Abstract
Unlike humans, certain adult vertebrates such as newts and zebrafish possess extraordinary abilities to functionally regenerate lost appendages and injured organs, including cardiac muscle. Here, we present new evidence that a remodeled extracellular matrix (ECM) directs cell activities essential for cardiac muscle regeneration. Comprehensive mining of DNA microarrays and Gene Ontology term enrichment analyses for regenerating newt and zebrafish hearts revealed that distinct ECM components and ECM-modifying proteases are among the most significantly enriched genes in response to local injury. In contrast, data analyses for mammalian cardiac injury models indicated that inflammation and metabolic processes are the most significantly activated gene groups. In the regenerating newt heart, we show dynamic spatial and temporal changes in tenascin-C, hyaluronic acid, and fibronectin ECM distribution as early as 3 days postamputation. Linked to distinct matrix remodeling, we demonstrate a myocardium-wide proliferative response and radial migration of progenitor cells. In particular, we report dramatic upregulation of a regeneration-specific matrix in the epicardium that precedes the accumulation and migration of progenitor cells. For the first time, we show that the regenerative ECM component tenascin-C significantly increases newt cardiomyocyte cell cycle reentry in vitro. Thus, the engineering of nature-tested extracellular matrices may provide new strategic opportunities for the enhancement of regenerative responses in mammals.
Keywords: heart, regeneration, extracellular matrix, newt, myocardial infarction
INTRODUCTION
The regenerative ability of the mammalian heart is very limited, with the default response to damage characterized by formation of a collagen-rich scar, causing significant myocardial stiffness with consequent decreased pumping and relaxation capacity (Burgess et al., 1996; Seccia et al., 1999; Thomas et al., 1992; Weber et al., 1994). Though some studies suggest that mammalian cardiomyocytes can undergo limited proliferation in response to cardiac damage or disease, the proliferative activity is not sufficient to overcome the fibrotic scarring associated with ischemia (Beltrami et al., 2001; Kajstura et al., 1998; Poss et al., 2002; Senyo et al., 2012). While stem cell-based therapies hold promise for functional tissue restoration after acute MI and ischemic cardiomyopathy, there are several unsolved problems, including the possible failure of cell homing/migration to the damaged myocardial tissue and inappropriate or incomplete differentiation and synchronization of transplanted cells (Ballard and Edelberg, 2007; Reffelmann et al., 2003; Strauer and Kornowski, 2003). Considering both regeneration-competent and incompetent species, we have taken a novel approach to understand how regenerative responses could be induced in damaged cardiac tissue.
The adult newt Notophthalmus viridescens possesses extraordinary abilities to regenerate lost or injured structures, including the limbs, lenses, tail, spinal cord, and heart, without scarring or impairment in the functional architecture of the tissue (Brockes, 1997; Del Rio-Tsonis et al., 1997; Lo et al., 1993; Makarev et al., 2007; Witman et al., 2011; Zukor et al., 2011). In particular, following surgical removal of approximately 20% of the ventricular apex, bleeding is arrested by the formation of a blood clot and minor myocardial contraction. Within 2 – 3 days postamputation (dpa), a dense fibrin matrix replaces the blood clot (Borchardt and Braun, 2007), and dedifferentiation of a subset of cardiomyocytes is thought to begin during this early regenerative period, as emphasized by downregulation of cardiac markers, such as α-myosin heavy chain and cardiac troponins (Bettencourt-Dias et al., 2003; Laube et al., 2006; Witman et al., 2011). Mounting evidence suggests that this cellular dedifferentiation facilitates cardiomyocyte cell cycle reentry beginning at 7 – 14 dpa with significant mitotic activity adjacent to the damaged myocardium (Bader and Oberpriller, 1979; Oberpriller and Oberpriller, 1971; Witman et al., 2011). Cell proliferation gradually subsides and the expression of mature cardiac muscle markers returns, with functional restoration of the heart ventricle by 60 dpa (Laube et al., 2006; Witman et al., 2011).
A remodeled extracellular environment is a well-known component of epimorphic limb regeneration. Previous research concerning the extracellular environment during newt limb regeneration revealed astounding similarities with ECMs critical in embryonic development (Calve et al., 2010; Gulati et al., 1983; Onda et al., 1991; Repesh et al., 1982; Tassava et al., 1996; Toole and Gross, 1971). In particular, the upregulation of hyaluronic acid (HA), tenascin-C (TNC), and fibronectin (FN) provide essential biochemical and mechanical cues that cooperatively regulate skeletal muscle regeneration (Calve et al., 2010; Calve and Simon, 2012). Differential matrix metalloproteinase (MMP) distribution and activity further contributes to the dynamics of this regenerative ECM and was shown to be essential for the regeneration of damaged limb tissue (Vinarsky et al., 2005). However, in vertebrate heart regeneration, a functional role for the ECM remained elusive. To better understand how the extracellular environment affects cardiac muscle regeneration, we performed interspecies microarray and Gene Ontology analyses and specifically investigated the composition and distribution of the ECM at the gene and protein level during newt heart regeneration, including testing the functional properties of ECM components on cardiomyocyte explants.
RESULTS
Evolutionarily conserved ECM remodeling segregates cardiac regeneration from repair
To discover regeneration-specific gene activities in cardiac muscle, we analyzed a temporal set of microarrays for regenerating newt hearts. These custom oligonucleotide arrays represent preselected cDNAs from mRNA differential display screens to enrich for regeneration-associated gene activities (Mercer et al., 2012). Approximately 20% of the distal ventricular tip was amputated, and at 3, 7, and 14 dpa the lower third of the ventricle containing the amputation plane was harvested to cover the time window thought to represent the most dynamic cell reprogramming events (Laube et al., 2006; Lepilina et al., 2006; Poss et al., 2002). Ventricular tissue of 8-10 intact and regenerating heart samples was pooled for probe making and competitive hybridization. Expression levels at each time point were normalized to the corresponding intact, unamputated tissue. We note that the Agilent microarrays contained between 7 and 23 replicate printings of the oligonucleotide set, generating expression profiles with very low average coefficients of variation (2-3% of each gene’s mean activity level; (Marx et al., 2007) and high reproducibility as determined by qRT-PCR (Mercer et al., 2012).
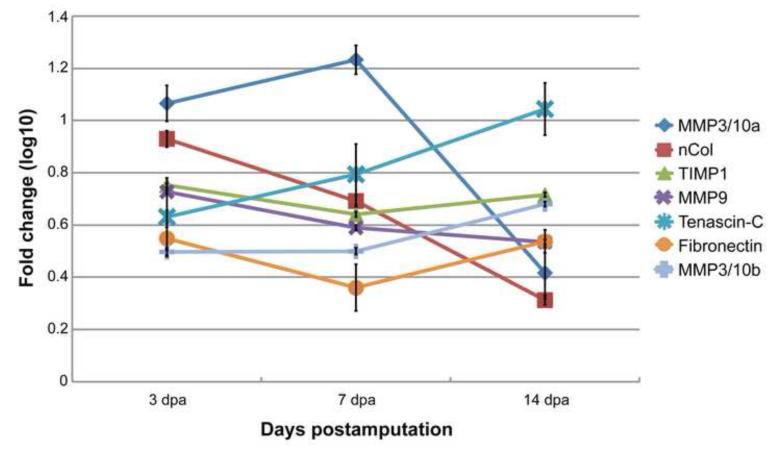
GeneSpring bioinformatics software was employed to identify 349 unique genes with differential expression (≥2-fold change relative to control) for at least one time point during heart regeneration. Pairwise comparisons revealed that regenerating hearts at 3 dpa displayed the highest number of differentially expressed genes while at 7 and 14 dpa comparable levels of differential gene expression were detected. Overall, gene activation was more common among the differentially expressed gene set, with 198 significantly upregulated genes versus 159 genes significantly downregulated for at least one time point. Global analysis identified MMPs to be among the first and most highly upregulated in response to tissue amputation (Figure 1). In particular, the stromelysin MMP3/10a displayed an 11-fold upregulation by 3 dpa, and the newt specific collagenase nCol (Vinarsky et al., 2005) was similarly increased 8-fold relative to control. Additionally, tissue inhibitor of metalloproteinases 1 (TIMP1) was significantly upregulated throughout this early time window (≥4-fold), potentially indicating that the degradation of cell debris and matrix proteins is tightly controlled, as it appears to be during newt limb regeneration (Stevenson et al., 2006). In line with the activity of MMPs, RNA encoding the ECM components TNC and FN was upregulated shortly after ventricular amputation (Figure 1). While FN expression remained mostly constant during the first 2 weeks of cardiac muscle regeneration, TNC gene expression further increased to a 11-fold upregulated level by 14 dpa. qRT-PCR analyses with a subset of ECM-associated genes confirmed the expression profiles and demonstrated the robustness of our microarray data (Table S1). The complete data set for our multi-tissue microarray screen that highlights regenerative molecular programs for a range of gene classes including immune response, cytoskeletal rearrangements, pattern regulation, and signaling cascades is openly available (Mercer et al., 2012; Vinarsky et al., 2005).
Figure 1. Genes controlling the extracellular environment are activated following ventricular resection.
Microarray analyses of newt heart regeneration reveal upregulation of genes encoding matrix metalloproteinases (MMPs) and their inhibitor (TIMP1), indicating a fine-tuned degradation of cell debris and ECM. The induction of the ECM components tenascin-C and fibronectin further indicate a substantial reorganization of the extracellular environment. Each time point was normalized to intact (day 0) expression levels and log10 transformed. Error bars represent ± standard deviation for each gene at each indicated time point.
To gain a deeper understanding of the biological context of the observed differential gene activities, we performed Gene Ontology (GO) term enrichment analyses. As the newt genome lacks GO annotation, all differentially expressed genes with known homology were assigned an official mouse gene symbol. GO term enrichment analysis for those annotated genes with ≥2-fold differential expression for at least one time point revealed 45 significantly enriched terms (p≤0.01). In line with differential expression measures, GO terms associated with the extracellular matrix represent the most significantly enriched terms in the data set (Table 1; Dataset S1). The redundancy in returned GO terms is a common limitation of GO analysis. In order to extract deeper and more specific biological information in our data set, we developed a filtering algorithm to remove GO terms that shared over 90% of their associated genes with a more significantly enriched GO term (Code S1). Applying this new filtering algorithm to the raw GO output, we were able to unmask significantly enriched GO terms associated with muscle function and cytoskeletal structure during newt heart regeneration (Table 1; Dataset S1).
Table 1.
