Abstract
Long-term diet influences the structure and activity of the trillions of microorganisms residing in the human gut1–5, but it remains unclear how rapidly and reproducibly the human gut microbiome responds to short-term macronutrient change. Here, we show that the short-term consumption of diets composed entirely of animal or plant products alters microbial community structure and overwhelms inter-individual differences in microbial gene expression. The animal-based diet increased the abundance of bile-tolerant microorganisms (Alistipes, Bilophila, and Bacteroides) and decreased the levels of Firmicutes that metabolize dietary plant polysaccharides (Roseburia, Eubacterium rectale, and Ruminococcus bromii). Microbial activity mirrored differences between herbivorous and carnivorous mammals2, reflecting trade-offs between carbohydrate and protein fermentation. Foodborne microbes from both diets transiently colonized the gut, including bacteria, fungi, and even viruses. Finally, increases in the abundance and activity of Bilophila wadsworthia on the animal-based diet support a link between dietary fat, bile acids, and the outgrowth of microorganisms capable of triggering inflammatory bowel disease6. In concert, these results demonstrate that the gut microbiome can rapidly respond to altered diet, potentially facilitating the diversity of human dietary lifestyles.
There is growing concern that recent lifestyle innovations, most notably the high-fat/high-sugar “Western” diet, have altered the genetic composition and metabolic activity of our resident microorganisms (the human gut microbiome)7. Such diet-induced changes to gut-associated microbial communities are now suspected of contributing to growing epidemics of chronic illness in the developed world, including obesity4,8 and inflammatory bowel disease6. Yet, it remains unclear how quickly and reproducibly gut bacteria respond to dietary change. Work in inbred mice shows that shifting dietary macronutrients can broadly and consistently alter the gut microbiome within a single day7,9. By contrast, dietary interventions in human cohorts have only measured community changes on timescales of weeks10 to months4, failed to find significant diet-specific effects1, or demonstrated responses among a limited number of bacterial taxa3,5.
Here, we examined if dietary interventions in humans can alter gut microbial communities in a rapid, diet-specific manner. We prepared two diets that varied according to their primary food source: a “plant-based diet”, which was rich in grains, legumes, fruits, and vegetables; and an “animal-based diet”, which was composed of meats, eggs, and cheeses (Supplementary Table 1). We picked these sources to span the global diversity of modern human diets, which includes exclusively plant-based and nearly exclusively animal-based regimes11 (the latter being the case among some high-latitude and pastoralist cultures). Each diet was consumed ad libitum for five consecutive days by six male and four female American volunteers between the ages of 21–33, whose body mass indices ranged from 19–32 kg/m2 (Supplementary Table 2). Study volunteers were observed for four days before each diet arm to measure normal eating habits (the baseline period) and for six days after each diet arm to assess microbial recovery (the washout period; Extended Data Fig. 1). Subjects’ baseline nutritional intake correlated well with their estimated long-term diet (Supplementary Table 3). Our study cohort included a lifetime vegetarian (see Supplementary Discussion, Extended Data Fig. 2, and Supplementary Table 4 for a detailed analysis of his diet and gut microbiota).
Each diet arm significantly shifted subjects' macronutrient intake (Fig. 1a–c). On the animal-based diet, dietary fat increased from 32.5±2.2% to 69.5±0.4% kcal and dietary protein increased from 16.2±1.3% to 30.1±0.5% kcal (p<0.01 for both comparisons, Wilcoxon signed-rank test; Supplementary Table 5). Fiber intake was nearly zero, in contrast to baseline levels of 9.3±2.1 g/1,000kcal. On the plant-based diet, fiber intake rose to 25.6±1.1 g/1,000kcal, while both fat and protein intake declined to 22.1±1.7% and 10.0±0.3%, respectively (p<0.05 for all comparisons). Subjects’ weights on the plant-based diet remained stable, but decreased significantly by day 3 of the animal-based diet (q<0.05, Bonferroni-corrected Mann-Whitney U test; Extended Data Fig. 3). Differential weight loss between the two diets cannot be explained simply by energy intake, as subjects consumed equal numbers of calories on the plant- and animal-based diets (1,695±172 kcal and 1,777±221 kcal, respectively; p=0.44).
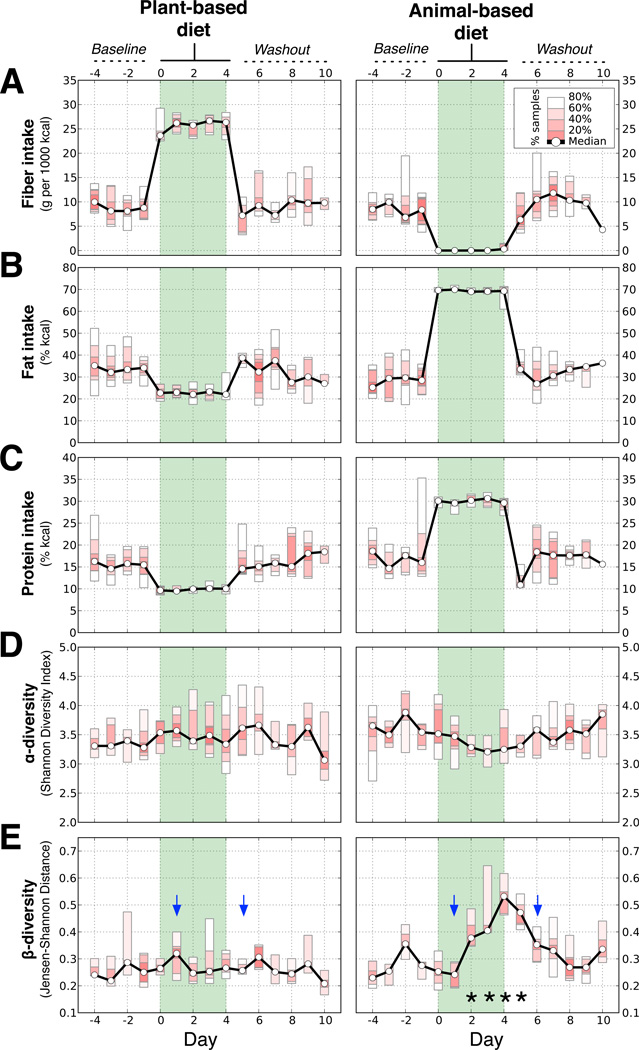
Fig. 1. Short-term diet alters the gut microbiota.
Ten subjects were tracked across each diet arm. (A) Fiber intake on the plant-based diet rose from a median baseline value of 9.3±2.1 to 25.6±1.1 g/1,000kcal (p=0.007; two-sided Wilcoxon signed-rank test), but was negligible on the animal-based diet (p=0.005). (B) Daily fat intake doubled on the animal-based diet from a baseline of 32.5±2.2% to 69.5±0.4% kcal (p=0.005), but dropped on the plant-based diet to 22.1±1.7% (p=0.02). (C) Protein intake rose on the animal-based diet to 30.1±0.5% kcal from a baseline level of 16.2±1.3% (p=0.005) and decreased on the plant-based diet to 10.0±0.3% (p=0.005). (D) Within-sample species diversity (α-diversity, Shannon’s Diversity Index), did not significantly change during either diet. (E) The similarity of each individual’s gut microbiota to their baseline communities (β-diversity, Jensen-Shannon distance) decreased on the animal-based diet (dates with q<0.05 identified with asterisks; Bonferroni-corrected, two-sided Mann-Whitney U test). Community differences were apparent one day after a tracing dye showed the animal-based diet reached the gut (blue arrows depict appearance of food dyes added to first and last diet day meals; Extended Data Fig. 3a).
To characterize temporal patterns of microbial community structure, we performed 16S rRNA gene sequencing on samples collected each day of the study (Supplementary Table 6). We quantified the microbial diversity within each subject at a given time-point (α-diversity) and the difference between each subjects' baseline and diet-associated gut microbiota (β-diversity) (Fig. 1d,e). Although no significant differences in α-diversity were detected on either diet, we observed a significant increase in β-diversity that was unique to the animal-based diet (q<0.05, Bonferroni-corrected Mann-Whitney U test). This change occurred a single day after the diet reached the distal gut microbiota (as indicated by the food tracking dye; Extended Data Fig. 3a). Subjects’ gut microbiota reverted to their original structure 2 days after the animal-based diet ended (Fig. 1e).
