Abstract
Next-Generation Sequencing (NGS) technologies are yielding ever-higher volumes of human genome sequence data. Given this large amount of data, it has become both a possibility and a priority to determine how disease-causing single nucleotide polymorphisms (SNPs) detected within gene regulatory regions (rSNPs) exert their effects on gene expression. Recently, several studies have explored whether disease-causing polymorphisms have attributes that can distinguish them from those that are neutral, attaining moderate success at discriminating between functional and putatively neutral regulatory SNPs. Here, we have extended this work by assessing the utility of both SNP-based features (those associated only with the polymorphism site and the surrounding DNA) and Gene-based features (those derived from the associated gene in whose regulatory region the SNP lies) in the identification of functional regulatory polymorphisms involved in either monogenic or complex disease. Gene-based features were found to be capable of both augmenting and enhancing the utility of SNP-based features in the prediction of known regulatory mutations. Adopting this approach, we achieved an AUC of 0.903 for predicting regulatory SNPs. Finally, our tool predicted 225 new regulatory SNPs with a high degree of confidence, with 105 of the 225 falling into linkage disequilibrium blocks of reported disease-associated GWAS SNPs.
Keywords: Regulatory mutations, Machine learning, Monogenic disease, Complex disease, Single Nucleotide Polymorphisms, SNP
Introduction
Single nucleotide polymorphisms (SNPs) occur approximately every 300 base-pairs along human chromosomes and represent the most common form of sequence variation [International HapMap Consortium, 2003]. Although it is likely that most SNPs lack functional significance, they are widely used as genetic markers throughout the genome [Kruglyak 1997; Sachidanandam et al., 2001]. However, some SNPs, depending upon their location, can influence gene transcription, transcript processing or protein synthesis, and a proportion of these may in turn be associated with human genetic disease [Buckland et al., 2004; Campino et al., 2008; Pastinen and Hudson, 2004; Prokunina and Alarcon-Riquelme, 2004; Savinkova et al., 2009]. Considerable efforts have been made to identify and characterize functional SNPs in human genes [Buckland, 2006; Chorley et al., 2008; Khan et al., 2006; Mottagui-Tabar et al., 2005; Pampin and Rodriguez-Rey, 2007]. However, given the large number of SNPs that exist in the human genome, it is currently impractical to investigate each of them individually in vitro. Computational approaches to the prediction of functional SNPs therefore provide an alternative means to address this problem [Mooney, 2005].
SNPs located within promoter regions can exert a functional effect by altering the regulation of gene transcription. For this reason, a number of promoter SNP prediction studies have focused exclusively on transcription factor binding sites (TFBS) [Andersen et al., 2008; Lapidot et al., 2008; Ponomarenko et al., 2002]. However, such studies are limited by our current rather incomplete knowledge of all existing TFBS. With the aim of improving our ability to predict functional SNPs, Montgomery et al. [2007] evaluated a number of allele- and sequence-based features for the prediction of functional regulatory polymorphisms. The most important features were found to be the distance from the transcriptional start site (TSS), the presence of a CpG island and local sequence repetitiveness. Torkamani and Schork [2008] have reported that the integration of Encyclopedia of DNA Elements (ENCODE) annotations improved the prediction of functional polymorphisms. Although it is a challenging task, and despite the need to address several outstanding methodological considerations pertaining to the analytical approach (e.g. biased features, imbalanced training sets and the means of evaluation), these initial results suggested that, with an appropriate feature set and machine learning method, functional regulatory polymorphisms ought to be inherently predictable.
Here, we have attempted to distinguish functional SNPs from likely neutral SNPs within putative transcription regulatory regions (defined here as 2500bp upstream of the TSS and 500 bp downstream of the TSS) of human genes. To this end, we employed a supervised machine learning method using a set of 445 known functional regulatory SNPs from the Human Gene Mutation Database (HGMD) together with a set of putatively neutral SNPs. By incorporating a series of novel features from each associated gene, we were able to demonstrate that functional regulatory SNPs are indeed predictable (our method achieved an area under the ROC curve (AUC) value of 0.903). Interestingly, features from the associated gene (as opposed to features pertaining solely to the SNP) were found to be highly predictive in this study. These findings promise to guide the development of better training data, a prerequisite not only for the improvement of our ability to predict disease-related polymorphisms but also, more fundamentally, for the prediction of those genes likely to play a role in genetic disease.
Materials and Methods
Data preparation
RefSeq sequences [Pruitt et al., 2007] which mapped ambiguously to multiple genomic positions were excluded from the analysis. This yielded a set of 20,826 non-redundant gene transcripts. Similarly, a set of 16,872,794 unambiguously mapped SNPs, derived from dbSNP version 130 [http://www.ncbi.nlm.nih.gov/SNP/index.html; Sherry et al., 2001], were employed in this analysis.
In order to evaluate features (attributes) that had the potential to be useful in identifying polymorphic sites responsible for altered gene expression, two datasets were collected. First, bona fide annotated functional SNPs were retrieved from the Human Gene Mutation Database (HGMD) [http://www.hgmd.org; Stenson et al., 2009] as a ‘positive set’. Second, a dataset of 241,465 SNPs (not present in the positive set of functional SNPs) was obtained from dbSNP as a ‘negative control dataset’. While a large proportion of this negative control SNP dataset is likely to be neutral, some of the SNPs could nevertheless exert a functional effect (hence we refer hereafter to this dataset as being ‘putatively neutral’). Both datasets were filtered to ensure that they mapped uniquely to the UCSC Human Genome Database Hg18 [Karolchik et al., 2008]. RefSeq transcripts in the UCSC database were used to define the locations of the SNPs. All SNPs in both the positive and negative sets were filtered so as to include only those promoter polymorphisms with the potential to directly impact upon the expression of their associated transcripts; hence, we confined our analysis to the putative transcriptional regulatory region of each gene (defined for the purposes of this study as the region spanning 2500bp upstream and 500bp downstream of the corresponding major transcriptional start site). A 3000bp region was selected in order to allow direct comparison with previously published methods [Andersen et al., 2008; Kim et al., 2008; Montgomery et al., 2007].
