Abstract
OBJECTIVE
To determine the source of a cluster of Klebsiella oxytoca isolates cultured from peritoneal fluid of three patients with cirrhosis on a single day.
DESIGN
Outbreak investigation and before-after study.
SETTING
A Veterans Affairs Medical Center.
METHODS
Epidemiologic investigation, analysis of antimicrobial susceptibility testing results and molecular typing of K. oxytoca isolates with repetitive sequence-based PCR (rep-PCR), review of microbiology laboratory procedures for processing peritoneal fluid cultures, and comparison of peritoneal fluid contamination rates 18 months before and after modification of laboratory procedures for culturing peritoneal fluid.
RESULTS
Each of the peritoneal fluid samples that grew K. oxytoca was inoculated into blood culture bottles by different clinicians at different hospital locations. None of the patients had clinical findings suggestive of peritonitis or elevated polymorphonuclear cell counts in peritoneal fluid (range, 3 to 25 cells/ µL). Molecular typing with rep-PCR demonstrated that the K. oxytoca isolates were genetically related (>95% similarity). Laboratory procedures included the routine addition of a culture medium supplement of yeast extract and dextrose from a multidose vial into blood culture bottles with peritoneal fluid. After discontinuing use of the culture medium supplement, there was a marked reduction in the number of peritoneal fluid cultures deemed as contaminants (14.3 % versus 0.9%, respectively; P<0.001).
CONCLUSION
A pseudo-outbreak of K. oxytoca peritonitis and high rates of contamination of peritoneal fluid were attributable to contamination of a multidose culture medium supplement. This report highlights the importance of discouraging the use of multidose vials in all clinical settings.
Keywords: multidose-vials, pseudo-outbreak, contamination
INTRODUCTION
Correct identification and early treatment of bacterial infection is crucial for the survival of patients with cirrhosis1. The diagnosis of spontaneous bacterial peritonitis (SBP), resulting from intestinal bacterial translocation in patients with cirrhosis, is often clinically elusive and rests on the white blood cell count and microbiologic analysis of peritoneal fluid. The presence of polymorphonuclear (PMN) white blood cells has demonstrated great sensitivity in establishing SBP. Conversely, the growth of bacteria from peritoneal fluid with normal PMN counts, especially when obtained from asymptomatic patients, can be interpreted as a contaminant2.
Contamination of cultures may occur at any point, including processing in the microbiology laboratory. The systematic occurrence of contamination may lead to a pseudo-outbreak, or a related cluster of false infections3. Bacterial pseudo-outbreaks due to contamination of cultures are often linked to the environment or equipment in the microbiology laboratory, although in some instances it has been difficult to identify the exact source for the contamination of cultures4–6.
Here, we describe a pseudo-outbreak of spontaneous bacterial peritonitis due to Klebsiella oxytoca that we suspect was caused by contamination of multidose vials of a supplement added to blood culture bottles to increase the yield of peritoneal fluid cultures.
METHODS
Setting
The Louis Stokes Cleveland Veterans Affairs Medical Center is a 225-bed acute-care facility, associated with a 165-bed long-term care facility and thirteen outpatient clinics, serving >100,000 patients/yearly. Approximately 300 peritoneal fluid specimens from patients with cirrhosis or with intra-abdominal infections are analyzed every year in the microbiology laboratory. Since 2001, only ten cultures of peritoneal fluid have demonstrated growth of K. oxytoca. On February 8, 2011, we were alerted by the microbiology laboratory about peritoneal fluid specimens from three different patients growing Gram-negative rods, eventually identified as K. oxytoca. Given that unusual coincidence, we initiated an outbreak investigation.
