Abstract
RNA localization is a regulated component of gene expression of fundamental importance in development and differentiation. Several RNA binding proteins involved in RNA localization during development in Drosophila have been identified, of which Y14, Mago, Pumilio, and IMP-1 are known to be expressed in adult mammalian intestine. The present study was undertaken to define the developmental and regional expression of these proteins, as well as Staufen-1, in mouse intestinal cells and in other tissues and cell lines using RT-PCR, and localization using in situ hybridization and immunohistochemistry. Staufen-1, Y14, Mago-m, and Pumilio-1 were expressed in intestinal epithelial cells of both villus and crypt and in Caco-2 and IEC-6 cells. In contrast, expression of IMP-1 was age- and region-specific, showing clear expression in distal fetal and newborn intestine, but very low or no expression in adult. The mRNAs were cytosolic, with more apical than basal expression in enterocytes. Staufen protein showed a similar localization pattern to that of its cognate mRNA. Overall, the data suggest an essential role for these proteins in intestinal cells. Age and regional expression of IMP-1 may indicate a role in regulation of site-specific translation of intestinal genes or in RNA localization.
Keywords: intestine, enterocytes, RNA binding proteins, RNA localization, differentiation
RNA localization is a conserved mechanism for the establishment of cellular asymmetries during both embryogenesis and adult life, facilitating protein function at final destinations within the cell. The phenomenon of RNA localization has been observed in embryos and oocytes in Drosophila and Xenopus (Bashirullah et al. 1998; Mowry and Cote 1999), in yeast (Long et al. 1997), and in somatic cells such as fibroblasts and neurons (Lawrence and Singer 1986; Steward 1997). We have previously demonstrated that mRNAs encoding proteins with enzymatic function are localized to specific regions in the cytoplasm of absorptive enterocytes in fetal rat (Rings et al. 1992) and mature human (Li et al. 1998) intestine, and most of these mRNAs colocalize with their encoded proteins.
Localization of mRNAs by RNA binding proteins is emerging as an important feature in development. The first RNA binding protein proven to play a role in RNA localization was Staufen, responsible for both anterior and posterior patterning of Drosophila embryos (Schupbach and Wieschaus 1986). Human Staufen and mouse Staufen are members of a large family of proteins involved in the transport and/or localization of mRNAs (Wickham et al. 1999; Brizard et al. 2000). Y14 and Mago are conserved eukaryotic proteins that associate with spliced mRNAs in the nucleus and remain associated during and after nuclear export. In the cytoplasm, Y14 is involved in mRNA quality control via the nonsense-mediated mRNA decay pathway and, together with Mago, is involved in localization of oskar mRNA in Drosophila (Fribourg et al. 2003). Mouse homologs of Staufen, Y14, and Mago have been identified in a gastric epithelial progenitor cell compartment (Mills et al. 2002).
The Drosophila Pumilio gene is a founding member of a novel family of evolutionarily conserved RNA binding proteins that are present in many eukaryotic organisms, from yeast to mammals and plants (Mac-Donald 1992; Souza et al. 1999; Spassov and Jurecic 2003); it has a role in translational repression during development and differentiation (Zhang et al. 1997; Tadauchi et al. 2001).
The IMPs (IGF-II mRNA binding proteins) have been implicated in posttranscriptional processes, such as mRNA localization and turnover, and translational control (reviewed in Nielsen et al. 2001; Yaniv and Yisraeli 2002). Vertebrate IMPs are coexpressed with IGF-II mRNA and H19 RNA in developing epithelia, muscle, and placenta in both mouse and human embryos (Nielsen et al. 1999). It has been demonstrated recently in mice that IMP-1 is essential for normal embryonal and postnatal growth, in particular for adaptation to extrauterine life through control of intestinal development (Hansen et al. 2004).
The information available for the localization of RNA-binding proteins in mammals is very limited, and it remains generally unclear whether there are any developmental or regional differences in their gene expression in intestine. We sought to study the developmental and regional expression of mRNA binding proteins in mouse intestinal cells and their expression in a variety of other tissues and cells. We find evidence for the ubiquitous expression of Staufen-1, Mago-m, Y14, and Pumilio-1 genes, as well as striking age-dependent and regional expression of IMP-1. In addition, Staufen-1, Y14, Mago-m, Pumilio-1, and IMP-1 mRNAs are cytosolic, but appear to be expressed more apically than basally. Staufen protein is expressed in a pattern similar to that of its cognate mRNA.
Materials and Methods
Reagents
All restriction enzymes and DNA-dependent RNA polymerases were purchased either from New England BioLabs (Beverly, MA), Promega Corporation (Madison, WI), or Invitrogen Life Technologies (Carlsbad, CA). RNase A was purchased from Sigma Chemical Company (St. Louis, MO). All other chemicals and reagents were purchased from Sigma, Gibco-BRL (Grand Island, NY), or Fisher Scientific (Fair Lawn, NJ).
DIG RNA labeling kit (Sp6/T7), alkaline phosphatase-conjugated anti-DIG antibody Fab fragments, Escherichia coli tRNA, 4-nitroblue tetrazolium chloride, and 5-bromo-4-chloro-3-indolyl-phosphate were purchased from Roche Diagnostics Corporation (Indianapolis, IN).
Tissue Preparation
Mouse duodenum and jejunum were obtained from normal adults. For fetal (days 15–18) and newborn samples, the whole small intestine was used. Tissues were fixed in 10% buffered formalin for 24 hr. After embedding in paraffin, the tissue was sectioned at 5 μm for in situ hybridization (Histology Core; Harvard Digestive Disease Center, Department of Pathology, Children's Hospital Boston, Boston, MA), and 6 μm for immunohistochemistry (Imaging Core; Harvard Digestive Disease Center, Beth Israel Deaconess Medical Center, Boston, MA). The integrity of the tissue sections was confirmed by staining with hematoxylin and eosin and reviewed by standard light microscopy.