For each HGMD functional SNP, ±30bp flanking sequences were obtained. The flanking sequences were aligned against the RefSNP sequences using BLAST. Where the flanking ±15bp sequences (deemed sufficient for the human genome) around the SNP positions were identical between the HGMD functional SNP and RefSNP, they were matched with the appropriate RefSNP id. By comparing the recorded genomic positions between dbSNP and the RefSeq sequences, a total of 445 functional SNPs and 241,465 background SNPs were obtained from putative transcriptional regulatory regions.
Disease-associated SNPs from published genome-wide association studies (GWAS) were downloaded from http://genome.gov/gwastudies/. CEU genotype data for non-redundant SNP assays from phases 1, 2 and 3 of the HapMap project were downloaded from HapMap website (http://hapmap.ncbi.nlm.nih.gov/). In order to determine linkage disequilibrium (LD) blocks, genotype information from relatives (i.e., children) was excluded from the original data. Haploview software was used to calculate the LD blocks with default settings.
Features
Features used in this study were split into two distinct sets: those directly relating to the SNP under consideration (SNP-based) and those pertaining to the gene in whose transcription regulatory region the SNP lies (Gene-based). SNP-based features included SNP distance to TSS, flanking nucleotide GC-content, flanking nucleotide conservation, SNP diversity, derived SNP frequency and SNP occurrence within known functional elements. Gene-based features were the same for each SNP lying within the regulatory region of a given gene. Gene-based features were further split into two sets: those pertaining to the function of the associated gene (Function-based) and those relating to the mRNA expression of the associated gene (Expression-based). For Function-based features, a set of prediction scores for GO biological process (1,788) and molecular function (344) terms were generated using the FANN-GO predictor of protein GO term annotations [Clark et al., 2011]. The use of predicted functions instead of experimentally determined functional annotations allowed us to obtain values for all data points and a set of features that is less likely to be biased towards genes frequently studied by biomedical researchers (which could result in an overestimation of performance accuracy). We also included interaction complexity (node degree in a protein-protein interaction network) which is derived from high-throughput experiments in this subset of function features. Expression-based features were generated using microarray platforms GPL1074 and GPL96 [Su et al., 2004]. A set of 158 features were generated that represent the normalized expression levels of each gene across 79 tissues. Features pertaining to the mean, standard deviation, coefficient of variation, maximum and minimum expression level of each gene across tissues were also generated. Finally, we generated 2 Codon usage features that were not classified as being either Expression-based or Function-based [see Table 1 for the complete list of SNP-based features and how these features were constructed].
Table 1. Features investigated in this study.
| Feature | Type | Source | Description |
|---|---|---|---|
| Individual tissue expression feature set | Gene-based | http://wombat.gnf.org/index.html | 158 expression data for 79 different types of human tissue/cell were retrieved from the GPL96 and GPL1074 data-sets. Expression values from all probe sets corresponding to the same gene were averaged. The raw expression values were log2 transformed. |
| Mean expression level | Gene-based | (same as above) | (same as above) |
| Minimum expression level | Gene-based | (same as above) | (same as above) |
| Maximum expression level | Gene-based | (same as above) | (same as above) |
| Coefficient of variation for expression level | Gene-based | (same as above) | (same as above) |
| Standard deviation for expression level | Gene-based | (same as above) | (same as above) |
| Frequency of optimal codons | Gene-based | ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot | Frequency of Optimal Codons(Fop) the ratio of optimal codons to synonymous codons. The reported values lie between 0 (where optimal codons were not used) and 1 (where only optimal codons were used). The Effective Number of Codons(ENC) is a measure of overall codon bias and is analogous to the Effective Number of Alleles measure used in population genetics. The reported value lies between 20 (when only one codon effectively was used for each amino acid) and 61 (when codons were used randomly). Fop and ENC values were calculated for human transcript coding sequences by means of CondonW. |
| Effective number of codons | Gene-based | (as above) | (as above) |
| FANN-GO feature set | Gene-based | Clark et al, 2011 | 2,132 FANN-GO features were generated using the FANN-GO predictor of Gene Ontology Function. FANNGO employs multi-output artificial neural networks that naturally incorporate the structure of the ontology in probabilistic inference. For a given data-point, each of the 2132 features represents an output score from FANN-GO generated using the protein sequence associated with the particular SNP. |
| Protein-protein interaction complexity | Gene-based | http://www.reactome.org/download/index.html; http://www.thebiogrid.org/downloads.php | Number of proteins recorded as interacting with a given protein. |
| Distance to transcription start site | SNP-based | http://genome.ucsc.edu/cgibin/hgTables | The Distance to transcription start site refers to the distance between a given SNP and the transcriptional start site of the transcript in the vicinity of each SNP. |
| GC content | SNP-based | ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/rs_fasta | Number of nucleotides that are either guanine or cytosine within the 21 bases flanking of a given SNP (i.e. 10 bp upstream and 10 bp downstream). |
| Sequence conservation | SNP-based | http://hgdownload.cse.ucsc.edu/goldenPath/hg18/phastCons28way | Average PhastCons scores for multiple alignments of 28 vertebrate genomes for the 21 base-pair sequence flanking a given SNP (10 bp upstream and 10 bp downstream). |
| Derived allele frequency | SNP-based | http://haplotter.uchicago.edu; //ftp.hapmap.org/genotypes/latest_ncbi_build36/forward/nonredundant | Derived alleles were identified, based on the estimation of the ancestral state for HapMap SNPs by alignment with the chimpanzee genome sequence. The frequency was then calculated using HapMap genotype data. SNP diversity was defined as 1-fA*fA-fB*fB, where fA and fB are the frequencies of the respective SNP allele, respectively. |
| SNP diversity | SNP-based | (same as above) | (same as above) |
| In CpG island | SNP-based | http://genome.ucsc.edu/cgi-bin/hgTables | Whether or not the given SNP is located in the pre-defined/validated functional region |
| In enhancer | SNP-based | http://www.dcode.org/EI | (same as above) |
| In insulator | SNP-based | http://insulatordb.utmem.edu | (same as above) |
| In RNA polymerase II-enriched region | SNP-based | http://dir.nhlbi.nih.gov/papers/lmi/epigenomes/hgtcell.html | (same as above) |
| In nuclease hypersensitive site | SNP-based | http://research.nhgri.nih.gov/DNaseHS/May2005; http://genome.ucsc.edu/cgi-bin/hgTables | (same as above) |
| In conserved non-coding sequences | SNP-based | http://www.bx.psu.edu/∼ross/dataset/DatasetHome.html | (same as above) |
| In transcription factor binding site | SNP-based | http://genome.ucsc.edu/cgi-bin/hgTables | (same as above) |
All data are based on UCSC Genome Database hg18 coordinates (where these were not available, data coordinates were converted to hg18).
