 0.90 | 95.25 | 94.92 | 95.09 | 0.12 | |
| Joint_CRF | 90.26 | 86.66 | 88.42 | 1.60 | 95.27 | 95.13 | 95.20 | 0.23 | |
| Joint_DD | 91.50 | 86.11 | 88.72 | 1.90 | 95.30 | 95.25 | 95.27 | 0.30 | |
| Ftr. Set 2 | Independent | 89.48 | 85.18 | 87.28 | 95.31 | 95.05 | 95.18 | ||
| Incremental | 89.59 | 86.03 | 87.77 | 0.49 | 95.26 | 95.10 | 95.18 | 0.00 | |
| Joint_CRF | 89.84 | 87.11 | 88.45 | 1.17 | 95.31 | 95.31 | 95.31 | 0.13 | |
| Joint_DD | 91.25 | 86.92 | 89.03 | 1.75 | 95.31 | 95.38 | 95.35 | 0.17 | |
| Ftr. Set 3 | Independent | 91.03 | 87.50 | 89.23 | 95.38 | 95.30 | 95.34 | ||
| Incremental | 90.96 | 88.10 | 89.51 | 0.28 | 95.48 | 95.56 | 95.55 | 0.21 | |
| Joint_CRF | 91.52 | 88.67 | 90.07 | 0.84 | 95.52 | 95.57 | 95.59 | 0.25 | |
| Joint_DD | 92.15 | 88.41 | 90.24 | 1.01 | 95.64 | 95.53 | 95.55 | 0.21 | |
| Problem | Test | Treatment | Medication | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F | P | R | F | P | R | F | P | R | F | ||
| Ftr. Set 1 | Independent | 90.4 | 87.9 | 89.1 | 90.5 | 82.8 | 86.5 | 79.9 | 66.0 | 72.3 | 91.2 | 71.1 | 79.9 |
| Incremental | 90.6 | 89.1 | 89.9 | 90.3 | 84.7 | 87.4 | 79.3 | 66.8 | 72.5 | 93.8 | 75.5 | 83.7 | |
| Joint_CRF | 91.2 | 89.8 | 90.5 | 90.9 | 85.6 | 88.1 | 81.0 | 67.9 | 73.9 | 89.9 | 80.4 | 84.9 | |
| Joint_DD | 92.3 | 89.8 | 91.0 | 92.4 | 84.1 | 88.0 | 81.9 | 69.3 | 75.1 | 93.7 | 72.5 | 81.8 | |
| Ftr. Set 2 | Independent | 90.4 | 88.7 | 89.5 | 89.0 | 82.6 | 85.7 | 79.9 | 67.1 | 72.9 | 92.5 | 75.9 | 83.4 |
| Incremental | 90.4 | 89.3 | 89.9 | 89.3 | 84.4 | 86.8 | 79.8 | 68.0 | 73.5 | 93.8 | 77.2 | 84.7 | |
| Joint_CRF | 90.8 | 90.0 | 90.4 | 89.6 | 85.6 | 87.6 | 81.2 | 69.3 | 74.8 | 89.5 | 82.8 | 86.0 | |
| Joint_DD | 92.0 | 90.5 | 91.3 | 92.1 | 84.1 | 87.9 | 81.5 | 69.7 | 75.1 | 94.0 | 75.9 | 84.0 | |
| Ftr. Set 3 | Independent | 92.0 | 90.5 | 91.2 | 91.1 | 85.5 | 88.2 | 80.0 | 67.1 | 73.0 | 93.6 | 86.4 | 89.9 |
| Incremental | 92.0 | 91.0 | 91.5 | 91.0 | 86.0 | 88.5 | 79.7 | 68.3 | 73.6 | 92.8 | 86.8 | 89.7 | |
| Joint_CRF | 92.6 | 91.3 | 91.9 | 91.5 | 87.0 | 89.2 | 81.3 | 70.1 | 75.3 | 92.6 | 88.7 | 90.6 | |
| Joint_DD | 93.1 | 91.2 | 92.1 | 92.2 | 86.3 | 89.2 | 82.4 | 69.7 | 75.5 | 93.7 | 87.6 | 90.6 | |
CRF, conditional random field; DD, dual decomposition. Bold indicates the highest values.
Discussion
Annotation of segmentation
In segmentation annotation, the general rule is ‘if the English translation of a group of Chinese characters is only one word, then the corresponding group of characters is labeled as a single word’. We employed this rule mainly to keep a one-to-one correspondence between Chinese and English medical terminologies (eg, ‘球结膜水肿’ and ‘chemosis’), which lays the groundwork for detecting pair links of the same objects between English and Chinese medical terminologies.