Review of cases
Spontaneous bacterial peritonitis (SBP) was defined as a peritoneal fluid analysis with a polymorphonuclear (PMN) cell count > 250 cells/µL2, with or without bacterial growth in peritoneal fluid cultures, and no evidence of secondary peritonitis, defined as an intra-abdominal infection extending to the peritoneal space due to perforation of a hollow viscus, bowel necrosis, or associated with the presence of a peritoneal dialysis catheter7. Contamination of peritoneal fluid cultures was defined as growth of bacteria in peritoneal fluid culture with peritoneal fluid PMN cell count < 250 cells/µL, and the absence of documented fever, abdominal pain, nausea, or vomiting at the time of the procedure. Demographic characteristics, reason for hospital admission, and nature of underlying chronic liver disease were extracted from the medical record.
Characterization of bacterial isolates
The VITEK2 system (bioMerieux, Inc., Durham, NC) was used for bacterial identification and antimicrobial susceptibility testing. Results were interpreted according to breakpoints defined by the Clinical and Laboratory Standards Institute (CLSI)8. Molecular typing was performed using repetitive sequence-based PCR (rep-PCR) in four isolates from peritoneal fluid (one each from the first two patients, and two from the third patient), as well as in two isolates from urine cultures from unrelated patients, all obtained during the same week in February 2011. Genomic DNA was extracted from bacterial isolates using the UltraClean™ Microbial DNA Isolation Kit (MoBio Laboratories, Carlsbad, CA). PCR amplification was performed using the DiversiLab® (bioMérieux, Athens, GA) Klebsiella fingerprinting kit, and rep-PCR amplicons were separated by electrophoresis on microfluidic chips and analyzed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The resulting band patterns were compared by Pearson’s correlation, and isolates that were >95% similar were considered genetically related9.
Investigation of microbiology laboratory procedures
To detect potential mechanisms of contamination, the location and form of collection of peritoneal fluid cultures was noted. Procedure notes were reviewed and, when necessary, the practitioner was contacted. Microbiology laboratory procedures pertaining to peritoneal fluid cultures were reviewed in detail and laboratory personnel were interviewed. To assess the possibility of an environmental source, we cultured medium supplements used in the processing of peritoneal fluid cultures.
Frequency of peritoneal fluid contamination before and after discontinuation of the multidose culture medium supplement
We reviewed the microbiology laboratory records to determine the frequency of occurrence of K. oxytoca in peritoneal fluid. We compared the results of peritoneal fluid cultures coupled with cell count analysis in the 18-month periods before and after discontinuation of the multidose culture medium supplement. The proportion of positive peritoneal cultures, discriminated between SBP, secondary peritonitis, and contamination (as defined above) was noted. Information collected on each specimen included hospital location, date and time of collection, result of peritoneal fluid culture, peritoneal fluid white blood cell count, associated symptoms, intra-abdominal infection, and underlying liver disease.
Statistical analysis
Fisher’s exact test was used to compare proportions of categorical variables. Analyses were performed using R version 2.15.2. P values <0.05 were considered significant.
RESULTS
Review of cases
In all three cases, peritoneal fluid was obtained in patients with underlying cirrhosis and ascites. SBP was considered unlikely based upon their clinical features and peritoneal fluid parameters (Table 1).
TABLE 1.
Cell count & differential and Gram stain of peritoneal fluid for the 3 case patients with cultures positive for Klebsiella oxytoca
| Case 1 | Case 2 | Case 3 | |
|---|---|---|---|
| Red blood cell count - cells/µl | 1000 | >1000 | 540 |
| White blood cell count - cells/µl | 159 | 162 | 75 |
| Polymorphonuclear cell percentage - cells/µl (%) | 3 (2) | 8 (5) | 22 (29) |
| Gram stain | No cells or organisms | No cells or organisms | No cells or organisms |
Patient 1, a 61 year-old man, was admitted to the hospital because of maroon stools and coffee ground emesis. He did not have fever or abdominal pain. He had hepatitis C virus infection and a history of alcohol abuse, complicated by cirrhosis, portal hypertension and ascites. On presentation, he was afebrile and hemodynamically stable. He was anemic and had leukocytosis. He underwent paracentesis on the day of the admission in the intensive care unit (February 7, 2011). He was found to have bleeding due to esophageal varices, which were banded. The patient died a month later after recurrence of bleeding.
