Abstract
Objectives
To understand the impact of GeneInsight Clinic (GIC), a web-based tool designed to manage genetic information and facilitate communication of test results and variant updates from the laboratory to the clinics, we measured the use of GIC and the time it took for new genetic knowledge to be available to clinicians.
Methods
Usage data were collected across four study sites for the GIC launch and post-GIC implementation time periods. The primary outcome measures were the time (average number of days) between variant change approval and notification of clinic staff, and the time between notification and viewing the patient record.
Results
Post-GIC, time between a variant change approval and provider notification was shorter than at launch (average days at launch 503.8, compared to 4.1 days post-GIC). After e-mail alerts were sent at launch, providers clicked into the patient record associated with 91% of these alerts. In the post period, clinic providers clicked into the patient record associated with 95% of the alerts, on average 12 days after the e-mail was sent.
Discussion
We found that GIC greatly increased the likelihood that a provider would receive updated variant information as well as reduced the time associated with distributing that variant information, thus providing a more efficient process for incorporating new genetic knowledge into clinical care.
Conclusions
Our study results demonstrate that health information technology systems have the potential effectively to assist providers in utilizing genetic information in patient care.
Keywords: Genetics, Medical Informatics Applications, Laboratory Software
Background and significance
The sequencing of the human genome in 2003 brought personalized medicine to the forefront of medicine and biomedical research. Since then, genetic information has revolutionized the diagnosis and management of an increasing number of diseases, and the rate of discovery in the fields of genetics and genomics has been accelerating.1 2 One of the results has been that clinicians and health systems are challenged to keep pace with the constantly evolving science of genetic medicine. Molecular diagnostic tests on germline DNA are increasingly used to help diagnose hereditary diseases such as hypertrophic cardiomyopathy and to distinguish them from diseases with similar clinical presentations.3 As genetic knowledge evolves, clinical interpretations of genetic variants may change thereby affecting treatment strategies and familial risk calculations, so it is essential for clinicians who see patients with genetic variants to stay current on the latest genetic understanding.4–9
In 2007, the American College of Medical Genetics and Genomics (ACMG) published a paper containing numerous recommendations regarding sequence variations, including the recommendation that ‘as more information becomes available allowing the interpretation of a new sequence variant, it is recommended that the laboratory amend previous reports and provide updated results to the physician’.10 This is a challenging task and clinics and laboratories must work together to provide optimal care for patients, embracing the most up-to-date genetic information. Personalized medicine will not reach its full potential if health systems are unable to connect providers to genetic information in clinically meaningful ways. Health information technology (HIT) infrastructure is poised to help ease the burden of this task, enhancing communication between the laboratory and the clinic and helping bridge the gap between laboratory science and clinical medicine.
In 2010, the Partners HealthCare Center for Personalized Genetic Medicine (PCPGM) launched the Patient Genome Explorer, later renamed GeneInsight Clinic (GIC). GIC is a web-based tool for clinics designed to manage genetic information and to facilitate communication of test results and variant updates from the laboratory to the clinics. GIC is paired with GeneInsight Lab (GIL) to form the GeneInsight Suite. GIL has been in use to support clinical molecular diagnostic laboratory practices since 2005. Study staff conducted usability and user workflow analyses in order to refine and enhance the GeneInsight tools. These efforts, as well as the early development of the suite, are described in detail elsewhere.11–13
Technology is only helpful if it is used and meets the needs of those who interact with it. Therefore, we conducted a study in which we measured the use of GIC by treating clinicians, and the impact on efficiency of new genetic knowledge being available to support clinical care as a result of GeneInsight's genetic information technology infrastructure.
Methods and materials
Study population
The data reported below were collected as part of a sub-analysis within a multi-aim investigation. PCPGM engaged with clinical practices interested in adopting GIC. Once practices had decided to implement GIC, study information was provided and they were invited to participate. Study staff contacted the practice management to assess interest before official study recruitment e-mails were sent to individual clinic staff. We recruited clinical staff who would, in the course of patient care, receive e-mails from GIC regarding patient variant changes. We enrolled staff from four clinics in the USA and Canada that were sending genetic tests related to cardiomyopathy to the PCPGM's Laboratory for Molecular Medicine (LMM) for processing. These clinics adopted GIC between April 2010 and June 2011. Clinics A and B went live in May and July of 2010, respectively, and clinics C and D went live in June 2011.