Intestinal Cell Extraction
The isolation of villus to crypt gradients of intact isolated intestinal epithelial cells from adult mouse small intestine was performed exactly as previously described (Weiser 1973). Briefly, isolated intestinal cell preparations were made from mouse small intestine using citrate to dissociate the epithelial cells; no proteases or other enzymes were used. The method isolates only epithelial cells and excludes serosal and interstitial cells. By a series of incubations at 37C and washing of gut loops, sequential fractions of isolated epithelial cells were obtained with a gradient of cells from villus tip to midvillus to crypt.
Cell Culture
IEC-6 and Caco-2 cells were obtained from the American Type Culture Collection (Manassas, VA) and routinely maintained in DMEM containing 10% FBS. The cells were harvested after 50% confluence, at confluence, day 11 or 60 postconfluence, and were then used for RNA isolation and subsequent gene expression studies.
Developmental and Regional Expression of Genes by RT-PCR Analysis
Mouse Staufen-1, Mago-m, Y14, Pumilio-1, IMP-1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin (as a control) mRNA expression was determined by RT-PCR analysis. For one-step RT-PCR, mouse intestinal total RNA from fetal (days 15–17), newborn, and adult was isolated by homogenization of intestinal tissues according to the manufacturer's instructions (RNeasy kit; Qiagen Inc., Valencia, CA). Total RNA from epithelial cells was also isolated following manufacturer's (Qiagen) specifications. RT-PCR was performed by using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) according to the manufacturer's protocol. Briefly, 100 ng of total RNA from different stages and regions (proximal = duodenum, middle = jejunum, and distal = ileum) of mouse intestine or other tissues and cell lines was reverse transcribed by SuperScript II H-reverse transcriptase and Platinum Taq in a reaction volume of 25 μl. The amplification reaction was carried out for 25 cycles with denaturation for 30 sec at 94C. Annealing was for 1 min at 54C for Staufen-1, Mago-m, and Y14; 51C for IMP-1, sucrase-isomaltase (SI), β-actin, and GAPDH; 57C for Pumilio-1; and extension for 1 min at 72C, followed by a final extension at 72C for 7 min. Primers for the amplification of Staufen-1, Mago-m, Y14 (Mills et al. 2002), and Pumilio-1 (Spassov and Jurecic 2003) genes were as described. Primers for IMP-1, SI, GAPDH and β-actin were designed by the vector NTI program (Infor-Max Inc.; Bethesda, MD). The sequences of the primers used for the amplification were as follows: for IMP-1, 5'-GGGGAAAGTAGAACTGCAAGG-3’ and 5'-TCCAAGATCATCTTGCACGC-3'; for SI, 5'-GGATTCCAACTATGTCGTTATGG-3’ and 5'-CTCTGGAAGCGTTAACAGCTTC-3'; for β-actin, 5'-TTGAACATGGCATTGTTACCAACTG-3' and 5'-CTGCTTGCTGATCCACATCTGCTG-3'; and for GAPDH, 5'-GTAGACAAAATGGTGAAGGTCGG-3’ and 5'-GTTGAATTTGCCGTGAGTGG-3'. The sizes of the amplicons are listed in Table 1.
Table 1.
Characteristics of RNA probe templates
| Template | Accession no. | Insert size | Location in full-length cDNA∗ (base no.) | Reference |
| Staufen-1 | NM_011490 | 151 | 753-903 | Wickham et al. 1999 |
| Mago-m | NM_010760 | 87 | 3-89 | Zhao et al. 1998 |
| Y14 | XM_129618 | 163 | 71-233 | NCBI Annotation Project 2003 |
| Pumilio-1 | AF321909 | 305 | 2996-3300 | Spassov and Jurecic 2003 |
| IMP-1 | BC051679 | 612 | 468-1079 | Strausberg et al. 2002 |
| SI | XM_143333 | 400 | 1078-1477 | NCBI Annotation Project 2002 |
| β-actin | NM_007393 | 869 | 292-1160 | Tokunaga et al. 1986 |
| GAPDH | XM_132897 | 175 | 42-216 | NCBI Annotation Project 2002 |
Start and all other coordinates are from beginning of coding sequence, where applicable.
Aliquots of the amplified products were analyzed on 1.2-2.0% agarose gel (Tris-borate-EDTA containing ethidium bromide) electrophoresis. The PCR products on the gels were scanned (wherever necessary) and analyzed using the program Quantity One (Bio-Rad Laboratories; Hercules, CA). For each analysis, a negative control prepared using all reagents except SuperScript II H— reverse transcriptase and an aliquot of the matching RNA yielded no detectable products, indicating that all RNAs were free of DNA contamination.
In cases in which a two-step reaction was preferred for the RT-PCR, first-strand cDNA synthesis was performed using ∼1.0-2.0 μg of total RNA, and either the SuperScript III first-strand synthesis system (Invitrogen) or the iScript cDNA synthesis kit (Bio-Rad) in a final reaction volume of 25 μl according to the manufacturer's specifications. After the completion of the reaction, the cDNA was further diluted four to five times with RNase-free water and ∼2–3 μl diluted cDNA was used for PCR amplification.
Subcloning, Sequence Analysis, and Preparation of Probe Templates
The RT-PCR amplified gene products were subcloned into the TOPO TA cloning system (Invitrogen), which contains Sp6 and T7 promoters. All constructs were confirmed by PCR and DNA sequencing (DNA Sequencing Core Facility; Children's Hospital Boston; Boston, MA). Database homology searches were made using the National Center for Biotechnology Information BLAST server (Altschul et al. 1997). The DNA was linearized with Spel or Xbal restriction endonucleases for the generation of template probes.