Classification method and identification of optimum predictive features
We evaluated several different machine learning methods including Support Vector Machines (SVMs), Bayesian networks and decision trees. Decision trees were selected on the basis of their interpretability, ease of use, and comparable performance with other methods. Evaluation of our model was performed using 10-fold cross-validation. The dataset was initially randomly split into 10 non-overlapping partitions, each containing 10% positive and 10% negative data points. In each step i∈ {1, 2, …, 10} of the 10 cross-validation steps, the ith fold was used as the test set whereas the remaining data were used to train classification models.
Predictors for each fold comprised an ensemble of 1000 trees. For each tree, training data were balanced by randomly sampling negative data points in order to have a balanced number of positive and negative data points in the training set. Missing values were replaced with the mean values from the respective feature with the null hypothesis (i.e. assuming no difference between the functional SNPs and the putatively neutral SNPs). Each testing data point's final prediction score was an average of all scores' output by the ensemble of 1000 decision trees. After completing the cross-validation steps, each data point contained exactly one predicted and one class value and the performance accuracy was estimated.
Classification performance was measured by calculating the Area Under Receiver Operator Characteristics curves (AUC). AUC provides a measure of the true positive rate (sensitivity) as a function of the false positive rate (1–specificity) over the entire [0, 1] interval. Given a set of data points and a decision threshold, sensitivity was defined as the fraction of positive data points correctly predicted (a data point was counted as a positive prediction if its predicted class value was greater than the decision threshold). Similarly, specificity was defined as the fraction of negative data points correctly predicted [Hastie et al., 2001].
We evaluated the performance of each individual feature by employing both the Wilcoxon test and by calculating the AUC (AUC only for individual tissue expression feature set and FANN-GO feature set) on prediction scores derived using the individual features as the sole feature when building an ensemble of trees. For the Wilcoxon test, statistical repeatability (defined as the frequency of statistical significance detected for all 1000 trees) was reported. The best performing features were reported, assuming that both threshold criteria were met (i.e. higher AUC value and higher statistical repeatability, as defined in the Tables). Training dataset and functional SNPs used in this study can be found at: http://www.mooneygroup.org/yiqiang/rSNP_data/. Prediction scores for each SNP investigated are also available in the Supporting Information (see Supp. Table S1).
Results
Model performance
With respect to the task of discriminating between functional SNPs and putatively neutral SNPs, we achieved an AUC of 0.903, sensitivity of 0.818 and specificity of 0.837 (decision threshold that maximizes the sum of sensitivity and specificity was used, and hereafter), using all features. Since some studies have suggested that selecting only informative features to train the classifier (feature selection) can improve prediction performance [Guyon, 2003; Saeys et al., 2007], we applied correlation-based feature selection (CFS) to ascertain a subset of features that would be the most informative for classification. Using a subset of the most relevant features decreased performance by 2-6% (data not shown), indicating that the ensemble method (1000 decision trees) was robust with respect to the noise introduced by less important features. We also evaluated the final classification model by constructing a random classifier for which the positive set was randomly selected from the putatively neutral SNPs. Consistent with our expectation, the random classifier achieved an AUC of 0.529.
Because the types of diseases associated with SNPs used in this study differ very considerably, the HGMD regulatory variants were subdivided into three categories: functional SNPs reported to cause monogenic disease (MS, n = 48), functional SNPs associated with complex disease (CS, n = 214) and SNPs with demonstrated functional significance but without any currently reported disease association (FS, n = 183). The analysis was then performed separately for these three categories of regulatory variants (MS, CS and FS). Prediction performance on the MS dataset was found to be the most accurate and yielded an overall performance AUC of 0.958, sensitivity of 0.896 and specificity of 0.941. We obtained comparable prediction performances for CS (AUC: 0.889, sensitivity: 0.799 and specificity: 0.821) and FS (AUC: 0.905, sensitivity: 0.809 and specificity: 0.870). AUC values were calculated on these subsets of SNPs by excluding prediction values for all other subclasses during evaluation. It should be noted that these values do not reflect how well a predictor would perform when built to identify specifically these SNPs; instead they indicate how well these subclasses of SNPs are identified by a general predictor.