In addition, we labeled body parts as independent words and separate location expressions as independent words. This is because body parts and location expressions are very important information in Chinese discharge summaries: even if information related to body parts and locations is slightly wrong, doctors will have trouble curing patients’ diseases. Besides, this rule is useful and effective for building a mapping from expressions of body parts to actual anatomies from each discharge summary. Such a mapping can greatly help physicians to combine all the anatomical information instead of being limited to independent organs or tissues to acquire a comprehensive knowledge of patients’ diseases.
Annotation of named entities
Slightly different from the method in I2B2 Challenge 2010 tasks, our method includes two more categories (medication and anatomy) in addition to the three used in the I2B2 Challenge tasks (medical problem, treatment, and test). Dividing medications from treatments will benefit future research on the usage and effectiveness of medications. Marking anatomy in problem and test phrases is also beneficial because it enables us to locate the exact positions of symptoms or medical tests.
However, our annotation method has limitations. For instance, allowing verb phrases and post-entity modifiers in an entity may cause difficulties in the design of automatic named entity recognition systems. Furthermore, there is some important information in clinical narratives that will be missed by our annotation guideline. For example, words indicating the changes in patients’ conditions are often missed, such as ‘恶化 (worsen)’, because these words are often unnecessary verb phrases.
Comparison between Chinese and English corpora
The most significant difference between Chinese and English corpora is the lack of a space between two Chinese words, which requires word segmentation for Chinese text. It is clear that the performance of word segmentation will greatly affect the accuracy of a named entity recognition system, as in the example phrase ‘停用新赛斯平 (stop using ciclosporin soft capsules)’. If the drug name ‘新赛斯平 (ciclosporin)’ is not segmented as a single word, then it is very difficult for the named entity recognition system to identify the word boundary correctly and the character ‘新 (new)’ may even be identified as an adjective by mistake.
Chinese word segmentation is not an easy task in general, but some properties of Chinese clinical reports make the segmentation and named entity recognition task even harder. For example, most drug names used in Chinese clinical reports are translations of their English counterparts and as Chinese is not an alphabetic language, there are a number of ways to translate one particular medicine, like ‘阿司匹林,’ ‘阿斯匹林,’ and ‘阿思匹林’ (all referring to aspirin). This additional ambiguity makes the Chinese named entity recognition task much harder than in English. Furthermore, there is no capitalization in the Chinese writing system, which is another challenge because we cannot recognize medication entities based on capitalized letters.
Another interesting difference between Chinese and English named entities in the medical domain is the formation of Chinese words for diseases. Many Chinese disease names contain anatomies, like ‘肺炎 (pneumonia, lung inflammation literally)’ and ‘肝炎 (hepatitis, liver inflammation literally)’. This shows that the identification of anatomy entities can help the recognition of some Chinese disease entities, whereas in English such an improvement is rare because the English words for pneumonia and hepatitis do not have any part for the corresponding anatomies (ie, lung and liver).
Methods
We first use an example to demonstrate why the method using independent models suffers from its lack of correlation information between word segmentation and named entity recognition, which eventually lowers its performance on both tasks. In the sentence ‘患者在医院输液治疗数天 (the patient was given infusion therapy in the hospital for several days)’, the CRF model for word segmentation correctly segmented the above sentence into ‘患者 (patient)/在 (in)/医院 (hospital)/输液 (infusion)/治疗 (therapy)/数天 (several days)’. However, the CRF model for named entity recognition gave an incorrect named entity ‘输液治疗数’, which is mistakenly considered by the model to mean ‘the number of infusion therapies’. Clearly, this mistake can be avoided if we consider word segmentation and named entity recognition jointly, because ‘数天 (several days)’ is correctly segmented as a single word and hence the phrase ‘输液治疗数’ should never be allowed to be a named entity because it contains a fraction of a word. As expected, incremental models and the two joint models gave correct answers for both word segmentation and named entity recognition.
Although incremental models can capture some correlation between the segmentation and entity recognition tasks, they suffered from the problem of error propagation because incorrect answers of one task will be likely to misguide the CRF model to give incorrect answers for the other task. We have observed this phenomenon when we were trying to process the phrase ‘还原性谷胱甘肽 (reduced glutathione)’. The independent models gave incorrect segmentation results for this phrase but gave correct named entity segmentation results, mainly because this phrase is enclosed with commas at both ends and the CRF model for named entity recognition correctly identified the named entity. However, when the (incorrect) initial segmentation results were used as features to train a CRF model incrementally for entity recognition, the newly trained model suffers from the incorrect segmentation and extracted an incorrect entity (‘原性谷胱甘肽’, missing the leading character ‘还’).