Preparation of Digoxigenin-labeled RNA Probes
Digoxigenin-labeled RNA probes were prepared using the DIG RNA Labeling kit (Sp6/T7) according to the manufacturer's instructions (Roche). Briefly, linearized templates were incubated with the buffered labeling mix containing DIG-UTP, Sp6 or T7 RNA polymerase, and RNase inhibitor for 2 hr at 37C. The transcription reactions were terminated by the addition of EDTA to a final concentration of 0.2 M. Probes greater than 612 bp (Table 1) were subjected to alkaline hydrolysis according to the protocol (Roche) to generate smaller fragments.
The sizes of the probes were verified by gel electrophoresis. The digoxigenin-labeled probes were purified using an RNeasy spin column (Qiagen). A dot-blot assay was used to estimate the digoxigenin incorporation into the probe using the standard (Roche), followed by the immunological detection assay. Aliquots in concentrations of 10 μg/ml were frozen at −80C until use.
In Situ Hybridization
In situ hybridization was carried out essentially as described (Barth et al. 1998), with some modifications. All steps were performed at room temperature unless indicated otherwise. Briefly, paraffin embedded sections (serial) were deparaffinized in xylene, rehydrated in graded treatments in ethanol, and then permeabilized with 0.2 N HCl, followed by digestion with prewarmed proteinase K (10 μg/ml) for 45 min at 37C. After digestion, sections were washed in 0.1% glycine, postfixed in 4% paraformaldehyde (pH 7.4), and washed with PBS. Sections were then incubated in 0.1 M triethanolamine (pH 8.0), acetylated with 0.25% acetic anhydride, washed in 2 × standard saline citrate (SSC), and dehydrated in graded treatments of ethanol. Hybridization was carried out overnight at 50–55C in a solution containing 50% formamide/10% dextran sulfate, 4 × SSC, 1 × Denhardt's, 1.0 mg/ml E. coli tRNA, 10 mM dithiothreitol (DTT), and 0.2 −1.0 ng/ml of digoxigenin-labeled RNA probe in a humidified chamber. Sections were then washed in 4 × SSC (four times), incubated in prewarmed RNase A (10 μg/ml) for 45 min at 37C, and rinsed twice for 5 min in RNase buffer (0.5 M NaCl; 0.01 M Tris, pH 8.0; 0.001 M EDTA) at 37C, followed by sequential washings of increasing stringencies in a buffer containing 0.01 M DTT: 2 × SSC, 15 min; 1 × SSC, 15 min; 0.5 × SSC, 15 min; and 0.1 × SSC, 30 min at 50–55C. Sections were blocked in a blocking buffer (1% BSA; 0.1% Triton-X100; 0.1 M Tris, pH 7.5; 0.15 M NaCl) for 45 min at 37C. Immunological detection of the hybridized probes was performed with alkaline phosphatase-conjugated anti-DIG antibody (1:500 dilutions) at 37C for 1 hr in a humidified chamber. After washing in TBS (0.1 M Tris, pH 7.5; 0.15 M NaCl) twice for 15 min, the sections were treated with 4.0 mM levamisole to block activity of any residual endogenous alkaline phosphatase. Slides were incubated in 0.5 ° chromogen solution containing levamisole, 4-nitroblue tetrazolium chloride, and 5-bromo-4-chloro-3-indolyl-phosphate in the dark. After stopping the reaction, when appropriate, sections were lightly counterstained with methyl green. Slides were mounted with aqua polymount (Polysciences Inc.; Warrington, PA), and visualized by light microscopy. Controls consisted of pretreatment with RNase A before hybridization to anti-sense probes, a nonspecific probe from the DIG RNA labeling kit (Roche), or treatment of sections with the sense probe. SI was used as an internal control.
Immunohistochemistry
Immunohistochemistry was performed as previously reported (Silberg et al. 2000) with slight modifications. Briefly, paraffin embedded tissues from prenatal, postnatal, and adult mice were serially sectioned. The deparaffinized and rehydrated sections on glass slides were immersed in 10 mM sodium citrate buffer at pH 6.0 and boiled in a microwave oven for 10 min, cooled, and gently washed with water for 10 min, followed by quenching with 2.25% hydrogen peroxide for 15 min. After washing in PBS (two times for 5 min), the slides were blocked with protein-blocking agent (Coulter-Immunotech; Miami, FL) and incubated for 3 hr at 37C with anti-Staufen antibody (1:1000 dilutions; a gift from J. Ortin, Centro Nacional de Biotecnologia, CSIC, Madrid, Spain). The slides were subsequently washed with PBS (two times for 5 min). The primary antibody was visualized with biotinylated secondary antibody (1:200 dilutions; 37C for 1 hr) and preformed avidin and biotin horseradish peroxidase macromolecular complex (Vectastain ABC kit; Vector Laboratories, Burlingame, CA) according to the protocol. The slides were developed with 3, 3'-diaminobenzidine tetrahydrochloride (Sigma). The tissue was lightly counterstained with methyl green, mounted with permount (Fisher), and viewed by light microscopy.
Microscopy
Images of mouse sections from the in situ hybridization experiments or immunohistochemistry were captured under brightfield illumination and digitized for computer visualization by a Nikon E800 microscope (Diagnostic Instruments Inc.; Sterling Heights, MI) equipped with SPOT RT camera and image capture software.
Statistics
Means were compared by one-way ANOVA. For statistically significant ANOVA, specific differences between or among groups were determined by the Tukey-Kramer multiple comparison analysis. All analyses were conducted using InStat software (GraphPad Software; San Diego, CA).
