Abstract
Background
Active clinical decision support (CDS) delivered through an electronic health record (EHR) facilitates gene-based drug prescribing and other applications of genomics to patient care.
Objective
We describe the development, implementation, and evaluation of active CDS for multiple pharmacogenetic test results reported preemptively.
Materials and methods
Clinical pharmacogenetic test results accompanied by clinical interpretations are placed into the patient's EHR, typically before a relevant drug is prescribed. Problem list entries created for high-risk phenotypes provide an unambiguous trigger for delivery of post-test alerts to clinicians when high-risk drugs are prescribed. In addition, pre-test alerts are issued if a very-high risk medication is prescribed (eg, a thiopurine), prior to the appropriate pharmacogenetic test result being entered into the EHR. Our CDS can be readily modified to incorporate new genes or high-risk drugs as they emerge.
Results
Through November 2012, 35 customized pharmacogenetic rules have been implemented, including rules for TPMT with azathioprine, thioguanine, and mercaptopurine, and for CYP2D6 with codeine, tramadol, amitriptyline, fluoxetine, and paroxetine. Between May 2011 and November 2012, the pre-test alerts were electronically issued 1106 times (76 for thiopurines and 1030 for drugs metabolized by CYP2D6), and the post-test alerts were issued 1552 times (1521 for TPMT and 31 for CYP2D6). Analysis of alert outcomes revealed that the interruptive CDS appropriately guided prescribing in 95% of patients for whom they were issued.
Conclusions
Our experience illustrates the feasibility of developing computational systems that provide clinicians with actionable alerts for gene-based drug prescribing at the point of care.
Keywords: pharmacogenetics, electronic health record, clinical decision support, personalized medicine
Background and significance
As implementation of pharmacogenetics into routine clinical practice progresses, the use of computational clinical decision support (CDS) delivered through the electronic health record (EHR) will be essential for effective application of pharmacogenetic data to patient care. CDS provides clinicians, patients, or others with knowledge and person-specific information, intelligently filtered and presented at appropriate times to enhance health and health care.1 There are many ways to implement CDS within a clinical environment, which can be classified on the basis of their effect on clinical workflow. Passive CDS includes order sets, patient data reports, and documentation templates while active CDS includes rules and alerts usually delivered through computerized provider order entry (CPOE) or other functions of the EHR.2–4 Active rules and alerts are among the most recognized and widely used types of CDS.5 CDS has been shown to improve healthcare processes and provider performance but its implementation can be challenging.6 7
Unique aspects of pharmacogenetic data make the ability to actively deliver information through decision support warnings to clinicians at the point of care crucial.8 Genetic test results differ from other laboratory test results because they remain relevant over a patient's entire lifetime. Without effective CDS, pharmacogenetic results collected in the remote past could easily be forgotten or lost within a patient's medical record by the time a patient with a high-risk phenotype is prescribed a high-risk drug affected by that phenotype. Additionally, consensus guidelines detailing the use of pharmacogenetic data to guide prescribing are now available from the Clinical Pharmacogenetics Implementation Consortium (CPIC) of the National Institutes of Health’s Pharmacogenomics Research Network (PGRN) and the Pharmacogenomics Knowledge Base (PharmGKB).9 As additional clinically relevant genes are discovered and incorporated into clinical practice guidelines, it will become increasingly difficult, even impossible, for clinicians to remember all high-risk genes and associated drugs and to apply this information to a specific patient's genetic data and drug therapy. Clearly, computational support will be needed to help guide clinical decisions.
CDS that enables use of pharmacogenetic data each time a relevant medication is prescribed is especially important as preemptive pharmacogenetic testing is embraced. With the preemptive approach, genotyping is performed and results are placed in the patient's EHR before a relevant high-risk drug is prescribed. This approach is also advantageous as therapy does not have to be delayed while clinicians are awaiting the return of test results. The preemptive approach to implementing pharmacogenetics is compelling because over half of all primary care patients are exposed to pharmacogenetically relevant drugs10 and the approach is increasingly feasible with the availability of cost-effective array-based genotyping of hundreds of pharmacogenes from a single sample.11 Because the intent of preemptive pharmacogenetics is to perform the test once and use the results repeatedly to improve drug therapy over a patient's lifetime, active CDS can also play a role in making certain that pharmacogenetic tests have been performed before a drug is prescribed and that genetic tests are not unnecessarily duplicated.