Multi-species GO analysis identifies molecular signatures of regeneration vs. repair.
| GO Term | P-Value | Fold Change |
Example Contributing Genes (with maximum log10 fold change value) |
|---|---|---|---|
| Newt heart regeneration | |||
| Extracellular region | 1.8E-08 | 3.3 | MMP9 (0.73), TIMP1 (0.75), TNC (1.04) |
| Contractile fiber part | 2.4E-06 | 17.6 | MYH4 (0.92), MYH13 (0.36), TCAP (−0.42) |
| Structural molecule activity | 3.4E-05 | 5.2 | COL2A1 (0.30), KRT5 (0.63), KRT8 (0.43) |
| Muscle contraction | 1.3E-04 | 18.9 | ACTA2 (0.51), MAP2K6 (−0.32), TTN (−0.59) |
| Muscle organ development | 9.8E-04 | 7.7 | GSC (0.32), MSX1 (0.41), TNC (1.04) |
| Zebrafish heart regeneration | |||
| Extracellular matrix | 3.4E-04 | 3.3 | FGF3 (−0.31), MMP9 (0.35), TIMP2B (1.01) |
| Homeostatic process | 1.6E-03 | 2.8 | PDCD8 (−0.37), PDIA3 (0.38), TTK (0.33) |
| Cytoskeletal protein binding | 4.8E-03 | 2.5 | ACTN1 (0.44), SDC4 (0.35), VMHC (0.37) |
| Hydrolase activity | 2.2E-02 | 1.5 | EDEM1 (0.43), FUCA1 (0.30), LYZ (−0.43) |
| Transition metal ion transport | 2.5E-02 | 3.6 | SLC39A7 (0.55), TFA (0.54) |
| Mouse heart post-MI | |||
| Mitochondrion | 5.1E-37 | 2.5 | COX6A2 (−0.70), GATM (0.76), TXN1 (0.47) |
| Organelle envelope | 4.0E-14 | 2.6 | EIF6 (0.46), HCCS (−0.49), RAC2 (0.31) |
| Generation of precursor metabolites and energy |
8.7E-13 | 3.7 | CYBB (0.51), ERO1L (0.58), ETFA (−0.61) |
| Cofactor metabolic process | 1.2E-10 | 3.1 | COQ3 (−0.44), CS (−0.57), FPGS (0.30) |
| Immune system process | 1.1E-07 | 2.0 | CXCL5 (1.26), IL6 (1.42), TLR6 (0.40) |
| Human heart post-MI | |||
| Inflammatory response | 4.7E-13 | 8.1 | CXCL2 (0.32), IL8 (0.37), ITGB2 (0.32) |
| Myofibril | 1.4E-11 | 12.9 | ACTC1 (−0.31), ENO1 (0.30), MYH7 (−0.40) |
| Immune system process | 9.8E-09 | 3.1 | CCL2 (0.40), LYN (0.37), PTX3 (0.41) |
| Regulation of biological quality | 1.9E-08 | 3.1 | CYP1B1 (0.42), DAB2 (0.38), NPPA (−0.31) |
| Heart development | 7.4E-08 | 6.3 | ADAP2 (0.33), CSRP3 (−0.36), PLN (−0.33) |
Gene Ontology (GO) term enrichment analysis revealed the most significantly enriched biological activities associated with heart regeneration in newt and zebrafish, compared to cardiac muscle repair following MI in mouse and human. Shown here are the top 5 most significantly enriched, non-redundant GO terms. Redundancy reducing algorithm available in Code S1. Full GO term enrichment results available in Dataset S1.
Given the conservation of a regenerative ECM between different regenerating tissue types in the newt (Calve et al., 2010; Mercer et al., 2012), we wanted to further investigate whether changes in the extracellular environment are an evolutionarily conserved mechanism in heart regeneration between species. In order to do so, we utilized available microarray data for zebrafish heart regeneration (Lien et al., 2006). Comparable to the newt, in that study the lower third of regenerating ventricles were harvested at 3, 7 or 14 dpa and compared to unamputated control tissue. Considering genes with a differential expression profile for at least one time point, we again found that the most significantly enriched GO terms in the zebrafish data set were those associated with the ECM (Table 1; Dataset S1). Interestingly, of the non-redundant GO terms, those associated with cytoskeletal structure are significantly enriched for both the newt and zebrafish, potentially suggesting that a change in cytoarchitecture supports regenerative cell behaviors such as proliferation and/or migration.
In contrast to the regeneration-competent newt and zebrafish, adult mammals including mice and humans are unable to mount a regenerative response to cardiac muscle injury. To gauge the significance of an evolutionarily conserved regenerative matrix in relation to the typical scar formation response in mammals, we obtained microarray data from the GEO online database for re-evaluation. The mouse post-MI data set comprises a time series (1, 4, 24, and 48 hours; 1 and 8 weeks) that compares sham operated left ventricles with infarcted left ventricles. Infarcted tissue samples were taken from the region between the left anterior descending artery and the apex (Tarnavski et al., 2004). The human heart array data came from a study that determined how left ventricular assist devices alter genomic signaling (Hall et al., 2004). We have taken a different perspective on this data set and evaluated only the pre-implantation microarrays. Specifically, we have analyzed gene expression for non-ischemic hearts (clinically classified as no evidence of coronary artery disease) versus hearts that have suffered an MI less than 10 days prior to tissue harvest. To extract the major biological themes of wound healing in mammalian cardiac tissue, we performed GO term enrichment analyses for all genes with ≥2-fold differential expression for at least one time point. The filtering algorithm (Code S1) proved especially useful in analyzing the human and mouse data, as each returned over 100 significantly enriched GO terms. In stark contrast to the newt and zebrafish, enriched non-redundant GO terms for the mammalian data set largely revealed immune and inflammatory responses, as well as energy conservation and production (Table 1; Dataset S1). These distinct responses provide important, novel insights into divergent molecular pathways that are activated in regeneration-competent and -incompetent species following cardiac injury.
Tenascin-C, Hyaluronic Acid and Fibronectin form a regenerative matrix in the newt heart
The interspecies differential expression and GO analyses suggested that a rapidly changing matrix environment may have critical functions early in heart regeneration, a role reminiscent of our previous findings in the regenerating newt limb (Calve et al., 2010; Calve and Simon, 2012). Detailed expression patterns of ECM components in the regenerating newt heart were not available, and we consequently determined the spatiotemporal distribution of TNC, HA, and FN during the first 10 weeks after ventricle resection. Antibodies for TNC and FN have been previously successfully employed in the newt model, as well as the use of biotinylated hyaluronic acid binding protein as a probe to specifically detect HA (Calve et al., 2010; Calve and Simon, 2012; Ripellino et al., 1985).
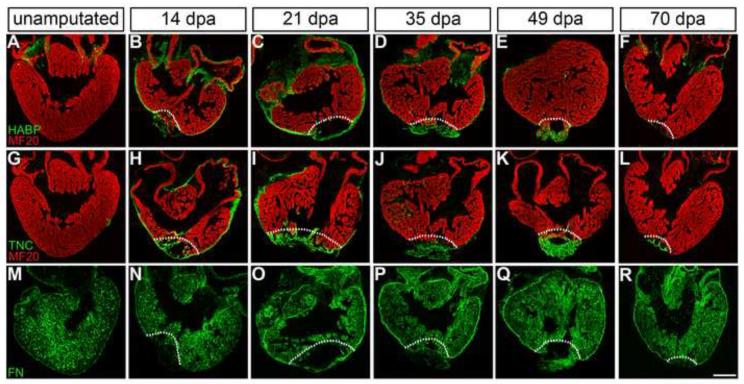
Immunohistochemistry of regenerating heart sections performed at defined time points (0, 3, 7, 14, 21, 35, 49, 70 dpa) revealed essentially no expression of TNC in the normal, unamputated heart, while HA distribution was confined to the outflow tract and atrioventricular canal. However, after ventricular tip amputation, we could demonstrate a dynamic upregulation of all three selected matrix components (overview provided in Figure 2; representative images; n≥3 for each time point). Immediately after amputation, the wound is closed by a fibrin-rich clot that is rapidly infiltrated by mononuclear cells (Witman et al., 2011). Near the injury site, tissue morphology changed dramatically (Figure 3). As early as 3 dpa, TNC became substantially activated in areas immediately adjacent to the amputation plane surrounding myocardial cells, with additional modest expression in the epicardium (Figure S1). By 1 week postamputation, both TNC and HA were expressed in the epicardium. At this time, HA also surrounded the nucleated clot at the regenerating apex (Figure S1). In contrast, while TNC remained in direct contact with the damaged myocardium, it was not robustly expressed around the perimeter of the fibrous clot until approximately 2 weeks postamputation (Figure 3), potentially indicating that TNC and HA play distinct roles in the spatial and/or temporal signals that support the early regenerative response. Counter-stain with the striated muscle-specific MF20 antibody revealed very little expression of both TNC and HA in myocardial muscle distant from the regenerate during this early time window (Figure S1). The epicardium-specific expression of both TNC and HA continued through 35 dpa, when these matrices were gradually downregulated and became restricted to the regenerating ventricular apex through 49 dpa (Figure 2). A substantial reduction of expression occurred as regeneration progressed through 70 dpa, at which point TNC and HA returned to near normal expression levels (Figure 2). In contrast, FN is expressed throughout the normal newt myocardium and epicardium. During the regenerative process, FN maintained a more subtle expression profile with delayed epicardial upregulation beginning at 21 dpa and lasting through 49 dpa (Figure 2). This modest FN protein upregulation aligns well with the 2–3 fold increased mRNA expression during the first two weeks of regeneration, in comparison to a 4–11 fold higher level for TNC relative to controls (Figure 1). For the first time in cardiac regeneration, we demonstrate a regeneration-specific dynamic temporal and spatial enrichment of TNC, HA and FN, which suggests a conserved function for the ECM in regenerative cell behaviors.
Figure 2. Cardiac injury induces the deposition of a regeneration-specific transitional ECM at the epicardium and injury site.
Spatiotemporal distribution of ECM components (green) at 14, 21, 35, 49, and 70 days postamputation (dpa) in comparison to the normal, unamputated heart, reveals dramatic extracellular matrix remodeling. Hyaluronic acid (HA, indirectly labeled with hyaluronic acid binding protein, HABP) and tenascin-C (TNC) are highly upregulated in the epicardium early in regeneration (B, C; H, I), with subsequent downregulation in all areas except the regenerating apex (D, E; J, K). In contrast, fibronectin (FN) is expressed throughout the normal myocardium (M), but shows a delayed upregulation in the epicardium of the regenerating heart (O – Q). Cardiac muscle is labeled with MF20 (red); white dotted lines demarcate the amputation site. Scale bar = 400 μm; dpa = days postamputation.
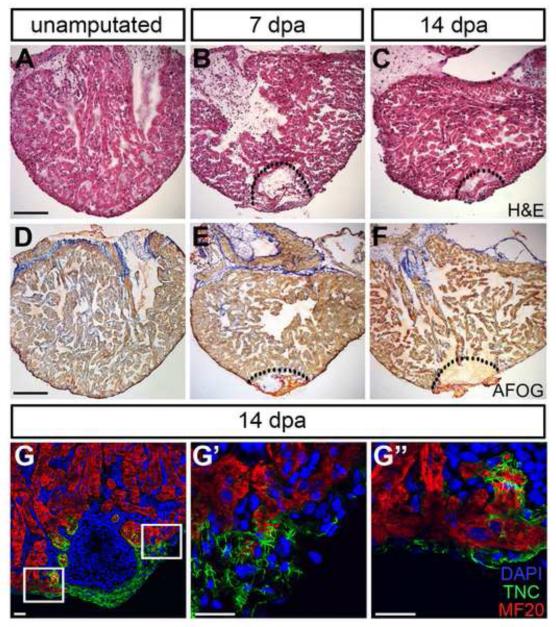
Figure 3. The regeneration-specific matrix surrounds the fibrous clot and adjacent myocardium.
Following ventricular resection, a fibrin-rich clot closes off the wound. Over the following two weeks, gross changes in tissue architecture are apparent, as visualized by Hematoxylin & Eosin stain (H&E; A – C) and Acid Fuchsin Orange G stain (AFOG; D – F). Immunohistochemistry reveals rapid influx of cells into the clot; by 14 dpa, a TNC-rich transitional matrix surrounds the injury site (G; nuclei, DAPI = blue; cardiomyocytes, MF20 = red; transitional matrix, TNC = green), and infiltrates the myocardial muscle directly adjacent to the amputation plane (G’, G’’). Scale bars for A – F = 400 μm; scale bars for G – G’’ = 50 μm. G’ represents the top and G’’ represents the lower box outline in G.
Ventricular amputation triggers a myocardium-wide proliferative response
Complete and functional restoration of the damaged myocardium depends on an appropriate proliferative response to generate progenitor cells (Bettencourt-Dias et al., 2003; Jopling et al., 2010; Laube et al., 2006). To determine the number and location of cells that enter the cell cycle after removing the ventricular tip, we employed EdU, a nucleotide analog that is incorporated into DNA during the synthesis (S) phase of the cell cycle. Because of its permeable dye-azide and click chemistry reaction, EdU labeling is also compatible with most antibody reactions to co-stain tissues (Salic and Mitchison, 2008). Newts were injected intraperitoneally with EdU every 24 hours for 3 days before tissue harvest. We found that at least a 3-day EdU pulse was necessary to label an adequate number of cells for visualization and statistical analysis, suggesting that in the newt heart a lower percentage of cells are cycling at any single time point compared to the regenerating limb and spinal cord, where substantial EdU incorporation was observed in as little as 3 hours (Calve et al., 2010; Mercer et al., 2012).
