Abstract
To date, there is no commercially available Y chromosome probe that can be used for fluorescence in situ hybridization (FISH) for the male rhesus monkey. We have recently generated a probe for FISH with high specificity to the short arm of the rhesus monkey Y chromosome. In this study, we further describe a method that keeps the integrity of tissue-specific antigenic structures for immunofluorescence staining subsequent to FISH on paraffin-embedded rhesus monkey tissues. We have examined this technique in combination with an epithelial cell—specific marker, cytokeratin 8/18 (CK8/18), on various tissues, including jejunum, liver, kidney, and pancreas. CK8/18 and Y chromosome signals were distinctly seen simultaneously on epithelial cells from the same tissue section from male but not female monkeys. These studies indicate that our FISH immunofluorescence technique can be reliably used to identify and phenotype male cells in paraffin-embedded rhesus monkey tissues.
Keywords: rhesus monkey, fluorescence in situ hybridization, Y chromosome immunofluorescence
Cell-tracking studies often require a label that is uniquely distinguishable and biologically stable. Donor cell tracking has been achieved through genetic labeling using either a gene reporter such as β-galactosidase (Alvarez-Dolado et al. 2003), a fluorescent protein such as green fluorescent protein (Weimann et al. 2003b) and its spectral variants (Feng et al. 2000), or detection of DNA-specific sequences by fluorescence in situ hybridization (FISH) (Eglitis and Mezey 1997; Mezey et al. 2000, 2003).
Y chromosome—specific probes for FISH have been used to detect male cells transplanted into female recipients in both mice and humans (Mezey et al. 2000, 2003; Weimann et al. 2003a). When FISH for the Y chromosome is combined with immunohistochemistry (IHC) or immunofluorescence staining, it allows a more reliable method for following the path taken by transplanted male stem cells and their differentiation into mature cells (Donadoni et al. 2004; Trotman et al. 2004). The combination of FISH with IHC has been used to identify male cells that have differentiated into tissue-specific cell types such as hepatocytes and cholangiocytes in humans (Theise et al. 2000; Korbling et al. 2002), as well as epithelial cells of liver, lung, gastrointestinal tract, and skin in mouse mixed-gender studies (Krause et al. 2001). Although these techniques are available for human and rodent models, the ability to discriminate male cells from female cells in rhesus monkey tissues has been very difficult, if not impossible, because no commercial probe(s) specific to the Y chromosome have been available.
The rhesus monkey is physiologically and phylogenetically similar to humans, and therefore is a clinically relevant animal model for biomedical research (Bontrop 2001; Pau and Wolf 2004). We have recently generated a DNA probe, by a chromosome microdissection technique, from the Y chromosome of the rhesus monkey (Taguchi et al. 2003). This probe has high specificity to the short arm of the Y chromosome; however, it has been successfully used only on metaphase cells to identify the Y chromosome using FISH.
In this study, we describe for the first time the combination of Y chromosome FISH and immunofluorescence staining on rhesus monkey paraffin-embedded tissues. This technique allows phenotyping of the same cells that show very distinct Y chromosome painting on jejunum, kidney, liver, and pancreas of only male rhesus monkeys.
Materials and Methods
Tissue Samples
Tissue samples from male and female rhesus monkeys were obtained according to the Institutional Animal Care and Use Committee (IACUC) approval at necropsy from the Division of Comparative Pathology, Tulane National Regional Primate Center. The tissue samples collected included jejunum, kidney, liver, pancreas, and bone marrow (not shown). The tissues were fixed by formalin and embedded in paraffin under standard conditions. Three-μm sections were placed on silanized slides and baked overnight at 56C before the FISH procedure.