There is relatively little published information about the development and implementation of active CDS for pharmacogenetics. A recent systematic review of CDS for personalized medicine identified only six primary research articles on CDS for pharmacogenomics.12 Three of these focused on CDS that was not implemented within the primary EHR, and one analyzed the feasibility of the computerized use of existing pharmacogenomic knowledge for CDS. In 2008, the Dutch Pharmacogenetics Working Group published their experience in developing computerized pharmacogenetic CDS that is accessible during electronic prescribing and automated medication surveillance throughout the nation.13 One recent article outlined a pilot study of the feasibility of incorporating preemptive pharmacogenetics into clinical care by creating a separate, genomics-based prescribing system used to report test results and related consultations and to provide links to additional information.14 Additionally, the Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT) project at Vanderbilt University Medical Center provides another example of preemptive implementation of pharmacogenetics.15 As part of an overall illustration of the implementation process, they describe their active CDS at the point of care to guide clopidogrel prescribing in cardiac patients genotyped for CYP2C19.
Objective
Here we describe the development, implementation, and evaluation of active CDS for multiple pharmacogenetic test results reported preemptively at St. Jude Children's Research Hospital (St. Jude).
Methods
Setting
The comprehensive pharmaceutical services at St. Jude Children's Research Hospital provide all medications to approximately 4200 patients per year, serving inpatients, the outpatient clinic, and patients requiring prescriptions at home; most patients are seen on an outpatient basis. While we have extensive clinical experience with single-gene pharmacogenetic tests,16 in May 2011 we implemented array-based pre-emptive genotyping through a clinical research protocol, St. Jude PG4KDS (http://www.stjude.org/pg4kds). In this protocol, genotyping for 225 genes is performed in a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory using the Affymetrix DMET Plus array supplemented with a CYP2D6 copy number assay.17 A subset of those 225 results are placed in the EHR, only after clear recommendations can be made for a gene–drug pair, such as through the CPIC guidelines.9 We designed an automated system to incorporate genetic results and their clinical interpretations directly into the EHR.18
Test results for each gene are reported as diplotypes using the nomenclature conventions for each gene, and these diplotypes are translated into probable phenotypes as defined by published CPIC guidelines (see online supplemental table S1). Each genotypically-determined phenotype is characterized as either routine or high-risk. High-risk phenotypes are defined as phenotypes that would require a change in drug therapy, such as dose modification or use of an alternate drug. As described below, specific CDS rules were written to link each high-risk phenotype with specific prescribing instructions for each affected drug.
Clinical decision support development and design
In 1997, St. Jude committed to transitioning to an EHR with CPOE (Millennium system, Cerner Corporation, Kansas City, Missouri, USA).19 As of 2011, St. Jude has fully implemented the EHR for all aspects of inpatient and outpatient care, including orders, documentation, laboratory, and pharmacy. CDS is used for multiple purposes throughout our EHR. Our focus has been to both optimize vendor-provided CDS, such as refining active CDS to limit alert fatigue,20 and to design and implement customized advanced passive and active decision support to prevent harm and improve care for St. Jude patients.
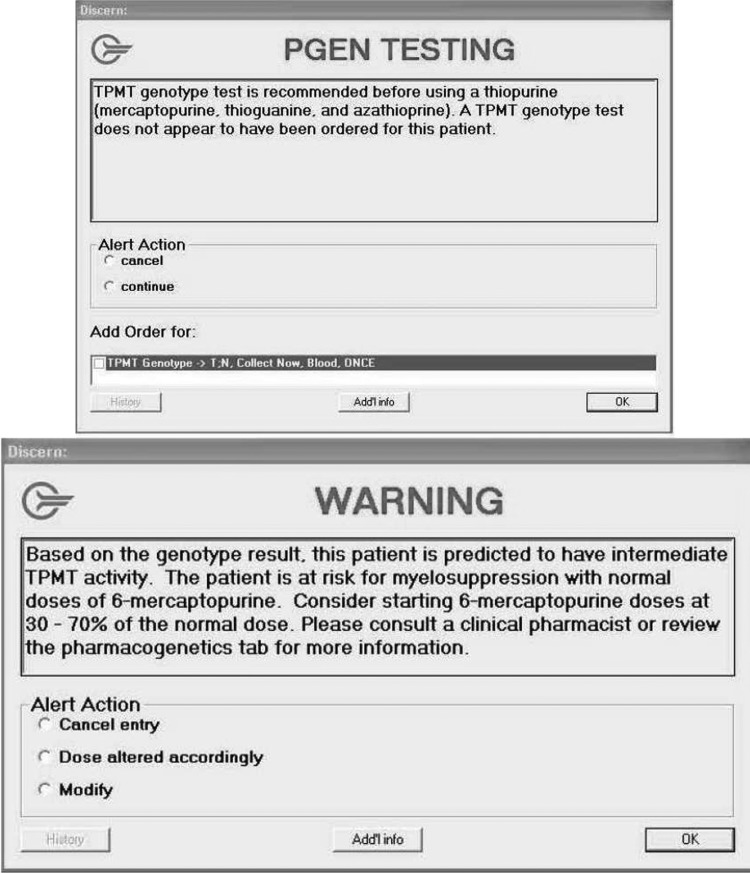