Gene-based features are important for prediction
Interestingly, by ranking features using the AUC of the ROC, we found that many of the informative features corresponded to those that were derived from the associated gene (i.e. Gene-based features) (Table 2). To validate this finding, we retrained the model so as to exclude all the Gene-based features; the overall performance decreased by approximately 13 percentage points (from an AUC of 0.903 to an AUC of 0.785). All three categories of regulatory variant displayed a deterioration of classification performance after removing Gene-based features (data not shown). In order to assess how likely the increased performance of our predictor when using Gene-based features was due to potential bias in the sample of genes associated with discovered bona fide annotated functional SNPs, we created a paired dataset, with no Gene-based features included. In this paired dataset, we selected only negative data points whose SNPs lie within the regulatory region of a transcript that also has a bona fide annotated functional SNP in its regulator region. The performance of the paired sets was found to be comparable to that of the original sets without Gene-based features (AUC of 0.785 versus AUC of 0.774 respectively). The difference in performance should therefore be attributed solely to the incorporation of Gene-based features in the original set.
Table 2. Optimal features for the prediction of all functional SNPs (MS, CS & FS).
| Feature | AUCa | Statistical repeatability | Directionb |
|---|---|---|---|
| FANN-GO feature set | 0.869 | NAc | NA |
| Individual tissue expression feature set | 0.775 | NA | NA |
| Maximum expression level of assoc. gene | 0.767 | 1 | + |
| Coefficient of variation for gene expression level | 0.763 | 0.994 | + |
| Standard deviation for gene expression level | 0.740 | 0.986 | + |
| Protein-protein interaction complexity | 0.705 | 0.998 | + |
| Distance to transcription start site in gene | 0.705 | 1 | − |
Using the maximum AUC value from random classifier (0.591) and statistical repeatability >0.6 as a threshold.
(+) indicates that the functional SNPs (MS, CS & FS) have higher median values than neutral SNPs; (−) indicates that the functional SNPs (MS, CS & FS) have lower median values than the neutral SNPs.
Wilcoxon tests is not done, because this is a feature set instead of a single feature.
Both Function-based and Expression-based features contributed greatly to prediction accuracy with the Function-based features performing slightly better than Expression-based features (Supp. Table S2). For the monogenic disease-related functional SNPs (MS), the importance of features used for classification (functional versus neutral) was found to share some similarities, but also some differences, when identifying functional complex disease-associated variants (CS). We found that 4 features pertaining to gene expression, codon usage and sequence conservation performed well only for MS prediction, whereas the protein-protein interaction complexity feature performed well only for CS prediction (Tables 3 and 4).
Table 3. Optimal features for the prediction of monogenic disease-causing SNPs (MS).
| Feature | AUCa | Statistical repeatability | Directionb |
|---|---|---|---|
| FANN-GO feature set | 0.931 | NAc | NA |
| Maximum expression level of assoc. gene | 0.918 | 1 | + |
| Individual tissue expression feature set | 0.884 | NA | NA |
| Coefficient of variation for gene expression level | 0.918 | 0.978 | + |
| Standard deviation for gene expression level | 0.904 | 1 | + |
| Mean gene expression level | 0.878 | 0.932 | + |
| Effective number of codons in assoc. gene | 0.860 | 0.988 | − |
| Distance to transcription start site of gene | 0.825 | 1 | − |
| Sequence conservation of ±10bp flanking SNP | 0.648 | 0.986 | + |
Using the maximum AUC value from random classifier (0.591) and statistical repeatability >0.6 as a threshold.
(+) indicates that the MS have higher median values than neutral SNPs; (−) indicates that the MS have lower median values than the neutral SNPs.
Wilcoxon tests is not done, because this is a feature set instead of a single feature.
Table 4. Optimal features for prediction of SNPs associated with complex disease (CS).
| Feature | AUCa | Statistical repeatability | Directionb |
|---|---|---|---|
| FANN-GO feature set | 0.841 | NAc | NA |
| Individual tissue expression feature set | 0.757 | NA | NA |
| Protein-protein interaction complexity | 0.749 | 0.986 | + |
| Coefficient of variation for gene expression level | 0.740 | 0.616 | + |
| Mean gene expression level | 0.721 | 0.622 | − |
| Distance to transcription start site of gene | 0.677 | 1 | − |
Using the maximum AUC value from random classifier (0.591) and statistical repeatability >0.6 as threshold.
(+) indicates that the CS have higher median values than neutral SNPs; (−) indicates that the CS have lower median values than the neutral SNPs.
Wilcoxon tests is not done, because this is a feature set instead of a single feature.