Y Chromosome FISH Probe
The DNA sequence used as the template to generate our FISH probe was originally obtained by a chromosome microdissection technique and then microcloned using the p-GEM-T Easy-Vector (Promega; Madison, WI) (Taguchi et al. 2003). The resultant microclone plasmids were extracted using the S.N.A.P Midi Prep Kit (Invitrogen; Carlsbad, CA) according to the manufacturer's instructions. The inserted DNA fragments (MMY#4) were sequenced with T7 RNA polymerase primers using ABI Prism (Model 3100; Version 3.7) software. Computer-assisted DNA alignments and comparisons were made using BLAST (PubMed) to search the GenBank database. The gene sequence was confirmed to be an exact match to the Y chromosome—specific repeated sequence MMDYZ1 (accession number AB100455), a partial sequence of Macaca mulatta (rhesus monkey) DNA. The MMY#4 DNA fragment was then excised from the plasmid DNA with NotI restriction endonucleases (New England Biolabs; Ipswich, MA) and extracted by standard methods. The FISH Y probe was generated by a random primer labeling technique using the MMY#4 DNA fragment. Digoxygenin (DIG)-labeled DNA labeling mix, hexanucleotide mix, and Klenow enzyme (Roche Applied Science; Penzberg, Germany) were used according to the manufacturer's protocol. The concentration of the DIG-labeled probe was determined by dot blot comparison to a known concentration of control DIG-labeled RNA (Roche Applied Science).
FISH Y Chromosome Painting
All protocol steps were carried out at room temperature unless otherwise specified. Tissue sections were deparaffinized and rehydrated according to standard protocols. After washing with PBS, the slides were treated with 0.2 N hydrochloric acid for 20 min, rinsed with double-distilled water (ddH2O) once, then washed twice for 5 min with 2× standard saline citrate solution (SSC). The slides were immersed in 0.1 M citric buffer (pH 6), steamed for 20 min at ≥96C, allowed to cool for 15 min, then washed twice for 5 min with 2× SSC. After prehybridization with hybridization buffer (50% formamide, 10% dextran sulfate, 2× SSC, 1% salmon sperm DNA, 1% Denhardt's solution) for 30 min at 42C, 12 μl digoxigenin-labeled Y probe (20 ng/section diluted with hybridization buffer) was applied to each tissue section and coverslipped. The edges of the coverslips were sealed with rubber cement. The probe and target DNA were denatured simultaneously by heating the slides for 8 min at 90C in a slide moat. Slides were then incubated in a humidified chamber at 42C overnight. Following incubation, the rubber cement was removed. The slides were washed twice for 15 min in 50% formamide/2× SSC at 42C in a water bath to remove excess unbound probe, then washed in 0.2× SSC two times for 5 min at 42C. One hundred μl blocking reagent (Roche Applied Science) was added to each section, and sections were incubated for 30 min at 37C. The anti-DIG-alkaline phosphatase (AP) Fab (Roche Applied Science) antibody was diluted 1:500 in blocking reagent, 100 μl was added to each section, and sections were incubated for 1 hr at 37C in the dark. The signals were then amplified by HNPP-Fast Red (Roche Applied Science) according to manufacturer's protocol. The slides were briefly checked with a fluorescence microscope under a red filter for adequate FISH signals prior to immunofluorescence staining.