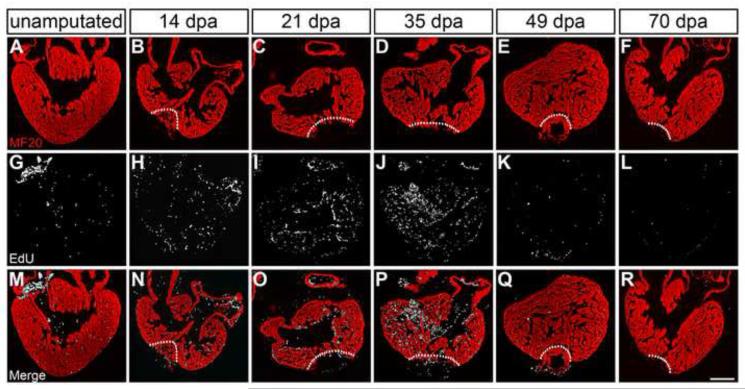
As expected, we found very few EdU+ cells within the normal adult newt heart (Figure 4; representative images; n≥3 for each time point). Similarly, we detected very limited proliferation during the first week following ventricular resection (Figure S2). Increased cycling began around 14 dpa, with EdU+ cells visible throughout the myocardium. The number of EdU+ cells increased substantially over the first 3 weeks of cardiac muscle regeneration, with a peak in cell cycle entry evident between 3-5 weeks postamputation (Figure 4). While most EdU+ cells were not cardiomyocytes, by 14 dpa we found that 15.5% (±1.2) of EdU+ cells in the myocardium were also MF20+, and this number increased to a maximum of 21.5% (±6.2) by 35 dpa (Figure S3; Table 2). Moreover, phospho-histone H3 (PH3) staining further revealed that cardiomyocytes are stimulated to enter the cell cycle and progress through mitosis, as we found multiple examples of PH3+/MF20+ cells during this peak proliferative period (Figure S3). As regeneration progressed, cell cycle entry slowed notably by 49 dpa, with near normal background levels by 70 dpa (Figure 4). Thus, our data suggest that ventricular distal tip amputation triggers a proliferative response throughout the myocardium in a subset of cardiomyocytes and other as yet unknown cell types (Figures 4 and S3).
Figure 4. Cell proliferation increases throughout the myocardium during heart regeneration.
Cardiac cells enter the cell cycle as determined by EdU incorporation at 14, 21, 35, 49, and 70 days postamputation (dpa). In the unamputated heart, only a few cells actively synthesizing DNA (EdU = white) were detected in the myocardium (MF20 = red) (G, M). The number of EdU+ cells increased by 14 dpa (H, N) and a substantial proliferative response throughout the myocardium was evident by 21 and 35 dpa (I, J; O, P). Cell cycling gradually subsided towards 70 dpa (L, R). White dotted lines demarcate the amputation site. Scale bar = 400 μm; dpa = days postamputation.
Table 2.
Cardiomyocyte cell cycle reentry during newt heart regeneration.
| 0 dpa | 14 dpa | 21 dpa | 35 dpa | 49 dpa | 70 dpa | |
|---|---|---|---|---|---|---|
|
% EdU+MF20+ cells from total
EdU+ cell population |
12.2 | 15.5 | 20.3 | 21.5 | 14.8 | 14.9 |
| Standard deviation | ±4.7 | ±1.2 | ±3.0 | ±6.2 | ±4.8 | ±3.9 |
To determine the portion of cardiomyocytes that contribute to the regenerative response, EdU+ cells and EdU+MF20+ double-labeled cells were counted throughout the regenerating newt heart at multiple time points. An observed increase in the percent of cardiomyocytes entering the cell cycle coincided with the peak proliferative response at 3-5 weeks after amputation (Figure 4). However, this increased EdU incorporation among cardiomyocytes did not reach statistical significance across the regenerative time course (n = 3 for each time point).
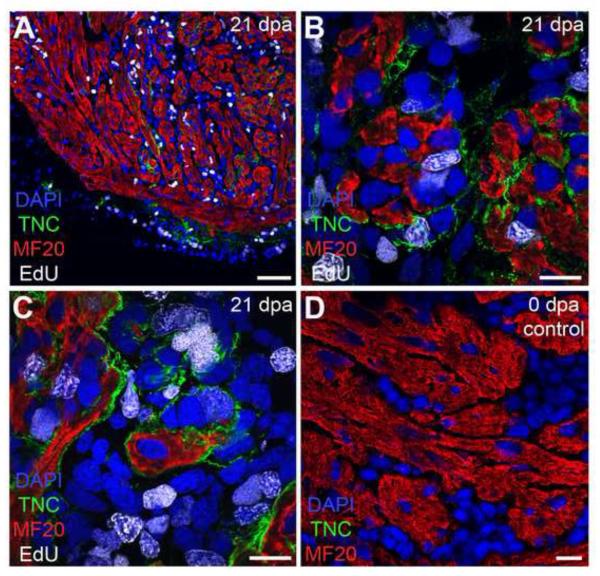
In an effort to understand how the regenerative matrix might instruct cell proliferation, we examined the expression of TNC and HA in cardiac muscle. Though we could not demonstrate a tissue-wide upregulation of HA or TNC in the myocardium, using high power confocal imaging we were able to detect local TNC distribution around the myocardial muscle in approximately 8% of our samples during the peak proliferative response at 21 -35 dpa (Figure 5). We found direct contact of TNC with MF20+ muscle and EdU+ nuclei; however, this TNC expression appeared transient and/or temporally variable between animals, such that we could not draw strict correlations between myocardial TNC distribution and EdU-labeling events.
Figure 5. Transient distribution of a TNC-rich matrix in the myocardium of the regenerating heart.
Tenascin-C is detected around myocardial muscle, contemporaneous with the appearance of proliferative activity at 21 dpa (A; nuclei, DAPI = blue; cardiomyocytes, MF20 = red; transitional matrix, TNC = green; EdU = white). High power confocal imaging demonstrated direct contact of TNC with MF20+ muscle and EdU+ nuclei (B, C). The regenerative matrix is not expressed in the myocardial muscle of unamputated hearts (D). Scale bar for A = 100 μm; scale bar for B – D = 20 μm. dpa = days postamputation.
The regenerative ECM serves as a path for proliferative progenitor cells
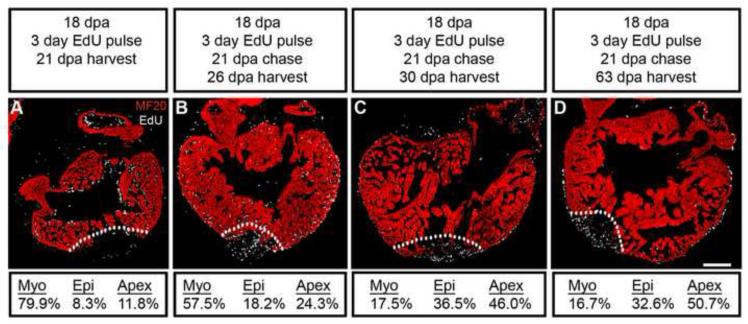
To gain insight into how myocardium-wide cell cycle entry can ultimately replace local tissue loss, we performed a series of pulse-chase (PC) EdU labeling assays in an effort to mark proliferating cells and monitor their location. Newts were intraperitoneally injected every 24 hours with 1mM EdU at the onset of the peak proliferative response period (18 dpa, pulse). After the 3-day labeling period, a second injection with 100-fold molar excess of thymidine served to effectively dilute the EdU and largely prevent further incorporation (21 dpa, chase). In this way, a substantial number of proliferative cells were permanently labeled with EdU, allowing us to follow their spatial distribution across the regenerative time course. Harvest at 21dpa revealed, as expected, an even distribution of EdU+ cells throughout the myocardium (Figure 6). However, after PC completion and harvest at 26, 30, and 63 dpa, the EdU+ cells displayed a successive enrichment toward the proximity of the epicardium, with eventual localization at the epicardial sheath and regenerating ventricular apex (Figure 6; representative images, n≥3 for each time point). The intermediate time point indicated that EdU+ cells gradually reach the transitional matrix-enriched epicardium and regenerating apex around 1 week after PC completion. Because we did not find substantial labeling of cells in the epicardium and regenerate during the initial EdU pulse, our data suggest that the EdU+ cells present in the epicardium and apex after PC largely originated in the myocardium. Of note, we observed a decrease in EdU+ cell numbers at later PC time points, which suggested either increased proliferative activity and signal dilution or cell death of the EdU-labeled cells. To test for the possibility of apoptosis, we performed TUNEL assays, but detected essentially no apoptotic activity within the investigated PC time window (Figure S4). We therefore reject the hypothesis that loss of EdU signal results from programmed cell death.
Figure 6. Proliferative cells born within the myocardium migrate to the epicardium and wound site.
EdU pulse-chase labeling identified cells actively synthesizing DNA (EdU = white) that were distributed throughout the ventricular myocardium at 21 dpa (A). Cells marked by EdU incorporation gradually accumulated near the epicardium and the regenerating ventricular apex around 26 dpa (B), ultimately migrating to the epicardium and continuing their accumulation at the ventricular apex by 30 dpa (C). EdU+ cells remain enriched in the epicardium and accumulate at the regenerating apex through 63 dpa (D). The dynamic change of progenitor cell location (myocardium vs. epicardium vs. regenerating apex) during the PC time course is quantified at the panel bottom (localization of EdU+ cells / total EdU+ cell population). MF20 identified striated muscle myosin heavy chain in the myocardium (red) and white dotted lines demarcate the amputation site. Scale bar = 400 μm; dpa = days postamputation.
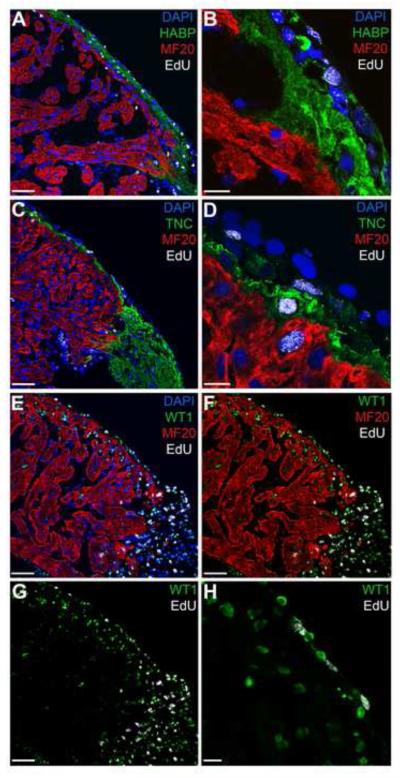
To further elucidate the behavior of cycling cells born within the myocardium, we performed an additional PC assay focusing on the time period following the peak proliferative response (39 dpa pulse, 42 dpa chase). As before, we found the majority of EdU+ cells located throughout the myocardium immediately after PC completion (42 dpa; Figure S5). However, within 1 week of PC, EdU-labeled cells were significantly enriched in the regenerating apex; this trend continued at 2 weeks after PC completion, at which point a substantial majority of EdU+ cells were located in the regenerate (Figure S5). In this PC study, the absolute numbers of EdU+ cells also increased at the apex over the time course (average of 3 biological samples: 55 EdU+ cells at 42 dpa harvest [Figure S5A], 119 EdU+ cells at 49 dpa harvest [Figure S5B], 152 EdU+ cells at 56 dpa harvest [Figure S5C]), strongly suggesting that the labeled cells are migrating to the injury site. Interestingly, different from the PC performed at the onset of peak proliferative activity, this late PC time course did not reveal the epicardial localization of EdU+ cells. We note that during the early PC (18 dpa pulse, 21 dpa chase; Figure 6), the regenerative ECM is robustly expressed at both the epicardium and wound site (Figure 2), while expression of TNC and HA is confined to the apex during the time window of the late PC series (39 dpa pulse, 42 dpa chase; Figure S5). In line with the differential spatiotemporal expression of the regenerative matrix, it seems possible that EdU+ cells accumulate in the epicardium at earlier time points to replace cells in the damaged epicardial layer. In support of this notion, using high power confocal imaging, we found that EdU+ cells are integrated into and located immediately exterior to the TNC and HA-rich ECM in the epicardium at 28 dpa (Figure 7A-D). At later time points, the epicardium is likely fully restored, such that migratory EdU+ cells localize predominantly to the regenerating apex to replace the lost ventricular myocardial tissue. Altogether, the PC labeling studies indicate that, after amputation, cells located throughout the myocardium are stimulated to enter the cell cycle and then migrate radially to the epicardium and the regenerating apex.
