Abstract
Only 20–25% of families screened for BRCA1/2 mutations are found positive. Because only a positive result is informative, we studied the role of BRCA1/2 immunohistochemistry as an additional method for patient selection. From 53 high-risk-affected probands, 18 (34%) had available paraffin blocks of their tumors and were selected for this study. Mutation screening was done by conformation-sensitive gel electrophoresis and multiplex ligation-dependent probe amplification. For immunohistochemistry, 21 neoplastic specimens (15 breast carcinomas, 5 ovary neoplasms, and 1 rectal adenocarcinoma) were analyzed with BRCA1 (monoclonal antibody, Ab-1, oncogene) and BRCA2 (polyclonal antibody, Ab-2, oncogene) antibodies. Absence of the BRCA1 protein was confirmed in negative tumors by Western blotting. Seven patients were positive for BRCA1/2 mutations: 5 for BRCA1 and 2 for BRCA2. Four out of five positive patients had tumors negative for BRCA1 immunostaining, and the remaining 13 BRCA1-negative patients had positive BRCA1 immunostaining in all tumor samples. Sensitivity to predict for BRCA1 mutation carriers was 80%, and specificity was 100%, with a positive predictive value of 100% and a negative predictive value of 93%. This correlation was statistically significant (p=0.001). No correlation was observed for BRCA2. If larger studies confirm these results, high-risk patients with BRCA1-negative tumors should be screened first for this gene.
Keywords: BRCA1/2, genetic screening, hereditary breast cancer, immunohistochemistry
Breast cancer is hereditary in 10% of cases, the majority related to mutations in the BRCA1 and BRCA2 genes (Ford et al. 1998). BRCA1 and BRCA2 are tumor suppressor genes (Crook et al. 1997; Scully et al. 1997) that span over 100 kb of genomic DNA each and encode proteins of 1863 and 3418 amino acids, respectively. Mutations in these genes predispose carriers mainly to breast cancer but also to other cancers (Easton et al. 1995; Breast Cancer Linkage Consortium 1999; Liede et al. 2004).
BRCA1/2 mutation detection is complex because of the large size of both genes and the absence of hot spots. Besides positive and negative tests, indeterminate results of this screening pose particular problems in the management of high-risk families. Selection of patients is then crucial and relies mainly on family history and phenotype ascertainment (Frank et al. 1998; Parmigiani et al. 1998; American Society of Clinical Oncology 2003), but only 20–25% of screened families have positive mutation results (Shih et al. 2002). This is due to the disparity of selection criteria used in different breast evaluation clinics, to the different molecular methodologies available, and also because other genes may be involved in hereditary breast cancer (Walsh et al. 2006). Screening is also complicated by the fact that mutation detection in one family must start in an affected relative, and in many families, all affected relatives are deceased.
More specific selection for genetic screening, taking into account characteristics of BRCA1/2 tumors, is essential. Histopathology of BRCA1/2 hereditary breast cancer may be useful, because a basal epithelial phenotype appears to be associated with germline BRCA1 mutations (Foulkes et al. 2003) and other pathological features distinguish BRCA1 tumors from BRCA2 and sporadic breast cancers (Lakhani et al. 2002). Immunohistochemistry of BRCA1/2 proteins in tumor cells could be useful as an additional method in patient selection for genetic screening. If the cancer tissue from the genetic proband of a high-risk breast/ovarian cancer family (the first person to be tested is a cancer-affected individual) shows an absence of labeling for either BRCA1 or BRCA2 protein in tumor cells, that could predict a BRCA mutation carrier. Tumorigenesis in individuals with germline BRCA mutations requires somatic inactivation of the wild-type allele, and BRCA deficiency is critical to the development of disease (Welcsh and King 2001). With a reliable immunohistochemical method, in most cases, only the mutated gene would be screened. This methodology has recently been demonstrated to be useful in the identification of carriers of mutations in the mismatch repair genes associated with hereditary non-polyposis colorectal cancer (Stormorken et al. 2005).
Several antibodies for BRCA proteins are available, but conflicting results have been published in the literature concerning their diagnostic usefulness. Concerning BRCA1 protein, different types of staining have been described: nuclear, cytoplasmic, or both (Chen et al. 1995; Scully et al. 1996; Coene et al. 1997; Perez-Valles et al. 2001). Although these different observations could be explained by differences in the specificity of the antibodies (Perez-Valles et al. 2001), different fixation methods (Scully et al. 1996; Coene et al. 1997), or by the existence of splice variant isoforms of the BRCA1 protein (Wilson et al. 1997), the use of monoclonal antibodies and fixation in neutral buffered formalin after antigen exposure in a microwave demonstrates a predominantly nuclear labeling (Scully et al. 1996). This localization is consistent with the role of BRCA1 in the maintenance of genome integrity: cell cycle control, apoptosis, and DNA repair. BRCA2 is also found in the nucleus, as expected for a key component of cell cycle control and DNA repair pathways (Marmorstein et al. 1998), although truncated forms have been observed in the cytoplasm (Spain et al. 1999). Because nuclear entry for both BRCA1 and BRCA2 is dependent on nuclear localization signals (Spain et al. 1999; Henderson 2005), it is expected that mutations in these domains may affect subcellular localization of BRCA proteins (Henderson 2005).
