Abstract
Fungi are exposed to broadly fluctuating environmental conditions, to which adaptation is crucial for their survival. An ability to respond to a wide pH range, in particular, allows them to cope with rapid changes in their extracellular settings. PacC/Rim signaling elicits the primary pH response in both model and pathogenic fungi and has been studied in multiple fungal species. In the predominant human pathogenic fungi, namely, Candida albicans, Aspergillus fumigatus, and Cryptococcus neoformans, this pathway is required for many functions associated with pathogenesis and virulence. Aspects of this pathway are fungus specific and do not exist in mammalian cells. In this review, we highlight recent advances in our understanding of PacC/Rim-mediated functions and discuss the growing interest in this cascade and its factors as potential drug targets for antifungal strategies. We focus on both conserved and distinctive features in model and pathogenic fungi, highlighting the specificities of PacC/Rim signaling in C. albicans, A. fumigatus, and C. neoformans. We consider the role of this pathway in fungal virulence, including modulation of the host immune response. Finally, as now recognized for other signaling cascades, we highlight the role of pH in adaptation to antifungal drug pressure. By acting on the PacC/Rim pathway, it may therefore be possible (i) to ensure fungal specificity and to limit the side effects of drugs, (ii) to ensure broad-spectrum efficacy, (iii) to attenuate fungal virulence, (iv) to obtain additive or synergistic effects with existing antifungal drugs through tolerance inhibition, and (v) to slow the emergence of resistant mutants.
INTRODUCTION
Opportunistic fungal infections have emerged as a major cause of morbidity and mortality in immunocompromised patients, including those with AIDS, hematological malignancies, and stem cell and organ transplant recipients. Collectively, invasive candidiasis, cryptococcal meningitis, invasive aspergillosis, and pneumocystis pneumonia are estimated to cause at least as many deaths worldwide as tuberculosis or malaria (1, 2). However the treatment of these life-threatening infections caused by eukaryotic pathogens still lags far behind that of infections due to other microorganisms, such as bacteria. Only four classes of drugs are currently available for treating invasive fungal infections: polyenes, azoles, pyrimidines, and echinocandins (1). This limited arsenal of effective drugs highlights the urgent need for the development of novel therapeutic agents, particularly in light of the emergence of drug-resistant strains, the growing number of immunocompromised patients to be treated, and the toxicity, high cost, and narrow activity spectra of the drugs currently available (1, 3, 4). Two nonexclusive approaches may be considered in the field of antifungal drug discovery: finding new inhibitors with a direct effect on specific targets in fungal cells or the improvement of existing drugs. Stress signaling in fungi is increasingly being recognized as important in adaptation to drug pressure, and its components are therefore being identified as potential targets (5, 6). The inhibition of some fungal cascades may be effective for both direct fungal and indirect synergistic approaches.
Ambient pH is one of the extracellular stresses to which these microorganisms must adapt rapidly. The major human pathogenic fungi, Candida albicans, Aspergillus fumigatus, and Cryptococcus neoformans, can survive in several anatomically distinct sites and can, therefore, provoke deep-seated infections in susceptible patients. In these settings, adaptation to pH is crucial and enables human pathogens to invade the bloodstream and tissues and to cause disseminated infections (7). Ambient pH is thought to be stable in the human body, and somewhat alkaline (around 7.4), but extreme variations occur. For example, spatial changes are observed along the digestive tract, together with temporal changes in the vaginal cavity with changes in hormone pressure, and the skin is acidic. PacC/Rim signaling is the best-studied pH response pathway. It has been extensively described in model organisms such as Aspergillus nidulans, Saccharomyces cerevisiae, and Yarrowia lipolytica (8–17). In pathogenic fungi, this pathway elicits the primary pH response and has been shown to be required not only for survival and growth in the host but also for invasive progression in tissues and for virulence (7, 18).
In this review, we focus on the way in which the PacC/Rim pathway is processed in the main fungal species, highlighting both conserved and divergent mechanisms. The role of this pathway in virulence, pathogenesis, and modulation of the host immune response is also discussed, together with its involvement in adaptation to drug pressure, all of which make this fungus-specific system a promising target for innovative antifungal strategies.
pH SIGNAL PROCESSING IN FUNGI
Fungi can grow at a wide range of pH values. The signaling pathways mediating responses to pH therefore play a key role in the cell biology of these organisms. Neutral-alkaline sensing relies mainly on the pathway called PacC in filamentous fungi and Rim101 in yeasts (7, 13–15, 18–21). This pathway is functional in deuteromycetes, ascomycetes, and basidiomycetes and is well conserved, although marked differences have been noted between these three groups of fungi (20, 22, 23). The regulation of pH has been studied most extensively in the nonpathogenic species A. nidulans, but studies of the yeasts Saccharomyces cerevisiae and Yarrowia lipolytica have also contributed to our current understanding. Thus, after a brief description of pH signal transduction and its multiple sequential steps in model organisms, we will focus on the situation in pathogenic fungi. We will concentrate on recent advances and refer to reviews for a comprehensive analysis of previous data (7, 13, 14, 24).