Passive CDS in the form of concise clinical pharmacogenetic test interpretations (or consultations) that are present statically in the EHR has been previously described.18 All pharmacogenetic tests at St. Jude are accompanied by a written consultation that provides a diplotype-specific interpretation, communicates the predicted phenotype, and offers general gene-based prescribing recommendations. Active CDS utilizes automated alerts to intercept the clinician at the point of care and includes both pre- and post-test alerts. Pre-test alerts are presented to clinicians when an order is placed for a high-risk drug (eg, a medication that is the subject of a published or planned CPIC guideline), but the patient does not yet have the appropriate pharmacogenetic test result in the EHR (figure 1). The wording and recommendations contained in the pre-test alerts, as well as the pre-test alert recipients, were customized depending on the medication (see online supplemental table S2). For example, pre-test alerts for thiopurines are presented on-screen to the prescriber and via email to clinical pharmacists strongly recommending testing prior to prescribing. For codeine, no on-screen pre-test alerts are delivered to prescribers to reduce alert fatigue. Instead, the CYP2D6 pre-test alert sends an email to a clinician who can order a CYP2D6 genetic test. Additionally, a pharmacogenetics pharmacist reviews all of the CYP2D6 pre-test alerts and follows up with the clinician as needed. At St. Jude, email is used for numerous other CDS alerts.
Figure 1.
Example of pre-test (top) and post-test (bottom) on-screen clinical decision-support alerts for TPMT.
Post-test alerts interrupt clinicians with prescribing authority to prompt a change in prescribing only when a high-risk drug is ordered for a patient with a high-risk phenotype (figure 1). These on-screen alerts are also presented to pharmacists on verification of the prescription. Post-test alerts are specific to each medication and provide concise pharmacotherapy recommendations for the phenotype and refer the prescriber to the consultation in the EHR or clinical pharmacist for additional information.
The text of interruptive alerts is drafted by the PG4KDS team, drawing from CPIC guidelines and other sources. The alert language is crafted to be concise and include only the most important information needed to make the clinical decision at hand. The resulting alert language is reviewed extensively by the PG4KDS team before final approval by the Pharmacogenetics Oversight Committee (see Clinical oversight and governance section below). This committee approves standard operating procedures to move a new gene–drug pair into the EHR and create CDS, and a checklist provides documentation ensuring that all steps are completed.
High-risk phenotypes are automatically posted as discrete entries in the EHR problem list based on the posting of a high-risk diplotype test result in the EHR (see online supplemental table S1). The problem list entry is the unambiguous trigger that drives the post-test alerts when high-risk drugs are prescribed. Because clinicians at our hospital can order single-gene tests for selected genes outside the PG4KDS protocol, the CDS is configured to apply to all clinical genetic test results and intercept possible duplicate genetic tests. Due to the lack of readily available Systematized Nomenclature of Medicine (SNOMED) or ICD-9 codes for pharmacogenetic test results, we developed custom problem list entries using a standardized naming convention. Our convention in building these custom codes was to list the gene (Human Genome Organization nomenclature)21 followed by the assigned phenotype (eg, CYP2D6 poor metabolizer; online supplemental table S1). Wherever possible, phenotype designation is consistent with CPIC guidelines.22–29 Automated emails are sent to each affected patient's primary physician and nurse practitioner when high-risk phenotypes are added to the problem list.
Clinical oversight and governance
To provide formal governance of pharmacogenetic CDS and other aspects of clinical pharmacogenetics at St. Jude, a multidisciplinary pharmacogenetics oversight committee was established in 2011. The committee comprises physicians of different specialties, clinical pharmacists, pathology representatives including lab personnel responsible for much of the genotyping, clinical informatics personnel, and an external advisor. The Pharmacogenetics Oversight Committee is a subcommittee of the hospital's Pharmacy and Therapeutics Committee, which reports to the Medical Executive Committee. To date, implementation of pharmacogenetic CDS has progressed without controversy, but this reporting structure ensures notification of the hospital's top committee for patient care policy (the Medical Executive Committee).
When adequate evidence is available through publication of CPIC guidelines or evaluation of primary literature using the criteria outlined by CPIC,9 decisions to migrate gene–drug pairs into the EHR and the corresponding use of CDS are made by the Pharmacogenetics Oversight Committee.