The use of BRCA1 and BRCA2 immunohistochemistry in the prediction of BRCA mutation carriers has been more extensively studied for BRCA1, but with contradictory results. The absence of BRCA1 in 4% of 50 tissue sections was proposed to represent familial cases (Chen et al. 1995), and more recent studies have considered the use of BRCA1 staining to be a good indicator for the presence of mutations in this gene (Kashima et al. 2000; Schofield et al. 2000). However, the opposite has also been reported (Perez-Valles et al. 2001). In this study, we analyzed the correlation between the immunoexpression for BRCA1/2 and the results of genetic screening for BRCA1/2 mutations in a group of high-risk women with available tumor paraffin blocks. We also established the sensitivity, specificity, and predictive values of the method.
Materials and Methods
Case Selection
Between July 2000 and July 2002, we identified 18 women selected for BRCA1/2 mutation screening with available paraffin-embedded tissue blocks of their tumors. These women belonged to a group of 53 affected individuals at high risk for BRCA mutations based on personal and family history [combined probability of BRCA1/2 mutation over 25% (Frank et al. 1998; Parmigiani et al. 1998) or female breast cancer under 30 years of age] included in a research study for the analysis of these genetic events in the Portuguese population. All patients were counseled and consented to genetic screening, according to procedures approved by the ethics committee of our center. In one case, BRCA1 genetic screening results were correlated with immunochemistry data from an obligate carrier in the same family.
Clinicopathological Data
Information about age and other neoplasias whose specimens were not available for review was obtained from patients' medical records. A pathologist blinded to the results of BRCA1/2 mutation testing reviewed the cases. Histological grade of differentiation was estimated using the Elston and Ellis system (Elston and Ellis 1991).
BRCA1/2 Mutation Screening
DNA was extracted from whole blood using the QIAmp blood Midi Kit (Qiagen; Hilden, Germany) and aliquots of patients' DNA were each subjected to PCR amplification using primers designed to amplify whole exonic sequences and intronic/exonic boundaries of BRCA1 and BRCA2 (The Breast Cancer Information Core Database 1999; http://research.nhgri.nih.gov/bic/). The amplified products were analyzed by conformation-sensitive gel electrophoresis (CSGE) (Ganguly et al. 1993) and positive samples were sequenced using an automated fluorescence-based cycle sequencer (ABI Prism 310; Applied Biosystems, Foster City, CA). Mutations were classified according to current guidelines (den Dunnen and Antonarakis 2000).
Detection of BRCA1 Rearrangements
All samples negative for BRCA1/2 mutations by CSGE analysis were tested for the presence of large rearrangements in the BRCA1 gene with the use of the multiplex ligation-dependent probe amplification (MLPA) (MRC Holland; Amsterdam, The Netherlands), according to the instructions provided by the manufacturer. Separation and relative quantification of the amplification products were obtained with an ABI Prism 310 automatic sequencer (Applied Biosystems) using the Genescan software.
Immunohistochemistry
Formalin-fixed, paraffin-embedded blocks of the 21 specimens available were analyzed using a monoclonal antibody for the BRCA1 protein [anti-BRCA1 (Ab-1), human (mouse); epitope: 1–304 N-terminal amino acids of BRCA1] and a polyclonal antibody for the BRCA2 protein [anti-BRCA2 (Ab-2), rabbit polyclonal IgG; epitope: 3245–3418 of human BRCA2] (Oncogene Research Products/Calbiochem; Darmstadt, Germany) at a dilution of 1:300 and 1:100, respectively. Antigen retrieval was performed by heating the specimens in a pressure cooker for 6 min in citrate buffer (pH 6). The Envision system (Dako; Copenhagen, Denmark) was used for detection. Sections from positive breast invasive ductal carcinoma were used as positive controls, and negative controls were obtained by omitting the primary antibodies. The reaction was considered positive if more than 10% of the cells showed distinctive nuclear staining. Specific staining was evaluated by two investigators who had no knowledge of the patients' family histories or the results of mutation analysis.
Protein Extraction
Total protein extracts were obtained from formalin-fixed paraffin-embedded tissue sections. Three 50-μm-thick sections were deparaffinized, and exclusively cancer tissues were collected and further cut into small pieces. Proteins were extracted from tissue sections with 50 μl of radioimmunoprecipitation (RIPA) buffer, pH 7.6 [50 mM Tris, 5 mM EDTA, 1% Triton X-100, 0.4% sodium deoxycholate, 150 mM NaCl, and protease inhibitors (Complete Mini, EDTA-free; Roche, Mannheim, Germany)], followed by incubation at 100C for 20 min and incubation at 60C for 2 hr. Next, the tissue lysates were centrifuged at 14,000 × g for 20 min at 4C, and the supernatants were collected (Ikeda et al. 1998). Protein concentration of the lysates was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories; Munich, Germany). This reaction is similar to the well-documented Bradford assay (Bradford 1976).