PacC PROCESSING IN ASPERGILLUS NIDULANS
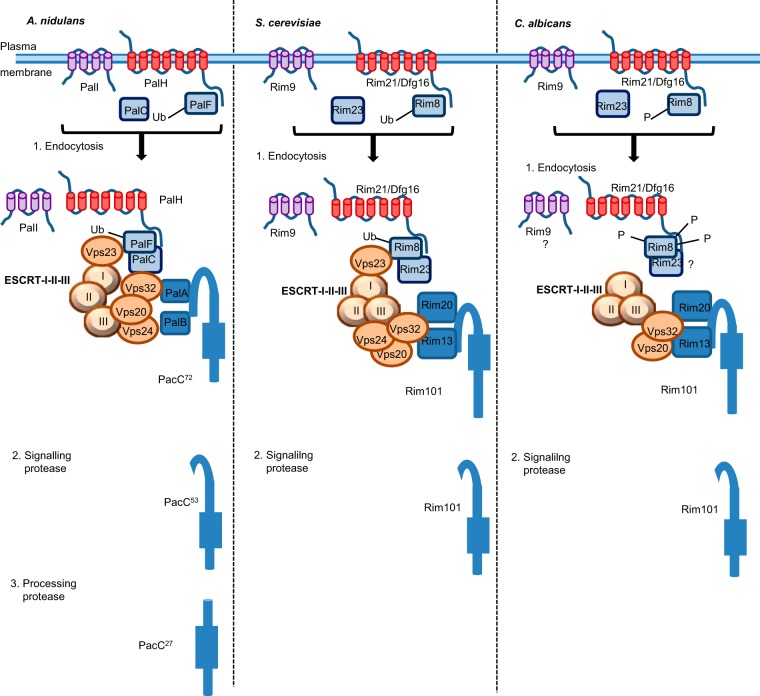
There are seven dedicated proteins involved in the PacC signaling pathway: PacC, the transcription factor, PalH, PalI, PalF, PalC, PalA, and PalB, which relay changes in environmental pH to PacC (Fig. 1). External pH sensing involves a cortical membrane complex of three proteins: the putative pH sensor PalH and its assistant PalI, together with an arrestin-like protein PalF. PalF binds to the C terminus of PalH and becomes phosphorylated and ubiquitinated at neutral to alkaline pH in a PalH-dependent manner (25–28). The N-terminal arrestin domain of PalF has been shown to be crucial for the PalF-PalH interaction and further activation of this pathway. This domain is also highly conserved in pathogenic fungi, such as A. fumigatus and yeasts (25, 28). Ubiquitinated PalF then binds the Bro1 domains of PalC and Vps23, a component of endosomal-sorting complex I required for transport (ESCRT-I) (27, 29, 30). This triggers the sequential recruitment, to the cortical region located at the plasma membrane, of additional Vps proteins that are normally part of the ESCRT complexes of the multivesicular body of the endosome, together with the two remaining Pal proteins (PalA and PalB). The Vps32/Vps20 subcomplex, a key component of ESCRT-III, interacts with PalC and PalA, whereas Vps24 recruits and possibly activates PalB, the signaling protease (29, 31, 32). In response to neutral to alkaline conditions, PalB mediates the cytoplasmic proteolysis of the full-length 72-kDa PacC72 precursor, generating the N-terminal 53-kDa fragment PacC53 (33, 34). PacC53 is the substrate of a second, possibly proteasomal, proteolytic event yielding the 27-kDa final product, PacC27. PacC27 is then translocated to the nucleus, where it turns on genes responsive to alkaline conditions and switches off genes responsive to acidic conditions (35). In summary, the current model suggests that the PacC pathway is triggered at alkaline pH by cortical structures at the plasma membrane that recruit all Pal proteins and components of the ESCRT endosomal machinery for the activation of PacC by proteolysis (Fig. 1 and Table 1) (30).
FIG 1.
Pal/Rim signaling cascades in Aspergillus nidulans, Saccharomyces cerevisiae, and Candida albicans. External pH sensing involves the plasma membrane complex, comprising PalH/Rim21-Dfg16 and its assistant, PalI/Rim9, an arrestin-like protein PalF/Rim8, and PalC/Rim23. At alkaline-neutral pH, the plasma membrane complex is endocytosed through PalF/Rim8 ubiquitination, which triggers interactions with the ESCRT complexes involved in the multivesicular body endocytic pathway. Vps32, a key component of ESCRT-III, interacts with PalC/Rim23 and PalA/Rim20. PalA/Rim20 in conjunction with PalB/Rim13 (the signaling protease) are members of the endosomal signaling complex and recruit PacC/Rim101. PalB/Rim13 mediates the cytoplasmic proteolysis of the full-length PacC/Rim101. In A. nidulans, a second processing event occurs independently of PalB. In all species, whether PalI/Rim9 is endocytosed together with PalC/Rim23, PalH/Rim21, and PalF/Rim8 has not been demonstrated. In C. albicans, the role of Vps23 has not been described. Also in C. albicans, Rim8 is not ubiquitinated but hyperphosphorylated and it associates with Rim21 and Rim101, suggesting a role in bridging the cortical and the endosomal signaling complexes.