Results
Expression of Genes in Staged Developmental Mouse Series by RT-PCR Analysis
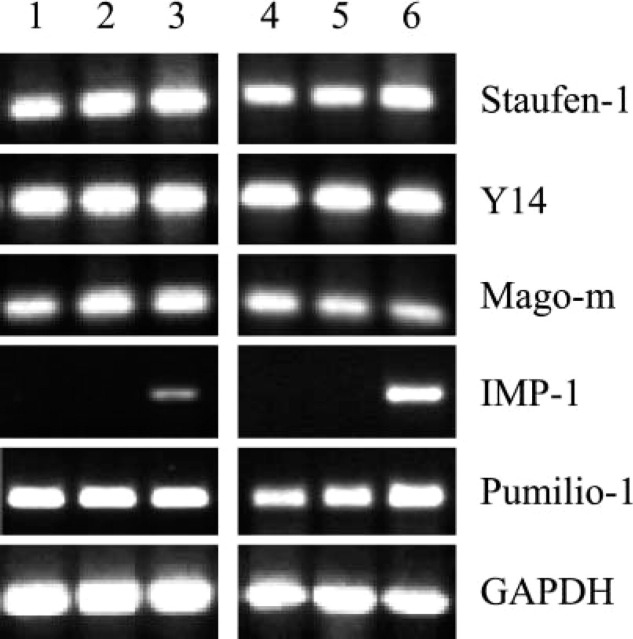
We initially assessed expression of the mRNAs for the RNA binding proteins in adult mouse small intestine using RT-PCR. Staufen-1, Y14, Mago-m, and Pumilio-1 were highly expressed along the horizontal axis from proximal to distal intestine (Figure 1). Furthermore, they were also expressed in liver, spleen, and testis. In contrast, IMP-1 mRNA expression was undetectable or very low in adult proximal and mid-intestine, but was detectable at a low level in distal intestine. In adult mouse liver and spleen, IMP-1 expression was not detected, whereas in the testis there was a high level of IMP-1 expression.
Figure 1.
Comparative RT-PCR analysis of RNA binding protein(s) expression in adult mouse tissues. Lanes 1–3: intestinal segments: proximal (Lane 1), middle (Lane 2), distal (Lane 3), liver (Lane 4), spleen (Lane 5), and testis (Lane 6). This is a representative of n = 3 (Staufen-1, Y14, Mago-m, and Pumilio-1) and n = 4 (IMP-1) identical experiments.
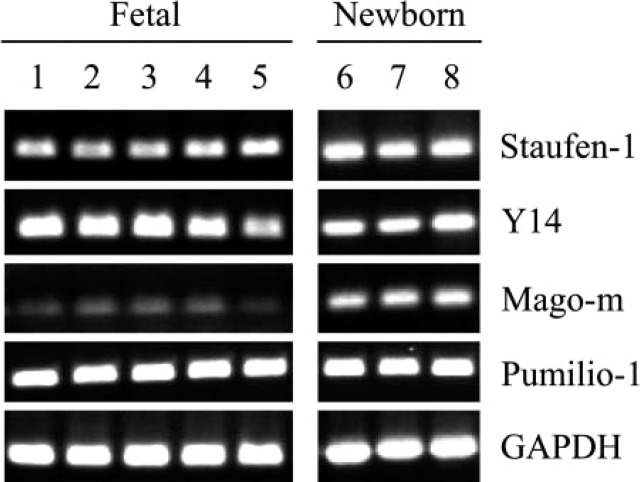
We next examined the expression of these mRNAs in developing mouse small intestine. Our results show that mRNAs for Staufen-1, Y14, Mago-m, and Pumilio-1 are expressed constitutively from fetal stages through the newborn stage (Figure 2), suggesting a role for these proteins during development. Moreover, the genes are expressed abundantly in all regions of fetal 17-day-old and newborn intestine.
Figure 2.
Developmental expression of Staufen-1, Y14, Mago-m, and Pumilio-1 in mouse small intestine using RT-PCR analysis. Fetal, 15 days (Lane 1); fetal, 16.5 days (Lane 2); fetal, 17 days (Lanes 3–5); and newborn (Lanes 6–8). Intestinal segments: proximal (Lanes 3 and 6), middle (Lanes 4 and 7), and distal (Lanes 5 and 8). This is a representative of n = 3 identical experiments.
Expression of IMP-1 in Developing and Newborn Mouse
Because IMP-1 was expressed at a minimal level in adult small intestine, we next determined its expression in developing and newborn stages using RT-PCR analysis. As shown in Figure 3, earlier fetal stages seem to express higher levels of IMP-1 than later stages of intestinal development. Moreover, there was an unequal distribution of IMP-1 expression along the horizontal axis of the intestine.
Figure 3.
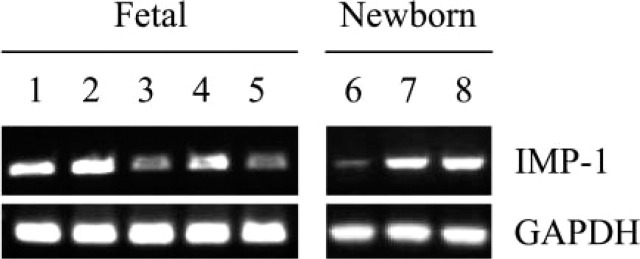
Expression of IMP-1 in developing and newborn mouse intestine using RT-PCR analysis. Fetal, 15 days (Lane 1); fetal, 16.5 days (Lane 2); fetal, 17 days (Lanes 3–5); and newborn (Lanes 6–8). Intestinal segments: proximal (Lanes 3 and 6), middle (Lanes 4 and 7), and distal (Lanes 5 and 8). This is a representative of identical experiments (fetal 15 days, n = 2; fetal 16.5 days, n = 6; fetal 17 days, n = 3; newborn, n = 8 for proximal and distal, n = 7 for middle).
The relative abundance of the IMP-1 mRNA in developing and adult mouse intestine was assessed by densitometric scanning of the signals obtained by RT-PCR (Figure 4) and compared with the signal intensities obtained for GAPDH mRNA. The abundance of IMP-1 mRNA showed dramatic differences in the different regions of fetal and adult intestine. Proximally, it was expressed at lower levels than distally; levels were, however, consistently higher in early stages compared with adult (p<0.05). Taken together, data from Figure 1, Figure 3, and Figure 4 suggest that IMP-1 mRNA expression is temporally and spatially regulated.