Alert process outcomes
Our CDS system provides event logging of each alert occurrence for prescribing or dispensing attempts. Custom retrievals for all alerts during the 18-month period of May 19, 2011 to November 25, 2012 were written to assemble the alert event data and the following corresponding data elements: alert timing (pre-test vs post-test), problem list entry, high-risk medication, role of the personnel to whom the alert was presented, and the patient's medical service. The data were examined to detect patterns in alert firing and handling, describe the volume of alerts, and assess alert effectiveness. Additionally, the data were used to identify prescribing attempts per patient in relation to the first post-test alert for TPMT and thiopurines and for CYP2D6 and codeine. Medical records were subsequently reviewed to determine whether the dose was modified or the medication was changed in compliance with on-screen alert recommendations.
Results
As of November 2012, 35 customized rules had been built and activated for two genes and eight drugs: TPMT and thiopurines (azathioprine, thioguanine, and mercaptopurine) and CYP2D6 and codeine, tramadol, amitriptyline, fluoxetine, and paroxetine (see online supplemental tables S2–S6). These drugs are commonly used in our patients, especially codeine and mercaptopurine. For example, in 2011, 18% of the 4245 patients who received medications at St. Jude received codeine and 7.5% of patients received a thiopurine. As of November 2012, 885 patients had TPMT genetic test results in the EHR (99 of whom had high-risk results, and 609 patients had CYP2D6 results (67 of whom had high-risk results). Between May 2011 and November 2012, 2658 alerts were presented to clinical personnel through either on-screen alerts or email (table 1).
Table 1.
Number of alerts presented to clinical personnel
| No. of alerts | No. of alerts | ||
|---|---|---|---|
| Pre-test alerts | 1106 | Post-test Alerts | 1552 |
| CYP2D6 | 1030 | CYP2D6 | 31 |
| Amitriptyline | 40 | Amitriptyline | 6 |
| Codeine | 953 | Codeine | 9 |
| Fluoxetine | 14 | Fluoxetine | 0 |
| Paroxetine | 1 | Paroxetine | 16 |
| Tramadol | 8 | Tramadol | 0 |
| Unknown CYP2D6* | 14 | Other CYP2D6 | 0 |
| TPMT | 76 | TPMT | 1521 |
| Azathioprine | 2 | Azathioprine | 0 |
| Mercaptopurine | 64 | Mercaptopurine | 1517 |
| Thioguanine | 10 | Thioguanine | 4 |
Evaluation period: May 19, 2011 to Nov 25, 2012.
*Pre-test alerts where the specific triggering drug (amitriptyline, codeine, fluoxetine, paroxetine, or tramadol) could not be recorded by the alert logging system. These cases were evaluated manually.
Of those 2658 total alerts, 1628 were presented on-screen to providers during prescribing or to pharmacists when the orders were processed for dispensing. The pre-test alerts were issued 1106 times (76 times for thiopurines and 1030 times for various drugs affected by CYP2D6), and the post-test alerts were issued 1552 times (1521 times for TPMT and 31 times for CYP2D6). Most pre-test alerts for CYP2D6 involved orders for codeine (93%) and almost all post-test alerts (99%) for TPMT were prompted by prescription of mercaptopurine. Most on-screen post-test alerts were viewed by attending physicians (n=651 alerts), followed by pharmacists (n=459) and nurse practitioners and physician assistants (n=454 alerts) (table 2).
Table 2.
Type of provider presented with on-screen clinical decision support (CDS) alerts
| Type of Provider | No. of Alerts | No. of Providers | Mean no. of Alerts per Provider | Median no. of Alerts per Provider |
|---|---|---|---|---|
| Attending physician | 651 | 17 | 38.3 | 37 |
| Oncology fellow/medical resident | 62 | 13 | 4.8 | 3 |
| Dentist | 2 | 2 | 1 | 1 |
| Nurse practitioner/physician assistant | 454 | 16 | 28.4 | 10.5 |
| Pharmacist | 459 | 34 | 13.5 | 6.5 |
Evaluation period: May 19, 2011 to Nov 25, 2012.
Most alerts (n=1732, 65%) were electronically issued to clinicians of patients on the leukemia/lymphoma service, likely related to the heavy use of mercaptopurine as a backbone for the treatment of acute lymphoblastic leukemia (table 3).30–33
Table 3.
Clinical decision support (CDS) alerts categorized by patient medical service at time of alert
| Service | Pre-Test | Post-Test | Total (% of total) |
|---|---|---|---|
| Hematology | 76 | 0 | 76 (2.9) |
| Leukemia/lymphoma | 223 | 1509 | 1732 (65.2) |
| Neuro-oncology | 298 | 15 | 313 (11.8) |
| Radiation oncology | 90 | 0 | 90 (3.4) |
| Solid tumor | 119 | 0 | 119 (4.5) |
| Surgery | 113 | 27 | 140 (5.3) |
| Transplant | 164 | 0 | 164 (6.2) |
| Other* | 23 | 1 | 24 (0.9) |
| Total | 1106 | 1552 | 2658 |
Evaluation period: May 19, 2011 to Nov 25, 2012.