Patient 2, a 57 year-old man with a history of alcohol abuse, cirrhosis and portal hypertension, required paracentesis approximately every 2 weeks to alleviate ascites refractory to diuretics. He underwent paracentesis on February 7, 2011 as an outpatient in the gastroenterology procedure suite and returned home. When he was contacted to discuss the results of peritoneal fluid cultures, he denied any fever, abdominal pain or other symptoms.
Patient 3, a 42 year-old man, was admitted to the hospital because of worsening ascites, in the setting of alcohol abuse, cirrhosis and chronic portal vein thrombosis. He had previously experienced alcohol withdrawal and delirium tremens, and was actively drinking until five days prior to admission. He had leucocytosis and elevation of transaminases, but was afebrile. He had negative urine and blood cultures. In order to rule out spontaneous bacterial peritonitis, he underwent paracentesis on February 7, 2011 while hospitalized on a medical ward. He was initially treated with ceftriaxone, but it was discontinued after PMN count was found to be inconsistent with SBP.
In all cases, peritoneal fluid was directly inoculated into blood culture bottles at the bedside immediately following paracentesis in order to increase the diagnostic yield of cultures10; in each case, a different practitioner was involved. Therefore, it was deemed unlikely that contamination with K. oxytoca occurred during specimen collection. Furthermore, the patients were hospitalized in different wards, and no overlapping stays in the same inpatient wards, emergency room, imaging facilities and outpatient clinics occurred in the previous month.
Investigation of microbiology laboratory procedures
Upon review of procedures for culture processing, we learned that in all cases the same microbiology laboratory technician added BD Difco Supplement B (Becton Dickinson, Franklin Lakes, NJ) to blood culture bottles inoculated with peritoneal fluid. The supplement contains hematin, yeast extract, L-glutamine and dextrose, and its purpose is to increase the yield for fastidious organisms such as Hemophilus influenzae and Neisseria gonorrhoeae. The supplement was in a 10 mL multidose vial kept in the refrigerator at 37 °F between doses. According to the laboratory technician, a sterile syringe and needle was used to aspirate each dose of the supplement from the multidose vial after cleaning the cap with an alcohol pad. The same supplement bottle was used to inoculate each of the positive peritoneal fluid cultures. Unfortunately, by the time the outbreak was noted, the vial had been discarded and was not available for culture. Cultures of three unopened supplement bottles from the same lot were negative.
Characterization of bacterial isolates
All isolates had the same antimicrobial susceptibility profile; they were resistant to ampicillin, but susceptible to gentamicin, cephalosporins, beta-lactam/beta-lactamase inhibitor combinations, ciprofloxacin, and trimethoprim/sulfamethoxazole. Molecular typing with rep-PCR revealed that K. oxytoca isolates from peritoneal fluid from all three patients shared >95% similarity among them, and with one of the two contemporary urine isolates (Figure 1).
FIGURE 1.

Results of molecular typing with rep-PCR. P1-Kox and P2-Kox are Klebsiella oxytoca isolated from peritoneal fluid in patients 1 and 2, respectively. P3-Kox and P4-Kox were isolated from patient 3. U1-Kox and U2-Kox are K. oxytoca isolated from contemporary urine samples of unrelated patients. Based on Pearson’s correlation, K. oxytoca from all three patients are >99% similar, and >95% similar to U1-Kox.
Frequency of peritoneal fluid contamination before and after discontinuation of the multidose culture medium supplement
In the 18-month period prior to the discovery of the pseudo-outbreak, 328 peritoneal fluid samples from 146 unique patients were cultured and bacteria grew in 58 specimens from 35 different patients; Staphylococcus spp., often considered a skin contaminant, was cultured from 33 specimens. In the 18-month period after discovery of the pseudo-outbreak and discontinuation of the use of the multidose culture medium supplement, 425 peritoneal fluid specimens from 170 patients were cultured and bacteria grew in only 18 specimens; Staphylococcus spp. was cultured from one specimen. Figure 2 shows the percentage of positive cultures in 6-month intervals before versus after discontinuation of the multidose supplement, separated into those deemed to represent SBP, secondary peritonitis, and contaminants. The percentage of contaminants decreased from 47 of 328 (14%) to only 4 of 425 (0.9%) after discontinuation of the supplement (P<0.001).