This study was approved by the Partners Human Research Committee (PHRC) and registered with ClinicalTrials.gov (ID: NCT01225978). The PHRC granted a waiver of informed consent for study components relating to clinic provider GIC usage data, as well as for patient clinical data. In total across the four study sites, we captured usage data for 34 clinicians over 24 months. At clinic A we captured data for 10 users, at clinic B for six users, and we captured data for nine users each at clinics C and D.
Intervention
Before GIC, there were no systematic processes in place to notify providers at study sites of gene variant changes, to identify easily all patients with the same variant, or to track variant changes. When new knowledge was identified based on an enquiry from a clinician regarding a specific patient, there was also no consistent process in place to record communications with clinicians regarding new information about their patients. With GIC, clinics have access to patient reports from the LMM and the history of genetic tests ordered for their patients. In addition, GIC contains an alerting system, whereby clinicians receive e-mail alerts when the LMM re-categorizes a variant in a way that may be relevant to one or more of their patients. As of March 2013, the LMM had information stored for 45 850 variants in 244 genes, evaluated by 150 tests in their GIL.
There are three ways in which providers can learn about a patient variant change via GIC: e-mail alert, patient search, or generating a list of unreviewed patients. First, providers may be contacted directly by e-mail. During the course of the study period, two different versions of alert e-mails were sent to study participants. These e-mail alerts did not contain protected health information. The original e-mail alerts sent from GIC to providers indicated the variant change and the number of patients affected. In order to view individual patient records, the e-mail recipient needed to click on the link, which brought them to the search results page, from where he/she could click into individual patient records. The e-mail did not make clear whether or not the patients in the list were the recipient's own patients, or other patients within his/her practice cared for by other providers. After usability testing and provider feedback, the GeneInsight team developed a more streamlined e-mail alert, in which links were patient specific and identified the associated provider. Moreover, the individual link brought the viewer directly to the patient view.
In addition to e-mail, providers can go directly into GIC to review variant changes in one of two ways. First, providers can log into GIC and generate a list of their patients who need to be reviewed. Patients are added to this list whenever their variant information is updated, and they are not removed from the list until a clinic provider clicks the button that indicates the alerts have been reviewed. Providers can also log into GIC to access the search page, where they can search for data about specific patients one at a time by entering a patient name and pulling up the corresponding patient record (see supplementary figures S1–S3, available online only).
Before implementation of GIC, clinicians learned of genetic variant updates either by proactively calling the LMM to check for new information on genetic test results on any of their patients, or if an individual within the LMM contacted the provider with updated variant information (via a report amendment, phone call, etc.). With the GIC alerting system, treating clinicians automatically receive genetic variant updates relevant to their patients whenever the LMM releases changes to variant classifications. Providers no longer have to request them nor does the LMM need to contact providers directly. Moreover, they have a reliable online site to review available genetic information on one or several patients or variants, without needing to call the laboratory for this information.
Study design and data collection
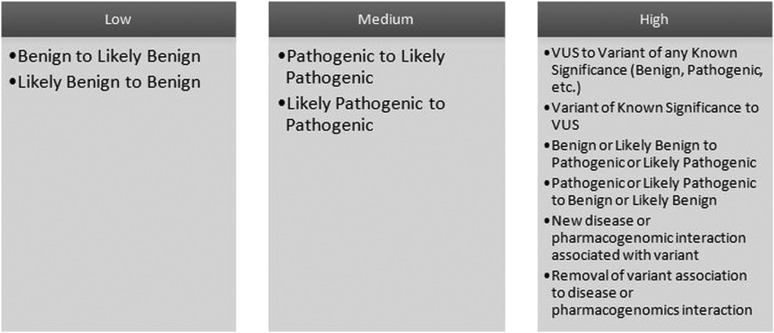
The primary outcome measures were the time (average number of days) between a variant change approval event and notification of clinic staff, and the time between notification and when the patient record was viewed. The categories employed to define gene variants included benign, likely benign, pathogenic, likely pathogenic, and variant of unknown significance. In turn, alert scenarios were defined based on changes between these categories, and the relative importance of each change (figure 1). Our system for variant classification is consistent with the most recent guidelines published by the ACMG in 2008.10 While these guidelines do not specify specific terms for variant classification, an ACMG working group is currently developing a revision of guidelines published in 2008, which has agreed to adopt the five terms used by the LMM laboratory that are part of this study, as well as the majority of clinical sequencing laboratories. These terms were also recommended by Plon et al14 and Kearney et al15 for classifying cancer and copy number variants, respectively.