Figure 7. The epicardium-associated regenerative matrix supports the accumulation of progenitor cells.
Dynamic remodeling at the epicardial sheath results in robust expression of a regenerative matrix by 28 dpa. Distribution of HA (indirectly labeled with HA binding protein HABP; A) and TNC (C) fosters the accumulation of EdU+ cells (18 dpa pulse, 21 dpa chase) following radial migration in the myocardium (Figure 6). High magnification (B, D) reveals that EdU+ cells are immersed in the regenerative matrix and/or interact with EdU- cells in the outer epicardial layer. WT1 is expressed in the epicardium and outer myocardium (E, F, G) and labels the majority of EdU+ cells as they reach the epicardial ECM (E – H). Scale bars for A, C, and E – G = 100 μm; scale bars for B, D and H = 20 μm; dpa = days postamputation.
To clarify the relationship between the epicardium and proliferative cells during heart regeneration, we co-stained tissue with an antibody directed against Wilms Tumor 1 (WT1), a recognized epicardial marker for studies of vertebrate cardiac development and zebrafish heart regeneration (Kikuchi et al., 2011; Perez-Pomares et al., 2002; Schnabel et al., 2011; Zhou et al., 2008). In the newt heart, WT1 identifies the majority of cells that are immediately superficial to the MF20+ myocardium, likely marking the epicardial lineage. Interestingly, we also found nuclear WT1 expression extending from the epicardium into the exterior portions of the myocardial muscle (WT1+ signal extends from the epicardial sheath ≈ 3 – 5 cell layers deep into the adjacent myocardium), labeling both cardiomyocytes and non-MF20+ cells (Figure 7E-H). We observed this epicardial and exterior myocardial expression pattern in normal, non-regenerating hearts with unaltered expression in regenerating hearts across the investigated time course. Contemporaneous with the accumulation of EdU+ cells near the epicardium around 4 weeks postamputation, we detected an increase in the population of EdU+ cells that were also WT1+ (Figure 7E-H). Of note, at 21 dpa (18 dpa EdU pulse), approximately 21.5% (±5.6) of the EdU+ cells were WT1+. When we followed this cell population to 28 dpa (18 dpa EdU pulse, 21 dpa chase), the number of double-labeled EdU+WT1+ cells increased to 80.7% (± 6.1). Thus, a majority of these proliferative cells upregulate WT1 expression as they approach the epicardium. Previous research indicated that WT1 can function in a variety of roles in multiple tissue types, including supporting alterations in differentiation status (Hohenstein and Hastie, 2006). It seems possible that the specific TNC and HA-rich regenerative ECM in the epicardium serves as a signaling center to attract myocardial progenitor cells to the epicardium where they begin to express WT1, enabling a change in their differentiation state. Thus, the WT1 labeling studies suggest that a subset of migratory EdU+ cells may replenish the regenerating epicardium, while direct interaction of EdU+ cells with the regenerative ECM could facilitate the migration of these progenitor cells to the injury site.
Regenerative ECMs support cell cycle reentry in cardiomyocytes
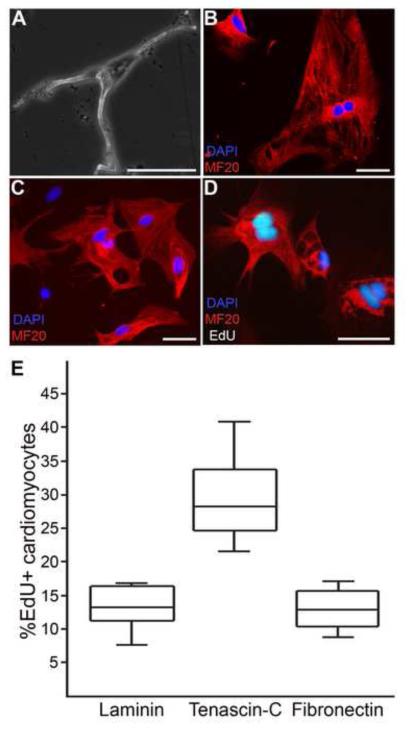
The temporal regulation of TNC, HA and FN matrix components not only indicates that they play important roles in regeneration, but also suggests that their distinct spatial distributions may be involved in instructing different regenerative cell behaviors. Therefore, in order to directly test whether the extracellular environment has instructive abilities on cardiac cells, we developed an in vitro culture system that utilizes primary newt cardiomyocytes plated on regeneration-specific ECMs. Following published protocols (Bettencourt-Dias et al., 2003), adult newt cardiomyocytes were isolated and displayed an elongated shape with the typical branched morphology in culture (Figure 8A). The cardiomyocytes adhered and spread slowly, displaying spontaneous beating after 5 – 7 days, and more robust synchronous beating when they formed intricate junctions 10 – 12 days after plating. During culture, cardiomyocytes maintained their differentiated state for at least 1 month, as indicated by positive reaction with the MF20 antibody that detects myosin heavy chain of mature muscle (Figure 8B, C).
Figure 8. The regeneration-specific ECM directs cardiomyocyte cell cycle entry.
Explanted newt ventricular cardiomyocytes maintain their differentiated status in vitro, as verified by the bifurcated structure and robust staining with the muscle myosin heavy chain MF20 antibody (A, phase contrast; B, C, MF20 = red). Cardiomyocytes were cultured under defined conditions for 7 days and the percentage of cells actively synthesizing DNA was determined by EdU incorporation. Cells plated on TNC-coated substrates incorporated EdU with visible nuclear division (D, MF20 = red; DAPI = blue, EdU = white) and demonstrated a significantly higher proliferative activity as compared to matrices associated with mature, differentiated cardiac muscle, including laminin and FN (E, One-way ANOVA, p<0.001). Box-and-whisker plots: center lines represent the median, surrounding box represents upper and lower quartiles, respectively; outside bars represent upper and lower extremes, respectively (n≥3 independent experiments for each ECM coating). Scale bar for A – D = 50 μm.
Having succeeded in establishing stable cultures of newt adult cardiomyocytes, we tested whether these explanted cells would differentially enter the cell cycle in response to distinct ECM environments. We chose to assess proliferative activity 7 days after plating, before the cells began to form complex networks that prevented proper observation and counting. EdU was added to the culture medium and cell cycle entry monitored. We note that, in our hands, newt cardiomyocytes did not adhere well to HA-coated plastic dishes, likely due to the hygroscopic properties of HA (Chen and Abatangelo, 1999). In line with a previous report that investigated newt cardiomyocyte behavior on laminin substrates (Bettencourt-Dias et al., 2003), we observed a comparable low, but highly reproducible proliferative response for laminin (12.8% ± 3.2) and FN (12.9% ± 2.8; Figure 8E). In contrast, TNC revealed a significantly higher proliferative index (29.4% ± 6.2), clearly supporting cell cycle entry and nuclear division (Figure 8D, E). Thus, a TNC-rich regenerative matrix can induce cell cycle entry in newt cardiomyocytes. However, as we could only identify variable and transient expression of TNC within the myocardial muscle (Figure 5), it seems likely that other factors besides TNC may also be responsible for cardiomyocyte proliferation.
DISCUSSION
Novel hypotheses for cardiac muscle regeneration in the newt
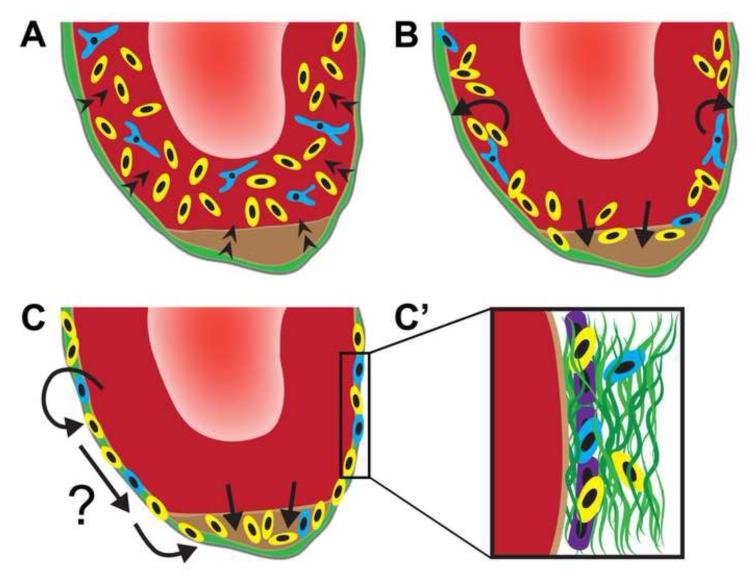
Extracellular matrix remodeling is a critical aspect of development, wound healing and regeneration (Cox and Erler, 2011). For the first time, we report the expression of a regeneration-specific transitional matrix composed of TNC, HA and FN during newt heart regeneration. The dynamic temporal and spatial distribution of these matrix components in combination with distinct physiologic activities of cardiac progenitor cells allowed for the development of a new working model for vertebrate cardiac muscle regeneration (Figure 9). Our model integrates existing knowledge from embryogenesis, as well as skeletal and cardiac muscle regeneration, implicating a coordinated function of these matrix components in the induction and maintenance of the regenerative response. We hypothesize that a TNC-rich regeneration-specific matrix directly or indirectly regulates the generation of proliferative progenitor cells throughout the myocardium. In addition, an epicardium- and apex-associated transitional ECM attracts the progenitor cells and supports their radial migration to the heart surface and wound site. As progenitor cells accumulate near the epicardium and regenerate, developmental gene programs, including WT1, are activated and promote a change in differentiation status. While some progenitors may take on an epicardial fate, it is possible that direct interaction with the regenerative matrix at the epicardium also facilitates the passage of progenitor cells to the distantly located site of tissue loss.
Figure 9. Model for newt cardiac muscle regeneration.
An epicardium- and apex-associated regenerative ECM serves as a reservoir for factors that signal inward (arrowheads in A) to promote cell cycle entry throughout the myocardium (yellow cells), including a subset of cardiomyocytes (blue cells; A). Proliferative cells subsequently migrate radially toward the epicardium and apex, likely undergoing a change in differentiation status (B). Embedded within the regeneration-specific matrix (C), the cells could integrate into the epicardium or migrate to the apex and contribute to the regenerating myocardium (C).
ECM remodeling is an evolutionarily conserved mechanism in tissue regeneration
Our previous work has shown that extracellular remodeling is a conserved process in newt forelimb, hindlimb, tail, spinal cord and heart regeneration (Mercer et al., 2012). Moreover, in vivo and in vitro studies of newt regenerating skeletal muscle have demonstrated that TNC, HA and FN instruct all regeneration-relevant cell behaviors, including dedifferentiation, proliferation and migration (Calve et al., 2010; Calve and Simon, 2012). This ECM remodeling is not only conserved across different regenerating tissue types in the newt, but also seems to be an evolutionarily conserved process among vertebrates capable of heart regeneration. By mining microarray data for zebrafish heart regeneration after ventricular resection, we detected differential expression of genes that are significantly enriched for GO terms associated with the extracellular matrix.