*Comprising after-completion-of-therapy, immunology, and infectious disease services.
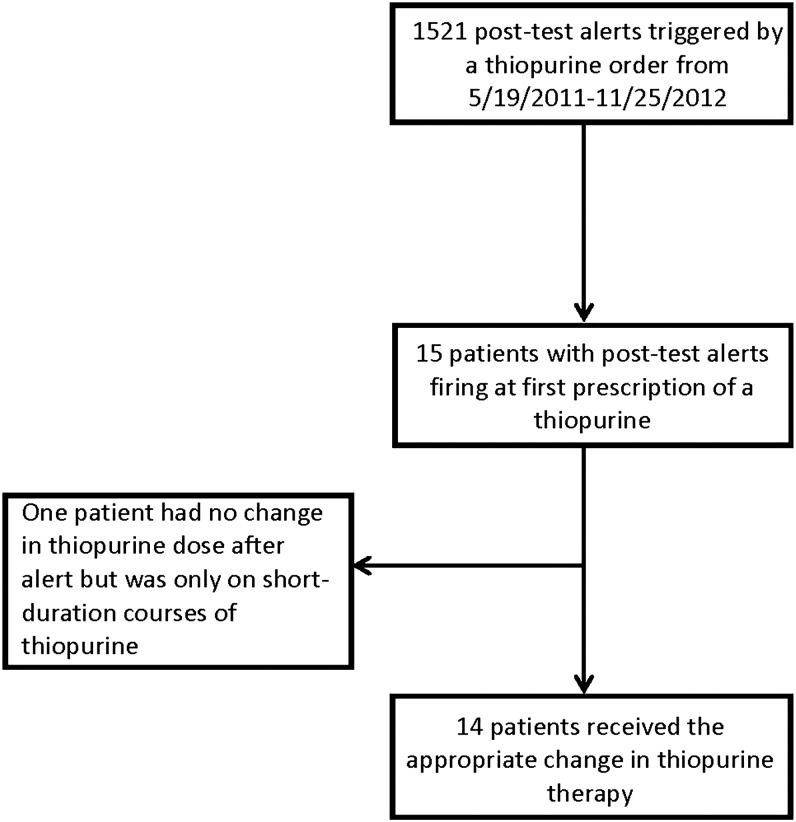
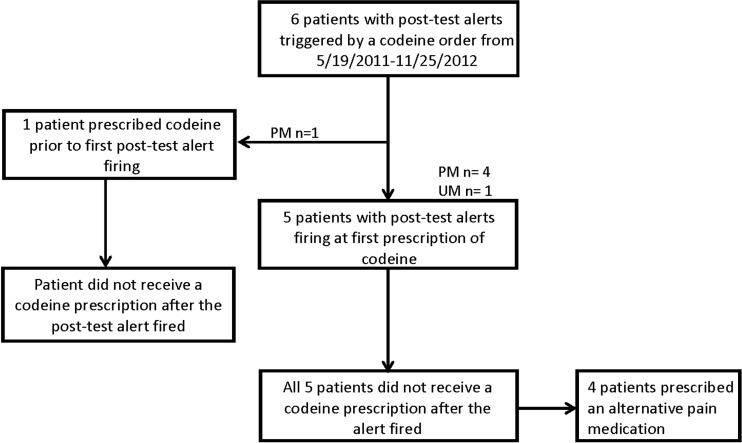
Fifteen patients had a TPMT post-alert issued at the first prescription of a thiopurine during the evaluation period (figure 2). All of the patients were being treated for either leukemia or lymphoma and had a high-risk phenotype of intermediate TPMT activity. Of those, 14 received the appropriate change in thiopurine therapy. The one patient who did not have a thiopurine dose reduction after the first interruptive alert was on a non-St. Jude protocol in which a thiopurine was used for a very short duration (thioguanine 50 mg/m2 per dose twice a day for 4 days). For the codeine post-test alerts, none of the six patients who were reported as either poor or ultrarapid metabolizers for CYP2D6 received codeine after the post-test alert (figure 3). When the alert outcome data for codeine and thiopurines were combined, 19 out of 20 (95%) patients who had a post-test alert at the time of the first prescription received the appropriate change in therapy as guided by the on-screen alert.
Figure 2.
Thiopurine prescribing outcomes after interruptive clinical decision support alerts during an 18-month period (May 2011 to November 2012).
Figure 3.
Codeine prescribing outcomes after interruptive clinical decision support alerts during an 18-month period (May 2011 to November 2012). UM, ultrarapid metabolizer; PM, poor metabolizer.