Figure 1.
Variant change alert levels at time of study. Please note the information and associated alerting algorithms presented in this figure represent those in effect at the time of the study. They have subsequently been refined to accommodate a higher degree of detail since the completion of our study.
Usage data were collected across four study sites at the time of GIC launch (when GIC was implemented at each practice) and post-GIC (after GIC implementation). Variant changes that were approved before 5/1/2012 are included in our data and we collected GIC access entries to 6/1/2012. It is possible that alerts were accessed after this date but these access entries would not be recorded in our data. In our analyses, if multiple patients were affected by the same variant change, the change would be counted multiple times—once for each patient. These are referred to as ‘patient variant changes’.
Alert release timing varied based on level of significance. High alerts were sent immediately after variant change approvals occurred. Medium and low alerts were only sent once a week in a summary e-mail. However, for all alerts, new variant information was available in GIC as soon as the variant change was approved. As a result, if clinic users proceeded directly into the system rather than waiting for alerts they would have been able to see medium and low level changes before receiving the weekly summary alert. In these situations, the number of days from e-mail notification to when a clinic provider clicked into the patient record would be a negative number.
At GIC launch, alerts were generated based on whether there was a difference between the most recently approved category of the individual variant and the most recently reported category for that patient, that is, the last reported category on their final or amended genetic test report. At launch, GIC records were brought up to date to create a historical patient record in GIC, thus triggering numerous alerts even though some of the patient variant changes sent as alerts may have been previously communicated to providers for individual patients. We consider notification date in this period to be the date the data were migrated into the system and alerts were generated. In addition, in order to support the system catch-up, the alert logic for GIC launch differed from current GIC alert logic. GIC launch therefore represents a unique point in time for our data collection that is less relevant to current practice.
Once GIC was launched and variant changes were reported consistently, variant changes generated alerts based on the change from the previously approved category to the most recently approved category for that variant. The time between variant approval and notification is based on the average number of days between when the variant category was approved and when the alert e-mail was sent to a clinic provider.
Pre-GIC data include patient variant changes in which a clinician did not get notified and some in which a clinician did get notified (either through an amendment, e-mail, or phone call from the laboratory, or an eventual alert at GIC launch). The data on notification of the clinician pre-GIC were not formally captured. In order to relate GIC alert logic to variant changes that occurred before GIC implementation, LMM analysts pulled data specific to each unique patient and his/her unique variant(s), indicating when and what type of change was made and therefore when an alert could have been triggered. Post-GIC, a system audit log was maintained that included when certain components of GIC were clicked on or accessed. The first click on a link in an e-mail alert or on a patient in GIC itself defined the first access to the patient record. Finally, after usage data were collected, we spoke with a primary user at the three clinics with the highest volume of patient variant changes in the study period to understand their unique workflows.
Results
Time from variant change approval to provider notification
Before GIC implementation there were 357 patient variant changes, affecting 266 unique patients; using GIC alert logic, 48% of these would have been in the high alert category in the GIC alerting system. In the post-GIC period there were 312 patient variant changes, affecting 242 unique patients. Of these, 33% were in the high alert category.
Of the patient variant changes that occurred pre-GIC, 222 generated alerts at launch. We compared the average number of days between a variant change approval and provider notification at the launch and post-GIC periods. In comparison to launch, the average number of days between a variant change approval and provider notification was dramatically shorter in the post-GIC period (average days at launch 503.8 days compared to 4.1 days post-GIC) (table 1). Similar changes were identified when moderating the influence of outliers on the data by calculating the median number of days (median days at launch 340 days compared to 4 days post-GIC) (table 1).
Table 1.
Average number of days from variant change approval to notification of a clinic provider
| Alert level | Launch | Post-GIC | ||||
|---|---|---|---|---|---|---|
| n | Average | Median | n | Average | Median | |
| Low | 87 | 708.3 | 844 | 152 | 5.0 | 4 |
| Medium | 39 | 254.5 | 153 | 56 | 8.4 | 6 |
| High | 96 | 419.6 | 320.5 | 104 | 0.5 | 0 |
| Total | 222 | 503.8 | 340 | 312 | 4.1 | 4 |
GIC, GeneInsight Clinic.