Analysis of the relative abundance of bacterial taxonomic groups supported our finding that the animal-based diet had a greater impact on the gut microbiota than the plant-based diet (Fig. 2). We hierarchically clustered species-level bacterial phylotypes by the similarity of their dynamics across diets and subjects (see Supplementary Methods and Supplementary Tables 7 and 8). Statistical testing identified 22 clusters whose abundance significantly changed while on the animal-based diet, while only 3 clusters showed significant abundance changes while on the plant-based diet (q<0.05, Wilcoxon signed-rank test; Supplementary Table 9). Notably, the genus Prevotella, one of the leading sources of inter-individual gut microbiota variation12 and hypothesized to be sensitive to long-term fiber intake1,13, was reduced in our vegetarian subject during consumption of the animal-based diet (see Supplementary Discussion). We also observed a significant positive correlation between subjects’ fiber intake over the past year and baseline gut Prevotella levels (Extended Data Fig. 4 and Supplementary Table 10).
Fig. 2. Bacterial cluster responses to diet arms.
Cluster log2 fold-changes on each diet arm were computed relative to baseline samples across all subjects and are drawn as circles. Clusters with significant fold-changes on the animal-based diet are colored in red, and clusters with significant fold-changes on both the plant- and animal-based diets are colored in both red and green. Uncolored clusters exhibited no significant fold-change on either the animal or plant-based diet (q<0.05, two-sided Wilcoxon signed-rank test). Bacterial membership in the clusters with the three largest positive and negative fold-changes on the animal-based diet are also displayed and colored by phylum: Firmicutes (purple), Bacteroidetes (blue), Proteobacteria (green), Tenericutes (red), and Verrucomicrobia (gray). Multiple OTUs with the same name are counted in parentheses.
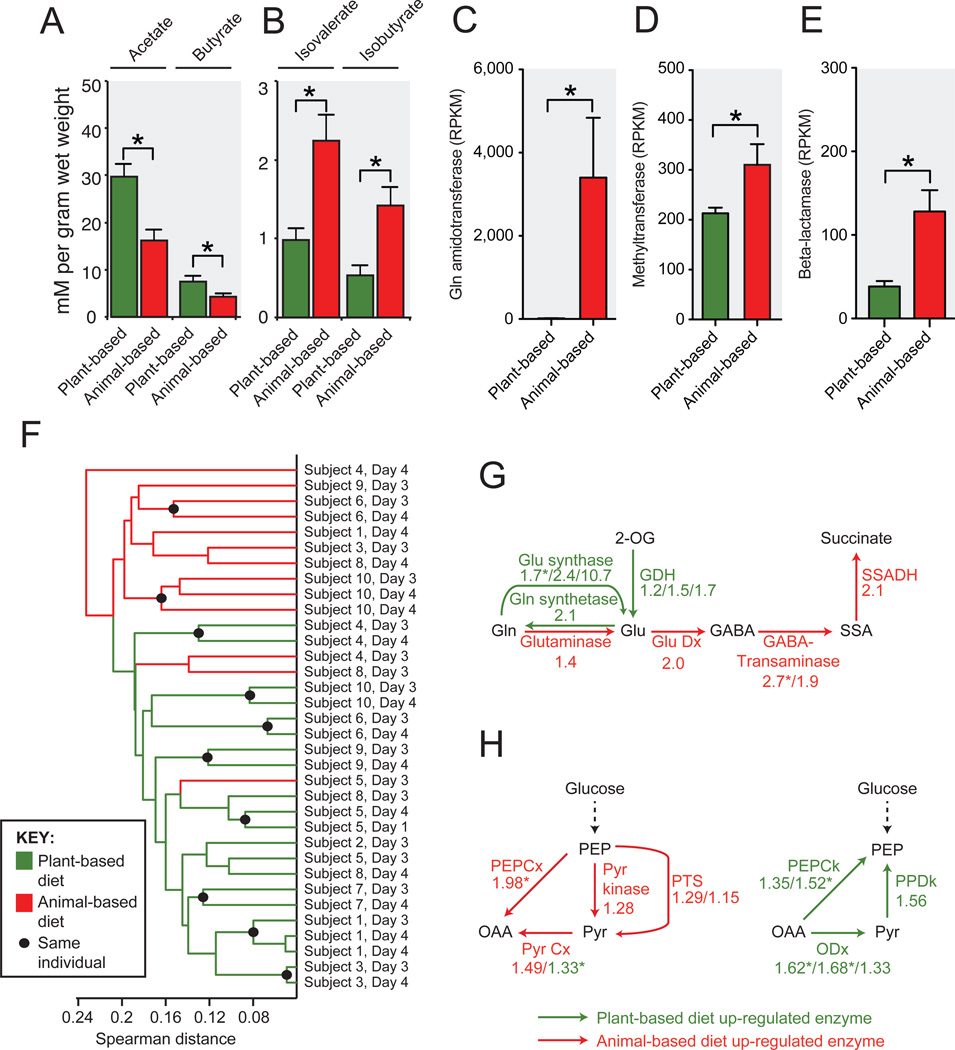
To identify functional traits linking clusters that thrived on the animal-based diet, we selected the most abundant taxon in the three most-enriched clusters (Bilophila wadsworthia, Cluster 28; Alistipes putredinis, Cluster 26; and a Bacteroides sp., Cluster 29), and performed a literature search for their lifestyle traits. That search quickly yielded a common theme of bile-resistance for these taxa, which is consistent with observations that high fat intake causes more bile acids to be secreted14.
Analysis of fecal SCFAs and bacterial clusters suggests that macronutrient shifts on both diets also altered microbial metabolic activity. Relative to the plant-based diet and baseline samples, the animal-based diet resulted in significantly lower levels of the products of carbohydrate fermentation and a higher concentration of the products of amino acid fermentation (Fig. 3a,b; Supplementary Table 11). When we correlated subjects’ SCFA concentrations with the same-day abundance of bacterial clusters from Fig. 2, we found significant positive relationships between clusters composed of putrefactive microbes15,16 (i.e. Alistipes putredinis and Bacteroides spp.) and SCFAs that are the end products of amino acid fermentation (Extended Data Fig. 5). We also observed significant positive correlations between clusters comprised of saccharolytic microbes3 (e.g. Roseburia, E. rectale, and F. prausnitzii) and the products of carbohydrate fermentation.
Fig. 3. Diet alters microbial activity and gene expression.
Fecal concentrations of SCFAs from (A) carbohydrate and (B) amino acid fermentation (*p<0.05, two-sided Mann-Whitney U test; n=9–11 fecal samples/diet arm; Supplementary Table 11). The animal-based diet was associated with significant increases in gene expression (normalized to reads per kilobase per million mapped, or RPKM; n=13–21 datasets/diet arm) among (C) glutamine amidotransferases (K08681, vitamin B6 metabolism), (D) methyltransferases (K00599, polycyclic aromatic hydrocarbon degradation), and (E) beta-lactamases (K01467). (F) Hierarchical clustering of gut microbial gene expression profiles collected on the animal-based (red) and plant-based (green) diets. Expression profile similarity was significantly associated with diet (p<0.003; two-sided Fisher’s exact test excluding replicate samples), despite inter-individual variation that preceded the diet (Extended Data Figs. 6a,b). Enrichment on animal-based diet (red) and plant-based diet (green) for expression of genes involved in (G) amino acid metabolism and (H) central metabolism. Numbers indicate the mean fold-change between the two diets for each KEGG orthologous group assigned to a given enzymatic reaction (Supplementary Table 17). Enrichment patterns on the animal- and plant-based diets agree perfectly with patterns observed in carnivorous and herbivorous mammals, respectively2 (p<0.001, Binomial test). Note: Pyr Cx is represented by two groups, which showed divergent fold-changes. Asterisks in panels C-E and G,H indicate p<0.05, Student’s t test. Values in panels A-E are mean±sem. Abbreviations: glutamate dehydrogenase (GDH), glutamate decarboxylase (Glu Dx), succinate-semialdehyde dehydrogenase (SSADH), phosphoenolpyruvate carboxylase (PEPCx), pyruvate carboxylase (Pyr Cx), phosphotransferase system (PTS), PEP carboxykinase (PEPCk), oxaloacetate decarboxylase (ODx), pyruvate, orthophosphate dikinase (PPDk).