Immunoprecipitation and Western Blot Analysis
Total protein extracts were immunoprecipitated with the Ab-1 monoclonal antibody. Samples were precleared with protein G-Sepharose beads (Sigma-Aldrich Chemie; Steinheim, Germany) for 1 hr at 4C in RIPA buffer, and supernatants were recovered and incubated overnight at 4C with the antibody at 2 μg/ml. The binding reactions were incubated for 1 hr 30 min at 4C with protein G-Sepharose beads. The immunoprecipitated proteins were obtained after three washes in RIPA buffer and boiling at 100C for 5 min in sample loading buffer (2-fold concentrate: 125 mM Tris, 20% glycerol, 4% SDS, 2% 2-mercaptoethanol, and 0.001% bromophenol blue). Proteins were analyzed by 8% SDS-PAGE gels and transferred onto nitrocellulose membranes (Hybond-C extra; Amersham Life Sciences, Buckinghamshire, UK). After incubation with the primary antibody (Ab-1) overnight at 2 μg/ml, the membranes were incubated for 2 hr with a goat anti-mouse IgG-horseradish peroxidase (Santa Cruz Biotechnology; Santa Cruz, CA) at a dilution of 1:5000. An enhanced chemiluminescence detection system and Kodak film (Amersham Pharmacia Biotech; Buckinghamshire, UK) were used to visualize the presence of proteins on the nitrocellulose blots.
Statistical Analysis
Sensitivity, specificity, and positive and negative predictive values were calculated for the results of immunohistochemistry to predict for BRCA1/2 mutations. The association between negative immunohistochemistry and mutation carrier status was analyzed using the two-tailed Fisher's exact test.
Results
Clinicopathological Data
Nine out of 18 women had been diagnosed with unilateral breast cancer, 2 with bilateral breast cancer, 3 with breast and ovarian cancer, 2 with ovarian cancer, 1 with breast and rectal cancer, and 1 with breast and lung cancer. Twenty-one specimens were available: 15 with breast cancer tissue, 5 with ovarian cancer, and 1 with rectal cancer. Two of the five ovarian cancer specimens were of surgically excised metastasis (epiploon and hepatic metastasis). The other three specimens included the primary tumor. Clinicopathological characteristics are described in Table 1.
Table 1.
Clinopathological data
| Case | Age (years) | Tumor specimen | Histological diagnosis | Histological grade | Pathological stage | Estrogen receptors |
| 1 | 28 | Breast | Invasive ductal carcinoma with intraductal component (<10%) | G2 | IIB | + |
| 2 | 69 | Breast | Invasive ductal carcinoma, NOS | G2 | IIA | + |
| 3 | 28 | Breast | Invasive ductal carcinoma, NOS | G1 | IIA | + |
| 4 | 46 | Breast | Invasive ductal carcinoma, NOS | G2 | IIB | − |
| 5 | 35 | Breast | (1) Invasive ductal carcinoma with predominant intraductal component | (1) G2 | (1) IIB | + |
| (2) Invasive ductal carcinoma, NOS | (2) G2 | (2) IIB | + | |||
| 6 | 41 | Ovary | Serous adenocarcinoma ∗ | G2 | IIIB | NA |
| 7 | 43 | Breast | Invasive ductal carcinoma, NOS | G3 | IIA | + |
| 8 | 49 | Breast | Osseous metastasis of invasive ductal carcinoma | G3 | IV | + |
| 9 | 52 | Breast | Invasive ductal carcinoma | G3 | I | − |
| 10 | 42 | Breast | Intraductal carcinoma | G2 | 0 | + |
| 11 | 58 | Ovary | Serous (60%) and endometrioid (40%) carcinoma | G3 | IB | NA |
| 12 | 34 | Breast | Invasive ductal carcinoma with intraductal component (<20%) | G2 | IIB | + |
| 13 | 67 | Breast | Invasive ductal carcinoma with predominant intraductal component | G3 | I | + |
| 14 | 37 | (1) Breast | (1) Invasive ductal carcinoma | (1) G3 | (1) IIB | (1)− |
| (2) Rectum | (2) Adenocarcinoma | (2) G2 | (2) IIIB | (2) NA | ||
| 15 | 32 | Breast | Invasive ductal carcinoma | G3 | IIA | + |
| 16 | 33 | Breast | Invasive ductal carcinoma with predominant intraductal component | G2 | I | + |
| 17 | 62 | Ovary | Liver metastasis of ovary serous carcinoma | G3 | IV | NA |
| 18 | 55 | Ovary | Clear cell carcinoma | G3 | IIIC | NA |
∗ Two samples reviewed, one of a surgically excised metastasis and the other including the primitive tumor.