Figure 4.
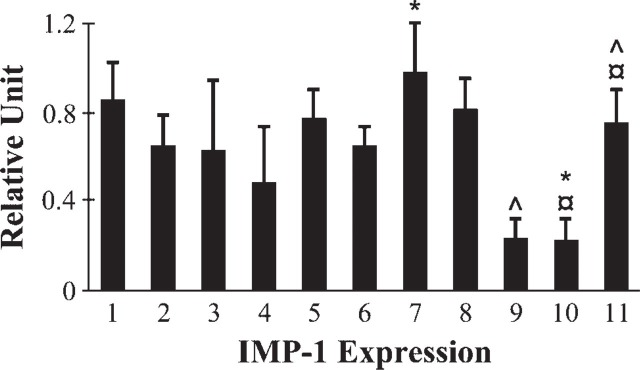
Quantification of IMP-1 mRNA expression. The level of IMP-1 expression was calculated after normalization to the glycer-aldehyde-3-phosphate dehydrogenase (GAPDH) level in each sample and presented as relative units. Fetal, 15 days (Lane 1); fetal, 16.5 days (Lane 2); fetal, 17 days (Lanes 3-5); newborn (Lanes 6-8); and adult (Lanes 9-11). Intestinal segments: proximal (Lanes 3, 6, and 9), middle (Lanes 4, 7, and 10), and distal (Lanes 5, 8, and 11). Data are means ± SE; (fetal 15 days, n = 2; fetal 16.5 days, n = 6; fetal 17 days, n = 3; newborn, n = 8 for proximal and distal, n = 7 for middle; adult proximal and distal, n = 7, middle n = 6). ^, ×, ∗ p<0.05 compared within or between the groups.
Histology
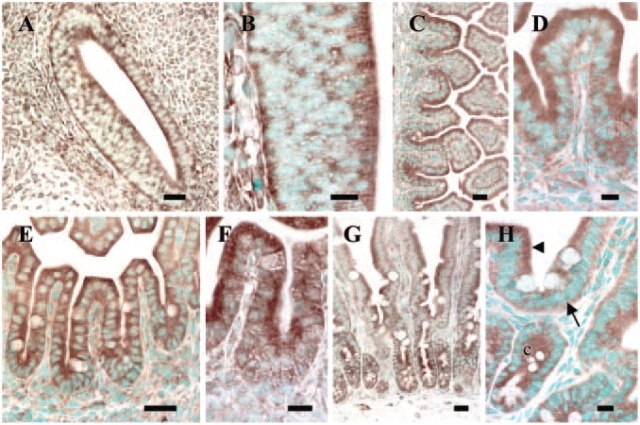
All fetal, newborn, and adult duodenal and jejunal sections were examined and demonstrated normal morphology and cellular architecture (Figures 5A-5D1).
Figure 5.
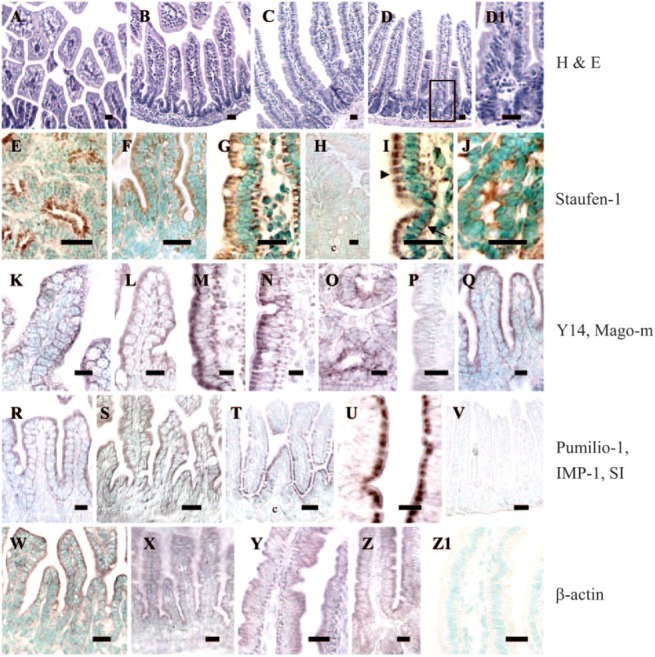
Intracellular localization of mRNAs for specific RNA-binding proteins in absorptive enterocytes. (A-D1) hematoxylin and eosin staining of sections demonstrating the integrity of the crypt-villus architecture. (D1) Magnified view of (D). (E-Z1) In situ hybridization assays of small intestine using digoxigenin-labeled probes. Antisense probes for Staufen-1 (E-J; except H), Y14 (K-O), Mago-m (Q), Pumilio-1 (R), IMP-1 (S), and β-actin (W-Z) mRNAs revealed that these transcripts are cytosolic, but appear to be expressed more apically (arrowhead) than basally (arrow). In adult tissues, crypt cells also showed expression of these transcripts (J, O). In contrast, SI mRNA was localized apically (T, U). There was no expression of SI in the basal region of enterocytes, or in the crypt cells (c). No consistent signal was detected when we used sense probes (P, sense for Y14; V, sense for SI), when sections pretreated with RNase A followed by antisense probes (H, for Staufen-1), or when nonspecific probes (Z1) were used. The brown or reddish brown color is the positive signal for the message. Where appropriate, nuclei are counterstained with methyl green. (Fetal 17 days: A, E, K, W; newborn 1 day: B, F, L, Q-S, X; adult duodenum: C, G, H, M, Y, Z1; adult jejunum: D, D1, I, J, N-P, T-V, Z). Bars: T, V = 50 μm; A-G, I, J, P, S, U, W-Z1 = 20 μm; H, K-O, Q, R = 10 μm. Shown here are sections representative of identical experiments (Staufen-1, n = 10; Y14, n = 5; Mago-m, n = 5; Pumilio-1, n = 3; IMP-1, n = 2; SI, n = 8; β-actin, n = 5).