In order to test whether the observed changes in microbial community structure and metabolic end products were accompanied by more widespread shifts in the gut microbiome, we measured microbial gene expression using RNA sequencing (RNA-Seq). A subset of samples was analyzed, targeting the baseline periods and the final 2 days of each diet (Extended Data Fig. 1, Supplementary Table 12). We identified several differentially-expressed metabolic modules and pathways during the plant- and animal-based diets (Supplementary Tables 13 and 14). The animal-based diet was associated with increased expression of key genes for vitamin biosynthesis (Fig. 3c); the degradation of polycyclic aromatic hydrocarbons (Fig. 3d), which are carcinogenic compounds produced during the charring of meat17; and the increased expression of β-lactamase genes (Fig. 3e). Metagenomic models constructed from our 16S rRNA data18 suggest that the observed expression differences are due to a combination of regulatory and taxonomic shifts within the microbiome (Supplementary Tables 15 and 16).
We next hierarchically-clustered microbiome samples based on the transcription of KEGG orthologous groups19, which suggested that overall microbial gene expression was strongly linked to host diet. Nearly all of the diet samples could be clustered by diet arm (p<0.003, Fisher’s exact test; Fig. 3f), despite the pre-existing inter-individual variation we observed during the baseline diets (Extended Data Fig. 6a,b). Still, subjects maintained their inter-individual differences on a taxonomic level on the diet arms (Extended Data Fig. 6c). Of the three RNA-Seq samples on the animal-based diet that clustered with samples from the plant-based diet, all were taken on day 3 of the diet arm. In contrast, all RNA-Seq samples from the final day of the diet arms (day 4) clustered by diet (Fig. 3f).
Remarkably, the plant- and animal-based diets also elicited transcriptional responses that were consistent with known differences in gene abundance between the gut microbiomes of herbivorous and carnivorous mammals, such as the tradeoffs between amino acid catabolism versus biosynthesis, and in the interconversions of phosphoenolpyruvate (PEP) and oxaloacetate2 (Fig. 3g,h). The former pathway favors amino acid catabolism when protein is abundant2, and we speculate that the latter pathway produces PEP for aromatic amino acid synthesis when protein is scarce20. In all 14 steps of these pathways, we observed fold-changes in gene expression on the plant- and animal-based diets whose directions agreed with the previously reported differences between herbivores and carnivores (p<0.001, Binomial test). Notably, this perfect agreement is not observed when the plant- and animal-based diets are only compared to their respective baseline periods, indicating that the expression patterns in Fig. 3g,h reflect functional changes from both diet arms (Supplementary Table 17).
Our findings that the human gut microbiome can rapidly switch between herbivorous and carnivorous functional profiles may reflect past selective pressures during human evolution. Consumption of animal foods by our ancestors was likely volatile, depending on season and stochastic foraging success, with readily available plant foods offering a fallback source of calories and nutrients21. Microbial communities that could quickly, and appropriately, shift their functional repertoire in response to diet change would have subsequently enhanced human dietary flexibility. Examples of this flexibility may persist today in the form of the wide diversity of modern human diets11.
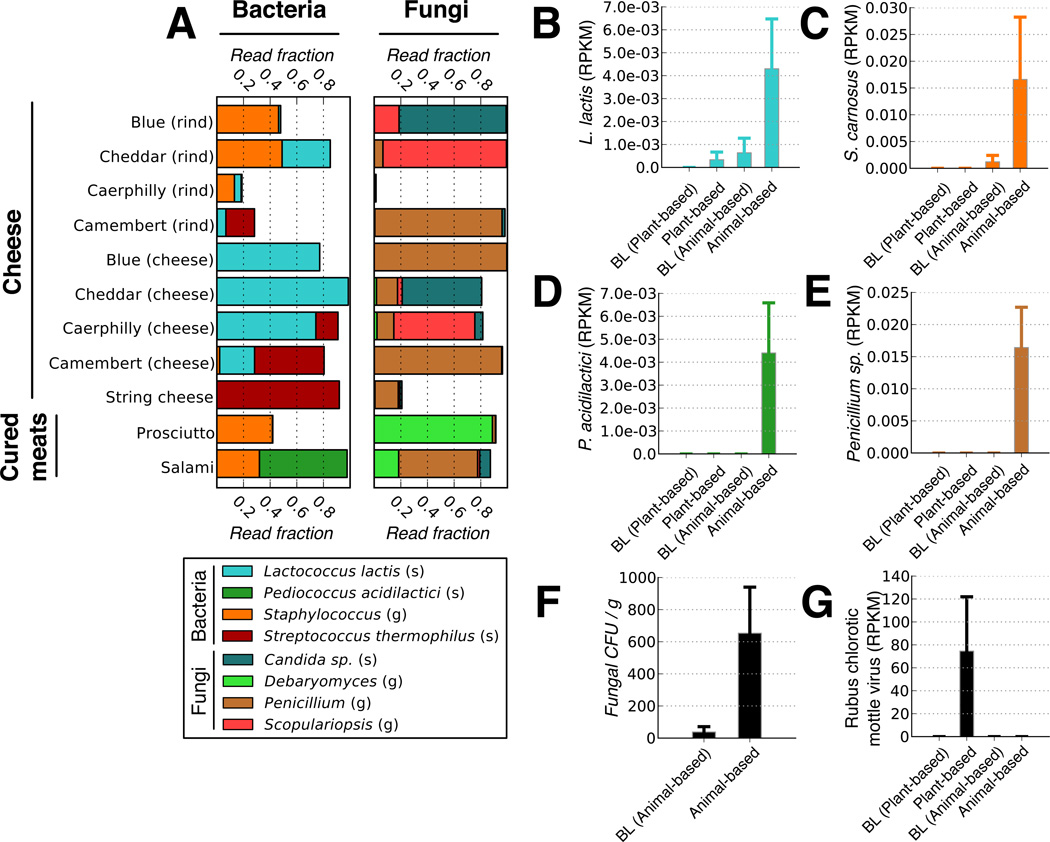
We next examined if, in addition to affecting the resident gut microbiota, either diet arm introduced foreign microorganisms into the distal gut. We identified foodborne bacteria on both diets using 16S rRNA gene sequencing. The cheese and cured meats included in the animal-based diet were dominated by lactic acid bacteria commonly used as starter cultures for fermented foods22,23: Lactococcus lactis, Pediococcus acidilactici, and Streptococcus thermophilus (Fig. 4a). Common non-lactic acid bacteria included several Staphylococcus taxa; strains from this genus are often used when making fermented sausages23. During the animal-based diet, three of the bacteria associated with cheese and cured meats (L. lactis, P. acidilactici, and Staphylococcus) became significantly more prevalent in fecal samples (p<0.05, Wilcoxon signed-rank test; Extended Data Fig. 7c), indicating that bacteria found in common fermented foods can reach the gut at abundances above the detection limit of our sequencing experiments (on average 1 in 4×104 gut bacteria; Supplementary Table 6).
Fig. 4. Foodborne microbes are detectable in the distal gut.
(A) Common bacteria and fungi associated with the animal-based diet menu items, as measured by 16S rRNA and ITS gene sequencing, respectively. Taxa are identified on the genus (g) and species (s) level. A full list of foodborne fungi and bacteria on the animal-based diet can be found in Supplementary Table 21. Foods on the plant-based diet were dominated by matches to the Streptophyta, which derive from chloroplasts within plant matter (Extended Data Fig. 7a). (B-E). Fecal RNA transcripts were significantly enriched (q<0.1, Kruskal-Wallis test; n=6–10 samples/diet arm) for several food-associated microbes on the animal-based diet relative to baseline (BL) periods, including (B) Lactococcus lactis, (C) Staphylococcus carnosus, (D) Pediococcus acidilactici, and (E) a Penicillium sp. A complete table of taxa with significant expression differences can be found in Supplementary Table 22. (F) Fungal concentrations in feces before and 1–2 days after the animal-based diet were also measured using culture media selective for fungal growth (plate count agar with milk, salt, and chloramphenicol). Post-diet fecal samples exhibit significantly higher fungal concentrations than baseline samples (p<0.02; two-sided Mann-Whitney U test; n=7–10 samples/diet arm). (G) Increased RNA transcripts from the plant-derived Rubus chlorotic mottle virus transcripts increase on the plant-based diet (q<0.1, Kruskal-Wallis test; n=6–10 samples/diet arm). Barplots (B-G) all display mean±sem.