Immunofluorescence: Cytokeratin 8/18 and E-Cadherin
The controls for cytokeratin 8/18 (CK8/18) immunofluorescence staining of tissue sections included jejunum, kidney, liver, and pancreas from one male and one female rhesus monkey. Another epithelial cell marker E-cadherin (E-cad) was also used to double stain the epithelial cells. The tissues were deparaffinized and rehydrated according to standard protocol. All protocol steps were carried out at room temperature unless otherwise specified. Slides were washed with PBS for 5 min, antigen retrieval was performed by immersing slides in 0.1 M citric buffer (pH 6), and slides were steamed for 20 min at ≥96C. The slides were allowed to cool for 15 min and were then washed for 5 min with PBS supplemented with 0.2% fish skin gelatin (FSG) and 0.1% Triton-X-100 buffer, followed by washing twice with PBS with 0.2% FSG only. From this point on, all tissue sections for FISH for the Y chromosome and control tissue sections for immunofluorescence staining were treated with a serum-free protein blocking reagent (DakoCytomation; Carpinteria, CA) for 30 min, prior to mouse anti-human CK8/18 LMW (clone 5D3, prediluted; Biocare Medical, Concord, CA). The tissues were then incubated for 1 hr at 37C in a humidified chamber. The control slides were not treated with the primary antibody. After incubation, all slides were washed three times with PBS for 5 min. The primary antibody was detected using Alexa 488—labeled goat anti-mouse antibody (Molecular Probes; Eugene, OR) diluted 1:1000 in antibody diluent (DakoCytomation). One hundred μl of diluted secondary antibody was added to each section, and sections were incubated for 30 min in the dark. At this point, slides used for FISH for the Y chromosome were rinsed as described above in PBS and further incubated for 10 min with To-pro-3 (nuclear marker, Molecular Probes), diluted 1:1000 in antibody diluent for nuclear staining. Slides used as controls with only immunofluorescence staining were further treated with an additional primary mouse monoclonal antibody against a fragment of human E-cad (amino acids 735–883) (clone 36/E-cadherin; BD Transduction Laboratories, San Diego, CA) diluted 1:100 in antibody diluent and incubated for 1 hr at room temperature in a humidified chamber. After three washings with PBS for 5 min, Alexa 568—labeled goat anti-mouse antibody (Molecular Probes) diluted 1:1000 in antibody diluent was used for 30 min in the dark to detect the E-cad. To-pro-3 was then used as described above. After rinsing in ddH2O for 5 min, the slides were mounted with anti-quenching solution and stored at 4C until confocal imaging.
Confocal Microscopy
Confocal microscopy was performed using a Leica TCS SP2 confocal microscope equipped with three lasers (Leica Microsystems; Exton, PA). NIH Image (version 1.62) and Adobe Photoshop (version 7.0) were used to assign colors to the four channels collected: HNPP/Fast Red substrate that fluoresces red when exposed to a 568-nm wavelength laser appears red; Alexa 488 is green; Alexa 568 is red; To-pro-3 appears blue; and the differential interference contrast (DIC) image appears in the gray scale. The four channels were collected simultaneously.
Results
Tissue samples, including jejunum, kidney, liver, and pancreas from male and female rhesus monkeys, were used to test the specificity and sensitivity of our FISH probe for the Y chromosome. The female rhesus monkeys were selected on the basis of gravidity to prevent the possibility of detecting lingering fetal male cells in female tissues. Hematoxylin and eosin staining confirmed that all tissues were free of disease and appeared normal.
For the FISH procedure, optimal pretreatment was by immersing the slides in 0.1 M citric buffer (pH 6) and steaming them for 20 min at ≥96C. Tissues were incubated with Y chromosome probe at 42C overnight for hybridization. The signals were amplified by anti-DIG-AP, Fab antibody with HNPP-Fast Red.
Following FISH, all tissues were stained with mouse anti-human CK8/18 and goat anti-mouse Alexa 488. To-pro-3 was also used to stain the nuclei. The tissues were examined by confocal microscopy. HNPP/Fast Red substrate is seen as red fluorescence, Alexa 488 is green, To-pro-3 appears blue, and the DIC image appears in the gray scale.