Only specimens available for review are shown. In case 3, the patient had contralateral breast cancer at age 35, and in case 5 a first diagnosis of invasive lobular carcinoma of the other breast had been made at age 33; Cases 9, 17, and 18 had both breast and ovarian cancer, and case 13 was treated for lung cancer before her breast cancer diagnosis. NA, not applicable; NOS, not otherwise specified.
Genetic Screening
Of the 18 women with available paraffin-embedded tumor blocks, 7 were found to be positive for BRCA1/2 mutations: 5 with BRCA1 mutations (c.536delA, g.Ex13ins6Kb, c.211A>G, and g.Ex11_Ex15del) and 2 with BRCA2 mutations (c.1369_1370ins2 and c.7208_ 7211del4). The remaining 11 women were negative for BRCA1/2 mutations, by CSGE and MLPA screening of BRCA1 rearrangements (Table 2).
Table 2.
Correlation between mutation testing and immunohistochemistry
| Case | BRCA1/2 mutation (designation in BIC database) | Codon | Exon | Coding effect | BRCA1 nuclear immunostaining | BRCA2 nuclear immunostaining |
| 1 | Negative | − | − | − | + | + |
| 2 | Negative | − | − | − | + | + |
| 3 | c.536delA | 179 | 8/1 | STOP 233 | − | − |
| 4 | Negative | − | − | − | + | − |
| 5 | c.1369_1370ins2 | 457 | 10/2 | STOP 460 | + | + ∗ |
| 6 | g.Ex13ins6Kb (exon13ins6Kb) | NA | 13/1 | STOP 1460 | − | − |
| 7 | c.7208_7211del4 (7436del4) | 2403 | 14/2 | STOP 2467 | + | − |
| 8 | Negative | − | − | − | + | − |
| 9 | Negative | − | − | − | + | − |
| 10 | Negative | − | − | − | + | + |
| 11 | g.Ex11_Ex15del | NA | 11–15/1 | ID | + | − |
| 12 | Negative | − | − | − | + | − |
| 13 | Negative | − | − | − | + | − |
| 14 | c.211A>G (R71G) | 71 | 5/1 | STOP 64 | − | − |
| 15 | Negative | − | − | − | + | − |
| 16 | Negative | − | − | − | + | + |
| 17 | Negative | − | − | − | + | + |
| 18 | c.211A>G (R71G) | 71 | 5/1 | STOP 64 | − | + |
∗ This patient relapsed in the breast 3 years latter and the specimen of invasive ductal carcinoma was negative for BRCA2 immunohistochemistry. BIC, Breast Cancer Information Core; ID, indeterminate; NA, not applicable.
Immunohistochemistry
Tumor samples from four of five women with BRCA1 mutations were BRCA1 negative, with absence of nuclear or cytoplasmic staining (Figure 1B). These cases included two breast cancer specimens (from cases 3 and 14), three ovarian cancer specimens (two from case 6 and one from case 18), and one rectal cancer specimen (case 14). In contrast, the other patient with a BRCA1 mutation (g.Ex11_Ex15del) had her ovarian cancer specimen stain positive for BRCA1 immunohistochemistry, with a clear nuclear immunoreactivity in tumor cells. All other tumor samples, from the 2 BRCA2-positive women and from the 11 patients negative for mutations in both genes, were positive for BRCA1 immunostaining (Table 2).
Figure 1.
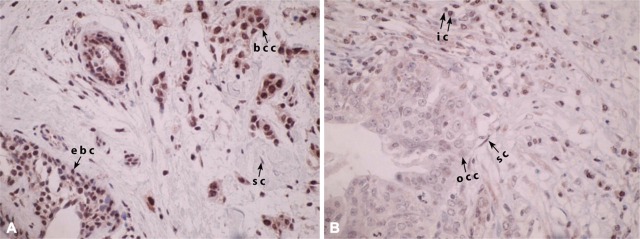
BRCA1 immunohistochemistry in cancer specimens. (A) Breast cancer specimen with clear nuclear BRCA1 labeling in cancer (bcc), epithelial (ebc), and stromal (sc) cells contrasts with an ovarian cancer sample (B), where cancer cells (occ) are negative for BRCA1, even if inflammatory (ic), and stromal cells (sc) keep the expected nuclear labeling.
BRCA2 staining was also optimized and nuclear staining observed in tumor samples. Cytoplasmic staining was not observed. One BRCA2-positive woman had her invasive breast cancer sample stain negative for BRCA2, whereas the opposite was observed in a sample of invasive ductal carcinoma with large areas of intraductal carcinoma of the other BRCA2 mutation carrier. In this last case, a relapse in the same location of the right breast was observed 3 years later, and the corresponding tumor sample, of invasive ductal carcinoma, was negative for BRCA2 staining. Most of the tumor samples from the 18 women were negative for BRCA2 labeling (Table 2).