Discussion
Although it is crucial to include pharmacogenetic test interpretation in the EHR as passive CDS,18 interruptive point-of-care alerts are necessary to ensure that high-risk pharmacogenetic patient characteristics are not overlooked when high-risk medications are prescribed or dispensed.34 Our experience illustrates the feasibility of developing computation-based systems that provide clinicians with actionable, real-time alerts for gene-based drug prescribing at the point of care. Alert language is designed to provide guidance on drug dosing or choice of drugs and the content reflects evidence-based CPIC guidelines that provide specific, peer-reviewed recommendations for drug–gene pairs. Each alert also refers clinicians to other resources: additional consultation with pharmacists, the Pharmacogenetics Tab in the EHR, or the PG4KDS web page (http://www.stjude.org/pg4kds), so that clinicians can learn more about the relevance of pharmacogenetic results for high-risk patients at the time of prescribing. Additionally, because our approach relies on core CDS functions from a widely used commercial EHR (Cerner), our methods to implement pharmacogenetic CDS can be transferred to other institutions using an EHR.
Alert fatigue is a well recognized dilemma with computerized medical systems, especially associated with active CDS.20 35 36 When too many alerts are presented to clinicians, alerts may be ignored or overridden, subsequently diminishing the patient safety benefits of CDS. In a review of 17 published studies, 49–96% of medical alerts were overridden, leading to inefficiency and frustration.20 In an effort to prevent alert fatigue, we targeted alerts only for the relatively rare event of prescribing a high-risk drug to a patient with a high-priority phenotype and we designed the alerts to present only the most crucial information (with concise dosing recommendations and links to additional educational resources). We created alerts that were very specific for each high-risk phenotype and associated high-risk drugs. For example, for TPMT, only approximately 10% of patients have a diplotype that triggers an interruptive alert if thiopurines are prescribed; without tailoring alerts to diplotype, the number of patients who would theoretically prompt an alert is increased 10-fold.
Also, instead of creating a single, generic warning for thiopurine dosing in patients with intermediate or low TPMT activity (as in the SNOMED diagnostic code of ‘TPMT deficiency’), we created different post-test alerts for each of the three thiopurines, offering dosing information specific for the drug and the predicted TPMT activity (intermediate, possible intermediate, and deficient (low) TPMT activity)—thus creating nine different alerts. This is more work for the informatics team, but less work for clinicians at the point of care who must process multiple alerts every day. While we carefully considered limiting the number of patient-specific alerts for high-risk TPMT phenotypes by individual prescriber to one per patient per specified time period (eg, one per day or per week), feedback from prescribers indicated a desire to continue receiving alerts with each prescribing attempt for these high-risk patients, and that these targeted warnings associated with high-risk pharmacogenetic results were considered the most highly valued of all CDS warnings.
We translated high-risk diplotypes into problem list entries and linked interruptive CDS to the problem list entry, rather than to the diplotype results themselves. This approach creates a high-risk diagnosis that serves as ‘shorthand’ to remind clinicians that the patient is at high risk of undesired drug effects with at least some medications. Second, it provides a logical way of summarizing multi-gene high-risk pharmacogenetic diagnoses (eg, the combination of CYP2C9 and VKORC1 inactivating variants could be summarized as ‘warfarin high sensitivity’ without having to deduce this from the raw genetic test results). Third, it will become more common for patients to have genetic test results generated from multiple laboratories, perhaps even from direct-to-consumer laboratories, as well as from phenotypic tests, and thus there must be a method for assigning pharmacogenetic diagnoses manually (if necessary) that synthesizes data from a variety of sources. It should be noted that this approach highlights the need for standardized pharmacogenetic diagnostic codes, and the time to develop such terms is now. In this regard, Logical Observation Identifiers Names and Codes (LOINC) codes have been developed (table 4) that should be considered as modifiers for relevant genes to create useful diagnostic codes using a standardized system.37–39
Table 4.
Potential derivation of standardized pharmacogenetic diagnostic codes from LOINC codes
| LOINC answer text | LOINC code | Possible pharmacogenetic problem list entry: CYP2D6* | Possible pharmacogenetic problem list entry: CYP2C19* |
|---|---|---|---|
| Ultrarapid metabolizer | LA10315-2 | CYP2D6 ultrarapid metabolizer | CYP2C19 ultrarapid metabolizer |
| Extensive metabolizer | LA10316-0 | CYP2D6 extensive metabolizer | CYP2C19 extensive metabolizer |
| Intermediate metabolizer | LA10317-8 | CYP2D6 intermediate metabolizer | CYP2C19 intermediate metabolizer |
| Poor metabolizer | LA9657-3 | CYP2D6 poor metabolizer | CYP2C19 poor metabolizer |
| Inconclusive | LA9663-1 | CYP2D6 inconclusive | CYP2C19 inconclusive |
*Combined LOINC code with HGNC gene name.