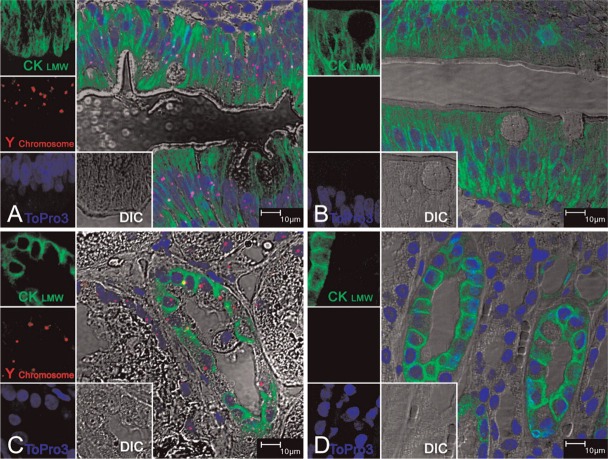
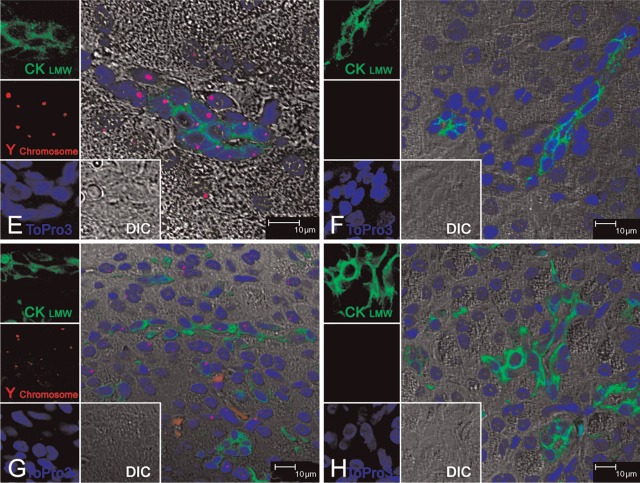
We found that the Y probe hybridized only with the majority of cells in representative tissue sections from male rhesus jejunum, kidney, liver, and pancreas. The Y labeling is specifically localized to the nucleus and is punctuated and discrete only in male tissues (Figures 1A, 1C, 1E, and 1G). The hybridization signal of the Y probe is absent in all of the corresponding control female tissue sections (Figures 1B, 1D, 1F, and 1H). The non-proteinase FISH protocol allowed the following immunofluorescence staining: Using mouse anti-human CK8/18, an epithelial cell—specific monoclonal antibody, we could identify the positive staining in the cytoplasm of all epithelial cells subsequent to the Y chromosome paint in all tissues. The morphology of the tissues was preserved very well by this protocol, as shown by DIC confocal imaging (Figures 1A–1H).
Figure 1.
Dual Y chromosome fluorescence in situ hybridization (FISH) with cytokeratin 8/18 (CK8/18) immunofluorescence staining on normal rhesus monkey tissues. Multilabel confocal microscopy in normal male rhesus monkey tissues: (A) jejunum, (C) kidney, (E) liver, and (G) pancreas; and female rhesus monkey tissues: (B) jejunum, (D) kidney, (F) liver, and (H) pancreas. Images for individual channels [epithelium marker CK8/18 with Alexa 488 is green, Y chromosome FISH with Fast Red is red, nucleus marker To-pro-3 is blue, and differential interference contrast (DIC) is gray] are shown on the left and bottom, and a large merged image containing three channels plus DIC showing double-positive cells is shown on the right. The pink/fuchsia spots (due to the combination of red and blue) indicate Y chromosome signals, which are located inside the blue-staining nucleus, as shown in A, C, E, and G. There were no Y chromosome paintings in the corresponding female tissues used as controls, as shown in B, D, F, and H. Immunofluorescence staining with CK8/18 was shown in all tissues used: the jejunum intestinal epithelium (A, B); the distal tubule epithelium in kidney (C, D); the bile duct epithelium of liver (E, F); and the intralobular duct epithelium in pancreas (G, H). The DIC images indicated that the morphology of all tissues was well preserved.
To further demonstrate that the green immunofluorescence staining of the CK8/18 was optimal and not interrupted by the FISH procedure in both male and female tissues (Figure 1), we used additional male and female tissues without FISH. We also used E-cad in combination with CK8/18 as an additional primary antibody that recognizes intercellular junction protein structures between epithelial cells in the same type of tissues (Figure 2). The intensity and specificity of the CK8/18 immunofluorescence was comparable for all tissues that were either treated or untreated for FISH (Figures 1 and 1). The red immunofluorescence and specificity of E-cad antibodies is seen overlapping with the green signals of CK8/18 as intense yellow immunofluorescence specifically between epithelial cells in the intestinal epithelium of jejunum (Figure 2A), the distal tubule of the kidney (Figure 2B), bile duct of liver (Figure 2C), and intralobular duct of pancreas (Figure 2D) and only where tight junctions are most prominent. In contrast, liver hepatocytes (Figure 2C) and ancinar cells of the pancreas (Figure 2D) show strong and distinct immunofluorescence signals only for E-cad (red) while maintaining a strong green immunofluorescence for CK8/18. The CK8/18 remained strong and distinct; no quenching of green fluorescence signals was observed when CK8/18 was used in combination with FISH or E-cad. Taken together, these results substantiate the validity of immunofluorescence staining of CK8/18 in all tissues in combination with FISH.