Recent work applying a cryoinjury to the zebrafish ventricle further supports our findings, demonstrating that TNC is specifically upregulated at the injury border throughout the first month of myocardial regeneration (Chablais et al., 2011). Interestingly, while both regeneration-competent species exhibit an activation of a TNC-enriched matrix during cardiac muscle regeneration, the spatiotemporal expression pattern of this ECM component differs between the species. In the newt, we find that both TNC and HA are specifically expressed in the epicardium and wound site early in the regeneration process (Figure S1), with a subsequent restriction to the injured apex tissue around 5 – 7 weeks postamputation (Figure 2). A combination of TNC and HA likely enhances the ability of cells to migrate in the regenerative matrix, as hyaluronic acid provides a hydrated environment resulting in a change of tissue stiffness that is known to support cell migration (Calve and Simon, 2012; Chen and Abatangelo, 1999). Therefore, in both the newt and zebrafish it seems possible that TNC counter-adhesive domains facilitate cell migration at the edges of the intact myocardium (Calve et al., 2010; Chablais et al., 2011; Poss et al., 2002). As cardiomyocyte migration was recently shown to be essential for complete and functional regeneration in the zebrafish heart (Itou et al., 2012), future studies with forced changes of ECM production in vivo and its influence on cardiomyocyte migration will be of great interest.
Local injury induces a global myocardial proliferative response
Our EdU incorporation and PH3 labeling data within the newt ventricular distal tip resection model reveal a proliferative response throughout the myocardium and support a previous report that used a different heart muscle resection scheme in combination with long-term BrdU labeling (Figures 4 and S3; (Witman et al., 2011). Such an organ-wide response has also been reported in the zebrafish and appears to be a conserved reaction in regeneration-competent vertebrates (Jopling et al., 2010; Witman et al., 2011). Interestingly, while the majority of proliferative cardiomyocytes are concentrated near the wound site in the regenerating zebrafish heart (Poss et al., 2002), a proportion can also be found in regions far from the amputated apex (Jopling et al., 2010). The same principle seems to hold true in the regenerating newt heart, as we find that up to 21.5% of cardiomyocytes are able to undergo cell cycle reentry within a 3-day labeling period during the regenerative response (Table 2), with a substantial portion of these cells near the ventricular apex (Figure S3). While we have demonstrated variable, transient expression of TNC in the myocardium in the proximity of EdU+ cells (Figure 5), these data were not conclusive for a direct role of the regenerative ECM in the proliferative response. However, our functional tests with myocardial explants under defined culture conditions provided clear first evidence that TNC has instructive properties on newt cardiomyocytes to engage in cell cycle entry (Figure 8). These findings are in line with our previous work on newt skeletal muscle cells, which also demonstrated a selective proliferative response to distinct ECM components, including TNC (Calve et al., 2010). Our data indicate that cardiomyocytes make up only a minority of proliferative cells in the regenerating newt heart. This finding is supported by an earlier report from Witman et al. (Witman et al., 2011), which also demonstrated that a subset of proliferative non-cardiomyocytes expresses the cardiogenic transcription factors GATA4 or Islet1, suggesting that a portion of progenitors could be derived from resident stem cells or cardiomyocyte dedifferentiation. Ongoing work in our laboratory is focused on determining other cell types that contribute to the proliferative response.
An epicardium-associated transitional matrix supports cardiac regeneration
The specific and strong upregulation of a regenerative matrix in the epicardium indicates robust activity and participation of this cellular sheath early in the regenerative response. We speculate that these specific ECMs serve as a reservoir for signaling factors. The local upregulation of MMPs could help release these factors and allow them to penetrate into the myocardium where they might be involved in regulating gene expression and cellular behaviors of newly arriving progenitor cells. In support of this notion, it has been shown that the ECM can provide soluble signaling factors, mediating signals to promote migration, proliferation and alterations in differentiation state (Kim et al., 2011; Rosso et al., 2004). In many ways, this mechanism could recapitulate aspects of cardiogenesis, where the epicardium is necessary for myocardial growth and function through inward signaling with a variety of factors, including FGFs, retinoic acid and cytokines (Chen et al., 2002; Kwee et al., 1995; Lavine et al., 2005).
The expression of the regenerative ECM likely coincides with the activation of molecular programs to support regenerative processes in the newt heart. While we have employed WT1 as a marker to discover the interactions of EdU+ cells in an epicardial regenerative environment (Figure 7), the constant expression of WT1 in the non-regenerating epicardium and adjacent myocardium suggests that WT1 might play multiple roles in the newt heart. Several recent reports have focused on the biological functions of WT1, ultimately discovering that this multi-faceted protein can support progenitor cell proliferation, as well as epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions (Hohenstein and Hastie, 2006; Smart et al., 2011; von Gise et al., 2011; Zhou et al., 2008). As a majority of EdU+ cells become WT1+ during their migration to the epicardium and apex, we hypothesize that the upregulation of this developmental gene program contributes to changes in the differentiation status of these recruited progenitor cells. In the alternative newt ventricular resection model, the cardiogenic transcription factors HAND2, GATA4 and GATA5 exhibit substantially higher expression levels at 23 dpa relative to control newts (Witman et al., 2011). This time frame is not only when we see robust epicardial expression of TNC, HA and FN, but is also near the peak of cell proliferation and migration in our apex resection model (Figures 2, 4, and 6). Thus, the transitional expression of a regeneration-specific matrix may provide cues to regulate a gene program that generates progenitor cells to functionally replace the lost apex tissue, either by cardiomyocyte dedifferentiation or stem cell activation.
The epicardial- and apex-enriched transitional ECM may also have a role in directing progenitor cells towards the wound site. Our data provide evidence that after cell cycle entry, the EdU+ cells that originate in the myocardium migrate radially towards the heart circumference. Specifically, the in vivo tracking of proliferative cells through long-term EdU pulse-chase indicate two distinct cell localization patterns. Progenitor cells born in the myocardium at the onset of the peak proliferative period migrate to the regenerative matrix-enriched apex and epicardium, where a substantial number apparently replace cells in the damaged epicardial layer (Figures 2 and 6). While not conclusive from our data, once in direct contact with the epicardial regenerative matrix, a portion of these cells may further migrate towards the ventricular apex, as both TNC and HA are substrates that support cell migration (Calve et al., 2010). Later in the regenerative response, when the epicardium is largely recovered and the TNC and HA epicardial matrix is downregulated, the vast majority of progenitor cells home instead to the regenerative matrix-rich apex tissue and contribute to the regenerative repair (Figure 2 and S5). In line with a recent report utilizing a crush injury model of newt heart regeneration (Piatkowski et al., 2013), after 49 days postinjury regeneration led to continuous improvement of tissue morphology, including differentiation of progenitors cells and muscle fiber repatterning. In this context we note that this group showed that the ECM component collagen type III serves as a scaffold in directing the integration of new cardiomyocytes into existing myocardial muscle. Therefore, it seems likely that additional matrices interact with the regeneration-specific matrix described here, contributing to the dynamic extracellular remodeling that facilitates a regenerative response in cardiac muscle.
Cardiac damage in mammals elicits a different matrix remodeling response
The regenerative response that occurs in damaged newt and zebrafish hearts is in stark contrast to the scar-forming repair response found in mammals. Our GO term enrichment analyses highlight these differences, and are supported by other reports on extracellular remodeling in mammals post-MI (Palojoki et al., 2001; Saraste et al., 1997; See et al., 2005; Weber et al., 1994). Mammalian cardiac tissue largely consists of a structural scaffold of interstitial collagens, a matrix network well suited to retain myocardial integrity and cardiac pump function. However, following MI, excessive accumulation of collagen in both infarcted and adjacent noninfarcted myocardium leads to tissue stiffness, ventricular diastolic and systolic dysfunction, and ultimately contributes to heart failure (Cleutjens and Creemers, 2002).
Alterations to the cardiac ECM following MI not only change the mechanical and functional properties of the heart, but are also thought to directly modulate the inflammatory response of the infarcted heart, a physiological activity that is also revealed in our GO analyses (Table 1). Immediately after MI, rapid activation of MMPs supports the degradation of the cardiac ECM (Dobaczewski et al., 2010), with the resulting matrix fragments exerting pro-inflammatory functions. Later, TIMP expression is induced to enhance new matrix deposition and preservation, while repressing inflammatory reactions. Different from this mammalian sequence of events, in the regenerating newt heart we see that MMPs and TIMPs are expressed at the same time early on in the regenerative response (Figure 1). This concurrent expression of MMPs and TIMPs is found in a number of other regenerating tissue types in the newt (Mercer et al., 2012; Stevenson et al., 2006), allowing the speculation that this dual activation supports the induction of matrix remodeling necessary to survive extensive organ damage, while also tempering and controlling inflammatory processes. In fact, studies using MMP inhibitors in experimental models of MI show promising results for attenuating adverse left ventricular remodeling (Creemers et al., 2001; Matsumura et al., 2005).
New strategies in cardiac regenerative medicine
Our model of newt heart regeneration implicates TNC, HA and FN in the induction and subsequent maintenance of the regenerative response. In this context, it is of interest that both regeneration-competent and -incompetent species undergo ECM remodeling and utilize similar matrix components while recovering from cardiac injury (Dobaczewski et al., 2006; Waldenstrom et al., 1991). However, the precise roles of these ECM components in the mammalian cardiac repair response remain unclear. We and others have shown that the ECM can differentially control cellular behavior during the regeneration process by mediating both growth factor availability and the specific binding of matrices to cell-membrane-localized receptors (Calve et al., 2010; Legate et al., 2009; Schultz and Wysocki, 2009). In this way, ECMs can trigger regeneration-specific gene pathways that are important in the recruitment, expansion, localization, and differentiation of cardiac progenitor cells.
The new data presented here indicate a critical role for the epicardium in regulating cardiac regenerative processes in the newt and are of great significance in light of recent studies in the mouse where the adult mammalian epicardium was shown to harbor dormant progenitor cells that can be activated and serve as a source for new cardiomyocytes (Riley, 2012). Moreover, it has been demonstrated that epicardial-derived signals can stimulate cardiomyocyte proliferation and coronary vasculogenesis within the myocardium (Kang et al., 2008; Olivey and Svensson, 2010; Stuckmann et al., 2003; Zhou et al., 2011). Thus, a deeper understanding of the differential functions of the epicardial transitional matrix in the regenerating newt heart may provide a template to engineer biomimetic matrices that promote regenerative rather than scar-forming responses in humans.
MATERIALS AND METHODS
Ethics Statement
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee at Northwestern University (IACUC animal welfare assurance number A-3995-01; IACUC approval number 99015). All surgery was performed under anesthesia, and every effort was made to minimize suffering.
Animal care and Surgical Procedures
Adult newts (Notophthalmus viridescens) were purchased from Connecticut Valley Biological Supply Company and housed at 22°C in tanks containing filtered dechlorinated water. Newts were fed bloodworms every 2-3 days. Before surgery, newts were anesthetized with 0.1% ethyl 3-aminobenzoate methanesulfonate salt (Sigma) in 0.04% Instant Ocean (Aquarium Systems). Following amputation of approximately 20% of the lower ventricle, animals were allowed to recover in 0.25% sulfamerazine sodium salt (Sigma) in 0.04% Instant Ocean. For proliferation studies, three days before tissue harvest, newts were injected intraperitoneally with 100 μL 1 mM 5-ethynyl-2′-deoxyuridine (EdU, Invitrogen) in 70% phosphate buffered saline (PBS, Cellgro) daily to allow for the identification of cells that had entered the cell cycle. For pulse-chase studies, newts were injected intraperitoneally with 100 μL 100 mM thymidine (Sigma) in 70% phosphate buffered saline (PBS, Cellgro) to dilute residual EdU and prevent further incorporation. Newts were re-anesthetized and regenerating hearts were harvested at designated time points, imbedded in OCT, frozen in an isopentane-dry ice slurry and stored at −80 °C.