Protein Analysis
For BRCA1, the absence of the 220-kDa BRCA1 protein in negative immunohistochemistry tumor samples was further confirmed with total protein extracts from paraffin tumor specimens, by Western blot. Normal breast tissue was used as control, and adequate total protein lysates were obtained from seven of our tissue-embeded samples. Total proteins were concentrated by immunoprecipitation, and in the experiment shown in Figure 2, the band corresponding to the BRCA1 protein was observed by Western blotting. BRCA1 protein was observed in normal breast tissue (positive control) and in a BRCA1-negative mutation case with positive stained BRCA1 specimens (case 1) but not in two BRCA1 mutation—positive cases with negative BRCA1 immunostaining (cases 6 and 14) and in the negative control (protein G-Sepharose beads with Ab-1 monoclonal antibody).
Figure 2.
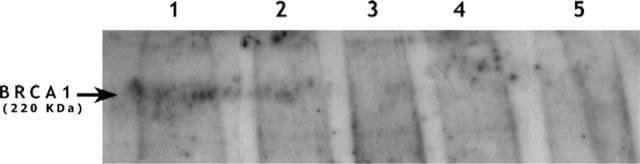
Absence of BRCA1 protein in negative breast cancer specimens. After Western blotting of tumor protein extracts with the Ab-1 antibody, BRCA1 protein was identified in normal breast, positive control (1) and in breast cancer from a BRCA1 mutation-negative woman but with positive Ab-1 immunostaining (2). In BRCA1 mutation-positive cases with negative immunostaining, the 220-kDa band corresponding to the BRCA1 protein was not observed (3 and 4). Negative control, protein G-Sepharose beads with Ab-1 monoclonal antibody (5).
Correlation Between Genetic Screening and Immunohistochemistry
Sensitivity of immunohistochemistry to detect BRCA1 mutation carriers was 80% and specificity 100%. Negativity for BRCA1 labeling in a tumor paraffin specimen was significantly correlated with BRCA1 carrier status (p=0.001); the positive predictive value of this test was 100%, and the negative predictive value 93%. For BRCA2, sensitivity of immunochemistry was 50% and specificity 38%. The results of immunohistochemistry for BRCA2 were not significantly correlated with BRCA2 carrier status (p=1).
Discussion
In this study, we observed that immunostaining with a monoclonal antibody against the N-terminal amino acids of the BRCA1 protein has a high specificity for the prediction of BRCA1 mutation carriers. These results suggest that BRCA1 immunohistochemistry, a rapid and easy test, can be used before the expensive mutation screening, to select which high-risk cases should be submitted to analysis of this gene; when BRCA1 staining shows integrity of the protein, BRCA2 screening should be done first. This methodology is helpful in case selection for subsequent mutation analysis and it can also be the only method to demonstrate inherited breast or ovarian cancer in deceased individuals belonging to high-risk breast/ovarian cancer families. However, the inclusion of BRCA1 immunohistochemistry in algorithms concerning patient selection for BRCA1/2 mutation screening is not recommended at this time, because the results of our study need confirmation in a larger, preferentially prospective sample. Confirmations of familiar cancers are not easy to obtain in breast cancer risk evaluation clinics, and cancer specimens are even more difficult to obtain. Collaborative studies are needed to overcome these difficulties.
Specificity of BRCA1 immunohistochemistry to predict for BRCA1 mutations was 100% and sensitivity 80%. In only one case was BRCA1 immunohistochemistry not concordant with the final mutation results: a woman negative for BRCA1/2 screening by CSGE was found to test positive for a large BRCA1 deletion after MLPA screening. This rearrangement included the deletion of exons 11–15 of BRCA1, and immunohistochemistry of this patient's ovarian tumor sample showed positive labeling of the BRCA1 protein. The most reasonable explanation for this observation is that besides this large deletion, the N-terminal portion of the BRCA1 protein is transcribed and is accessible to the Ab-1 antibody. A previous study had already shown that BRCA1 exon 11 mutations may not affect the immunostaining of both C-terminal and N-terminal antibodies (Kashima et al. 2000) (Table 3). In the present study, most of the BRCA1 mutations occurred downstream of exon 11; the only two exceptions were the large deletion encompassing exons 11–15 and Ex13ins6Kb, another large rearrangement occurring upstream of exon 11. In this last case, complete negativity of BRCA1 was observed, as in all other cases with mutations downstream of exon 11. These observations reinforce the notion that BRCA1 immunohistochemistry can be used as a prescreen for mutation testing but, because sensitivity is 80%, it cannot replace mutation testing. However, its inclusion in a decision process also considering phenotypic and pathological factors can rationalize the approach and the cost of BRCA1/2 mutations.
Table 3.