Figure 2.
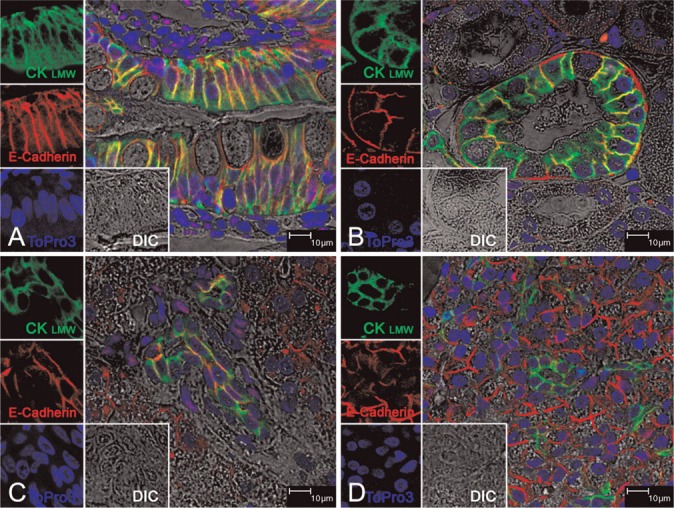
Immunofluorescence staining of CK8/18 on healthy rhesus monkey tissues. All panels show triple-labeled confocal microscopy (four channels). Images for individual channels [CK8/18 with Alexa 488 is green, E-cadherin (E-cad) with Alexa 568 is red, To-pro-3 is blue, and DIC is gray] are shown on the left and the main panel shows the merged image containing all three channels plus DIC. The areas of green and red fluorescence overlap are seen as yellow. (A) Jejunum epithelial cells; (B) kidney distal tubule epithelial cells; (C) bile duct epithelial cells of liver; (D) intralobular ductal epithelial cells of pancreas. Structures are all predominantly stained for CK8/18 and E-cad, whereas E-cad red fluorescence but not CK8/18 is seen on hepatocytes in C and acinar cells of pancreas in D. Taken together, CK8/18 is seen exclusively to stain epithelial cell structures of all of these tissues.
Discussion
In this study, we described an efficient and reliable way to combine FISH and immunofluorescence staining on male rhesus monkey paraffin-embedded tissue sections by using an antigen-retrieval step commonly employed in IHC. Our technique was validated by using the FISH probe alone or in combination with immunofluorescence using CK8/18, or conversely, comparing those results obtained by immunofluorescence technique without FISH, using instead CK8/18 in combination with E-cad, a second epithelial cell-specific protein marker.
We were able to generate a highly specific probe for FISH by using our template DNA previously obtained by microdissection of the Y chromosome from rhesus chromosomal smears (Taguchi et al. 2003). Our probe was specifically derived from micro-clone MMY#4, also previously shown to have a hybridized signal only to the short arm of the Y chromosome of rhesus monkeys, despite the sequence similarity to several satellite DNAs of many primates. Our goal now was to adapt our probe for FISH to be used on paraffin-embedded tissue sections and then use the FISH technique in combination with immunofluorescence or IHC so that we could simultaneously identify and phenotype cells bearing the Y chromosome in various tissues obtained from healthy male rhesus monkeys.
One of the major problems was related to the use of proteinase treatment. Although current FISH protocols usually require protein digestion to make nuclear DNA accessible to the probe, protein digestion often renders tissues unsuitable for subsequent IHC or immunofluorescence staining. To avoid potential limitations in the ability to visualize protein markers required to identify tissue-specific cell types, current studies have replaced the protein digestion step with a sodium citrate—based buffer to pretreat paraffin-embedded sections to make the cells permeable (Zaidi et al. 2000; Dundas et al. 2001; Trotman et al. 2004) We also found that immersing the tissue sections in 0.1 M citric acid buffer (pH 6) at 96C preserves all tissue integrity as detected by DIC and allows subsequent codetection of the Y chromosome by FISH and of epithelium antigenic structures by immunofluorescence. We used mouse anti-human CK8/18 monoclonal antibody to detect epithelial cells in jejunum, liver, kidney, and pancreas. The confocal images show that combining FISH with immunofluorescence allows the covisualization of CK8/18-positive cytoskeletal filaments localized in the cytoplasm and Y chromosome signals localized only in the nucleus of the same cells in all tissues examined.