In contrast to incremental models, our proposed joint model uses dual decomposition to coordinate results from the two tasks and avoid being misled by incorrect answers of one task. In fact, because the named entity recognition system correctly identified the phrase as a medication name and we have specified constraints requiring all named entities of type medication must be labeled as a single word when segmenting the text, the dual decomposition algorithm automatically changes the segmentation labels to the correct ones. This example clearly demonstrates the effectiveness and fitness of our proposed method for word segmentation and named entity recognition tasks.
Another important advantage of our proposed joint model based on dual decomposition is its smaller inference complexity. Unlike a complete joint CRF model, which combines labels for the two tasks altogether, our proposed method trains models for each task separately, which significantly reduces the amount of inference time. In our experiment, the joint CRF model requires an average of 10 h to finish training while our proposed dual decomposition method completes training in less than 3 h. Note that the number of labels used in the joint CRF model is proportional to the product of the number of labels used in each task; we expect the joint CRF model runs much slower than our proposed dual decomposition method as the number of tasks and their labels grow larger.
Errors in the treatment category
Noticeably, in all the four semantic categories (ie, problems, tests, medication and treatments), the performance of treatment identification (especially the recall) is significantly lower than the performance of the other three categories. This is due to the complicated structure of treatment entities: unlike the other three categories whose entities are mostly noun phrases (eg, ‘咳嗽 (cough; problem)’, ‘血压 (blood pressure; test)’, ‘阿司匹林 (aspirin; medication)’, etc.), many treatment entities involve verb phrases (eg, ‘扩张支气管’ (expansion of bronchus, literally ‘expand (扩张) bronchus (支气管)’) and ‘祛痰’ (expectorant, literally ‘remove (祛) sputum (痰)’)). As most current named entity recognition (NER) algorithms focus on entities that are noun phrases, the structure of treatment entities makes its recognition understandably harder.
Domain difference
The performance of state-of-the-art segmentation methods in common corpus is 96%/F or so36 while our best performance is 95.5% or so in Chinese discharge summaries. This clearly demonstrates that our annotation guideline is a practical one because both methods achieved comparable performance.
Conclusion and future work
In this paper, we created a new corpus of word segmentation and name entity recognition based on Chinese discharge summaries. Named entities are divided into four categories: problem, test, treatment, and medication. We described the annotation guideline of word segmentation in detail. Combining the correlation between named entity recognition and word segmentation, a new joint model using dual decomposition is presented to perform the two tasks. Experiments demonstrate that the joint model has evident advantages in terms of performance as well as running time, compared with other baseline models.
In future, dual decomposition as a framework will be designed for several primary and essential tasks in Chinese discharge summaries, such as POS tagging and dependency parsing.
Supplementary Material
Footnotes
Contributors: All authors focused on writing code, made valuable contributions to study design, and took part in the writing of the paper.
Funding: This work was supported by Microsoft Research Asia (MSR Asia). The work was supported by MSRA eHealth grant, Grant 61073077 from the National Science Foundation of China and Grant SKLSDE-2011ZX-13 from the State Key Laboratory of Software Development Environment in Beihang University in China.
Competing interests: None.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Chapman WW, Nadkarni PM, Hirschman L, et al. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions. J Am Med Inform Assoc 2011;18:540–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dogan RI, Neveol A, Lu ZY. A context-blocks model for identifying clinical relationships in patient records. BMC Bioinform 2011;12(Suppl. 3):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soualmia LF, Prieur-Gaston E, Moalla Z, et al. Matching health information seekers’ queries to medical terms. BMC Bioinform 2012;13(Suppl. 14):S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu H, Markatou M, Dimova R, et al. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues. BMC Bioinform 2006;7:334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriquez-Molinero A, Lopez-Diequez M, Tabuenca AI, et al. Joint segmentation and named entity recognition using dual decomposition in Chinese discharge summaries. BMC Geriatr 2006;6:1316952319 [Google Scholar]
- 6.Uzuner Ö, Bodnari A, Shen S, et al. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc 2012;19:786–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uzuner Ö, South B, Shen S, et al. 2010 i2b2/VA Challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011;18:552–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao JF, Li M, Wu AD, et al. Chinese word segmentation and named entity recognition: a pragmatic approach. Comput Linguist 2005:31:1–42 [Google Scholar]