Prediction of BRCA1 carrier status with immunohistochemistry for the BRCA1 protein
In cases negative for immunostaining, this finding was striking in that almost all tumor cells were negative. Although we indicated a cutoff at less than 10% of labeled tumor cells to consider the labeling negative, in only one of the tumors analyzed (one of the ovarian cancer specimens) very few cells (much less than 10%) were scarcely positive for BRCA1. This is in contrast with the results of another study in which only “islands” of negativity for BRCA1 immunohistochemistry were observed in the presence of BRCA1 mutations (Schofield et al. 2000) (Table 3). The results we present here, however, were confirmed by Western blotting: negative samples after BRCA1 immunohistochemistry were negative for the detection of the BRCA1 protein, and the opposite was observed for positive samples. No other study had previously done this type of assay, probably because of the technical difficulties in obtaining protein lysates from paraffin-embedded specimens. A recent study whose aim was to assess the specificity and sensitivity of immunohistochemistry as a screening method for demonstrating BRCA1 expression concluded that Ab-1 was the only BRCA1 antibody whose staining correlated significantly with loss of BRCA1 expression as determined by RT-PCR (Al-Mulla et al. 2005). Table 3 summarizes prior studies attempting to analyze the sensitivity and specificity of several BRCA1 antibodies and the prediction of BRCA1 mutation carrier status with immunohistochemistry.
Small sample size, one of the limitations of our study, is due to the difficulty in assembling tissue specimens from all patients tested for BRCA1/2 mutations. In spite of this, one of the strengths of our assay is that BRCA1 results were concordant not only in tumors of different origins (breast, ovarian, and one case of rectal cancer) but also in primitive and metastatic lesions. Rectal cancer has been an interesting although complicated issue since it was first associated with BRCA1 (Brose et al. 2002; Thompson and Easton 2002), but more recent studies did not confirm that association (Niell et al. 2004). BRCA1 loss is expected in a subgroup of colorectal cancers (Grabsch et al. 2006), and it may be that our specimen was negative because it was a sporadic rectal cancer in a confirmed BRCA1 carrier or because the specific splicing BRCA1 mutation that this patient inherited is truly associated with colorectal and other digestive tumors (Vega et al. 2001).
Exclusive nuclear staining could be considered to represent the normal phenotype and was observed for both BRCA1 and BRCA2 antibodies in all tumor specimens analyzed in our study. Some studies (Chen et al. 1995; Coene et al. 1997; Perez-Valles et al. 2001; Al-Mulla et al. 2005) also reported a cytoplasmatic staining. Several reasons may account for the observation of BRCA1/2 in the cytoplasm: cross-reaction between the target epitopes; the presence of BRCA1 splice variants (Wilson et al. 1997); BRCA2 truncated forms (Spain et al. 1999), or loss of the nuclear localization signals of the BRCA proteins. In fact, the ability to enter the nucleus may be compromised by germline mutations in the BRCT (Rodriguez et al. 2004) domain of BRCA1 and in the carboxyl terminus of BRCA2 (Marmorstein et al. 1998).
Prediction of BRCA2 mutation carriers remains a problem. We could not find a correlation with immunohistochemistry in our study, and several studies show that pathological or microarray tumor analysis of BRCA1 and BRCA2 tumors is more distinctive for BRCA1 than for BRCA2: BRCA1 tumors tend to be of higher grade and are also more frequently negative for hormone receptors and more p53-positive than are their BRCA2 counterparts (Lakhani et al. 2002); type D cyclins and their associated CDK4 and CDKIs (p16, p21, and p27) were found to be downregulated in BRCA1 in respect to BRCA2 carcinomas. Also, over 80% of triple-negative breast tumors (negative for estrogen, progestagen, and HER2 receptors) are “basal-like,” and basal markers have been shown to be specific to a subset of BRCA1 carcinomas (Palacios et al. 2004). The apparent relative excess of BRCA1 germinal mutations in association with triple negative tumors is under study (Kandel et al. unpublished data). The characterization of BRCA2 breast tumors and their distinction from sporadic breast cancers is an area of active research, and one recent study proposed that BRCA2 tumors could be identified through the use of an array of markers that included CHEK2 and RAD51: CHEK2 was found to be more frequently expressed in BRCA1 and BRCA2 tumors than in non-BRCA1/2, and BRCA2 was found to be necessary for the translocation of RAD51 (Honrado et al. 2005).
In conclusion, we observed a high specificity for the prediction of BRCA1 carriers with immunohistochemistry using a monoclonal BRCA1 antibody. Validation of this assay, using a larger sample, will allow the use of immunohistochemistry for deciding which high-risk patients should be screened first for the BRCA1 gene. This recommendation does not exclude the relevance of other known risk factors for mutations in this gene (family history, age at cancer diagnosis, histological characteristics of the tumors, triple negativity) but is intended to contribute to a more specific patient selection. Because only positive results are informative for probands of these families, better patient selection is likely to increase the possibilities of obtaining informative genetic results.