LOINC, Logical Observation Identifiers Names and Codes.
Because the field of pharmacogenetics is constantly evolving, the scalability of active CDS is important. In our system, CDS can easily be added for new drugs affected by already implemented genes. For example, we first implemented the gene–drug pair of CYP2D6 and codeine. After more evidence became available for amitriptyline, paroxetine, and fluoxetine with CYP2D6 genotype, we implemented CDS for those medications prospectively as well as retrospectively for patients who already had pharmacogenetic results in the EHR for CYP2D6. In the latter case, we performed CDS retrievals to assess whether any patient with high-risk CYP2D6 phenotypes had been prescribed one of the ‘new’ high-risk medications and notified physicians of any potential problems. We also have the ability to update any CDS language as new evidence or safety warnings evolve, such as the new Food and Drug Administration boxed warning for codeine in children.40 Due to this boxed warning, we also anticipate modifying our existing CYP2D6 pre-test alerts to include additional on-screen alerts to prescribers in the future for codeine and other opioids. Our approach is to implement new gene–drug pairs based on the level of evidence (generally as supported by CPIC guidelines), regardless of the frequency of the high-risk diplotype or the frequency of use of affected drugs. The strategies we developed for pharmacogenetic CDS could be readily applied at many institutions. As further data emerge in the future, our approach will also accommodate more complex pharmacogenetic CDS, such as alerts that incorporate multiple genes or drug interactions.41
Conclusion
We have implemented an extensible CDS system for delivering interruptive point-of-care pharmacogenetic alerts based on classifying genotype results into phenotypic diagnoses. The system is designed to minimize alert fatigue and to prevent inappropriate prescribing and dispensing decisions. Analysis of alert intervention outcomes revealed that the interruptive CDS appropriately changed prescribing per the alert recommendation in 95% of patients for whom they were issued during our 18-month review period.
Supplementary Material
Acknowledgments
The authors thank the participating clinical and laboratory staff, patients, and their parents, and especially our department research nurses, Sheri Ring, Lisa Walters, Terri Kuehner, Paula Condy, Margaret Edwards, Melinda Wood, and Shannon Gibbs.
Footnotes
Contributors: GCB, KRC, MRW, CEH, JKH, DKB, NMK, WY, SJC, SCH, WEE, UB, MVR, and JMH contributed to conception and design. GCB, JMH, MRW, KRC, RRF, and MVR were involved in analysis and interpretation of data. GCB, MVR, KRC, JMH, MRW, and RRF contributed to drafting the article or revising it critically for important intellectual content. All authors gave final approval of the version to be published.
Funding: Supported by NCI grants CA 36401, CA 21765, NIH/NIGMS Pharmacogenomics Research Network (U01 GM92666, U19 GM61388, U01 HL105918), and by the American Lebanese Syrian Associated Charities (ALSAC).
Competing interests: MVR and WEE receive a portion of the income St. Jude receives from licensing patent rights related to TPMT.
Ethics approval: St. Jude Children's Research Hospital Institutional Review Board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Osheroff JA, Teich JM, Middleton B, et al. A roadmap for national action on clinical decision support. J Am Med Inform Assoc 2007;14:141–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson JE, Ash JS, Sittig DF, et al. Multiple perspectives on the meaning of clinical decision support. AMIA Annual Symposium Proceedings; American Medical Informatics Association, 2010 [PMC free article] [PubMed] [Google Scholar]
- 3.Ash JS, Sittig DF, Guappone KP, et al. Recommended practices for computerized clinical decision support and knowledge management in community settings: a qualitative study. BMC Med Inform Decis Mak 2012;12:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Troiano D, Jones MA, Smith AH, et al. The need for collaborative engagement in creating clinical decision-support alerts. Am J Health-Syst Pharm 2013;70:150–3 [DOI] [PubMed] [Google Scholar]
- 5.Kaushal R, Shojania KG, Bates DW. Effects of computerized physician order entry and clinical decision support systems on medication safety: a systematic review. Arch Intern Med 2003;163:1409–16 [DOI] [PubMed] [Google Scholar]
- 6.Bright TJ, Wong A, Dhurjati R, et al. Effect of clinical decision-support systems: a systematic review. Ann Intern Med 2012;157:29–43 [DOI] [PubMed] [Google Scholar]
- 7.Jaspers MW, Smeulers M, Vermeulen H, et al. Effects of clinical decision-support systems on practitioner performance and patient outcomes: a synthesis of high-quality systematic review findings. J Am Med Inform Assoc 2011;18:327–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther 2011;89:379–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin Pharmacol Ther 2011;89:464–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans WE, Crews KR, Pui C-H. A healthcare system perspective on implementing genomic medicine: pediatric acute lymphoblastic leukemia as a paradigm. Clin Pharmacol Ther 2013;94:224–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crews KR, Hicks JK, Pui CH, et al. Pharmacogenomics and individualized medicine: translating science into practice. Clin Pharmacol Ther 2012;92:467–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welch BM, Kawamoto K. Clinical decision support for genetically guided personalized medicine: a systematic review. J Am Med Inform Assoc 2013;20:388–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swen J, Wilting I, De Goede A, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther 2008;83:781–7 [DOI] [PubMed] [Google Scholar]