Prediction of functional SNPs in GWAS studies
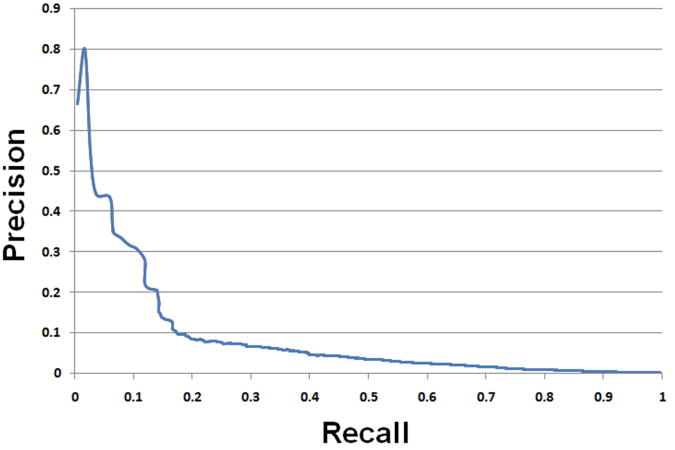
On the basis that functional SNPs are likely to be comparatively rare (as compared with neutral SNPs), a prediction tool to identify functional SNPs requires high specificity (i.e. the proportion of correctly identified neutral SNPs) to be useful in a research context. Applying a very conservative decision threshold to our method, we obtained a specificity of 99.9%. We then applied our method (with this conservative decision threshold) to all SNPs (n = 241,465) in the candidate regulatory region, thereby prospectively identifying 225 SNPs (not present in our positive training dataset) that represent good candidates for SNPs with functional significance (Supp. Table S3). By applying the 99.9% specificity threshold, the prediction precision (i.e. the proportion of true functional SNPs) reached 20.6%. Since regulatory SNPs are likely to be individually very rare (in the present case, 445 functional SNPs and 241,465 background SNPs, 0.18%), our method promises to greatly simplify the task of identifying a regulatory SNP in the genome (see Figure 1 for the overall recall-precision plot). With one exception (see Case Study below), no experimental evidence for the functional significance of these 225 SNPs has so far been reported in the literature. However, the recent increase in reported GWAS data provides us with an opportunity to establish post hoc the potential functional/clinical significance of these SNPs. Although not all disease-associated SNPs reported in GWAS studies are directly causative of the observed disease association, some will indeed be of functional significance and hence will also be likely to be causative of the reported disease association. Analysis of GWAS data and the 225 SNPs predicted to be functional, revealed that 105 of these 225 predicted functional SNPs (47%), distributed between 66 different genes, occurred within the same LD block as a reported disease-associated GWAS SNP. Although these 225 candidate functional regulatory SNPs still await in vitro validation by reporter gene assay, their frequent spatial coincidence within the same LD blocks as reported disease-associated GWAS SNPs suggests that a substantial proportion may eventually turn out to be bona fide functional regulatory SNPs. On the other hand, we believe that many of the remaining 120 SNPs could still be important in functional terms since having a regulatory role does not necessarily imply that it is also going to be of pathological significance.
Figure 1.
The recall-precision plot for the prediction model.
Case study
We retrospectively searched the literature for any experimental evidence of a functional effect for the 225 candidate regulatory SNPs identified in this study. Functional evidence was obtained for one candidate SNP (rs2280789,T/C) in the Chemokine (C-C motif) ligand 5 (CCL5) gene. This SNP occurs within an up-regulating intron 1 element; employing a luciferase reporter gene assay, it was shown that the ‘C’ allele of rs2280789 was associated with a highly significant 3-fold reduction in gene expression as compared to the ‘T’ allele (P < 0.001) [An et al., 2002]. The ‘C’ allele was also reported to be associated with rapid disease progression to AIDS for individuals with an HIV infection.
Discussion
Assessment of performance
In this study, we employed what is, to our knowledge, the most comprehensive functional regulatory SNP dataset available. Compared to previous studies that have used relatively small numbers of functional regulatory SNPs (about 100 regulatory SNPs) and an imbalanced training approach without special treatment [Montgomery et al., 2007; Torkamani and Schork, 2008], we have performed a robust analysis of the prediction of functional SNPs within promoter regions. We achieved this by incorporating biologically relevant features of the downstream genes and using a forest-like tree method that greatly improved prediction performance (AUC of 0.903, sensitivity of 0.818 and specificity of 0.837). Owing to the likely low prevalence (as compared to neutral SNPs) of functional regulatory SNPs in the human genome, the accurate prediction of functional regulatory SNPs is inherently very difficult. Our method nevertheless provides a high-throughput means to identify potentially functional regulatory SNPs. Employing this method, we report here 225 high-confidence candidates that we consider worthy of laboratory testing.
This study does however indicate that much work still remains to be done in order to improve the prediction of polymorphic sites of functional significance. Indeed, several major challenges lie ahead. First, available bona fide (i.e. experimentally supported) functional polymorphism data are still limited. Since millions of SNPs remain uncharacterized, we are currently working with only a very small proportion of the complete dataset of functional SNPs within regulatory regions. Second, although the definition of functional features is proceeding apace, it is hard to escape the conclusion that functional SNPs have been disproportionately derived from those genes which have been functionally well characterized [including, of course, disease genes; Osada et al., 2009]. With the features (both Gene-based and SNP-based) employed in this study, we were able to successfully identify functional SNPs with a high degree of confidence. However, in this study we can only predict rSNP by genome location. Our method would not be able to distinguish the direction of the nucleotide changes which would result in a functional effect. (i.e. A to T vs A to C). As more biological knowledge becomes available, improvements (e.g. discovery of new TFBS) to existing SNP-based features will increase classification performance, thereby reducing the dependency of classification methods on those Gene-based features that tend to be biased or suffer from sparseness.
In order to improve the prediction of disease-related SNPs, additional novel features still need to be identified. Previous studies have suggested that disease genes may possess specific properties that can serve to distinguish them from non-disease genes such as longer sequence length and a lower nucleotide substitution rate [Cooper and Mort, 2010; Khaitovich et al., 2004; Lopez-Bigas and Ouzounis, 2004]. These features were not included in the current analysis but the addition of evolutionary attributes and other disease gene-specific properties could easily be incorporated so as to improve the predictive performance in the context of the monogenic disease-causing SNPs. Similarly, the topological parameters of a gene within a network or pathway represent promising features for the prediction of CS [Hahn and Kern, 2005; Zhu et al., 2007].