Acknowledgments
This research was supported by grant 47320 from Serviço de Saúde e Desenvolvimento da Fundação Calouste Gulbenkian, Lisboa, and by the grant Investigação em Oncologia NRS/LPCC-Terry Fox “2001-2002,” from Liga Portuguesa Contra o Cancro, Lisboa, Portugal.
We are grateful to Fernanda Silva and Teresa Pereira for their technical assistance and to Dr. Cristina Casalou for technical assistance and review of the manuscript.
Literature Cited
- Al-Mulla F, Abdulrahman M, Varadharaj G, Akhter N, Anim JT. (2005) BRCA1 gene expression in breast cancer: a correlative study between real-time RT-PCR and immunohistochemistry. J Histochem Cytochem 53: 621–629 [DOI] [PubMed] [Google Scholar]
- American Society of Clinical Oncology (2003) American Society of Clinical Oncology policy statement update: genetic testing for cancer susceptibility. J Clin Oncol 21: 2397–2406 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Breast Cancer Linkage Consortium (1999) Cancer risks in BRCA2 mutation carriers. J Natl Cancer Inst 91: 1310–1316 [DOI] [PubMed] [Google Scholar]
- Brose MS, Rebbeck TR, Calzone KA, Stopfer JE, Nathanson KL, Weber BL. (2002) Cancer risk estimates for BRCA1 mutation carriers identified in a risk evaluation program. J Natl Cancer Inst 94: 1365–1372 [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen CF, Riley DJ, Allred DC, Chen PL, Von Hoff D, Osborne CK, et al. (1995) Aberrant subcellular localization of BRCA1 in breast cancer. Science 270: 789–791 [DOI] [PubMed] [Google Scholar]
- Coene E, Van Oosteveldt P, Willems K, Van Emmelo J, De Potter CR. (1997) BRCA1 is localized in cytoplasmic tube-like invaginations in the nucleus. Nat Genet 16: 122–124 [DOI] [PubMed] [Google Scholar]
- Crook T, Crossland S, Crompton MR, Osin P, Gusterson BA. (1997) P53 mutations in BRCA1-associated familial breast cancer. Lancet 350: 638–639 [DOI] [PubMed] [Google Scholar]
- Den Dunnen JT, Antonarakis SE. (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15: 7–12 [DOI] [PubMed] [Google Scholar]
- Easton DF, Ford D, Bishop DT. (1995) Breast and ovarian cancer incidence in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Am J Hum Genet 56: 265–271 [PMC free article] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow up. Histopathology 19: 403–410 [DOI] [PubMed] [Google Scholar]
- Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, et al. (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am J Hum Genet 62: 676–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Stefansson IM, Chappuis PO, Begin LR, Goffin JR, Wong N, Trudel M, et al. (2003) Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 95: 1482–1485 [DOI] [PubMed] [Google Scholar]
- Frank TS, Manley SA, Olopade OI, Cummings S, Garber JE, Bernhardt B, Antman K, et al. (1998) Sequence analysis of BRCA1 and BRCA2: correlation of mutations with family history and ovarian cancer risk. J Clin Oncol 7: 2417–2425 [DOI] [PubMed] [Google Scholar]
- Ganguly A, Rock MJ, Prockop DJ. (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90: 10325–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabsch H, Dattani M, Barker L, Maughan N, Maude K, Hansen O, Gabbert HE, et al. (2006) Expression of DNA double-strand break repair proteins ATM and BRCA1 predicts survival in colorectal cancer. Clin Cancer Res 12: 1494–1500 [DOI] [PubMed] [Google Scholar]
- Henderson BR. (2005) Regulation of BRCA1, BRCA2 and BARD1 intracellular trafficking. Bioessays 27: 884–893 [DOI] [PubMed] [Google Scholar]
- Honrado E, Osorio A, Palacios J, Milne RL, Sanchez L, Diez O, Cazorla A, et al. (2005) Immunohistochemical expression of DNA repair proteins in familial breast cancer differentiate BRCA2-associated tumors. J Clin Oncol 23: 7503–7511 [DOI] [PubMed] [Google Scholar]
- Ikeda K, Monden T, Kanoh T, Tsujie M, Izawa H, Haba A, Ohnishi T, et al. (1998) Extraction and analysis of diagnostically useful proteins from formalin-fixed, paraffin-embedded tissue sections. J Histochem Cytochem 46: 397–403 [DOI] [PubMed] [Google Scholar]
- Kashima K, Oite T, Aoki Y, Takakuwa K, Aida H, Nagata H, Sekine M, et al. (2000) Screening of BRCA1 mutation using immunohistochemical staining with C-terminal and N-terminal antibodies in familial ovarian cancers. Jpn J Cancer Res 91: 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhani SR, Van De Vijver MJ, Jacquemier J, Anderson TJ, Osin PP, McGuffog L, Easton DF. (2002) The pathology of familial breast cancer: predictive value of immunohistochemical markers, estrogen receptor, progesterone receptor, HER-2, and p53 in patients with mutations in BRCA1 and BRCA2. J Clin Oncol 20: 2310–2318 [DOI] [PubMed] [Google Scholar]