- 10.Zhao H, Huang CN, Li M. An improved Chinese word segmentation system with conditional random field. Proceedings of the Fifth SIGHAN Workshop on Chinese Language Processing; Sydney, 2006:162–5 [Google Scholar]
- 11.Xu W, Fu B, Fu L, et al. Domain extension of Chinese named entity recognition. The Ninth National Computational Linguistics Conference Proceedings; 2007:503–8 [Google Scholar]
- 12.Qiao W. Research on several key issues in Chinese word segmentation [PHD dissertation]. Tsinghua University, 2010 [Google Scholar]
- 13.Xu Y, Hong K, Tsujii J, et al. Feature engineering combined with machine learning and rule-based methods for structured information extraction from narrative clinical discharge summaries. J Am Med Inform Assoc 2012;19:824–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, Tsujii J, Chang EI. Named entity recognition of follow-up and time information in 20 000 radiology reports. J Am Med Inform Assoc 2012;19:792–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kruengkrai C, Uchimoto K, Kazama J, et al. An error-driven word-character hybrid model for joint Chinese word segmentation and POS tagging. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP; Association for Computational Linguistics, 2009:513–21 [Google Scholar]
- 16.Hatori J, Matsuzaki T, Miyao Y, et al. Incremental joint POS tagging and dependency parsing in Chinese. Proceedings of IJCNLP; 2011:1216–24 [Google Scholar]
- 17.Srikumar V, Roth D. A joint model for extended semantic role labeling. Proceedings of the Conference on Empirical Methods in Natural Language Processing; Association for Computational Linguistics, 2011:129–39 [Google Scholar]
- 18.Dantzig GB, Wolfe P. Decomposition principle for linear programs. Operations Research 1960;8:101–11 [Google Scholar]
- 19.Rush AM, Sontag D, Collins M, et al. On dual decomposition and linear programming relaxations for natural language processing. Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing; Association for Computational Linguistics, 2010:1–11 [Google Scholar]
- 20.Koo T, Rush AM, Collins M, et al. Dual decomposition for parsing with non-projective head automata. Proceedings of the 2010 Conference on Empirical Methods in Natural Language Processing; Association for Computational Linguistics, 2010:1288–98 [Google Scholar]
- 21.Rush AM, Collins M. Exact decoding of syntactic translation models through lagrangian relaxation. Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics; Human Language Technologies, 2011:72–82 [Google Scholar]
- 22.Chang YW, Collins M. Exact decoding of phrase-based translation models through lagrangian relaxation. Proceedings of the Conference on Empirical Methods in Natural Language Processing; Association for Computational Linguistics, 2011:26–37 [Google Scholar]
- 23.McClosky D, Surdeanu M, Manning CD. Event extraction as dependency parsing for bionlp 2011. Proceedings of the BioNLP Shared Task 2011 Workshop; Association for Computational Linguistics, 2011:41–5 [Google Scholar]
- 24.Riedel S, McClosky D, Surdeanu M, et al. Model combination for event extraction in BioNLP 2011. Proceedings of the BioNLP Shared Task 2011 Workshop; Association for Computational Linguistics, 2011:51–5 [Google Scholar]
- 25.Riedel S, McCallum A. Fast and robust joint models for biomedical event extraction. Proceedings of the Conference on Empirical Methods in Natural Language Processing; Association for Computational Linguistics, 2011:1–12 [Google Scholar]
- 26.McClosky D, Riedel S, Surdeanu M, et al. Combining joint models for biomedical event extraction. BMC Bioinform. BioMed Central Ltd 2012:13:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo Y, Asahara M, Matsumoto Y. Dual decomposition method for Chinese predicate-argument structure analysis. Natural Language Processing and Knowledge Engineering (NLP-KE), 2011 7th International Conference; IEEE, 2011: 409–14 [Google Scholar]
- 28.Hanamoto A, Matsuzaki T, Tsujii J. Coordination structure analysis using dual decomposition. Proceedings of the 13th Conference of the European Chapter of the Association for Computational Linguistics; 2012:430–8 [Google Scholar]
- 29.Lafferty J, McCallum A, Pereira FCN. Conditional random fields: probabilistic models for segmenting and labeling sequence data. Proceedings of the 18th International Conference on Machine Learning ACM, 2001:282–9 [Google Scholar]
- 30.Tseng H, Chang P, Andrew G, et al. A conditional random field word segmenter for sighan bakeoff 2005. Proceedings of the Fourth SIGHAN Workshop on Chinese Language Processing; Jeju Island, Korea, 2005:171 [Google Scholar]
- 31.Das D, Martins AFT, Smith NA. An exact dual decomposition algorithm for shallow semantic parsing with constraints. Proc. of *SEM; 2012 [Google Scholar]
- 32.Boyd S, Xiao L, Mutapcic A, et al. Notes on decomposition methods. Stanford University, 2008 [Google Scholar]
- 33.CRF++. http://code.google.com/p/crfpp/
- 34.Xu Y, Wang YN, Sun JT, et al. Building large collections of Chinese and English medical terms from semi-structured and encyclopedia websites. PLoS ONE 2013 Jul 9;8: doi:10.1371/journal.pone.0067526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ICTCLAS. http://ictclas.org/
- 36.Song Y, Cai DF, Zhang GP, et al. Approach to Chinese word segmentation based on character-word decoding. J Software 2009;20:2366–75 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.