We also sequenced the internal transcribed spacer (ITS) region of the rRNA operon from community DNA extracted from food and fecal samples to study the relationship between diet and enteric fungi, which to date remains poorly characterized (Supplementary Table 18). Menu items on both diets were colonized by the genera Candida, Debaryomyces, Penicillium, and Scopulariopsis (Fig. 4a and Extended Data Fig. 7a), which are often found in fermented foods22. A Penicillium sp. and Candida sp. were consumed in sufficient quantities on the animal- and plant-based diets to show significant ITS sequence increases on those respective diet arms (Extended Data Fig. 7b,c).
Microbial culturing and re-analysis of our RNA-Seq data suggested that foodborne microbes survived transit through the digestive system and may have been metabolically active in the gut. Mapping RNA-Seq reads to an expanded reference set of 4,688 genomes (see Supplementary Methods) revealed a significant increase on the animal-based diet for transcripts expressed by food-associated bacteria (Fig. 4b–d) and fungi (Fig. 4e; q<0.1, Kruskal-Wallis test). Many dairy-associated microbes remained viable after passing through the digestive tract, as we isolated 19 bacterial and fungal strains with high genetic similarity (>97% ITS or 16S rRNA) to microbes cultured from cheeses fed to the subjects (Supplementary Table 19). Moreover, L. lactis was more abundant in fecal cultures sampled after the animal-based diet, relative to samples from the preceding baseline period (p<0.1; Wilcoxon Signed-Rank test). We also detected an overall increase in the fecal concentration of viable fungi on the animal-based diet (Fig. 4f; p<0.02; Mann-Whitney U test). Interestingly, we detected RNA transcripts from multiple plant viruses Extended Data Fig. 8). One plant pathogen, Rubus chlorotic mottle virus, was only detectable on the plant-based diet (Fig. 4g). This virus infects spinach24, which was a key ingredient in the prepared meals on the plant-based diet. These data support the hypothesis that plant pathogens can reach the human gut via consumed plant matter25.
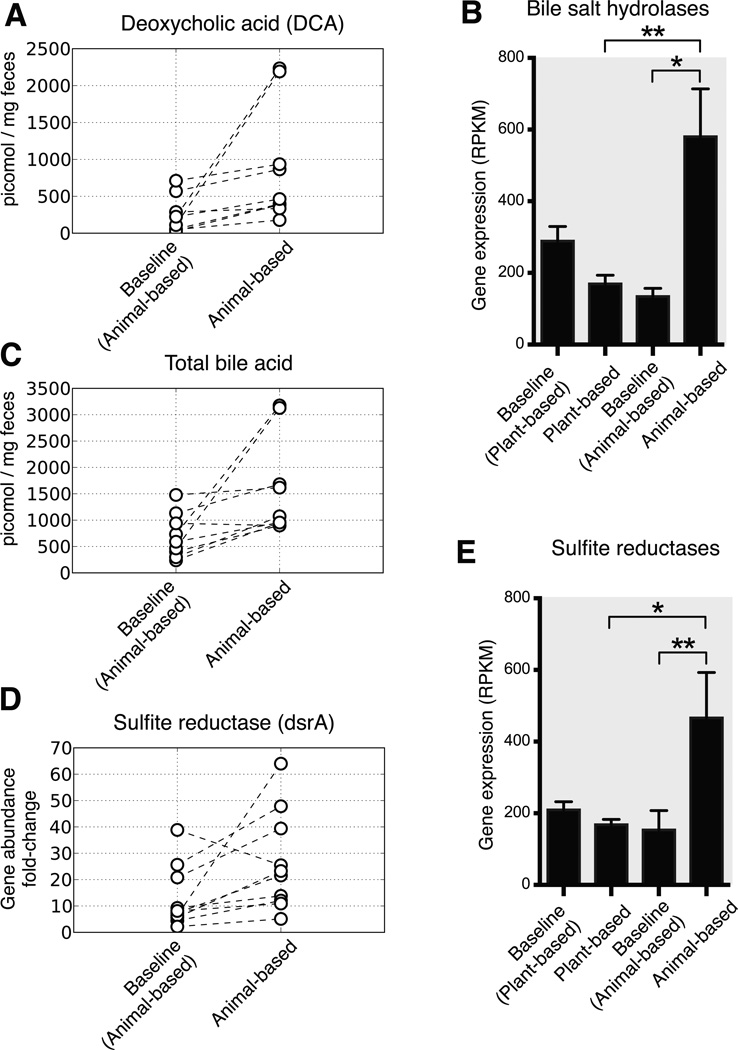
Finally, we found that microbiota changes on the animal-based diet could be linked to altered fecal bile acid profiles and the potential for human enteric disease. Recent mouse experiments have shown high-fat diets lead to increased enteric deoxycholic concentrations (DCA); this secondary bile acid is the product of microbial metabolism and promotes liver cancer26. In our study, the animal-based diet significantly increased the levels of fecal DCA (Fig. 5a). Expression of bacterial genes encoding microbial bile salt hydrolases, which are prerequisites for gut microbial production of DCA27, also exhibited significantly higher expression on the animal-based diet (Fig. 5b). Elevated DCA levels in turn, may have contributed to the microbial disturbances on the animal-based diet, as this bile acid can inhibit the growth of members of the Bacteroidetes and Firmicutes phyla28.
Fig. 5. Changes in the fecal concentration of bile acids and biomarkers for Bilophila on the animal-based diet.
(A) Deoxycholic acid, a secondary bile acid known to promote DNA damage and hepatic carcinomas26, accumulates significantly on the animal-based diet (p<0.01, two-sided Wilcoxon signed-rank test; see Supplementary Table 23 for the diet response of other secondary bile acids). (B) RNA-Seq data also supports increased microbial metabolism of bile acids on the animal-based diet, as we observe significantly increased expression of microbial bile salt hydrolases (K01442) during that diet arm (q<0.05, Kruskal-Wallis test; normalized to reads per kilobase per million mapped, or RPKM; n=8–21 samples/diet arm). (C) Total fecal bile acid concentrations also increase significantly on the animal-based diet, relative to the preceding baseline period (p<0.05, two-sided Wilcoxon signed-rank test), but do not change on the plant-based diet (Extended Data Fig. 9). Bile acids have been shown to cause IBD in mice by stimulating the growth of the bacterium Bilophila6, which is known to reduce sulfite to hydrogen sulfide via the sulfite reductase enzyme (dsrA; Extended Data Fig. 10). (D) Quantitative PCR showed a significant increase in microbial DNA coding for dsrA on the animal-based diet (p<0.05; two-sided Wilcoxon signed-rank test), and (E) RNA-Seq identified a significant increase in sulfite reductase expression (q<0.05, Kruskal-Wallis test; n=8–21 samples/diet arm). Barplots (B,E) display mean±sem.
Mouse models have also found evidence that inflammatory bowel disease can be caused by B. wadsworthia, a sulfite-reducing bacterium whose production of H2S is thought to inflame intestinal tissue6. Growth of B. wadsworthia is stimulated in mice by select bile acids secreted while consuming saturated fats from milk. Our study provides several lines of evidence confirming that B. wadsworthia growth in humans can also be promoted by a high-fat diet. First, we observed B. wadsworthia to be a major component of the bacterial cluster that increased most strongly while on the animal-based diet (C28; Fig. 2 and Supplementary Table 8). This Bilophila-containing cluster also showed significant positive correlations with both long-term dairy (p<0.05; Spearman correlation) and baseline saturated fat intake (Supplementary Table 20), supporting the proposed link to milk-associated saturated fats6. Second, the animal-based diet led to significantly increased fecal bile acid concentrations (Fig. 5c and Extended Data Fig. 9). Third, we observed significant increases in the abundance of microbial DNA and RNA encoding sulfite reductases on the animal-based diet (Fig. 5d,e). Together, these findings are consistent with the hypothesis that diet-induced changes to the gut microbiota may contribute to the development of inflammatory bowel disease. More broadly, our results emphasize that a more comprehensive understanding of diet-related diseases will benefit from elucidating links between nutritional, biliary, and microbial dynamics.