- Liede A, Karlan BY, Narod SA. (2004) Cancer risks for male carriers of germline mutations in BRCA1 or BRCA2: a review of the literature. J Clin Oncol 22: 735–742 [DOI] [PubMed] [Google Scholar]
- Marmorstein LY, Ouchi T, Aaronson SA. (1998) The BRCA2 gene product functionally interacts with p53 and RAD51. Proc Natl Acad Sci USA 95: 13869–13874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niell BL, Rennert G, Bonner JD, Almog R, Tomsho LP, Gruber SB. (2004) BRCA1 and BRCA2 founder mutations and the risk of colorectal cancer. J Natl Cancer Inst 96: 15–21 [DOI] [PubMed] [Google Scholar]
- Palacios J, Honrado E, Osorio A, Diez O, Rivas C, Benitez J. (2004) Re: Germline BRCA1 mutations and a basal epithelial phenotype in breast cancer. J Natl Cancer Inst 96: 712–714 [DOI] [PubMed] [Google Scholar]
- Parmigiani G, Berry DA, Aguilar O. (1998) Determining carrier probabilities for breast cancer susceptibility genes BRCA1 and BRCA2. Am J Hum Genet 62: 145–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Valles A, Martorell-Cebollada M, Nogueira-Vazquez E, Garcia-Garcia JA, Fuster-Diana E. (2001) The usefulness of antibodies to the BRCA1 protein in detecting the mutated BRCA1 gene. An immunohistochemical study. J Clin Pathol 54: 476–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Au WW, Henderson BR. (2004) Cytoplasmic mislocalization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp Cell Res 293: 14–21 [DOI] [PubMed] [Google Scholar]
- Schofield AC, Payne S, Ross VG, Miller ID, Heys SD, Haites NE. (2000) Immunohistochemical detection of a germline BRCA1 mutation in a breast and ovarian cancer family. Breast 9: 286–291 [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, et al. (1997) Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell 88: 265–275 [DOI] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Brown M, De Caprio JA, Cannistra SA, Feunteun J, Schnitt S, et al. (1996) Location of BRCA1 in human breast and ovarian cancer cells. Science 272: 123–126 [DOI] [PubMed] [Google Scholar]
- Shih HA, Couch FJ, Nathanson KL, Blackwood MA, Rebbeck TR, Armstrong KA, Calzone K, et al. (2002) BRCA1 and BRCA2 mutation frequency in women evaluated in a breast cancer risk evaluation clinic. J Clin Oncol 20: 994–999 [DOI] [PubMed] [Google Scholar]
- Spain BH, Larson CJ, Shihabuddin LS, Gage FH, Verma IM. (1999) Truncated BRCA2 is cytoplasmic: implications for cancer-linked mutations. Proc Natl Acad Sci USA 96: 13920–13925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormorken AT, Bowitz-Lothe IM, Noren T, Kure E, Aase S, Wijnen J, Apold J, et al. (2005) Immunohistochemistry identifies carriers of mismatch repair gene defects causing hereditary nonpolyposis colorectal cancer. J Clin Oncol 23: 4705–4712 [DOI] [PubMed] [Google Scholar]
- Thompson D, Easton DF. (2002) Cancer incidence in BRCA1 mutation carriers. Breast Cancer Linkage Consortium. J Natl Cancer Inst 94: 1358–1365 [DOI] [PubMed] [Google Scholar]
- Vaz FH, Machado PM, Brandão RD, Laranjeira CT, Eugénio JS, Fernandes AH, André SP. (In Press) Familial breast/ovarian cancer and BRCA1/2 genetic screening: the role of immunohistochemistry as an additional method in the selection of patients. J Histochem Cytochem. [DOI] [PMC free article] [PubMed]
- Vega A, Campos B, Bressac-de-Paillerets B, Bond PM, Janin N, Douglas FS, Domenech M, et al. (2001) The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum Mutat 17: 520–521 [DOI] [PubMed] [Google Scholar]
- Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J, Roach KC, et al. (2006) Spectrum of mutations in BRCA1, BRCA2, CHEK2, and TP53 in families at high risk of breast cancer. JAMA 295: 1379–1388 [DOI] [PubMed] [Google Scholar]
- Welcsh PL, King MC. (2001) BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum Mol Genet 10: 705–713 [DOI] [PubMed] [Google Scholar]
- Wilson CA, Payton MN, Elliot GS, Buaas FW, Cajulis EE, Grosshans D, Ramos L, et al. (1997) Differential subcellular localization, expression and biological toxicity of BRCA1 and the splice variant BRCA1-delta 11b. Oncogene 14: 1–16 [DOI] [PubMed] [Google Scholar]