Intracellular Localization of mRNAs Encoding Specific RNA-binding Proteins
The intracellular distribution of the mRNAs specific for RNA-binding proteins in absorptive enterocytes was studied using digoxigenin-labeled RNA probes. Previous studies from our laboratory have shown apical localization for human SI mRNA using a digoxigenin-labeled RNA probe for in situ hybridization (Barth et al. 1998; Li et al. 2000). In the present study, we have used a mouse SI probe as an internal control for studying mRNA expression patterns for specific RNA-binding proteins. Figures 5E-5Z1 show in situ hybridization results for fetal, newborn, or adult mouse small intestine. mRNA expression for Staufen-1 (Figures 5E-5J, except Figure 5H), Y14 (Figures 5K-5O), Mago-m (Figure 5Q), Pumilio-1 (Figure 5R), and IMP-1 (Figure 5S) was predominantly in the epithelial cells of the developing and adult villus. All mRNAs were detected both apically (arrowhead) and basally (arrow) relative to the nucleus (Figure 5I); in some sections, the mRNAs seemed to be localized more apically. The mRNA localization patterns showed no differences between developmental and adult stages. In the adult tissues, crypt cells also showed expression of these messages (Figures 5J and 5O, respectively). Because the expression patterns of these mRNAs under in situ hybridization conditions were so similar during development and in the adult, we have presented the data on localization only in the newborn stage for Mago-m, Pumilio-1, and IMP-1 mRNA. Our results in newborn for IMP-1 mRNA (Figure 5S) were also similar to those seen by Hansen et al (2004) in 17.5-day-old fetal intestine.
In control experiments, SI and β-actin mRNAs were imaged. As expected, SI mRNA was localized apically in villus absorptive cells with little or no detectable signal in crypt cells or any other region of the cells (Figures 5T and 5U, respectively), and β-actin mRNA was found both apically and basally (Figures 5W-5Z). There was no consistent expression of the messages for any of the RNA-binding proteins in the goblet cells. No significant reaction product was detected in tissue sections hybridized with nonspecific probes (Figure 5Z1), with antisense probes pretreated with RNase A (Figure 5H; for Staufen-1 probe), or with sense probes (Figures 5P and 5V, respectively; Figure 5P, sense probe for Y14; and Figure 5V, sense probe for SI). Data for other control probes are not shown.
Expression of mRNAs for RNA-binding Proteins in Cell Lines
Figure 6 shows the RT-PCR analysis of the expression pattern of the mRNAs in two intestinal epithelial cell lines, IEC-6 and Caco-2. Staufen-1, Y14, Mago-m, and Pumilio-1 mRNAs were expressed in both cell lines, and at each stage of confluence and postconfluence. However, IMP-1 mRNA was expressed in Caco-2 cells, but the expression was almost undetectable in IEC-6 cells, although a faint signal was consistently present.
Figure 6.
Expression of mRNAs specific for RNA-binding proteins in epithelial cell lines using RT-PCR analysis. IEC-6 cells (Lanes 1–3); Caco-2 cells (Lanes 4–6). 50% confluent (Lanes 1, 4); confluent (Lanes 2, 5); 60 days after confluence (Lane 3); 11 days after confluence (Lane 6). This is representative of n = 3 identical experiments.
Expression of Staufen-1, Y14, Mago-m, Pumilio-1, IMP-1, and SI in Enterocytes
To confirm the expression of these RNA-binding proteins in the villus and crypt compartments, isolated villus and crypt cells from adult mouse small intestine were separated and analyzed by RT-PCR. The data shown in Figure 7 confirm that Staufen-1, Y14, Mago-m, and Pumilio-1 mRNAs are expressed in villus and in crypt cells. IMP-1 had a very minimal or no expression in the adult mouse intestine (Figure 1), and we could not see any detectable level of IMP-1 in the isolated villus and crypt enterocytes (not shown). As expected, SI mRNA was undetectable in the crypt cells.
Figure 7.
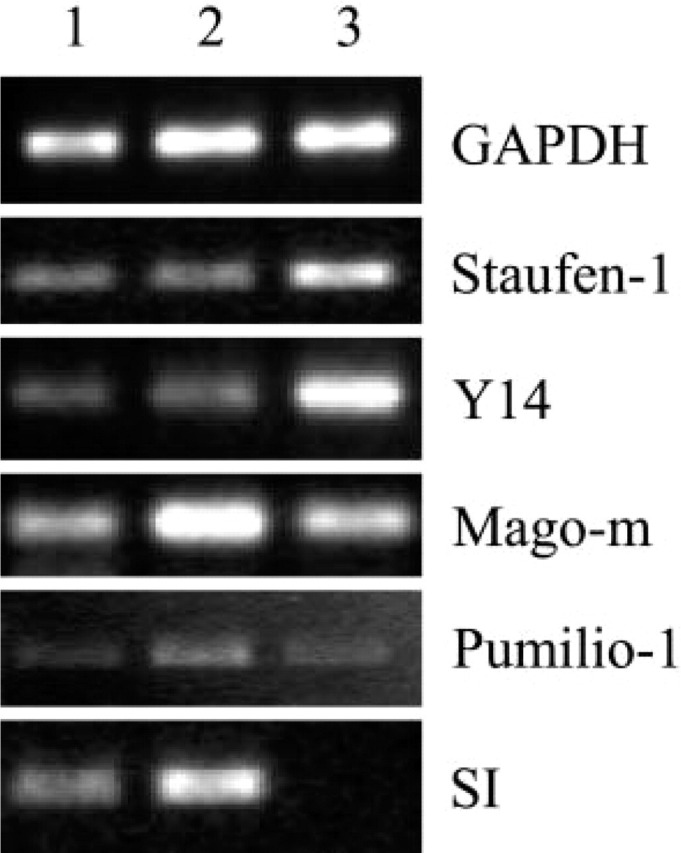
Expression of Staufen-1, Y14, Mago-m, Pumilio-1, and SI in villus and crypt compartments of mouse small intestine using RT-PCR analysis. Upper villus (Lane 1); mid-villus (Lane 2); crypt (Lane 3). This is representative of n = 3 identical experiments. Note that SI mRNA is expressed only in the villus cells (Lanes 1, 2) and not in crypt cells, consistent with previously published observations (Barth et al. 1998).