Microarray and Gene Ontology analysis
Custom microarrays were prepared by Agilent Technologies using data from both in-house DNA sequences and public databases. Each microarray represented 1,915 putative transcripts from 10 newt species with over 90% of the transcript sequences being derived from N. viridescens. The majority of N. viridescens genes were cloned from mRNA and sequenced. Three 60-mer probes were designed for each transcript using Agilent eArray software and each probe was represented 7 or 8 times on a grid containing 44,000 oligonucleotide probes. The apex of the heart (≈20% of the ventricle) was removed with iridectomy scissors and flash frozen. RNA was isolated from regenerating tissues at 3, 7, or 14 dpa as previously described (Mercer et al., 2012). An equivalent section of non-regenerating heart tissue was collected as a 0 dpa time point control. RNA probe preparation and hybridizations were performed at The Huntsman Cancer Institute Microarray Core Facility at the University of Utah. RNA probes were generated by amplifying the isolated RNA and labeling with Cy3 or Cy5. Microarrays were competitively hybridized with Cy5- and Cy3-labeled probes from regenerating and day 0 (intact) tissues. Using GeneSpring GX bioinformatics software (Agilent), microarray data were Lowess normalized and Cy5:Cy3 ratios (regenerating vs. intact tissues) were obtained for each hybridization. Data were log10 transformed and the geometric means and standard deviations were obtained for repetitive hybridizations. All microarray experiments conform to MIAME guidelines.
Microarray data for zebrafish heart regeneration were obtained from Dr. Ching-Ling Lien and previously reported (Lien et al., 2006). Mouse and human post-MI microarray data are publicly available on the GEO database and discussed elsewhere (Hall et al., 2004; Tarnavski et al., 2004). All interspecies Gene Ontology term enrichment analyses were performed in GeneSpring. BLAST searches using NCBI databases were performed to identify gene orthologs or most closely related homologs to newt sequences when possible. Subsequently, the corresponding mouse official gene symbols were obtained for all annotated newt genes that exhibited differential expression in the microarrays for GO analysis. If at least 2 genes were associated with a returned GO term and the corrected P-value was ≤0.01, a GO term was considered significantly enriched for the input data set. For the GO data presented in Table 1, a filtering algorithm (Code S1) was applied to the raw data set to remove essentially identical GO terms (those with ≥90% overlapping associated genes) to avoid unnecessary redundancies; the entire GO data set is available in Dataset S1.
Immunohistochemistry
Serial cryosections of 14 μm were collected. Sections were fixed in 4% paraformaldehyde (PFA; Sigma) for 5 min, rinsed in PBS, permeabilized with 0.5% Triton X-100 (Amersham Life Science) in PBS for 1 min, then rinsed with PBS. Sections were blocked for 30 min [blocking buffer: 20% goat serum (Invitrogen), 0.2% bovine serum albumin (BSA, Sigma), 50 mM ammonium chloride (Sigma), 25 mM glycine (Fisher), 25 mM lysine (Sigma), and 0.02% sodium azide (Sigma) in PBS]. Primary antibodies were applied for 1 h and sections rinsed with 0.1% Tween-20 (Sigma) in PBS. Slides were blocked again for 5 min before staining with the appropriate secondary detection reagents for 30 min. Slides were rinsed with 0.1% Tween-20 in PBS and mounted with FLUORO-GEL (Electron Microscopy Sciences).
To visualize hyaluronic acid distribution, biotinylated hyaluronic acid binding protein (HABP, Calbiochem) was used as an indirect probe. After fixation and permeabilization, and prior to labeling sections with HABP, endogenous avidin and biotin activity was inactivated with an Avidin/Biotin blocking kit (Zymed Labs). Specificity was confirmed by preincubating HABP with a solution of 1 mg/mL hyaluronic acid sodium salt (Calbiochem).
To identify cells that incorporated EdU, a cell-permeable dye-azide was conjugated to EdU via a copper catalyzed click-chemistry reaction (Salic and Mitchison, 2008). Briefly, after fixation, permeabilization and blocking, sections were incubated for 30 min with AF488 conjugated azide (Invitrogen) diluted 1:500 in 2 M Tris (pH 8.5, Fisher), 50 mM copper (II) sulfate (Sigma) and 0.5 M ascorbic acid (Sigma). Sections were either imaged using a Leica DMI6000 or Zeiss LSM 510 Meta Confocal microscope. Image-Pro Analyzer (Media Cybernetics) was utilized to assemble individual sections into a single composite image. Adobe Photoshop was used to quantify EdU+/MF20+/WT1+ cells; cell count data was subsequently processed in Excel.
For the TUNEL assay, tissue sections were reacted with TMR Red-dUTP using the in situ cell death detection kit (Roche) following the manufacturer’s protocol with minor modifications. The sections were fixed with 4% PFA for 10 minutes, permeabilized in 0.25% Triton-X 100 and 0.1% sodium citrate (Fisher), and incubated for 2 h at room temperature with TMR Red-dUTP reagent. Positive and negative controls were obtained according to manufacturer’s protocol.
Antibodies
The following primary antibodies were used: mouse monoclonal anti-chick myosin heavy chain (MF20; Developmental Studies Hybridoma Bank), rabbit polyclonal anti-chick tenascin-C (Millipore), mouse monoclonal anti-human fibronectin (Sigma), rabbit polyclonal anti-human Wilms tumor 1 (WT1; Calbiochem), rabbit monoclonal anti-human phospho-histone H3 (PH3; Cell Signaling Technology). All primary antibodies were tested for species specificity with appropriate controls. ECM antibodies have been previously verified in the newt model (Calve et al., 2010; Calve and Simon, 2012). For immunofluorescence studies, primary antibodies were detected with appropriate species-specific Alexa Fluor-conjugated secondary antibodies (Invitrogen) and nuclei were stained with DAPI (Roche).
Cell culture
Primary cultures of newt cardiomyocytes were prepared using a modified version of previously described methods (Bettencourt-Dias et al., 2003). In short, ventricles were amputated from adult newts and stored overnight in 70% L15 media (Gibco) supplemented with 1x Pen/Strep/Fungiezone (Hyclone). Hearts were subsequently digested in 70% PBS (Cellgro) with 0.5% trypsin (Sigma), 400 U/ml type II collagenase (Sigma), 0.15% BSA (Sigma), 0.3% glucose (Sigma) and 1x Pen/Strep/Fungiezone for 6 – 10 hours in a shaking water bath at 27°C, with a change of enzyme solution every 2 hours. The resulting cell suspension was added to 4 ml 70% minimum essential medium (AMEM; Cellgro) with 10% FBS (Gibco), passed through a 100 μm microsieve, and centrifuged to collect the cells. The sedimented cells were resuspended in AMEM with 10% FBS and preplated in 10-cm culture dishes for 3 days at 25°C in a humidi ed 2.0% CO2 incubator. Myocytes remained in the non-adherent fraction during this purification step and were subsequently plated onto ECM-coated dishes at a density of 3,000 cells/cm2.
Assessing DNA synthesis in cultured cells
Commercially available ECMs were diluted in serum-free AMEM and were allowed to passively adsorb to multi-well tissue culture polystyrene dishes for a minimum of 1 h, then rinsed with PBS. Wells were coated in triplicate for the DNA synthesis assay. Importantly, ECM-coated dishes were not allowed to dry at any time before plating with cells. Successful coating was confirmed via IHC with respective antibodies. As described above, cardiomyocytes were cultured in AMEM for 7 days before EdU was added to the media of each well (final concentration of 1 μM). After 12 hours cells were fixed, permeabilized and stained with AF488 azide and other antibodies as described above. ≥10 non-overlapping images were acquired for each well and cell counting was performed in Adobe Photoshop. One-way ANOVA was used in PAST software to statistically assess the effects of different matrices on in vitro DNA synthesis.
Supplementary Material
Figure S1. Dynamic spatial expression of the transitional matrix early in the regenerative response. At 3 dpa, hyaluronic acid (HA, indirectly labeled with hyaluronic acid binding protein, HABP) is absent from the regenerating apex (A), but displays weak epicardial expression (B, C); in contrast, tenascin-C (TNC) infiltrates the myocardial muscle surrounding the resection (D) and is moderately expressed in the epicardium at this early time point (E, F). By 7 dpa, HA has enclosed the nucleated clot at the wound site (G) and is expressed throughout the epicardial sheath (H, I). In the regenerating newt heart, TNC remains localized to the epicardium at 7 dpa (K, L), with additional robust expression surrounding cardiac cells adjacent to the resection (J, L). Scale bars for A, B, D, E, G, H, J and K = 100 μm; scale bars for C, F, I and L = 50 μm. dpa = days postamputation.
Figure S2. Limited proliferative activity during the first week of newt heart regeneration. Cell cycle entry as determined by EdU incorporation at 3 and 7 dpa. At these early time points, only very few cells at the amputation plane (A, C) and in the adjacent myocardium (B, D; MF20 = red) are actively synthesizing DNA (EdU = white). Scale bar = 100 μm; dpa = days postamputation.
Figure S3. Existing cardiomyocytes contribute to the proliferative response. Confocal fluorescent microscopy reveals that after ventricular amputation both cardiac muscle cells (cardiomyocytes, MF20 = red) and non-cardiomyocytes (nuclei, DAPI = blue) reenter the cell cycle and synthesize DNA (EdU = white) at various time points and locations (A = 49 dpa near regenerating apex, B = 21 dpa near central myocardium). Double labeling for EdU (white) and phospho-histone H3 (PH3 = green) at 21 dpa further demonstrates that mature cardiomyocytes progress into mitosis and divide in support of tissue regeneration (C – F). Scale bar for A and C = 100 μm; scale bar for B and D – F = 50 μm.
Figure S4. Minimal apoptotic activity during the later stages of newt heart regeneration. Apoptotic cells as identified by TUNEL assay at 42 dpa. Six weeks after ventricular resection only extremely low numbers of apoptotic cells (red) were detected at the wound site (A – C) and in the myocardium/epicardium (D – F). Cycling cells (white) identified by EdU incorporation were found throughout the heart at this time point (A, D), with no obvious spatial correlation to apoptotic activity. Treatment of comparable tissue sections with DNase I causes extensive TUNEL labeling and serves as a positive reference for assay function (G); conversely, the TUNEL assay reveals no cell staining in the enzyme-free labeling reaction (H). Scale bars for A, B, D, E, G and H = 100 μm; scale bars for C and F = 50 μm.
Figure S5. Proliferative cells migrate from the myocardium to the regenerating ventricular apex late in the regenerative process. Following the peak proliferative response, EdU pulse-chase labeling identified cycling cells (EdU = white) throughout the myocardial space at 42 dpa (A). 1 week after pulse-chase, cells marked by EdU incorporation were substantially enriched at the regenerating apex (B), with further accumulation of EdU+ cells within the regenerate after 2 weeks (C). The dynamic change of progenitor cell location (myocardium vs. epicardium vs. regenerating apex) during the PC time course is quantified at the panel bottom (localization of EdU+ cells / total EdU+ cell population). MF20 identified striated muscle myosin heavy chain in the myocardium (red) and white dotted lines demarcate the amputation site. Scale bar = 400 μm; dpa = days postamputation.
Code S1. Filtering algorithm for identifying redundancy in Gene Ontology term enrichment results. This algorithm identifies returned GO terms that exhibit ≥90% overlap in their contributing genes. The program is written in Python; the input is a CSV file using tabs as delimiters with categories: GO term, associated genes, p-value, and accession number. The program assumes the input GO terms are sorted in the desired order. GO terms were first sorted by p-value so that increasingly less significant GO terms would be removed based on the redundancy requirement. The program outputs two files named terms.csv and terms_full.csv. The terms.csv file lists the GO terms in order of significance without any term determined to be redundant for a higher ranked non-redundant term. The terms_full.csv file provides the ranked GO terms from the input with annotation to indicate redundancies and the ratio of overlap between associated genes that contribute to the redundancy.