In total, for 10% (31) of the post-GIC patient variant changes, the number of days between approval and notification of provider was longer than expected, based on GIC alert rules. For low and medium level alerts, GIC alert rules predict that the time between variant change approval and provider notification should be 7 days or less, given that a weekly summary report is generated. For high level alerts, the variant change approval and provider notification should occur on the same day (0 days in between approval and alert). Of the 10% that were delivered later than expected, the low alerts averaged 10.9 days to delivery in comparison to 4.6 days for those delivered on time, the medium averaged 29.3 days in comparison to 5.4 days, and the high alerts averaged 3.1 days to delivery in comparison to 0 days. Two factors contributed to the delay in delivery of these notifications. The majority of these alerts were temporarily delayed due to transmission or processing issues that put the alerts on hold. To reduce potential hold times, the GeneInsight team constructed functionality that proactively alerts them when an alert is held. This substantially reduced the hold time of future messages. A small percentage were delayed for quality assurance testing. For 18 days after launching one of the clinics, the GeneInsight team internally monitored all alerts to make sure the alerting mechanism was functioning properly. The accumulated alerts were then released to the clinic, which then began receiving alerts normally.
Time from provider notification to viewing patient record
At launch, once e-mail alerts were sent, clinic providers clicked into the patient record associated with 91% of these alerts overall—77% of low, and 100% of both medium and high alerts. Post-GIC, clinic providers clicked into the patient record associated with 95% of the alerts overall—91% of low, 100% of medium and 99% of high alerts (table 2). For 22 (7.4%) patient variant changes, clinicians clicked into the patient record before receiving the e-mail notification. For the remaining 275 patient variant changes, clinicians clicked on the patient record on average 14.4 days after receiving the e-mail notification for low alerts, 15.3 days for medium alerts and 11.9 days for high alerts.
Table 2.
Post GIC access to patient record
| Variant change alert level | No of alerts for which a clinic provider clicked into patient record | Average no of days from e-mail notification to when a clinic provider clicked into patient record | Median no of days from e-mail notification to when a clinic provider clicked into patient record |
|---|---|---|---|
| Low | 138 (91%) | 12.2 | 1 |
| Medium | 56 (100%) | 13.2 | 3 |
| High | 103 (99%)* | 11.9 | 4 |
| Total | 297 (95%) | 12.3 | 3 |
*The patient record for one high alert in the post-GIC period was not viewed. A user did click on the link in the e-mail for this record. Other patient records affected by this same variant change were all viewed the same day by one clinic provider.
GIC, GeneInsight Clinic.
Workflows differed by clinic. At the three clinics with the highest volume of patient variant changes, the time between notification and viewing patient records was calculated for the post-GIC period. For these clinics we also calculated the time between notification and viewing variant interpretations, which requires an additional step within GIC. The average for high alerts ranged from 1.1 to 18.1 days between notification and viewing the patient record, and 10.1 to 22.6 days between notification and viewing the variant interpretations (table 3). One high alert took 101 days between receipt of the alert and a provider clicking into the patient record.
Table 3.
Post-GIC average days from e-mail notification to viewing patient record and variant interpretation for the three clinics with the highest volume of patient variant changes
| Average no of days from e-mail notification to when a responsible provider clicked into patient record (N) | Average no of days from e-mail notification to when a responsible provider clicked into individual variant interpretation (N) | |||||
|---|---|---|---|---|---|---|
| Clinic A | Clinic B | Clinic C | Clinic A | Clinic B | Clinic C | |
| Low | 12.4 (74) | 21.3 (34) | −1.2 (25) | 21.0 (66) | 21.3 (34) | 1.0 (25) |
| Medium | 12.3 (11) | 20.3 (25) | −1.8 (12) | 86.9 (7) | 22.1 (25) | −0.8 (12) |
| High | 1.1 (33) | 18.1 (58) | 11.3 (9) | 10.1 (29) | 22.6 (58) | 11.3 (9) |
Negative numbers reflect cases in w hich a provider clicked into GIC before receiving the e-mail notification.
GIC, GeneInsight Clinic.
Discussion
We evaluated the impact of a tool designed to help providers learn about genetic variants and found that GIC greatly increased the likelihood that a provider would receive updated variant information, as well as reducing the time associated with distributing that variant information, thus providing a more efficient process for incorporating new genetic knowledge into clinical care.