E-cad was also used with CK8/18 without FISH to substantiate the CK8/18 staining when combined with FISH. We chose E-cad because it is an adhesion/tight junction protein that is exclusively expressed in epithelium (Eidelman et al. 1989). Both the villous and crypt epithelial layers showed uniformly strong positivity for E-cad in jejunum mucosa, as shown in Figure 2A (Jahnsen et al. 1998). In sections of normal kidney, E-cad was specifically detected within the distal convoluted tubules (Figure 2B) but not in proximal tubules (Krishnamachary et al. 2006). In liver, both hepatocytes and bile duct cells express E-cad (Figure 2C) (Ihara et al. 1996; Kozyraki et al. 1996), and pancreatic epithelium expresses high levels of E-cad (Figure 2D) (Weinel et al. 1996). Our dual-immunofluorescence staining with CK8/18 and E-cad confirms these previous reports and substantiates CK8/18 staining following the FISH procedure.
In conclusion, we have described for the first time a protocol enabling the combination of FISH and immunofluorescence that allows the simultaneous phenotype of Y-positive cells on paraffin-mounted male rhesus monkey tissue sections that include jejunum, kidney, liver, and pancreas. Our methodology could contribute to future advances in mixed-gender studies in rhesus monkeys and possibly other Old World monkeys.
Acknowledgments
This study was supported by a grant from the State of Louisiana Board of Regents, Health Excellence Fund, HEF No. (2001-06)-03 (to VFLR).
We thank Dr. Charles S. Hemenway for assistance with the microcloning technique. We also thank Dr. Juan Borda for assistance with histology.
Literature Cited
- Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, Fike JR, Lee HO, Pfeffer K, Lois C, et al. (2003) Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature 425: 968–973 [DOI] [PubMed] [Google Scholar]
- Bontrop RE. (2001) Non-human primates: essential partners in biomedical research. Immunol Rev 183: 5–9 [DOI] [PubMed] [Google Scholar]
- Donadoni C, Corti S, Locatelli F, Papadimitriou D, Guglieri M, Strazzer S, Bossolasco P, et al. (2004) Improvement of combined FISH and immunofluorescence to trace the fate of somatic stem cells after transplantation. J Histochem Cytochem 52: 1333–1339 [DOI] [PubMed] [Google Scholar]
- Dundas SR, Boyle S, Bellamy CO, Hawkins W, Garden OJ, Ross JA, Bickmore W. (2001) Dual Y-chromosome painting and immunofluorescence staining of archival human liver transplant biopsies. J Histochem Cytochem 49: 1321–1322 [DOI] [PubMed] [Google Scholar]
- Eglitis MA, Mezey E. (1997) Hematopoietic cells differentiate into both microglia and macroglia in the brains of adult mice. Proc Natl Acad Sci USA 94: 4080–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidelman S, Damsky CH, Wheelock MJ, Damjanov I. (1989) Expression of the cell-cell adhesion glycoprotein cell-CAM 120/80 in normal human tissues and tumors. Am J Pathol 135: 101–110 [PMC free article] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, Wallace M, Nerbonne JM, et al. (2000) Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28: 41–51 [DOI] [PubMed] [Google Scholar]
- Ihara A, Koizumi H, Hashizume R, Uchikoshi T. (1996) Expression of epithelial cadherin and alpha- and beta-catenins in nontumoral livers and hepatocellular carcinomas. Hepatology 23: 1441–1447 [DOI] [PubMed] [Google Scholar]