FIGURE 2.
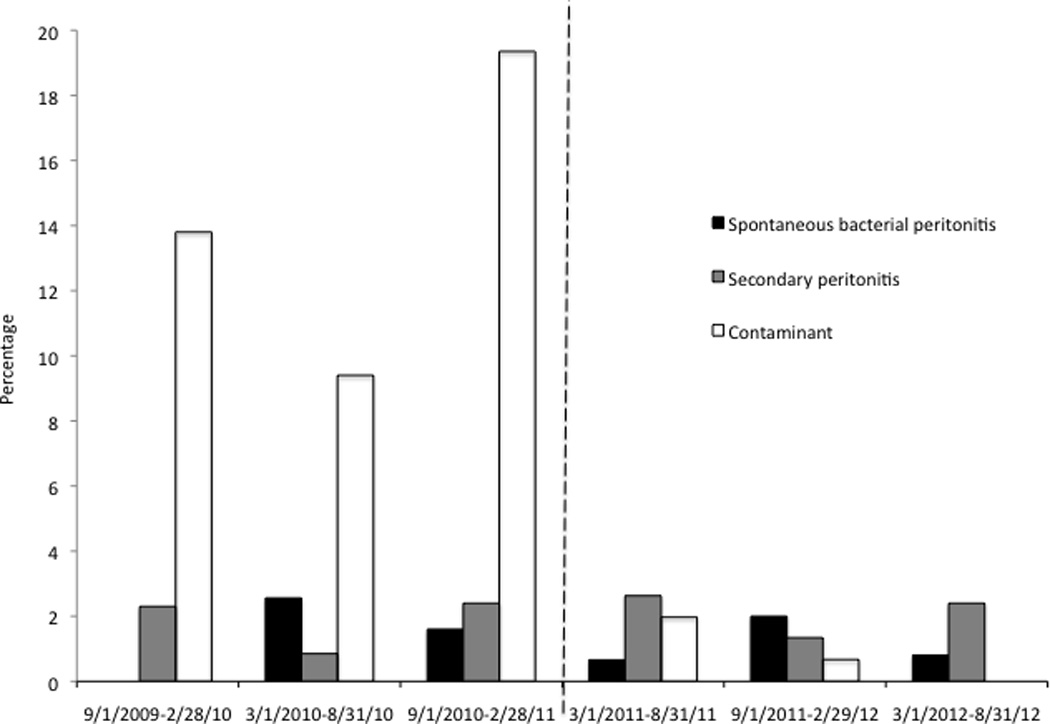
Percentage of peritoneal fluid cultures that grew bacteria, classified as spontaneous bacterial peritonitis, secondary peritonitis and contamination, from September 2009 through September 2012. The pseudo-outbreak of Klebsiella oxytoca was detected in February 2011, and the use of the culture medium supplement for fastidious organisms was discontinued in March 2011, signified by the interrupted line.
DISCUSSION
We describe a pseudo-outbreak of K. oxytoca SBP involving three patients with advanced liver disease and cirrhosis. Our findings suggest that none of the patients were truly infected, nor were they linked to a common hospital location or practitioner. Rather, their peritoneal fluid samples were inoculated in the microbiology laboratory with culture medium supplement from a multidose vial. All samples grew K. oxytoca isolates that were determined to be genetically similar by rep-PCR. We postulate that the multidose vial of culture medium supplements may have been accessed by a needle or syringe that was previously used on a sample from an infected patient, or that was inadvertently in contact with contaminated equipment or surfaces. After introduction of bacteria into the medium supplement, subsequent contamination of the three cultures with the same organism may have occurred. Unfortunately, we were unable to culture K. oxytoca from the vial, since it had been discarded.