We were unable to compare directly the average time between variant change approval and provider notification between pre-GIC and post-GIC time periods because there are inadequate records of when providers contacted the LMM for variant change updates in the pre-GIC period and not all changes generated an amended report. If providers knew any of these variant changes it was either due to the provider proactively contacting the LMM for this information, or a change being updated at the LMM and the laboratory staff informing the providers either through an amended report, phone call, or e-mail. From conversations with study clinics and the LMM staff, it is our understanding that reporting these variant changes to providers was inconsistent before GIC launch and alerting. However, the average number of days between variant approval and provider notification before implementation of GIC can be estimated by the average number of days between variant change approval and provider notification at launch, although the results do represent an overestimate. We understand from the LMM that before GIC implementation it was not unusual for variant changes to have been approved for over a year without providers being notified of these changes. This was the case because standard practice at the time was that providers were responsible for checking with the laboratory to see if there were any new variant updates. However, once GIC was established, it took, on average, fewer than 5 days for providers to be notified when a variant knowledge change had been identified.
Our data also support the hypothesis that GIC would impact the process of incorporating genetic knowledge into clinical care because of the high level of provider use of GIC. Providers viewed the patient record associated with an alert for 91% of alerts sent at launch and 95% of alerts sent post-GIC. This percentage was 100% for high level alerts in the launch period and 99% in the post-GIC period. This suggests that the alerts are not only being sent but also that the providers are reviewing the alerts and are able to consider their relevance in the care of their patients. Our data also show that users clicked into the patient record to view a variant change even before a weekly e-mail alert was sent on medium or low category changes.
Our findings are best understood in the context of individual clinic workflow regarding GIC. We previously found that clinicians responded well to the design of the tool and were positive with regard to its value and utility.13 However, there is more to learn about how providers are using the system, in order to continue to evolve the system to maximize its value to clinicians. In particular, it will be an important next step to evaluate the effect of GIC on clinical care, as well as how it affects clinical decision-making. Such a study would also help us evaluate the way GeneInsight is integrated with other systems that support clinical processes. It might also identify ways these integrations could be enhanced to optimize further the workflow that surrounds each patient.
Given clinic differences in workflow, a few of our results merit further examination. First, as reported above and in table 2, clinic providers clicked into the patient record for 99% of high alerts. The patient record for one high alert in the post-GIC period was not viewed. However, other patient records affected by this same variant change were all viewed the same day by one clinic provider. It may be that the clinician became aware of the variant change for the patient without having to access the record directly.
On average across all clinics, it took approximately 12 days after e-mail receipt of a high alert for providers to click into the patient record. Based on follow-up conversations with staff at the study clinics we understand that this average is higher than expected due to the workflow practice at clinic B and an outlier data point at clinic C. At clinic B, only one staff member accesses GIC. Typically this individual logs into GIC to view unreviewed items approximately once every 2–3 weeks. All low, medium, and high alerts are reviewed in batches. They review each patient affected and mark reviewed on the reports and variant changes, which means they view the patient record and the individual variant interpretation page for almost every alert and every patient. Therefore, although clinic B has developed a thorough workflow that meets its needs, this workflow contributes to a longer average number of days between e-mail alerts and clicks into the patient record.
Unlike clinic B, the staff member primarily responsible for reviewing alerts at clinic C often uses the ‘display un-reviewed items only’ checkbox in order to see all of the variants that have not yet been checked off as reviewed. This process can result in patient variant change information being viewed before the weekly summary alert e-mail of medium or low category changes is sent. However, the data from clinic C also include one outlier that took 101 days between receipt of the alert and a provider clicking into the patient record. We understand from clinic staff that the variant change that triggered this alert was at a splice site, and therefore its upgraded status was not a significant change to the clinic staff's knowledge and did not have the same urgency for review as other high alerts.
Finally, it is also important to note that subsequent to the conclusion of our data collection, GIC algorithms have been updated so that they are more refined for alerting. For example, in response to provider feedback, GIC is no longer alerting on low level variant changes, although these changes continue to be available in GIC. In addition, different clinical classifications have been incorporated into the algorithm for different disease areas, resulting in a more detailed alerting system. Therefore, the classifications in figure 1, while in place during the course of our study, have been refined for improved specificity in the GIC system that is currently in use.
Limitations
This study has several limitations. As there was no systematic process of recording communications about variant updates before the launch of GIC, the average length of time between when the most recently approved category was set and when the most recently reported category change occurred is an overestimate. However, even if notification data were consistently available and this average time was considerably reduced, it would still be far greater than the few days it takes with the use of GIC.