Staufen Protein Expression in Developing and Adult Mouse Intestine
Histological distribution of Staufen protein was detected by immunohistochemistry in the developing and adult mouse small intestine. At all stages of intestinal development, distinct immunostaining was observed (Figure 8). The staining was seen in the cytoplasm of epithelial cells, very similar to the mRNA expression pattern. Adult crypt cells also expressed Staufen (Figures 8G and 8H, respectively). As seen in the figure, there was no consistent expression of Staufen protein in the Paneth cells, goblet cells, or lamina propria. No immunolabeling was detected when primary antibody was omitted in each experiment (not shown).
Figure 8.
Localization of Staufen protein during developmental and adult stages using immunohistochemical staining. Fetal 15 days (A, B), fetal 18 days (C, D), newborn (E, F), and adult jejunum (G, H). The brown color is positive signal for the protein. Nuclei are counterstained with methyl green. Bars: A, C, E, G = 20 μm; B, D, F, H = 10 μm. Staufen protein staining was detected in the apical region (arrowhead) of mouse enterocytes, but was also visualized basally (arrow). The protein was also expressed in crypt cells (c) in adult jejunum (H). No consistent signals were detected in the goblet cells, Paneth cells, or in lamina propria in any section. This is representative of n = 3 identical experiments.
Discussion
In this study, we demonstrate that the mRNAs for the RNA binding proteins, Staufen-1, Y14, Mago-m, Pumilio-1, and IMP-1 are expressed in both proliferating and differentiated intestinal epithelial cells of fetal, newborn, and adult mice. These mRNAs are cytosolic, but appear to be expressed more apically than basally. Staufen protein is expressed in a pattern similar to that of its cognate mRNA. IMP-1 mRNA is also developmentally regulated, showing higher expression in fetal than in mature intestine, and exhibiting a striking increase in expression in distal compared with proximal intestine.
Using microarray technology, Mills et al. (2002) have reported expression of Staufen, Y14, and Mago-m in gastric progenitor cells, but only Y14 and Mago-m showed enhanced expression in crypt cells, and Staufen-1 could not be detected in intestinal cells isolated from putative progenitor compartments in adult mice. In contrast, our RT-PCR analysis demonstrates that Staufen-1, Y14, and Mago-m mRNAs are ubiquitously expressed and share a similar expression profile that remains primarily unchanged from fetal through postnatal development. Staufen mRNA is also present in multiple adult human tissues (Wickham et al. 1999), and we identified it in liver, spleen, and testis.
Based on conserved function in invertebrates and lower vertebrates, it was recently suggested that Pumilio proteins support proliferation and/or self-renewal of stem cells (Wickens et al. 2002). This raises the possibility that mammalian orthologs of Pumilio may control asymmetric division and cell fate specification in a variety of mammalian lineages, including stem cell maintenance and self-renewal in the intestine. This would predict higher levels of Pumilio expression in crypt cells. However, in the present study, the RT-PCR data demonstrated expression of Pumilio-1 mRNA in both crypt and villus compartments. Mouse Pumilio-1 is expressed constitutively in many tissues (Spassov and Jurecic 2003), and our RT-PCR data for Pumilio-1 are consistent with those results. No previous studies have examined Pumilio-1 during intestinal development. We have demonstrated constitutive expression in fetal, newborn, and adult enterocytes.
Mouse IMPs are expressed in a biphasic fashion, with elevated expression during the early stages of embryogenesis (Nielsen et al. 2001; Yaniv and Yisraeli 2002) and around embryonic day 12.5 followed by a decline toward birth (Nielsen et al. 1999). Our data show that IMP-1 mRNAs exhibit similar patterns with high expression in fetal intestine and a decline toward birth. In the present study, IMP-1 mRNA could not be detected in adult mouse tissues such as liver and spleen. Similar observations for liver (Leeds et al. 1997; Nielsen et al. 1999; Ioannidis et al. 2001) and spleen (Nielsen et al. 1999) have been reported.
A striking new finding in this study is the distinct pattern of expression of IMP-1 mRNA—low in the proximal and high in the distal small intestine. This pattern suggests that differential expression of IMP-1 genes may be involved in the proximal to distal progression of intestinal development. Thus the age and regional expression of IMP-1 mRNA may indicate a role in the regulation of site-specific translation of intestinal genes, such as IGF-II, or in RNA localization.
The patterns of expression of the mRNAs studied in native intestine agree with our data from IEC-6 cells, an undifferentiated intestinal cell line, and Caco-2 cells, a human colon carcinoma cell line that has characteristics of differentiated small intestinal cells. Both cell lines showed comparable expression of the mRNAs examined. The ubiquitous presence of most of these transcripts in villus and crypt cells, and in cell lines, suggests that their translated proteins are likely required for constitutive cellular function. However, the exact role of these proteins in intestinal cells remains to be defined.
Taken together, the evidence presented here is the first comparative report describing differential expression of mRNAs for RNA-binding proteins in mouse small intestine. More precisely, our data indicate widespread expression of Staufen-1, Y14, Mago-m, and Pumilio-1 mRNAs. IMP-1 gene expression is strictly controlled during fetal development and adult life in mouse intestine, and may be important in regulation of proximal/distal development of the gastrointestinal tract.
Acknowledgments
Supported by NIH Research Grant R37 DK-32658 (MERIT) and Harvard Digestive Disease Center Grant P30 DK-34854.