A survey of disease-related SNPs and disease genes
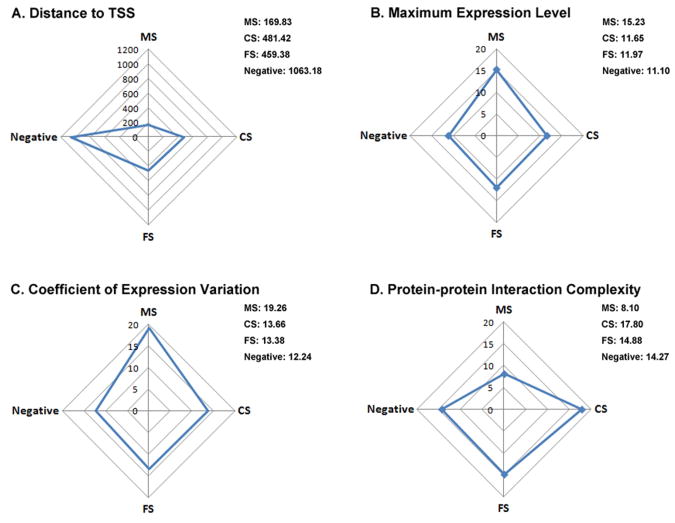
The functional SNPs investigated in this study will only be predicted to give rise to changes in gene expression rather than to protein structure or function. However, the consequences of an expression change may include either a deleterious gene dosage alteration [Anneren and Edman, 1993; Stayner et al., 2006; Toivonen et al., 2003] or a change in the functional role of the associated gene product in the context of a given biological pathway or protein interaction network [Cunningham et al., 2005; Tepper et al., 2005]. Our studies are suggestive of both these possibilities. The prediction of monogenic disease-related functional SNPs (MS) was most accurate, with the Expression-based features contributing highly to the performance (Table 3 and Figure 2; for complete statistical summary for each features, see Supp. Table S4). Thus, the gene expression level appears likely to exert an important (and direct) influence on the genotype-phenotype relationship in monogenic disease. The fact that the Codon usage feature works well only for MS prediction, taken together with the observation that MS were generally located within core promoter regions and hence were significantly closer to the transcriptional start sites than was the case for CS and putatively neutral SNPs (Wilcoxon tests, p<0.001, Bonferroni-corrected), also point in the same direction. However, compared to MS, the effect of Expression-based features is less pronounced for complex disease (CS) yet (although still good) protein-protein interaction complexity works well (Table 4 and Figure 2). This suggests that there may be underlying differences in the mechanism(s) by which a given SNP exerts its functional effect between monogenic and complex diseases. The disruption of protein-protein interactions and biological pathways induced by a change in gene expression may underlie a high proportion of complex disease regulatory SNPs.
Figure 2.
Features that exhibit differences between different data sets. MS: SNPs associated with, or causing, monogenic disease; CS: SNPs associated with complex disease; FS: SNPs with demonstrated functional significance but without any reported disease association; Negative: Neutral SNPs.
The evolutionary conservation of sequences flanking SNPs was shown to be an effective predictive feature for the MS set. Although not statistically significant owing to the small sample size, the SNP diversity (Table 1) of MS (median: 0.082) was lower by comparison to the putatively neutral SNPs (median: 0.367) and CS (median: 0.341). Taken together, this is indicative of MS being under strong negative selection. Although we could not rule out the possibility that CS are under balancing selection (either heterozygote advantage or environmental heterogeneity), based on the observation of a lower derived allele frequency and higher SNP diversity as compared to MS, CS appear more likely to have evolved neutrally because the CS flanking sequences were not evolutionarily conserved, consistent with previous analyses of gene promoter regions [Keightley et al., 2005; Khaitovich et al., 2004]. Detailed disease gene categorization is required to determine whether the paucity of evidence for selection was due to genetic drift, slightly deleterious conditions or to diseases with late onset.
Owing to the lower level of sequence conservation and greater distance to the core promoter exhibited by CS (in comparison to MS), SNP-based features are not as discriminating for CS as with MS. There are some good Gene-based features for CS prediction (e.g. protein-protein interaction complexity) but, we could only speculate that genes with certain attributes were more likely to harbor functional SNPs. Generally speaking, CSs appear much more difficult to predict. It is the tacit assumption of most promoter studies that the location of known transcription factor binding sites (TFBS) or other functional annotations would be useful in the identification of regulatory mutations and polymorphisms [Andersen et al., 2008; Conde et al., 2004; Lapidot et al., 2008; Mottagui-Tabar et al., 2005]. In this study, functional annotations such as TFBS actually display very limited predictive power (AUC=0.504) in terms of discriminating functional regulatory SNPs from putatively neutral SNPs. Possible reasons for this might be: (i) our knowledge of the structure and function of regulatory elements in our genome is still very inadequate (the information employed in this study might not be representative) due to data sparseness (small percentage of data points actually has been annotated), and/or (ii) more detailed positional information is required in relation to SNPs located within the regulatory elements since such elements can be redundant, and not every base within a given regulatory element is critical to its function. Consistent with previous studies [Buckland et al., 2005; Guo and Jamison, 2005; Montgomery et al., 2007], the distance to the transcriptional start site was one of the best performing features. Although the promoter was generally considered to be very important for gene regulation, the influence of a particular SNP may be quite complex because multiple regulatory elements can overlap and the effect of different promoter variants can be additive. To test if the distance to the transcriptional start site is a dominant feature in making rSNP predictions, we evaluated our model with the full feature set but excluding just this feature. The result showed that performance dropped only slightly from an AUC of 0.903 to an AUC of 0.895, suggesting that other features used in our model appear able to compensate for the information provided by this important feature.