Methods
Sample collection
We recruited 11 unrelated subjects (n=10 per diet; 9 individuals completed both arms of the study). One participant suffered from a chronic gastrointestinal disease, but all other volunteers were otherwise healthy. The volunteers’ normal bowel frequencies ranged from three times a day to once every other day. Three participants had taken antibiotics in the past year. Additional subject information is provided in Supplementary Table 2. Gut microbial communities were sampled from feces. Subjects were instructed to collect no more than one sample per day, but to log all bowel movements. No microbiota patterns were observed as a function of sampling time of day (data not shown). Subjects collected samples by placing disposable commode specimen containers (Claflin Medical Equipment, Warwick, RI) under their toilet seats before bowel movements. CultureSwabs™ (BD, Franklin Lakes, NJ) were then used to collect fecal specimens for sequencing analyses, and larger collection tubes were provided for harvesting larger, intact stool samples (~10g) for metabolic analyses. Each sample was either frozen immediately at −80°C or briefly stored in personal −20°C freezers before transport to the laboratory.
Diet design
We constructed two diet arms, each of which consisted mostly of plant- or animal-based foods (Extended Data Fig. 1). Subjects on the plant-based diet ate cereal for breakfast and precooked meals made of vegetables, rice, and lentils for lunch and dinner (see Supplementary Table 1 for a full list of diet ingredients). Fresh and dried fruits were provided as snacks on this diet. Subjects on the animal-based diet ate eggs and bacon for breakfast, and cooked pork and beef for lunch. Dinner consisted of cured meats and a selection of four cheeses. Snacks on this diet included pork rinds, cheese, and salami. Ingredients for the plant-based diet, dinner meats and cheeses for the animal-based diet, and snacks for both diets were purchased from grocery stores. Lunchmeats for the animal-based diet were prepared by a restaurant that was instructed to not add sauce to the food. On each diet arm, subjects were instructed to eat only provided foods or allowable beverages (water or unsweetened tea for both diets; coffee was allowed on the animal-based diet). They were also allowed to add 1 salt packet per meal, if desired for taste. Subjects could eat unlimited amounts of the provided foods. Outside of the five-day diet arms, subjects were instructed to eat normally.
Food logs, subject metadata, and dietary questionnaires
Subjects were given notepads to log their diet, health, and bowel movements during the study. Subjects transcribed their notepads into digital spreadsheets when the study ended. Each ingested food (including foods on the diet arm) was recorded, as well as data on time, location, portion size, and food brand. Subjects were provided with pocket digital scales (American Weigh, Norcross, GA) and a visual serving size guide to aid with quantifying the amount of food consumed. Each day, subjects tracked their weight using either a scale provided in the lab, or their own personal scales at home. While on the animal-based diet, subjects were requested to measure their urinary ketone levels using provided Ketostix strips (Bayer, Leverkusen, Germany; Extended Data Fig. 1). if subjects recorded a range of ketone levels (the Ketostix color key uses a range-based reporting system), the middle value of that range was used for further analysis. Subjects were encouraged to record any discomfort they experienced while on either diet (e.g. bloating, constipation). Subjects tracked all bowel movements, regardless of whether or not they collected samples, recording movement time, date, and location, and qualitatively documented stool color, odor, and type1. Subjects were also asked to report when they observed stool staining from food dyes consumed at the beginning and end of each diet arm (Extended Data Fig. 3a).
Diet quantification
We quantified subjects' daily nutritional intake during the study using CalorieKing and Nutrition Data System for Research (NDSR). The CalorieKing™ food database (La Mesa, CA) was accessed via the CalorieKing Nutrition & Exercise Manager software (version 4.1.0). Subjects' food items were manually transferred from digital spreadsheets into the CalorieKing software, which then tabulated each food's nutritional content. Macronutrient content per serving was calculated for each of the prepared meals on the animal- and plant-based diet using lists of those meals’ ingredients. Nutritional data was outputted from CalorieKing in CSV format and parsed for further analysis using a custom Python script. NDSR intake data were collected and analyzed using Nutrition Data System for Research software version 2012, developed by the Nutrition Coordinating Center (NCC), University of Minnesota, Minneapolis, MN. We estimated subjects' long-term diet using the National Cancer Institute’s Diet History Questionnaire II2 (DHQ). We used the DHQ to quantify subjects' annual diet intake, decomposed into 176 nutritional categories. Subjects completed the yearly, serving size-included version of the DHQ online using their personal computers. We parsed the survey's results using the Diet*Calc software (version 1.5; Risk Factor Monitoring and Methods Branch, NCI) and its supplied 'Food and Nutrient Database', and 'dhqweb.yearly.withserv.2010.qdd' QDD file.
There was good agreement between subjects’ diets as measured by CalorieKing, the NDSR, and the DHQ: 18 of 20 nutritional comparisons between pairs of databases showed significant correlations (Supplementary Table 3). Unless specified, nutritional data presented in this manuscript reflect CalorieKing measurements.
16S rRNA gene sequencing and processing
Temporal patterns of microbial community structure were analyzed from daily fecal samples collected across each diet (Extended Data Fig. 1). Samples were kept at −80°C until DNA extraction with the PowerSoil bacterial DNA extraction kit (MoBio, Carlsbad CA). The V4 region of the 16S rRNA gene was PCR amplified in triplicate, and the resulting amplicons were cleaned, quantified, and sequenced on the Illumina HiSeq platform according to published protocols3,4 and using custom barcoded primers (Supplementary Table 6). Raw sequences were processed using the QIIME software package (Quantitative Insights Into Microbial Ecology)5. Only full-length, high-quality reads (−r=0) were used for analysis. Operational taxonomic units (OTUs) were picked at 97% similarity against the Greengenes database6 (constructed by the nested_gg_workflow.py QiimeUtils script on 4 Feb 2011), which we trimmed to span only the 16S rRNA region flanked by our sequencing primers (positions 521–773). In total, we characterized an average of 43,589±1,826 16S rRNA sequences for 235 samples (an average of 0.78 samples per person per study day; Supplementary Table 6). Most of the subsequent analysis of 16S rRNA data, including calculations of α- and β-diversity, were performed using custom Python scripts, the SciPy Python library7, and the Pandas Data Analysis Library8. Correction for multiple hypothesis testing utilized the fdrtool9 R library, except in the case of small test numbers, in which case the Bonferroni correction was used.
OTU clustering
We used clustering to simplify the dynamics of thousands of OTUs into a limited number of variables that could be more easily visualized and manually inspected. Clustering was performed on normalized OTU abundances. Such abundances are traditionally computed by scaling each sample's reads to sum to a fixed value (e.g. unity); this technique is intended to account for varying sequencing depth between samples. However, this standard technique may cause false relationships to be inferred between microbial taxa, as increases in the abundance of one microbial group will cause decreases in the fractional abundance of other microbes (this artifact is known as a “compositional” effect10). To avoid compositional biases, we employed an alternative normalization approach, which instead assumes that no more than half of the OTUs held in common between two microbial samples change in abundance. This method uses a robust (outlier-resistant) regression to estimate the median OTU fold-change between communities, by which it subsequently rescales all OTUs.
To further simplify community dynamics, we only included in our clustering model OTUs that comprised 95% of total reads (after ranking by normalized abundance). Abundances for each included OTU were then converted to log-space and mediancentered.
We computed OTU pairwise distances using the Pearson correlation (OTU abundances across all subjects and time points were used). The resulting distance matrix was subsequently input into Scipy's hierarchical clustering function ('fcluster'). Default parameters were used for fcluster, with the exception of the clustering criterion, which was set to 'distance', and the clustering threshold, which was set to '0.7'. These parameters were selected manually so that cluster boundaries visually agreed with the correlation patterns plotted in a matrix of pairwise OTU distances.
Statistics on cluster abundance during baseline and diet periods were computed by taking median values across date ranges. Baseline date ranges were the 4 days preceding each diet arm (i.e. days −4 through −1). Date ranges for the diet arms were chosen so as to capture the full effects of each diet. These ranges were not expected to perfectly overlap with the diet arms themselves, due to the effects of diet transit time. We therefore chose diet arm date ranges that accounted for transit time (as measured by food dye; Extended Data Fig. 3a), picking ranges that began 1 day after foods reached the gut, and ended 1 day before the last diet arm meal reached the gut. These criteria led microbial abundance measurements on the plant-based diet to span days 2–4 of that study arm, and animal-based diet measurements to span days 2–5 of that diet arm.