We would like to thank Dr. Juan Ortin (Centro Nacional de Biotecnologia, CSIC; Madrid, Spain) for the Staufen antibody. We also thank Dr. Stephen D. Krasinski (Division of Gastroenterology and Nutrition, Department of Medicine, Children's Hospital Boston; Boston, MA) for the statistical analysis. We are grateful to Dr. Susan J. Hagen, Jessica S. Wagner, and Daniel A. Brown (Imaging Core, Harvard Digestive Disease Center, Beth Israel Deaconess Medical Center and Children's Hospital Boston) for microscopy support. Finally, we thank Lena T. Liu (Histology Core, Harvard Digestive Disease Center, Department of Pathology, Children's Hospital Boston) for help in preparation of tissue sections.
Literature Cited
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth JA, Li W, Krasinski SD, Montgomery RK, Verhave M, Grand RJ. (1998) Asymmetrical localization of mRNAs in enterocytes of human jejunum. J Histochem Cytochem 46: 335–343 [DOI] [PubMed] [Google Scholar]
- Bashirullah A, Cooperstock RL, Lipshitz HD. (1998) RNA localization in development. Ann Rev Biochem 67: 335–394 [DOI] [PubMed] [Google Scholar]
- Brizard F, Luo M, Desgroseillers L. (2000) Genomic organization of the human and mouse stau genes. DNA Cell Biol 19: 331–339 [DOI] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E. (2003) A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat Struct Biol 10: 433–439 [DOI] [PubMed] [Google Scholar]
- Hansen TVO, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, et al. (2004) Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 24: 4448–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis P, Trangas T, Dimitriadis E, Samiotaki M, Kyriazoglou I, Tsiapalis CM, Kittas C, et al. (2001) C-MYC and IGF-II mRNA-binding protein (CRD-BP/IMP-1) in benign and malignant mesenchymal tumors. Int J Cancer 94: 480–484 [DOI] [PubMed] [Google Scholar]
- Lawrence JB, Singer RH. (1986) Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45: 407–415 [DOI] [PubMed] [Google Scholar]
- Leeds P, Kren BT, Boylan JM, Betz NA, Steer CJ, Gruppuso PA, Ross J. (1997) Developmental regulation of CRD-BP, an RNA-binding protein that stabilizes c-myc mRNA in vitro. Oncogene 14: 1279–1286 [DOI] [PubMed] [Google Scholar]
- Li W, Krasinski SD, Verhave M, Montgomery RK, Grand RJ. (1998) Three distinct messenger RNA distribution patterns in human jejunal enterocytes. Gastroenterology 115: 86–92 [DOI] [PubMed] [Google Scholar]
- Li W, Wang J, Coluccio LM, Matsudaira P, Grand RJ. (2000) Brush border myosin I (BBMI): a basally localized transcript in human jejunal enterocytes. J Histochem Cytochem 48: 89–94 [DOI] [PubMed] [Google Scholar]
- Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. (1997) Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 277: 383–387 [DOI] [PubMed] [Google Scholar]
- MacDonald PM. (1992) The Drosophila Pumilio gene: an unusually long transcription unit and an unusual protein. Development 114: 221–232 [DOI] [PubMed] [Google Scholar]
- Mills JC, Andersson N, Hong CV, Stappenbeck TS, Gordon JI. (2002) Molecular characterization of mouse gastric epithelial progenitor cells. Proc Natl Acad Sci USA 99: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Cote CA. (1999) RNA sorting in Xenopus oocytes and embryos. FASEB J 13: 435–445 [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Nielsen J, Christiansen J. (2001) A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl 234: 93–99 [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. (1999) A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rings EH, Bueller HA, de Boer PA, Grand RJ, Montgomery RK, Lamers WH, Charles R, et al. (1992) Messenger RNA sorting in enterocytes: co-localization with encoded proteins. FEBS Lett 300: 183–187 [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E. (1986) Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol 113: 443–448 [DOI] [PubMed] [Google Scholar]
- Silberg DG, Swain GP, Suh ER, Traber PG. (2000) Cdx1 and Cdx2 expression during intestinal development. Gastroenterology 119: 961–971 [DOI] [PubMed] [Google Scholar]
- Souza GM, da Silva AM, Kuspa A. (1999) Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development 126: 3263–3274 [DOI] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. (2003) Mouse Pum1 and Pum2 genes, members of the Pumilio family of RNA-binding proteins, show differential expression in fetal and adult hematopoietic stem cells and progenitors. Blood Cells Mol Diseases 30: 55–69 [DOI] [PubMed] [Google Scholar]
- Steward O. (1997) mRNA localization in neurons: a multipurpose mechanism? Neuron 18: 9–12 [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, et al. (2002) Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA 99: 16899–16903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadauchi T, Matsumoto K, Herskowitz I, Irie K. (2001) Post-transcriptional regulation through the HO 3'-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J 20: 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga K, Taniguchi H, Yoda K, Shimizu M, Sakiyama S. (1986) Nucleotide sequence of a full-length cDNA for mouse cytoskeletal beta-actin mRNA. Nucleic Acids Res 14: 2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser MM. (1973) Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J Biol Chem 248: 2536–2541 [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. (2002) A PUF family portrait: 3'-UTR regulation as a way of life. Trends Genet 18: 150–157 [DOI] [PubMed] [Google Scholar]
- Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. (1999) Mammalian Staufen is a double-stranded RNA and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 19: 2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K, Yisraeli J. (2002) The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 287: 49–54 [DOI] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. (1997) A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390: 477–484 [DOI] [PubMed] [Google Scholar]
- Zhao XF, Colaizzo-Anas T, Nowak NJ, Shows TB, Elliott RW, Aplan PD. (1998) The mammalian homologue of Mago nashi encodes a serum-inducible protein. Genomics 47: 319–322 [DOI] [PubMed] [Google Scholar]