Finally, we observed that the MS-associated genes had (i) a higher level of gene expression and (ii) greater variance of gene expression than the putatively neutral SNPs. Initially, this seemed to be contradictory since these two attributes are generally negatively correlated. Genes exhibiting a high expression level are usually expressed less variably [Subramanian and Kumar, 2004] and are could be less likely to be involved in disease because of their essential nature (on the basis that mutations in such genes would have tended not to come to clinical attention [Cooper et al. 2010]). One explanation for their co-occurrence might be differences in the clinical severity of different monogenic diseases. Some monogenic diseases are very severe clinically (either because the gene is critically important to health or because the mutation might have a strong impact on gene function), while others may not be. However, a lower mean expression level and a higher expression variance were found for complex disease, consistent with the view that complex disease is generally less severe and has a tendency to be associated with tissue-specific expression [Winter et al., 2004].
In conclusion, we have developed a method for predicting disease-associated functional SNPs within gene regulatory regions. We found Gene-based features were useful in making such predictions, possibly because such features represent a proxy for the disease mechanism. Finally, we identify a number of putative regulatory SNPs that we believe are likely to be of potential functional/clinical significance and which therefore represent good candidates for in vitro analysis as well as inclusion in future GWAS studies.
Supplementary Material
Acknowledgments
We would like to acknowledge funding support from the National Library of Medicine [grants K22LM009135 (PI: Mooney), R01LM009722 (PI: Mooney)] and funds from INGEN. The Indiana Genomics Initiative (INGEN) is funded in part from a grant by endowment of Eli Lilly and Co.
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- An P, Nelson GW, Wang L, Donfield S, Goedert JJ, Phair J, Vlahov D, Buchbinder S, Farrar WL, Modi W, O'Brien SJ, Winkler CA. Modulating influence on HIV/AIDS by interacting RANTES gene variants. Proc Natl Acad Sci USA. 2002;99:10002–10007. doi: 10.1073/pnas.142313799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen MC, Engstrom PG, Lithwick S, Arenillas D, Eriksson P, Lenhard B, Wasserman WW, Odeberg J. In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput Biol. 2008;4:e5. doi: 10.1371/journal.pcbi.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anneren G, Edman B. Down syndrome--a gene dosage disease caused by trisomy of genes within a small segment of the long arm of chromosome 21, exemplified by the study of effects from the superoxide-dismutase type 1 (SOD-1) gene. APMIS Suppl. 1993;40:71–79. [PubMed] [Google Scholar]
- Buckland PR. The importance and identification of regulatory polymorphisms and their mechanisms of action. Biochim Biophys Acta. 2006;1762:17–28. doi: 10.1016/j.bbadis.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Buckland PR, Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O'Donovan MC. Strong bias in the location of functional promoter polymorphisms. Hum Mutat. 2005;26:214–223. doi: 10.1002/humu.20207. [DOI] [PubMed] [Google Scholar]
- Buckland PR, Hoogendoorn B, Guy CA, Coleman SL, Smith SK, Buxbaum JD, Haroutunian V, O'Donovan MC. A high proportion of polymorphisms in the promoters of brain expressed genes influences transcriptional activity. Biochim Biophys Acta. 2004;1690:238–249. doi: 10.1016/j.bbadis.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Campino S, Forton J, Raj S, Mohr B, Auburn S, Fry A, Mangano VD, Vandiedonck C, Richardson A, Rockett K, Clark TG, Kwiatkowski DP. Validating discovered cis-acting regulatory genetic variants: application of an allele specific expression approach to HapMap populations. PLoS ONE. 2008;3:e4105. doi: 10.1371/journal.pone.0004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorley BN, Wang X, Campbell MR, Pittman GS, Noureddine MA, Bell DA. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: current and developing technologies. Mutat Res. 2008;659:147–157. doi: 10.1016/j.mrrev.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WT, Radivojac P. Analysis of protein function and its prediction from amino acid sequence. Proteins. 2011;79(7):2086–2096. doi: 10.1002/prot.23029. [DOI] [PubMed] [Google Scholar]
- Conde L, Vaquerizas JM, Santoyo J, Al-Shahrour F, Ruiz-Llorente S, Robledo M, Dopazo J. PupaSNP Finder: a web tool for finding SNPs with putative effect at transcriptional level. Nucleic Acids Res. 2004;32(Web Server issue):W242–248. doi: 10.1093/nar/gkh438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Chen JM, Ball EV, Howells K, Mort M, Phillips AD, Chuzhanova N, Krawczak M, Kehrer-Sawatzki H, Stenson PD. Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Hum Mutat. 2010;31:631–655. doi: 10.1002/humu.21260. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Mort M. Do inherited disease genes have distinguishing functional characteristics? Genet Test Mol Biomarkers. 2010;14:289–291. doi: 10.1089/gtmb.2010.0033. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Swartzlander D, Liyanarachchi S, Davuluri RV, Herman GE. Changes in gene expression associated with loss of function of the NSDHL sterol dehydrogenase in mouse embryonic fibroblasts. J Lipid Res. 2005;46:1150–1162. doi: 10.1194/jlr.M400462-JLR200. [DOI] [PubMed] [Google Scholar]
- Guo Y, Jamison DC. The distribution of SNPs in human gene regulatory regions. BMC Genomics. 2005;6:140. doi: 10.1186/1471-2164-6-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon I. An introduction to variable and feature selection. J Machine Learning Res. 2003;3:1157–1182. [Google Scholar]
- Hahn MW, Kern AD. Comparative genomics of centrality and essentiality in three eukaryotic protein-interaction networks. Mol Biol Evol. 2005;22:803–806. doi: 10.1093/molbev/msi072. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman JH. The elements of statistical learning: data mining, inference, and prediction. New York, NY: Springer Verlag; 2001. [Google Scholar]