RNA-Seq sample preparation and sequencing
In order to test if the observed changes in community structure were accompanied by changes to the active subset of the human gut microbiome, we measured communitywide gene expression using meta-transcriptomics11–14 (RNA sequencing, RNA-Seq; Supplementary Table 12). Samples were selected based on our prior 16S rRNA gene sequencing-based analysis, representing 3 baseline days and 2 timepoints on each diet (n=5–10 samples/timepoint; Extended Data Fig. 1). Microbial cells were lysed by a bead beater (BioSpec Products, Bartlesville, OK), total RNA was extracted with phenol:chloroform:isoamyl alcohol (pH 4.5, 125:24:1, Ambion 9720) and purified using Ambion MEGAClear columns (Life Technologies, Grand Island, NY), and rRNA was depleted via Ambion MICROBExpress subtractive hybridization (Life Technologies, Grand Island, NY) and custom depletion oligos. The presence of genomic DNA contamination was assessed by PCR with universal 16S rRNA gene primers. cDNA was synthesized using SuperScript II and random hexamers ordered from Invitrogen (Life Technologies, Grand Island, NY), followed by second strand synthesis with RNaseH and E.coli DNA polymerase (New England Biolabs, Ipswich, MA). Samples were prepared for sequencing with an Illumina HiSeq instrument after enzymatic fragmentation (NEBE6040L/M0348S). Libraries were quantified by quantitative reverse transcriptase PCR (qRT-PCR) according to the Illumina protocol. qRT-PCR assays were run using ABsoluteTM QPCR SYBR® Green ROX Mix (Thermo Scientific, Waltham, MA) on a Mx3000P QPCR System instrument (Stratagene, La Jolla, CA). The size distribution of each library was quantified on an Agilent HS-DNA chip. Libraries were sequenced using the Illumina HiSeq platform.
Functional analysis of RNA-Seq data
We used a custom reference database of bacterial genomes to perform functional analysis of the RNA-Seq data12. This reference included 538 draft and finished bacterial genomes obtained from human-associated microbial isolates15, and the Eggerthella lenta DSM2243 reference genome. All predicted proteins from the reference genome database were annotated with KEGG16 orthologous groups (KOs) using the KEGG database (version 52; BLASTX e-value<10–5, Bit score>50, and >50% identity). for query genes with multiple matches, the annotated reference gene with the lowest evalue was used. When multiple annotated genes with an identical e-value were encountered after a BLAST query, we included all KOs assigned to those genes. Genes from the database with significant homology (BLASTN e-value<10–20) to non-coding transcripts from the 539 microbial genomes were excluded from subsequent analysis. High-quality reads (see Supplementary Table 12 for sequencing statistics) were mapped using SSAHA217, to our reference bacterial database and the Illumina adaptor sequences (SSAHA2 parameters: “-best 1 -score 20 -solexa”). The number of transcripts assigned to each gene was then tallied and normalized to reads per kilobase per million mapped reads (RPKM). To account for genes that were not detected due to limited sequencing depth, a pseudocount of 0.01 was added to all samples. Samples were clustered in Matlab (version 7.10.0) using a Spearman distance matrix (commands: pdist, linkage, and dendrogram). Genes were grouped by taxa, genomes, and KEGG orthologous groups (KOs) by calculating the cumulative RPKM for each sample. HUMAnN18 was used for metabolic reconstruction from metagenomic data followed by LefSe19 analysis to identify significant biomarkers. A modified version of the “SConstruct” file was used to input KEGG orthologous group counts into the HUMAnN pipeline for each RNA-Seq dataset. We then ran LefSe on the resulting KEGG module abundance file using the “-o 1000000” flag.
Taxonomic analysis of RNA-Seq data
We used Bowtie 2 read alignment program20 and the Integrated Microbial Genomes (IMG; version 3.5) database21 to map RNA-Seq reads to a comprehensive reference survey of prokaryotic, eukaryotic, and viral genomes. Our reference survey included all 2,809 viral genomes in IMG (as of version 3.5), a set of 1,813 bacterial and archaeal genomes selected to minimize strain redundancy22, and 66 genomes spanning the Eukarya except for the plants and non-nematode Bilateria. Reads were mapped to reference genomes using Bowtie, which was configured to analyze mated paired-end reads, and return fragments with a minimum length of 150bp and a maximum length of 600bp. All other parameters were left to their default values. The number of base pairs in the reference genome dataset exceeded Bowtie's reference size limit, so we split the reference genomes into four subsets. Each read was mapped to each of these four subreference datasets, and the results were merged by picking the highest-scoring match across the sub-references. We settled tied scores by randomly choosing one of the best-scoring matches. To more precisely measure the presence or absence of specific taxa, we next filtered out reads that mapped to more than reference sequence. Raw read counts were computed for each reference genome by counting the number of reads that mapped to coding sequences according to the IMG annotations; these counts were subsequently normalized using RPKM scaling. Our analysis pipeline associated several sequences with marine algae, which are unlikely to colonize the human gut. We also detected a fungal pathogen exclusively in samples from subjects consuming the animal-based diet (Neosartorya fischeri); this taxon was suspected of being a misidentified cheese fungus, due to its relatedness to Penicillium. We thus reanalysed protist and N. fischeri reads associated with potentially mis-annotated taxa using BLAST searches against the NCBI non-redundant database, and we assigned taxonomy manually based on the most common resulting hits (Extended Data Fig. 8).
Quantitative PCR
Community DNA was isolated with the PowerSoil bacterial DNA extraction kit (MoBio, Carlsbad CA). To determine the presence of hydrogen consumers, PCR was performed on fecal DNA using the following primer sets: (i) Sulfite reductase23 (dsrA), F-5’- CCAACATGCACGGYT CCA-3’, R-5’-CGTCGAACTTGAACTTGAACTTGTAGG-3’; and (ii) Sulfate reduction24,25 (aps reductase), F-5’-TGGCAGATMATGATYMACGG-3’, R-5’- GGGCCGTAACCGTCCTTGAA-3’. qPCR assays were run using ABsoluteTM QPCR SYBR® Green ROX Mix (Thermo Scientific, Waltham, MA) on a Mx3000P QPCR System instrument (Stratagene, La Jolla, CA). Fold-changes were calculated relative to the 16S rRNA gene using the 2-ΔΔCt method and the same primers used for 16S rRNA gene sequencing.
Short-chain fatty acid measurements
Fecal SCFA content was determined by gas chromatography. Chromatographic analysis was carried out using a Shimadzu GC14-A system with a flame ionization detector (FID) (Shimadzu Corp, Kyoto, Japan). Fused silica capillary columns 30m × 0.25 mm coated with 0.25um film thickness were used (Nukol™ for the volatile acids and SPB™-1000 for the nonvolatile acids (Supelco Analytical, Bellefonte, PA). Nitrogen was used as the carrier gas. The oven temperature was 170°C and the FID and injection port was set to 225°C. The injected sample volume was 2 µL and the run time for each analysis was 10 minutes. The chromatograms and data integration was carried out using a Shimadzu C-R5A Chromatopac. A volatile acid mix containing 10 mM of acetic, propionic, isobutyric, butyric, isovaleric, valeric, isocaproic, caproic, and heptanoic acids was used (Matreya, Pleasant Gap, PA). A non-volatile acid mix containing 10 mM of pyruvic and lactic and 5 mM of oxalacetic, oxalic, methy malonic, malonic, fumaric, and succinic was used (Matreya, Pleasant Gap, PA). A standard stock solution containing 1% 2-methyl pentanoic acid (Sigma-Aldrich, St. Louis, MO) was prepared as an internal standard control for the volatile acid extractions. A standard stock solution containing 50 mM benzoic acid (Sigma-Aldrich, St. Louis, MO) was prepared as an internal standard control for the non-volatile acid extractions.