Dataset S1. Full multi-species Gene Ontology term enrichment analysis results with identified and sorted redundancies. Provided are all significantly enriched GO terms (p≤0.01) for newt and zebrafish heart regeneration at 3, 7, and 14 days postamputation; mouse cardiac repair following MI at 1, 4, 24, and 48 hours, as well as 1 and 8 weeks; and human cardiac repair within 10 days of an acute MI. GO analyses were performed using GeneSpring software and are derived from genes exhibiting differential expression (≥2-fold change relative to control) for the respective microarray data set. A custom-made filtering algorithm (Code S1) was applied to the GO term enrichment analysis results. GO terms are ranked by their p-value and redundancies with preceding (more significantly enriched) terms identified based on ≥90% overlap in the associated gene lists that contribute to the GO term.
Table S1. qRT-PCR validation of microarrays.
HIGHLIGHTS.
ECM remodeling is a conserved process during vertebrate heart regeneration
Tenascin-C, Hyaluronic Acid, and Fibronectin compose a regeneration-specific matrix
Ventricular resection triggers a myocardium-wide proliferative response
Tenascin-C supports cell cycle entry in newt cardiomyocytes
Progenitor cells migrate radially to the regenerative ECM-rich epicardium and apex
ACKNOWLEDGEMENTS
The authors are very grateful to W.M. Siler for writing the filtering algorithm utilized in the GO analyses. We thank C. Guzman and Dr. S. Calve for expert technical support. We are indebted to J. Krcmery for graciously providing the cartoon features of the model figure. We thank all members of the Simon laboratory for helpful discussions and contributions to the manuscript.
FUNDING STATEMENT This work was supported by DARPA, Restorative Injury Repair BAA04-12 Addendum B (H. -G. Simon), Searle Funds at the Chicago Community Trust (H. -G. Simon), American Heart Association predoctoral fellowship 11PRE5570021 (S. Mercer), and National Institutes of Health (NIH) Cellular and Molecular Basis of Disease training program T-32 GM008061-27 (S. Mercer). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- Bader D, Oberpriller J. Autoradiographic and electron microscopic studies of minced cardiac muscle regeneration in the adult newt, notophthalmus viridescens. J. Exp. Zool. 1979;208:177–193. doi: 10.1002/jez.1402080206. [DOI] [PubMed] [Google Scholar]
- Ballard VL, Edelberg JM. Stem cells and the regeneration of the aging cardiovascular system. Circul. Res. 2007;100:1116–1127. doi: 10.1161/01.RES.0000261964.19115.e3. [DOI] [PubMed] [Google Scholar]
- Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami CA, Anversa P. Evidence that human cardiac myocytes divide after myocardial infarction. N. Engl. J. Med. 2001;344:1750–1757. doi: 10.1056/NEJM200106073442303. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Mittnacht S, Brockes JP. Heterogeneous proliferative potential in regenerative adult newt cardiomyocytes. J. Cell Sci. 2003;116:4001–4009. doi: 10.1242/jcs.00698. [DOI] [PubMed] [Google Scholar]
- Borchardt T, Braun T. Cardiovascular regeneration in non-mammalian model systems: what are the differences between newts and man? Thromb. Haemost. 2007;98:311–318. [PubMed] [Google Scholar]
- Brockes JP. Amphibian limb regeneration: rebuilding a complex structure. Science. 1997;276:81–87. doi: 10.1126/science.276.5309.81. [DOI] [PubMed] [Google Scholar]
- Burgess ML, Buggy J, Price RL, Abel FL, Terracio L, Samarel AM, Borg TK. Exercise- and hypertension-induced collagen changes are related to left ventricular function in rat hearts. Am. J. Physiol. 1996;270:H151–159. doi: 10.1152/ajpheart.1996.270.1.H151. [DOI] [PubMed] [Google Scholar]
- Calve S, Odelberg SJ, Simon HG. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev. Biol. 2010;344:259–271. doi: 10.1016/j.ydbio.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calve S, Simon HG. Biochemical and mechanical environment cooperatively regulate skeletal muscle regeneration. FASEB J. 2012;26:2538–2545. doi: 10.1096/fj.11-200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chablais F, Veit J, Rainer G, Jazwinska A. The zebrafish heart regenerates after cryoinjury-induced myocardial infarction. BMC Dev Biol. 2011;11:21. doi: 10.1186/1471-213X-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Chang TC, Kang JO, Choudhary B, Makita T, Tran CM, Burch JB, Eid H, Sucov HM. Epicardial induction of fetal cardiomyocyte proliferation via a retinoic acid-inducible trophic factor. Dev. Biol. 2002;250:198–207. doi: 10.1006/dbio.2002.0796. [DOI] [PubMed] [Google Scholar]
- Chen WY, Abatangelo G. Functions of hyaluronan in wound repair. Wound Repair Regen. 1999;7:79–89. doi: 10.1046/j.1524-475x.1999.00079.x. [DOI] [PubMed] [Google Scholar]
- Cleutjens JP, Creemers EE. Integration of concepts: cardiac extracellular matrix remodeling after myocardial infarction. J. Card. Fail. 2002;8:S344–348. doi: 10.1054/jcaf.2002.129261. [DOI] [PubMed] [Google Scholar]
- Cox TR, Erler JT. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Disease models & mechanisms. 2011;4:165–178. doi: 10.1242/dmm.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creemers EE, Cleutjens JP, Smits JF, Daemen MJ. Matrix metalloproteinase inhibition after myocardial infarction: a new approach to prevent heart failure? Circ. Res. 2001;89:201–210. doi: 10.1161/hh1501.094396. [DOI] [PubMed] [Google Scholar]
- Del Rio-Tsonis K, Jung JC, Chiu IM, Tsonis PA. Conservation of fibroblast growth factor function in lens regeneration. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13701–13706. doi: 10.1073/pnas.94.25.13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobaczewski M, Bujak M, Zymek P, Ren G, Entman ML, Frangogiannis NG. Extracellular matrix remodeling in canine and mouse myocardial infarcts. Cell Tissue Res. 2006;324:475–488. doi: 10.1007/s00441-005-0144-6. [DOI] [PubMed] [Google Scholar]
- Dobaczewski M, Gonzalez-Quesada C, Frangogiannis NG. The extracellular matrix as a modulator of the inflammatory and reparative response following myocardial infarction. J. Mol. Cell. Cardiol. 2010;48:504–511. doi: 10.1016/j.yjmcc.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati AK, Zalewski AA, Reddi AH. An immunofluorescent study of the distribution of fibronectin and laminin during limb regeneration in the adult newt. Dev. Biol. 1983;96:355–365. doi: 10.1016/0012-1606(83)90173-2. [DOI] [PubMed] [Google Scholar]
- Hall JL, Grindle S, Han X, Fermin D, Park S, Chen Y, Bache RJ, Mariash A, Guan Z, Ormaza S, Thompson J, Graziano J, de Sam Lazaro SE, Pan S, Simari RD, Miller LW. Genomic profiling of the human heart before and after mechanical support with a ventricular assist device reveals alterations in vascular signaling networks. Physiol Genomics. 2004;17:283–291. doi: 10.1152/physiolgenomics.00004.2004. [DOI] [PubMed] [Google Scholar]
- Hohenstein P, Hastie ND. The many facets of the Wilms’ tumour gene, WT1. Hum. Mol. Genet. 2006;15(Spec No 2):R196–201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- Itou J, Oishi I, Kawakami H, Glass TJ, Richter J, Johnson A, Lund TC, Kawakami Y. Migration of cardiomyocytes is essential for heart regeneration in zebrafish. Development. 2012;139:4133–4142. doi: 10.1242/dev.079756. [DOI] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajstura J, Leri A, Finato N, Di Loreto C, Beltrami CA, Anversa P. Myocyte proliferation in end-stage cardiac failure in humans. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8801–8805. doi: 10.1073/pnas.95.15.8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Gu Y, Li P, Johnson BL, Sucov HM, Thomas PS. PDGF-A as an epicardial mitogen during heart development. Dev. Dyn. 2008;237:692–701. doi: 10.1002/dvdy.21469. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Gupta V, Wang J, Holdway JE, Wills AA, Fang Y, Poss KD. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Turnbull J, Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 2011;209:139–151. doi: 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- Kwee L, Baldwin HS, Shen HM, Stewart CL, Buck C, Buck CA, Labow MA. Defective development of the embryonic and extraembryonic circulatory systems in vascular cell adhesion molecule (VCAM-1) deficient mice. Development. 1995;121:489–503. doi: 10.1242/dev.121.2.489. [DOI] [PubMed] [Google Scholar]
- Laube F, Heister M, Scholz C, Borchardt T, Braun T. Re-programming of newt cardiomyocytes is induced by tissue regeneration. J. Cell Sci. 2006;119:4719–4729. doi: 10.1242/jcs.03252. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Dev Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes Dev. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- Lepilina A, Coon AN, Kikuchi K, Holdway JE, Roberts RW, Burns CG, Poss KD. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo DC, Allen F, Brockes JP. Reversal of muscle differentiation during urodele limb regeneration. Proc. Natl. Acad. Sci. U. S. A. 1993;90:7230–7234. doi: 10.1073/pnas.90.15.7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarev E, Call MK, Grogg MW, Atkinson DL, Milash B, Odelberg SJ, Tsonis PA. Gene expression signatures in the newt irises during lens regeneration. FEBS Lett. 2007;581:1865–1870. doi: 10.1016/j.febslet.2007.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx KA, Sharko J, Grinstein GG, Odelberg S, Simon HG. Evidence for proximal to distal appendage amputation site effects from global gene expression correlations found in newt microarrays. Proceedings of the 7th Ieee International Symposium on Bioinformatics and Bioengineering.2007. pp. 131–136. [Google Scholar]
- Matsumura S, Iwanaga S, Mochizuki S, Okamoto H, Ogawa S, Okada Y. Targeted deletion or pharmacological inhibition of MMP-2 prevents cardiac rupture after myocardial infarction in mice. J. Clin. Invest. 2005;115:599–609. doi: 10.1172/JCI22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer SE, Cheng CH, Atkinson DL, Krcmery J, Guzman CE, Kent DT, Zukor K, Marx KA, Odelberg SJ, Simon HG. Multi-tissue microarray analysis identifies a molecular signature of regeneration. PLoS One. 2012;7:e52375. doi: 10.1371/journal.pone.0052375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberpriller J, Oberpriller JC. Mitosis in adult newt ventricle. J. Cell Biol. 1971;49:560–563. doi: 10.1083/jcb.49.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ. Res. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda H, Poulin ML, Tassava RA, Chiu IM. Characterization of a newt tenascin cDNA and localization of tenascin mRNA during newt limb regeneration by in situ hybridization. Dev. Biol. 1991;148:219–232. doi: 10.1016/0012-1606(91)90331-v. [DOI] [PubMed] [Google Scholar]
- Palojoki E, Saraste A, Eriksson A, Pulkki K, Kallajoki M, Voipio-Pulkki LM, Tikkanen I. Cardiomyocyte apoptosis and ventricular remodeling after myocardial infarction in rats. Am J Physiol Heart Circ Physiol. 2001;280:H2726–2731. doi: 10.1152/ajpheart.2001.280.6.H2726. [DOI] [PubMed] [Google Scholar]
- Perez-Pomares JM, Phelps A, Sedmerova M, Carmona R, Gonzalez-Iriarte M, Munoz-Chapuli R, Wessels A. Experimental studies on the spatiotemporal expression of WT1 and RALDH2 in the embryonic avian heart: a model for the regulation of myocardial and valvuloseptal development by epicardially derived cells (EPDCs) Dev. Biol. 2002;247:307–326. doi: 10.1006/dbio.2002.0706. [DOI] [PubMed] [Google Scholar]