- 14.O'Donnell PH, Bush A, Spitz J, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 2012;92:446–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulley J, Denny J, Peterson J, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 2012;92:87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crews KR, Cross SJ, McCormick JN, et al. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am J Health Syst Pharm 2011;68:143–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez C, Smith C, Yang W, et al. Concordance of DMET Plus genotyping results with those of orthogonal genotyping methods. Clin Pharmacol Ther 2012;92:360–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks JK, Crews KR, Hoffman JM, et al. A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther 2012;92:563–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffman JM, Baker DK, Howard SC, et al. Safe and successful implementation of CPOE for chemotherapy at a children's cancer center. J Natl Compr Canc Netw 2011;9:S-36–50 [DOI] [PubMed] [Google Scholar]
- 20.van der Sijs H, Aarts J, Vulto A, et al. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc 2006;13:138–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.HUGO gene nomenclature committee. http://www.genenames.org (accessed 29 Mar 2013)
- 22.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing. Clin Pharmacol Ther 2011;89:387–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott S, Sangkuhl K, Gardner E, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin Pharmacol Ther 2011;90:328–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther 2012;91:321–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin MA, Klein TE, Dong BJ, et al. Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin Pharmacol Ther 2012;91:734–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilke RA, Ramsey LB, Johnson SG, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin Pharmacol Ther 2012;92:112–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hershfield MS, Callaghan JT, Tassaneeyakul W, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for human leukocyte antigen-B genotype and allopurinol dosing. Clin Pharmacol Ther 2013;93:153–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther 2011;90:625–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hicks JK, Swen JJ, Thorn CF, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Tricyclic Antidepressants. Clin Pharmacol Ther 2013;93:402–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pui C-H, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med 2006;354:166–78 [DOI] [PubMed] [Google Scholar]
- 31.Pui C-H, Mullighan CG, Evans WE, et al. Pediatric acute lymphoblastic leukemia: where are we going and how do we get there? Blood 2012;120:1165–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmiegelow K, Forestier E, Hellebostad M, et al. Long-term results of NOPHO ALL-92 and ALL-2000 studies of childhood acute lymphoblastic leukemia. Leukemia 2010;24:345–54 [DOI] [PubMed] [Google Scholar]
- 33.Silverman LB, Stevenson KE, O'Brien JE, et al. Long-term results of Dana-Farber Cancer Institute ALL Consortium protocols for children with newly diagnosed acute lymphoblastic leukemia (1985–2000). Leukemia 2009;24:320–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teng K, Eng C, Hess CA, et al. Building an innovative model for personalized healthcare. Cleve Clin J Med 2012;79 (Suppl 1):S1–9 [DOI] [PubMed] [Google Scholar]
- 35.Isaac T, Weissman JS, Davis RB, et al. Overrides of medication alerts in ambulatory care. Arch Intern Med 2009;169:305–11 [DOI] [PubMed] [Google Scholar]
- 36.Lin CP, Payne TH, Nichol WP, et al. Evaluating clinical decision support systems: monitoring CPOE order check override rates in the Department of Veterans Affairs’ Computerized Patient Record System. J Am Med Inform Assoc 2008;15:620–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forrey AW, McDonald CJ, DeMoor G, et al. Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem 1996;42:81–90 [PubMed] [Google Scholar]
- 38.Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifier Names and Codes (LOINC) vocabulary. J Am Med Inform Assoc 1998;5:276–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem 2003;49:624–33 [DOI] [PubMed] [Google Scholar]
- 40.FDA Drug Safety Communication: Safety review update of codeine use in children; new Boxed Warning and Contraindication on use after tonsillectomy and/or adenoidectomy. http://www.fda.gov/Drugs/DrugSafety/ucm339112.htm (accessed 28 Mar 2013)
- 41.Johnson JA, Klein TE, Relling MV. Clinical implementation of pharmacogenetics: more than one gene at a time. Clin Pharmacol Ther 2013;93:384–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.