Samples were kept frozen at −80°C until analysis. The samples were removed from the freezer and 1,200µL of water was added to each thawed sample. The samples were vortexed for 1 minute until the material was homogenized. The pH of the suspension was adjusted to 2–3 by adding 50 µL of 50% sulfuric acid. The acidified samples were kept at room temperature for 5 minutes and vortexed briefly every minute. The samples were centrifuged for 10 minutes at 5,000g. 500 µL of the clear supernatant was transferred into two tubes for further processing. For the volatile extraction 50 µL of the internal standard (1% 2-methyl pentanoic acid solution) and 500 µL of ethyl ether anhydrous were added. The tubes were vortexed for 30 seconds and then centrifuged at 5,000g for 10 minutes. 1 µL of the upper ether layer was injected into the chromatogram for analysis. For the nonvolatile extraction 50 µL of the internal standard (50 mM benzoic acid solution) and 500 µL of boron trifluoride-methanol solution (Sigma-Aldrich St. Louis, MO) were added to each tube. These tubes were incubated overnight at room temperature. 1 mL of water and 500 µL of chloroform were added to each tube. The tubes were vortexed for 30 seconds and then centrifuged at 5,000g for 10 minutes. 1 µL of the lower chloroform layer was injected into the chromatogram for analysis. 500 µL of each standard mix was used and the extracts prepared as described for the samples. The retention times and peak heights of the acids in the standard mix were used as references for the sample unknowns. These acids were identified by their specific retention times and the concentrations determined and expressed as mM concentrations per gram of sample.
Bulk bile acid quantification
Fecal bile acid concentration was measured as described previously26. 100 mg of lyophilized stool was heated to 195°C in 1 mL of ethylene glycol KOH for 2 hours, neutralized with 1 mL of saline and 0.2 mL of concentrated HCl, and extracted into 6 mL of diethyl ether 3 times. After evaporation of the ether, the sample residues were dissolved in 6 mL of methanol and subjected to enzymatic analysis. Enzymatic reaction mixtures consisted of 66.5 mmol/L Tris, 0.33 mmol/L EDTA, 0.33 mol/L hydrazine hydrate, 0.77 mmol/L NAD (N 7004, Sigma-Aldrich, St. Louis, MO), 0.033U/mL 3〈- hydroxysteroid dehydrogenase (Sigma-Aldrich, St. Louis, MO) and either sample or standard (taurocholic acid; Sigma-Aldrich, St. Louis, MO) dissolved in methanol. After 90 minutes of incubation at 37°C, absorbance was measured at 340 nm.
Measurement of primary and secondary bile acids
Profiling of fecal primary and secondary bile acids was performed using a modified version of a method described previously27. To a suspension of ~100 mg of stool and 0.25 mL of water in a 1 dram Teflon-capped glass vial was added 200 mg of glass beads. The suspension was homogenized by vortexing for 60–90 seconds. Ethanol (1.8 mL) was added, and the suspension was heated with stirring in a heating block at 80°C for 1.5 h. The sample was cooled, transferred to a 2 mL Eppendorf tube, and centrifuged at 13500 rpm for 1–2 min. The supernatant was removed and retained. The pellet was resuspended in 1.8 mL of 80% aqueous ethanol, transferred to the original vial, and heated to 80°C for 1.5 h. The sample was centrifuged again, and the supernatant was removed and added to the first extraction supernatant. The pellet was resuspended in 1.8 mL of chloroform:methanol (1:1 v/v) and refluxed for 30–60 min. The sample was centrifuged, and the supernatant removed and concentrated to dryness on a rotary evaporator. The ethanolic supernatants were added to the same flask, the pH was adjusted to neutrality by adding aqueous 0.01N HCl, and the combined extracts were evaporated to dryness. The dried extract was resuspended in 1 mL of 0.01N aqueous HCl by sonication for 30 min. A BIO-RAD Poly-Prep chromatography column (0.8×4cm) was loaded with Lipidex 1000 as a slurry in MeOH, allowed to pack under gravity to a final volume of 1.1 mL, and washed with 10 mL of distilled water. The suspension was filtered through the bed of Lipidex 1000 and the effluent was discarded. The flask was washed with 3 × 1 mL of 0.01N HCl, the washings were passed through the gel, and the bed was washed with 4 mL of distilled water. Bile acids and sterols were recovered by elution of the Lipidex gel bed with 8 mL of methanol. A BIO-RAD Poly-Prep chromatography column (0.8×4cm) was loaded with washed SP-Sephadex as a slurry in 72% aqueous MeOH to a final volume of 1.1 mL. The methanolic extract was passed through the SP-Sephadex column, and the column was washed with 4 mL of 72% aqueous methanol. The extract and wash were combined, and the pH was brought to neutral with 0.04N aqueous NaOH. A BIO-RAD Poly-Prep chromatography column (0.8×4cm) was loaded with Lipidex-DEAP, prepared in the acetate form, as a slurry in 72% aqueous MeOH to a final volume of 1.1 mL. The combined neutralized effluent was applied to the column, and the solution was eluted using air gas pressure (flow rate ~25 mL/h). The flask and column were washed with 2 × 2 mL of 72% aqueous ethanol, and the sample and washings were combined to give a fraction of neutral compounds including sterols. Unconjugated bile acids were eluted using 4 mL of 0.1 M acetic acid in 72% (v/v) aqueous ethanol that had been adjusted to pH 4.0 by addition of concentrated ammonium hydroxide. The fraction containing bile acids was concentrated zto dryness on a rotary evaporator.
The bile acids were converted to their corresponding methyl ester derivatives by the addition of 0.6 mL of MeOH followed by 40 µL of a 2.0 M solution of (trimethylsilyl)diazomethane in diethyl ether. The solution was divided in half, and each half of the sample was concentrated to dryness on a rotary evaporator. The bile acids in the first half of the sample were converted to their corresponding trimethylsilyl ether derivatives by the addition of 35 µL of a 2:1 solution of N,Obis(trimethylsilyl)trifluoroacetamide and chlorotrimethylsilane and analyzed by GC-MS. The identities of individual bile acids were determined by comparison of retention time and fragmentation pattern to known standards. Both the ratio of cholest-3-ene to deoxycholic acid in the sample and the amount of internal standard to be added were determined by integrating peak areas. A known amount of the internal standard, 5®- cholestane-3®-ol (5®-coprostanol), was added to the second half of the sample (0.003– 0.07 mmol). The bile acids in the second half of the sample were converted to their corresponding trimethylsilyl ether derivatives by the addition of 35 µL of a 2:1 solution of N,O-bis(trimethylsilyl)trifluoroacetamide and chlorotrimethylsilane and analyzed by GCMS. Amounts of individual bile acids were determined by dividing integrated bile acid peak area by the internal standard peak area, multiplying by the amount of internal standard added, and then dividing by half of the mass of fecal matter extracted. In the event that the first half of the sample contained cholest-3-ene, the coprostanol peak area in the second half of the sample was corrected by subtracting the area of the cholest-3-ene peak, determined by applying the cholest-3-ene:deoxycholic acid ratio calculated from the first half of the sample.
ITS sequencing
Fungal amplicon libraries were constructed with primers that target the internal transcribed spacer (ITS), a region of the nuclear ribosomal RNA cistron shown to promote successful identification across a broad range of fungal taxa28. We selected primers (ITS1f29 and ITS230) focused on the ITS1 region because it provided the best discrimination between common cheese-associated fungi in preliminary in silico tests. Multiplex capability was achieved by adding Golay barcodes to the ITS2 primer. Due to relatively low concentrations, fungal DNA was amplified in three serial PCR reactions, with the first reaction using 1 ul of the PowerSoil DNA extract, and the subsequent two reactions using 1 ul of the preceding PCR product as the template. In each round of PCR, sample reactions were performed in triplicate and then combined. Barcoded amplicons were cleaned, quantified and pooled to achieve approximately equal amounts of DNA from each sample using methods identical to those used for 16S. We gel purified the pool, targeting amplicons between 150 bp and 500 bp in size, and submitted it for Illumina sequencing.
Preliminary taxonomic assignments of ITS reads using the 12_11 UNITE OTUs ITS database (see http://qiime.org) resulted in many unassigned reads. To improve the percentage of reads assigned, we created our own custom database of ITS1 sequences. We extracted ITS sequences from GenBank by targeting specific collections of reliable ITS sequences (e.g. AFTOL, Fungal Barcoding Consortium) and by searching for sequences of yeasts and filamentous fungi that have been previously isolated from dairy and other food ecosystems. We also retrieved a wider range of fungi for our database by searching GenBank with the query internal transcribed spacer[All Fields] AND fungi NOT ‘uncultured’. Sequences that did not contain the full ITS1 were removed. We also included reference OTUs that were identified as widespread cheese fungi in a survey of cheese rinds (Wolfe, Button, and Dutton, unpublished data), but were not in public databases.