- Piatkowski T, Muehlfeld C, Borchardt T, Braun T. Reconstitution of the myocardium in regenerating newt hearts is preceded by transient deposition of extracellular matrix components. Stem cells and development. 2013 doi: 10.1089/scd.2012.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Reffelmann T, Leor J, Muller-Ehmsen J, Kedes L, Kloner RA. Cardiomyocyte transplantation into the failing heart-new therapeutic approach for heart failure? Heart Fail Rev. 2003;8:201–211. doi: 10.1023/a:1024796912475. [DOI] [PubMed] [Google Scholar]
- Repesh LA, Fitzgerald TJ, Furcht LT. Changes in the distribution of fibronectin during limb regeneration in newts using immunocytochemistry. Differentiation. 1982;22:125–131. doi: 10.1111/j.1432-0436.1982.tb01236.x. [DOI] [PubMed] [Google Scholar]
- Riley PR. An epicardial floor plan for building and rebuilding the mammalian heart. Curr. Top. Dev. Biol. 2012;100:233–251. doi: 10.1016/B978-0-12-387786-4.00007-5. [DOI] [PubMed] [Google Scholar]
- Ripellino JA, Klinger MM, Margolis RU, Margolis RK. The hyaluronic acid binding region as a specific probe for the localization of hyaluronic acid in tissue sections. Application to chick embryo and rat brain. J. Histochem. Cytochem. 1985;33:1060–1066. doi: 10.1177/33.10.4045184. [DOI] [PubMed] [Google Scholar]
- Rosso F, Giordano A, Barbarisi M, Barbarisi A. From cell-ECM interactions to tissue engineering. J. Cell. Physiol. 2004;199:174–180. doi: 10.1002/jcp.10471. [DOI] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U. S. A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste A, Pulkki K, Kallajoki M, Henriksen K, Parvinen M, Voipio-Pulkki LM. Apoptosis in human acute myocardial infarction. Circulation. 1997;95:320–323. doi: 10.1161/01.cir.95.2.320. [DOI] [PubMed] [Google Scholar]
- Schnabel K, Wu CC, Kurth T, Weidinger G. Regeneration of cryoinjury induced necrotic heart lesions in zebrafish is associated with epicardial activation and cardiomyocyte proliferation. PLoS One. 2011;6:e18503. doi: 10.1371/journal.pone.0018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz GS, Wysocki A. Interactions between extracellular matrix and growth factors in wound healing. Wound Repair Regen. 2009;17:153–162. doi: 10.1111/j.1524-475X.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- Seccia TM, Bettini E, Vulpis V, Quartaroli M, Trist DG, Gaviraghi G, Pirrelli A. Extracellular matrix gene expression in the left ventricular tissue of spontaneously hypertensive rats. Vol. 8. Blood Press; 1999. pp. 57–64. [DOI] [PubMed] [Google Scholar]
- See F, Kompa A, Martin J, Lewis DA, Krum H. Fibrosis as a therapeutic target post-myocardial infarction. Curr. Pharm. Des. 2005;11:477–487. doi: 10.2174/1381612053382098. [DOI] [PubMed] [Google Scholar]
- Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2012 doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson TJ, Vinarsky V, Atkinson DL, Keating MT, Odelberg SJ. Tissue inhibitor of metalloproteinase 1 regulates matrix metalloproteinase activity during newt limb regeneration. Dev. Dyn. 2006;235:606–616. doi: 10.1002/dvdy.20654. [DOI] [PubMed] [Google Scholar]
- Strauer BE, Kornowski R. Stem cell therapy in perspective. Circulation. 2003;107:929–934. doi: 10.1161/01.cir.0000057525.13182.24. [DOI] [PubMed] [Google Scholar]
- Stuckmann I, Evans S, Lassar AB. Erythropoietin and retinoic acid, secreted from the epicardium, are required for cardiac myocyte proliferation. Dev. Biol. 2003;255:334–349. doi: 10.1016/s0012-1606(02)00078-7. [DOI] [PubMed] [Google Scholar]
- Tarnavski O, McMullen JR, Schinke M, Nie Q, Kong S, Izumo S. Mouse cardiac surgery: comprehensive techniques for the generation of mouse models of human diseases and their application for genomic studies. Physiol Genomics. 2004;16:349–360. doi: 10.1152/physiolgenomics.00041.2003. [DOI] [PubMed] [Google Scholar]
- Tassava RA, Nace JD, Wei Y. Extracellular matrix protein turnover during salamander limb regeneration. Wound Repair Regen. 1996;4:75–81. doi: 10.1046/j.1524-475X.1996.40113.x. [DOI] [PubMed] [Google Scholar]
- Thomas DP, McCormick RJ, Zimmerman SD, Vadlamudi RK, Gosselin LE. Aging- and training-induced alterations in collagen characteristics of rat left ventricle and papillary muscle. Am. J. Physiol. 1992;263:H778–783. doi: 10.1152/ajpheart.1992.263.3.H778. [DOI] [PubMed] [Google Scholar]
- Toole BP, Gross J. The extracellular matrix of the regenerating newt limb: synthesis and removal of hyaluronate prior to differentiation. Dev. Biol. 1971;25:57–77. doi: 10.1016/0012-1606(71)90019-4. [DOI] [PubMed] [Google Scholar]
- Vinarsky V, Atkinson DL, Stevenson TJ, Keating MT, Odelberg SJ. Normal newt limb regeneration requires matrix metalloproteinase function. Dev. Biol. 2005;279:86–98. doi: 10.1016/j.ydbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- von Gise A, Zhou B, Honor LB, Ma Q, Petryk A, Pu WT. WT1 regulates epicardial epithelial to mesenchymal transition through beta-catenin and retinoic acid signaling pathways. Dev. Biol. 2011;356:421–431. doi: 10.1016/j.ydbio.2011.05.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldenstrom A, Martinussen HJ, Gerdin B, Hallgren R. Accumulation of hyaluronan and tissue edema in experimental myocardial infarction. J. Clin. Invest. 1991;88:1622–1628. doi: 10.1172/JCI115475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KT, Sun Y, Tyagi SC, Cleutjens JP. Collagen network of the myocardium: function, structural remodeling and regulatory mechanisms. J. Mol. Cell. Cardiol. 1994;26:279–292. doi: 10.1006/jmcc.1994.1036. [DOI] [PubMed] [Google Scholar]
- Witman N, Murtuza B, Davis B, Arner A, Morrison JI. Recapitulation of developmental cardiogenesis governs the morphological and functional regeneration of adult newt hearts following injury. Dev. Biol. 2011;354:67–76. doi: 10.1016/j.ydbio.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Zhou B, Honor LB, He H, Ma Q, Oh JH, Butterfield C, Lin RZ, Melero-Martin JM, Dolmatova E, Duffy HS, Gise A, Zhou P, Hu YW, Wang G, Zhang B, Wang L, Hall JL, Moses MA, McGowan FX, Pu WT. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukor KA, Kent DT, Odelberg SJ. Meningeal cells and glia establish a permissive environment for axon regeneration after spinal cord injury in newts. Neural Dev. 2011;6:1. doi: 10.1186/1749-8104-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Dynamic spatial expression of the transitional matrix early in the regenerative response. At 3 dpa, hyaluronic acid (HA, indirectly labeled with hyaluronic acid binding protein, HABP) is absent from the regenerating apex (A), but displays weak epicardial expression (B, C); in contrast, tenascin-C (TNC) infiltrates the myocardial muscle surrounding the resection (D) and is moderately expressed in the epicardium at this early time point (E, F). By 7 dpa, HA has enclosed the nucleated clot at the wound site (G) and is expressed throughout the epicardial sheath (H, I). In the regenerating newt heart, TNC remains localized to the epicardium at 7 dpa (K, L), with additional robust expression surrounding cardiac cells adjacent to the resection (J, L). Scale bars for A, B, D, E, G, H, J and K = 100 μm; scale bars for C, F, I and L = 50 μm. dpa = days postamputation.
Figure S2. Limited proliferative activity during the first week of newt heart regeneration. Cell cycle entry as determined by EdU incorporation at 3 and 7 dpa. At these early time points, only very few cells at the amputation plane (A, C) and in the adjacent myocardium (B, D; MF20 = red) are actively synthesizing DNA (EdU = white). Scale bar = 100 μm; dpa = days postamputation.
Figure S3. Existing cardiomyocytes contribute to the proliferative response. Confocal fluorescent microscopy reveals that after ventricular amputation both cardiac muscle cells (cardiomyocytes, MF20 = red) and non-cardiomyocytes (nuclei, DAPI = blue) reenter the cell cycle and synthesize DNA (EdU = white) at various time points and locations (A = 49 dpa near regenerating apex, B = 21 dpa near central myocardium). Double labeling for EdU (white) and phospho-histone H3 (PH3 = green) at 21 dpa further demonstrates that mature cardiomyocytes progress into mitosis and divide in support of tissue regeneration (C – F). Scale bar for A and C = 100 μm; scale bar for B and D – F = 50 μm.
Figure S4. Minimal apoptotic activity during the later stages of newt heart regeneration. Apoptotic cells as identified by TUNEL assay at 42 dpa. Six weeks after ventricular resection only extremely low numbers of apoptotic cells (red) were detected at the wound site (A – C) and in the myocardium/epicardium (D – F). Cycling cells (white) identified by EdU incorporation were found throughout the heart at this time point (A, D), with no obvious spatial correlation to apoptotic activity. Treatment of comparable tissue sections with DNase I causes extensive TUNEL labeling and serves as a positive reference for assay function (G); conversely, the TUNEL assay reveals no cell staining in the enzyme-free labeling reaction (H). Scale bars for A, B, D, E, G and H = 100 μm; scale bars for C and F = 50 μm.
Figure S5. Proliferative cells migrate from the myocardium to the regenerating ventricular apex late in the regenerative process. Following the peak proliferative response, EdU pulse-chase labeling identified cycling cells (EdU = white) throughout the myocardial space at 42 dpa (A). 1 week after pulse-chase, cells marked by EdU incorporation were substantially enriched at the regenerating apex (B), with further accumulation of EdU+ cells within the regenerate after 2 weeks (C). The dynamic change of progenitor cell location (myocardium vs. epicardium vs. regenerating apex) during the PC time course is quantified at the panel bottom (localization of EdU+ cells / total EdU+ cell population). MF20 identified striated muscle myosin heavy chain in the myocardium (red) and white dotted lines demarcate the amputation site. Scale bar = 400 μm; dpa = days postamputation.
Code S1. Filtering algorithm for identifying redundancy in Gene Ontology term enrichment results. This algorithm identifies returned GO terms that exhibit ≥90% overlap in their contributing genes. The program is written in Python; the input is a CSV file using tabs as delimiters with categories: GO term, associated genes, p-value, and accession number. The program assumes the input GO terms are sorted in the desired order. GO terms were first sorted by p-value so that increasingly less significant GO terms would be removed based on the redundancy requirement. The program outputs two files named terms.csv and terms_full.csv. The terms.csv file lists the GO terms in order of significance without any term determined to be redundant for a higher ranked non-redundant term. The terms_full.csv file provides the ranked GO terms from the input with annotation to indicate redundancies and the ratio of overlap between associated genes that contribute to the redundancy.
Dataset S1. Full multi-species Gene Ontology term enrichment analysis results with identified and sorted redundancies. Provided are all significantly enriched GO terms (p≤0.01) for newt and zebrafish heart regeneration at 3, 7, and 14 days postamputation; mouse cardiac repair following MI at 1, 4, 24, and 48 hours, as well as 1 and 8 weeks; and human cardiac repair within 10 days of an acute MI. GO analyses were performed using GeneSpring software and are derived from genes exhibiting differential expression (≥2-fold change relative to control) for the respective microarray data set. A custom-made filtering algorithm (Code S1) was applied to the GO term enrichment analysis results. GO terms are ranked by their p-value and redundancies with preceding (more significantly enriched) terms identified based on ≥90% overlap in the associated gene lists that contribute to the GO term.
Table S1. qRT-PCR validation of microarrays.