- Jahnsen FL, Farstad IN, Aanesen JP, Brandtzaeg P. (1998) Phenotypic distribution of T cells in human nasal mucosa differs from that in the gut. Am J Respir Cell Mol Biol 18: 392–401 [DOI] [PubMed] [Google Scholar]
- Korbling M, Katz RL, Khanna A, Ruifrok AC, Rondon G, Albitar M, Champlin RE, et al. (2002) Hepatocytes and epithelial cells of donor origin in recipients of peripheral-blood stem cells. N Engl J Med 346: 738–746 [DOI] [PubMed] [Google Scholar]
- Kozyraki R, Scoazec JY, Flejou JF, D'Errico A, Bedossa P, Terris B, Fiorentino M, et al. (1996) Expression of cadherins and alphacatenin in primary epithelial tumors of the liver. Gastroenterology 110: 1137–1149 [DOI] [PubMed] [Google Scholar]
- Krause DS, Theise ND, Collector MI, Henegariu O, Hwang S, Gardner R, Neutzel S, et al. (2001) Multi-organ, multi-lineage engraftment by a single bone marrow-derived stem cell. Cell 105: 369–377 [DOI] [PubMed] [Google Scholar]
- Krishnamachary B, Zagzag D, Nagasawa H, Rainey K, Okuyama H, Baek JH, Semenza GL. (2006) Hypoxia-inducible factor-1-dependent repression of E-cadherin in von Hippel-Lindau tumor suppressor-null renal cell carcinoma mediated by TCF3, ZFHX1A, and ZFHX1B. Cancer Res 66: 2725–2731 [DOI] [PubMed] [Google Scholar]
- Mezey E, Chandross KJ, Harta G, Maki RA, McKercher SR. (2000) Turning blood into brain: cells bearing neuronal antigens generated in vivo from bone marrow. Science 290: 1779–1782 [DOI] [PubMed] [Google Scholar]
- Mezey E, Key S, Vogelsang G, Szalayova I, Lange GD, Crain B. (2003) Transplanted bone marrow generates new neurons in human brains. Proc Natl Acad Sci USA 100: 1364–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pau KY, Wolf DP. (2004) Derivation and characterization of monkey embryonic stem cells. Reprod Biol Endocrinol 2: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi T, Akimaru K, Hirai H, Hirai Y, Mwenda JM, Yuri K. (2003) A probe generated by chromosome microdissection, useful for analyzing Y chromosome evolution in Old World monkeys. Chromosome Res 11: 147–152 [DOI] [PubMed] [Google Scholar]
- Theise ND, Nimmakayalu M, Gardner R, Illei PB, Morgan G, Teperman L, Henegariu O, et al. (2000) Liver from bone marrow in humans. Hepatology 32: 11–16 [DOI] [PubMed] [Google Scholar]
- Trotman W, Beckett T, Goncz KK, Beatty BG, Weiss DJ. (2004) Dual Y chromosome painting and in situ cell-specific immunofluorescence staining in lung tissue: an improved method of identifying donor marrow cells in lung following bone marrow transplantation. Histochem Cell Biol 121: 73–79 [DOI] [PubMed] [Google Scholar]
- Weimann JM, Charlton CA, Brazelton TR, Hackman RC, Blau HM. (2003a) Contribution of transplanted bone marrow cells to Purkinje neurons in human adult brains. Proc Natl Acad Sci USA 100: 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimann JM, Johansson CB, Trejo A, Blau HM. (2003b) Stable reprogrammed heterokaryons form spontaneously in Purkinje neurons after bone marrow transplant. Nat Cell Biol 5: 959–966 [DOI] [PubMed] [Google Scholar]
- Weinel RJ, Neumann K, Kisker O, Rosendahl A. (1996) Expression and potential role of E-cadherin in pancreatic carcinoma. Int J Pancreatol 19: 25–30 [DOI] [PubMed] [Google Scholar]
- Zaidi AU, Enomoto H, Milbrandt J, Roth KA. (2000) Dual fluorescent in situ hybridization and immunohistochemical detection with tyramide signal amplification. J Histochem Cytochem 48: 1369–1375 [DOI] [PubMed] [Google Scholar]