Second, our study participants were self-selected individuals and clinics. Recruited and participating clinics were those that had already agreed to implement GIC. Therefore, our participant population may be biased in favor of clinics willing and able to integrate this tool into their practice for any reason, including familiarity with other HIT software, sufficient staff to manage the rollout, and interest in effectively integrating variant change information into their care processes.
Conclusions
As genetic knowledge continues to expand, healthcare providers will need streamlined means by which to learn of and integrate this information into the care of their patients. Our study results demonstrate that HIT systems, when well designed and implemented, have the potential to meet this important, growing need effectively, in particular for the management of variants, which is a complex problem. Further research needs to be done evaluating the impact of such tools on patient care, the effectiveness of these systems at more clinic sites, and evaluating their utility for different genetic conditions.
Supplementary Material
Acknowledgments
The authors would like to thank Sara Samaha and Samantha Baxter for their assistance in conducting this research study.
Footnotes
Collaborators: Sara Samaha and Samantha Baxter.
Contributors: ARW wrote the manuscript. PMN, LAV, SJA, HLR and DWB revised the manuscript. PMN, LAV, LPN, EHC, LJB, MV, SJA, HLR and DWB contributed to the concept and design. ARW, PMN and LAV analyzed and interpreted the data. EHC and LJB extracted the data and reviewed manuscript. All authors approved the final version.
Funding: The National Library of Medicine, National Institutes of Health (RC1LM010526) provided financial support for the conduct of the research and preparation of this manuscript. The content is the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
Competing interests: This research was conducted at Brigham and Women's Hospital, which is an affiliated entity of Partners HealthCare. Partners HealthCare licensed the GeneInsight technology involved in this study in November 2012, to a company in which it acquired 100% equity ownership. The Committee on Conflicts of Interest reviewed this research in light of Partners acquisition of this financial interest, and the committee, consistent with Partners policy, required that notice of Partners financial interest in the technology be provided to journals and in publications and presentations resulting from the research.
Ethics approval: This study was approved by the Partners Human Research Committee (PHRC) and registered with ClinicalTrials.gov (ID: NCT01225978). The PHRC granted a waiver of informed consent for study components relating to clinic provider GIC usage data, as well as for patient clinical data.
Provenance and peer review: Not commissioned; externally peer reviewed.
Contributor Information
Collaborators: Sara Samaha and Samantha Baxter
References
- 1.Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med 2008;359:2143–53 [DOI] [PubMed] [Google Scholar]
- 2.Hindorff LA, MacArthur J, Wise A, et al. A Catalog of Published Genome-Wide Association Studies 2012 http://www.genome.gov/gwastudies/ (accessed 18 Mar 2013).
- 3.Arad M, Maron BJ, Gorham JM, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med 2005;352:362–72 [DOI] [PubMed] [Google Scholar]
- 4.Bean LJ, Tinker SW, da Silva C, et al. Free the data: one laboratory's approach to knowledge-based genomic variant classification and preparation for EMR integration of genomic data. Hum Mutat 2013;34(9):1183-8. [DOI] [PubMed] [Google Scholar]
- 5.Bell CJ, Dinwiddie DL, Miller NA, et al. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 2011;3:65ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue Y, Chen Y, Ayub Q, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet 2012;91:1022–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norton N, Robertson PD, Rieder MJ, et al. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet 2012;5:167–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng L, Kavaslar N, Ustaszewska A, et al. Lack of MEF2A mutations in coronary artery disease. J Clin Invest 2005;115:1016–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunt KA, Smyth DJ, Balschun T, et al. Rare and functional SIAE variants are not associated with autoimmune disease risk in up to 66,924 individuals of European ancestry. Nature Genet 2012;44:3–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards CS, Bale S, Bellissimo DB, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet Med 2008;10:294–300 [DOI] [PubMed] [Google Scholar]
- 11.Aronson SJ, Clark EH, Babb LJ, et al. The GeneInsight suite: a platform to support laboratory and provider use of DNA-based genetic testing. Hum Mutat 2011;32:532–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aronson SJ, Clark EH, Varugheese M, et al. Communicating new knowledge on previously reported genetic variants. Genet Med 2012;14:713–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neri PM, Pollard SE, Volk LA, et al. Usability of a novel clinician interface for genetic results. J Biomed Inform 2012;45:950–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plon SE, Eccles DM, Easton D, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum Mutat 2008;29:1282–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kearney HM, Thorland EC, Brown KK, et al. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011;13:680–5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.