Contamination of multidose vials has been linked to numerous true outbreaks of infection with viral and bacterial pathogens11–13. Although multidose vials may be more efficiently stored and reduce waste and cost, the Centers for Disease Control and Prevention recommends against their use, given the high risk of contamination and transmission of infection14. In the microbiology laboratory, contamination of multidose vials of culture medium supplement has led to a pseudo-outbreak of bloodstream infections involving Mycobacterium chelonae in patients with AIDS15. Patients with cirrhosis who frequently undergo investigation for SBP may represent a group at particular risk for contamination of cultures if supplements are used, as illustrated by this report and by a pseudo-outbreak of Bordetella bronchoseptica linked to the use of rabbit blood to supplement culture media16.
We suspect that the use of culture medium supplement in our microbiology laboratory contributed to multiple previous instances of contamination of peritoneal fluid cultures. After the practice of supplementing cultures to enhance growth of fastidious organisms was abandoned, contamination decreased dramatically, while the proportion of true cases of SBP remained unchanged (Figure 2). Although culture medium supplements may enhance the growth of Neisseria gonorrhea and Hemophilus influenzae from cultures of other sterile body fluids, supplementation does not appear to increase the diagnostic yield of peritoneal fluid cultures17. On the other hand, increased rates of contamination have been observed with the use of medium supplements. For instance, other investigators found a high proportion of contaminated cerebrospinal fluid cultures supplemented with Difco Supplement B (12%), similar to what we observed18.
In summary, our report demonstrates the value of periodically reviewing contamination rates and handling practices for different types of specimens in order to detect pseudo-outbreaks and other systematic errors in the microbiology laboratory. We suggest avoiding unwarranted routine microbiological analyses and unnecessary supplementation of clinical samples as a strategy to prevent contamination, false positive results, and inappropriate administration of antimicrobials. Finally, our report emphasizes the importance of eliminating or reducing as much as possible the use of multidose vials in every clinical setting, including the microbiology laboratory.
ACKNOWLEDGMENTS
Financial support. Funds and/or facilities provided by the Cleveland Department of Veterans Affairs, the Veterans Affairs Merit Review Program and the Geriatric Research Education and Clinical Center VISN 10 supported this work. This work was also supported by funds from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, under award numbers R01AI072219, R01AI063517, and R01AI100560 to R. A. B. The Cleveland Translational Science Award (UL1TR000439) supports F. P. Funding organizations were not involved in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the Veterans Administration.
Footnotes
Potential conflicts of interest. F. P. has received grant funding from STERIS Corporation. R. A. B. has received grant funding from STERIS Corporation, AstraZeneca, Pfizer and Rib-X Pharmaceuticals. C. J. D. has received grant funding from STERIS Corporation, Pfizer, Cubist, and EcoLab. All authors agree that there are no other relationships or activities that readers could perceive to influence, or that give the appearance of potentially influencing this report. All authors submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest, and the conflicts that the editors consider relevant to this article are disclosed here.