- International HapMap Consortium TIH. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Kuhn RM, Baertsch R, Barber GP, Clawson H, Diekhans M, Giardine B, Harte RA, Hinrichs AS, Hsu F, Kober KM, Miller W, Pedersen JS, Pohl A, Raney BJ, Rhead B, Rosenbloom KR, Smith KE, Stanke M, Thakkapallayil A, Trumbower H, Wang T, Zweig AS, Haussler D, Kent WJ. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36(Database issue):D773–779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Lercher MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Pääbo S. A neutral model of transcriptome evolution. PLoS Biol. 2004;2(5):E132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IA, Mort M, Buckland PR, O'Donovan MC, Cooper DN, Chuzhanova NA. In silico discrimination of single nucleotide polymorphisms and pathological mutations in human gene promoter regions by means of local DNA sequence context and regularity. Silico Biol. 2006;6:23–34. [PubMed] [Google Scholar]
- Kim BC, Kim WY, Park D, Chung WH, Shin KS, Bhak J. SNP@Promoter: a database of human SNPs (single nucleotide polymorphisms) within the putative promoter regions. BMC Bioinformatics. 2008;9(1):S2. doi: 10.1186/1471-2105-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L. The use of a genetic map of biallelic markers in linkage studies. Nat Genet. 1997;17:21–24. doi: 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- Lapidot M, Mizrahi-Man O, Pilpel Y. Functional characterization of variations on regulatory motifs. PLoS Genet. 2008;4(3):e1000018. doi: 10.1371/journal.pgen.1000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Ouzounis CA. Genome-wide identification of genes likely to be involved in human genetic disease. Nucleic Acids Res. 2004;32:3108–3114. doi: 10.1093/nar/gkh605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SB, Griffith OL, Schuetz JM, Brooks-Wilson A, Jones SJ. A survey of genomic properties for the detection of regulatory polymorphisms. PLoS Comput Biol. 2007;3:e106. doi: 10.1371/journal.pcbi.0030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney S. Bioinformatics approaches and resources for single nucleotide polymorphism functional analysis. Brief Bioinform. 2005;6:44–56. doi: 10.1093/bib/6.1.44. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Faghihi MA, Mizuno Y, Engstrom PG, Lenhard B, Wasserman WW, Wahlestedt C. Identification of functional SNPs in the 5-prime flanking sequences of human genes. BMC Genomics. 2005;6:18. doi: 10.1186/1471-2164-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N, Mano S, Gojobori J. Quantifying dominance and deleterious effect on human disease genes. Proc Natl Acad Sci USA. 2009;106:841–846. doi: 10.1073/pnas.0810433106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampin S, Rodriguez-Rey JC. Functional analysis of regulatory single-nucleotide polymorphisms. Curr Opin Lipidol. 2007;18:194–198. doi: 10.1097/MOL.0b013e3280145093. [DOI] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Ponomarenko JV, Orlova GV, Merkulova TI, Gorshkova EV, Fokin ON, Vasiliev GV, Frolov AS, Ponomarenko MP. rSNP_Guide: an integrated database-tools system for studying SNPs and site-directed mutations in transcription factor binding sites. Hum Mutat. 2002;20:239–248. doi: 10.1002/humu.10116. [DOI] [PubMed] [Google Scholar]
- Prokunina L, Alarcon-Riquelme ME. Regulatory SNPs in complex diseases: their identification and functional validation. Expert Rev Mol Med. 2004;6:1–15. doi: 10.1017/S1462399404007690. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, Sherry S, Mullikin JC, Mortimore BJ, Willey DL, Hunt SE, Cole CG, Coggill PC, Rice CM, Ning Z, Rogers J, Bentley DR, Kwok PY, Mardis ER, Yeh RT, Schultz B, Cook L, Davenport R, Dante M, Fulton L, Hillier L, Waterston RH, McPherson JD, Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D International SNP Map Working Group. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- Savinkova LK, Ponomarenko MP, Ponomarenko PM, Drachkova IA, Lysova MV, Arshinova TV, Kolchanov NA. TATA box polymorphisms in human gene promoters and associated hereditary pathologies. Biochemistry (Mosc) 2009;74:117–129. doi: 10.1134/s0006297909020011. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stayner C, Iglesias DM, Goodyer PR, Ellis L, Germino G, Zhou J, Eccles MR. Pax2 gene dosage influences cystogenesis in autosomal dominant polycystic kidney disease. Hum Mol Genet. 2006;15:3520–3528. doi: 10.1093/hmg/ddl428. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1(1):13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Kumar S. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics. 2004;168:373–381. doi: 10.1534/genetics.104.028944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepper CG, Gregg JP, Shi XB, Vinall RL, Baron CA, Ryan PE, Desprez PY, Kung HJ, deVere White RW. Profiling of gene expression changes caused by p53 gain-of-function mutant alleles in prostate cancer cells. Prostate. 2005;65:375–389. doi: 10.1002/pros.20308. [DOI] [PubMed] [Google Scholar]
- Toivonen JM, Manjiry S, Touraille S, Alziari S, O'Dell KM, Jacobs HT. Gene dosage and selective expression modify phenotype in a Drosophila model of human mitochondrial disease. Mitochondrion. 2003;3:83–96. doi: 10.1016/S1567-7249(03)00077-1. [DOI] [PubMed] [Google Scholar]
- Torkamani A, Schork NJ. Predicting functional regulatory polymorphisms. Bioinformatics. 2008;24:1787–1792. doi: 10.1093/bioinformatics/btn311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter EE, Goodstadt L, Ponting CP. Elevated rates of protein secretion, evolution, and disease among tissue-specific genes. Genome Res. 2004;14:54–61. doi: 10.1101/gr.1924004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Gerstein M, Snyder M. Getting connected: analysis and principles of biological networks. Genes Dev. 2007;21:1010–1024. doi: 10.1101/gad.1528707. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.