Microbial culturing
Fecal samples were cultured under conditions permissive for growth of food-derived microbes. Fecal samples were suspended in a volume of phosphate-buffered saline (PBS) equivalent to ten times their weight. Serial dilutions were prepared and plated on brain heart infusion agar (BD Biosciences, San Jose, CA), supplemented with 100ug/ml cycloheximide, an antifungal agent, and plate count agar with milk and salt (per liter: 5g tryptone, 2.5g yeast extract, 1g dextrose, 1g whole milk powder, 30g NaCl, 15g agar) supplemented with 50ug/ml chloramphenicol, an antibacterial agent. Plates were incubated under aerobic conditions at room temperature for 7 days. Plates supplemented with chloramphenicol which yielded significant growth of bacteria, as determined by colony morphology, were excluded from further analysis. Plates were examined by eye for bacterial colonies or fungal foci whose morphological characteristics were similar to previously characterized food-derived microbes. Candidate food-derived microbes were isolated and identified by Sanger sequencing of the 16S rRNA gene (for bacteria; primers used were 27f, 5- AGAGTTTGATCCTGGCTCAG, and 1492r, 5-GGTTACCTTGTTACGACTT) or ITS region (for fungi; primers used were ITS1f, 5-CTTGGTCATTTAGAGGAAGTAA, and ITS4, 5-TCCTCCGCTTATTGATATGC). After select colonies had been picked for isolation, the surface of each plate was scraped with a razor blade to collect all remaining colonies, and material was suspended in PBS. Dilutions were pooled, and DNA was extracted from the resulting pooled material using a PowerSoil kit (MoBio, Carlsbad, CA). The remaining pooled material was stocked in 20% glycerol and stored at −80°C.
Supplementary Material
Acknowledgements
We would like to thank Andrew Murray, Guido Guidotti, Erin O’Shea, Jeffrey Moffitt, and Bodo Stern for insightful comments; Mary Delaney (Harvard Digestive Disease Core) for biochemical analyses; Christian Daly, Michele Clamp, and Claire Reardon for sequencing support; N. Fierer for providing ITS primers; An Luong and Kylynda Bauer for technical assistance; Jennifer Brulc and Ravi Menon (General Mills) for nutritional guidelines; Atiqur Rahman for menu suggestions; Aviva Must and Jeanette Queenan for nutritional analysis; and our diet study volunteers for their participation. This work was supported by the National Institutes of Health (P50 GM068763), the Boston Nutrition Obesity Research Center (DK0046200), and the General Mills Bell Institute of Health and Nutrition, Minneapolis, MN.
Footnotes
Supplementary Information
Supplementary information is linked to the online version of the paper at www.nature.com/nature.
Author contributions
LAD, RJD, and PJT designed the study, and developed and prepared the diets. LAD, CFM, RNC, DBG, JEB, BEW, and PJT performed the experimental work. AVL, ASD, YV, MAF, and SBB conducted bile acid analyses. LAD and PJT performed computational analyses. LAD and PJT prepared the manuscript.
RNA-Seq data are deposited in the Gene Expression Omnibus (GEO) database (accession number GSE46761); 16S and ITS rRNA gene sequencing reads are deposited in MG-RAST (project ID 6248). Reprints and permissions information are available at www.nature.com/reprints. The authors have no competing interests.
References
- 1.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duncan SH, et al. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Walker AW, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 9.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota's response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell WR, et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr. 2011;93:1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- 11.Cordain L, et al. Plant-animal subsistence ratios and macronutrient energy estimations in worldwide hunter-gatherer diets. Am J Clin Nutr. 2000;71:682–692. doi: 10.1093/ajcn/71.3.682. [DOI] [PubMed] [Google Scholar]
- 12.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy BS. Diet and excretion of bile acids. Cancer Res. 1981;41:3766–3768. [PubMed] [Google Scholar]
- 15.Smith EA, Macfarlane GT. Enumeration of amino acid fermenting bacteria in the human large intestine: effects of pH and starch on peptide metabolism and dissimilation of amino acids. FEMS Microbiol Ecol. 1998;25:355–368. [Google Scholar]
- 16.Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- 17.Sinha R, et al. High concentrations of the carcinogen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) occur in chicken but are dependent on the cooking method. Cancer Res. 1995;55:4516–4519. [PubMed] [Google Scholar]
- 18. PICRUSt ( http://picrust.github.com)
- 19.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pittard J, Wallace BJ. Distribution and function of genes concerned with aromatic biosynthesis in Escherichia coli. J Bacteriol. 1966;91:1494–1508. doi: 10.1128/jb.91.4.1494-1508.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawkes K, O'Connell JF, Jones NG. Hunting income patterns among the Hadza: big game, common goods, foraging goals and the evolution of the human diet. Philos Trans R Soc Lond B Biol Sci. 1991;334:243–250. doi: 10.1098/rstb.1991.0113. discussion 250–241. [DOI] [PubMed] [Google Scholar]
- 22.Bourdichon F, Berger B, Casaregola S. Safety Demonstration of Microbial Food Cultures (MFC) in Fermented Food Products. Bulletin of the International Dairy Federation. 2012;455 [Google Scholar]
- 23.Nychas GJ, Arkoudelos JS. Staphylococci: their role in fermented sausages. Soc Appl Bacteriol Symp Ser. 1990;19:167S–188S. doi: 10.1111/j.1365-2672.1990.tb01806.x. [DOI] [PubMed] [Google Scholar]
- 24.McGavin WJ, Macfarlane SA. Rubus chlorotic mottle virus, a new sobemovirus infecting raspberry and bramble. Virus Res. 2009;139:10–13. doi: 10.1016/j.virusres.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshimoto S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 27.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 28.Islam KB, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 29.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Methods
- 1.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol. 1997;32:920–924. doi: 10.3109/00365529709011203. [DOI] [PubMed] [Google Scholar]
- 2.Diet History Questionnaire, Version 2.0. National Institutes of Health, Applied Research Program, National Cancer Institute; 2010. [Google Scholar]
- 3.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.SciPy: Open source scientific tools for Python. http://www.scipy.org/
- 8.McKinney W. Proceedings of the 9th Python in Science Conference. 2010 [Google Scholar]
- 9.Strimmer K. fdrtool: a versatile R package for estimating local and tail areabased false discovery rates. Bioinformatics. 2008;24:1461–1462. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 10.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rey FE, et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J Biol Chem. 2010;285:22082–22090. doi: 10.1074/jbc.M110.117713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson KE, et al. A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11:1725–1729. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abubucker S, et al. Metabolic reconstruction for metagenomic data and its application to the human microbiome. PLoS Comput Biol. 2012;8:e1002358. doi: 10.1371/journal.pcbi.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markowitz VM, et al. IMG: the Integrated Microbial Genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin J, et al. Optimizing read mapping to reference genomes to determine composition and species prevalence in microbial communities. PLoS ONE. 2012;7:e36427. doi: 10.1371/journal.pone.0036427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deplancke B, et al. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl Environ Microbiol. 2000;66:2166–2174. doi: 10.1128/aem.66.5.2166-2174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart JA, Chadwick VS, Murray A. Carriage, quantification, and predominance of methanogens and sulfate-reducing bacteria in faecal samples. Lett Appl Microbiol. 2006;43:58–63. doi: 10.1111/j.1472-765X.2006.01906.x. [DOI] [PubMed] [Google Scholar]
- 26.Porter JL, et al. Accurate enzymatic measurement of fecal bile acids in patients with malabsorption. J Lab Clin Med. 2003;141:411–418. doi: 10.1016/S0022-2143(03)00040-4. [DOI] [PubMed] [Google Scholar]
- 27.Setchell KD, Lawson AM, Tanida N, Sjovall J. General methods for the analysis of metabolic profiles of bile acids and related compounds in feces. J Lipid Res. 1983;24:1085–1100. [PubMed] [Google Scholar]
- 28.Schoch CL, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi . Proc Natl Acad Sci U S A. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes--application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 30.White TJ, Bruns T, Lee S, Taylor J. Gelfand Ma Innis Dh, Shinsky Jj, White Tj., editors. PCR Protocols: A Guide to. Methods and Applications. 1990:315–322. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.