REFERENCES
- 1.Fernandez J, Gustot T. Management of bacterial infections in cirrhosis. J Hepatol. 2012;56(Suppl 1):S1–S12. doi: 10.1016/S0168-8278(12)60002-6. [DOI] [PubMed] [Google Scholar]
- 2.Wong CL, Holroyd-Leduc J, Thorpe KE, Straus SE. Does this patient have bacterial peritonitis or portal hypertension? How do I perform a paracentesis and analyze the results? JAMA. 2008;299(10):1166–1178. doi: 10.1001/jama.299.10.1166. [DOI] [PubMed] [Google Scholar]
- 3.Curran ET. Pseudo outbreaks and no-infection outbreaks. J Infect Prevention. 2013;14:108. [Google Scholar]
- 4.Aronoff DM, Thelen T, Walk ST, et al. Pseudo-outbreak of Clostridium sordellii infection following probable cross-contamination in a hospital clinical microbiology laboratory. Infect Control Hosp Epidemiol. 2010;31(6):640–642. doi: 10.1086/652774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pereira LA, Chan DS, Ng TM, et al. Pseudo-outbreak of Rhizobium radiobacter infection resulting from laboratory contamination of saline solution. J Clin Microbiol. 2009;47(7):2256–2259. doi: 10.1128/JCM.02165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blossom DB, Alelis KA, Chang DC, et al. Pseudo-outbreak of Mycobacterium abscessus Infection Caused by laboratory contamination. Infect Control Hosp Epidemiol. 2008;29(1):57–62. doi: 10.1086/524328. [DOI] [PubMed] [Google Scholar]
- 7.Solomkin JS, Mazuski JE, Bradley JS, et al. Diagnosis and management of complicated intra-abdominal infection in adults and children: guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2010;50(2):133–164. doi: 10.1086/649554. [DOI] [PubMed] [Google Scholar]
- 8.Clinical Laboratory Standard Institute. CLSI document M100-S20. Wayne, PA: 2010. Performance standards for antimicrobial susceptibility testing. [Google Scholar]
- 9.Endimiani A, Hujer AM, Perez F, et al. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother. 2009;63(3):427–437. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bobadilla M, Sifuentes J, Garcia-Tsao G. Improved method for bacteriological diagnosis of spontaneous bacterial peritonitis. J Clin Microbiol. 1989;27(10):2145–2147. doi: 10.1128/jcm.27.10.2145-2147.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macedo de Oliveira A, White KL, Leschinsky DP, et al. An outbreak of hepatitis C virus infections among outpatients at a hematology/oncology clinic. Ann Intern Med. 2005;142(11):898–902. doi: 10.7326/0003-4819-142-11-200506070-00007. [DOI] [PubMed] [Google Scholar]
- 12.Blossom DB, Kallen AJ, Patel PR, et al. Outbreak of adverse reactions associated with contaminated heparin. N Engl J Med. 2008;359(25):2674–2684. doi: 10.1056/NEJMoa0806450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattner F, Gastmeier P. Bacterial contamination of multiple-dose vials: a prevalence study. Am J Infect Control. 2004;32(1):12–16. doi: 10.1016/j.ajic.2003.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Siegel JD, Rhinehart E, Jackson M, Chiarello L. 2007 Guideline for Isolation Precautions: Preventing Transmission of Infectious Agents in Health Care Settings. American journal of infection control. 2007;35(10 Suppl 2):S65–S164. doi: 10.1016/j.ajic.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashford DA, Kellerman S, Yakrus M, et al. Pseudo-outbreak of septicemia due to rapidly growing mycobacteria associated with extrinsic contamination of culture supplement. J Clin Microbiol. 1997;35(8):2040–2042. doi: 10.1128/jcm.35.8.2040-2042.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calfee DP, Kornblum J, Jenkins SG. Pseudo-outbreak of Bordetella bronchiseptica infection associated with contaminated rabbit blood used as a broth culture supplement. Infect Control Hosp Epidemiol. 2007;28(6):758–760. doi: 10.1086/516796. [DOI] [PubMed] [Google Scholar]
- 17.Fuller DD, Davis TE, Kibsey PC, et al. Comparison of BACTEC Plus 26 and 27 media with and without fastidious organism supplement with conventional methods for culture of sterile body fluids. J Clin Microbiol. 1994;32(6):1488–1491. doi: 10.1128/jcm.32.6.1488-1491.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ajello GW, Feeley JC, Hayes PS, et al. Trans-isolate medium: a new medium for primary culturing and transport of Neisseria meningitidis, Streptococcus pneumoniae and Haemophilus influenzae. J Clin Microbiol. 1984;20(1):55–58. doi: 10.1128/jcm.20.1.55-58.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]