TABLE 1.
pH signal processing in fungal model organisms and in the main human fungal pathogens
| Processing step | Fungal species |
||||
|---|---|---|---|---|---|
| Aspergillus nidulans | Saccharomyces cerevisiae | Candida albicans | Aspergillus fumigatus | Cryptococcus neoformans | |
| Sensing | |||||
| Cell location | Plasma membrane | Plasma membrane | Plasma membrane | Plasma membrane | Not described |
| Proteins (orthologues) | PalH with 7 TMDs,a PalI with 4 TMDs, PalF arrestin like | Dfg16/Rim21(PalH), Rim9 (PalI), Rim8 (PalF) | Dfg16/Rim21, Rim9, Rim8 | PalH, PalF | Not described |
| Trigger | PalH-PalF interaction, PalF phosphorylation and ubiquitination (neutral-alkaline pH dependent) | Rim21-Rim8 interaction, Rim8 phosphorylation and ubiquitination (pH independent) | Rim21-Rim8 interaction, Rim8 hyperphosphorylation dependent on Rim101 activation | Not describedb | Not described |
| Signaling | |||||
| Cell location | Plasma membrane, endosome | Plasma membrane, endosome | Plasma membrane, endosome | Not describedb | Not described |
| Proteins (orthologues) | PalH, PalF, PalC, Vps23 (ESCRT-Ic), Vps32/Vps20, Vps24 (ESCRT-IIIc) | Rim21, Rim8, Rim23 (PalC), Vps23, Vps32/Vps20, Vps24 | Rim21, Rim8, Rim23, Vps32/Vps20 | Not describedb | Not described |
| Trigger | Binding of ubiquitinated PalF-PalH to PalC and Vps23, PalF-dependent endocytosis of PalH through Vps23 recruitment of Vps32/Vps20 | Binding of ubiquitinated Rim8-Rim21 to Rim23 and Vps23, Rim8-dependent endocytosis of Rim21 through Vps23 recruitment of Vps32/Vps20/Vps24 | Interaction of hyperphosphorylated Rim8 with Rim21and Rim101, Rim8-dependent endocytosis of Rim21 through Vps32/Vps20 binding | Not described | Not described |
| Processing of PacC/Rim101 | |||||
| Cell location | First proteolysis in endosome, second proteolysis in proteasome | Single proteolysis in endosome | Single proteolysis in endosome | Not describedb | Not described |
| Proteins | Interaction of Vps32/Vps20 with PalA, and Vps24 with PalB. First PalB-mediated proteolysis of PacC72 to generate PacC53. Second proteolysis mediated by the proteasome | Interaction of Vps32/Vps20/Vps24 with Rim20, Rim13, and Rim101, Rim13-mediated proteolysis of Rim101 | Interaction of Vps32 with Rim20, Rim13, and Rim101, Rim13-mediated proteolysis of Rim101 to generate Rim10165 | Not describedb | Not described, but two proteolysis steps dependent on Rim13, Rim20, and cAMP/PKA pathway |
| Trigger | PacC27 translocation to the nucleus | Translocation to nucleus of Rim101 truncated form corresponding to PacC53 | Translocation to the nucleus of Rim10165 | Not described | Not described |
| Activation | |||||
| Cell location | Nucleus | Nucleus | Nucleus | Not described | Not described |
| Proteins | Binding of PacC27 to pH responsive genes | Binding of truncated Rim101to pH-responsive genes | Binding of truncated Rim101to pH-responsive genes | Not described | Not described |
| Regulation | |||||
| Proteins | PacC | Not described | Rim8, Rim101 | Not described | Not described |
| Trigger | Autogenous PacC mediated activation | Not described | Rim101-mediated repression and degradation of Rim8 and autogenous Rim101-mediated activation | Not described | Not described |
TMD, transmembrane domain.
Thought to be highly similar to that in A. nidulans.
Endosomal sorting complex required for transport.
In Saccharomyces cerevisiae and Yarrowia lipolytica, the PacC/Rim101 pH transduction pathway is called Rim, because RIM101 was first identified as a regulator of IME2, a gene involved in meiosis initiation in S. cerevisiae. This pathway was subsequently studied mostly for its role in the pH response mediated by Rim101 (formerly Rim1, the PacC orthologue in yeasts) (12, 36–38). All components of the A. nidulans PacC pathway have homologues in yeasts. However, the selection of yeast mutants with impaired pH responses and the identification of proteins interacting with known Rim proteins led to the identification of additional players, mostly components of the endocytic pathway that were later shown to have similar, but not identical, functions in A. nidulans. Interestingly, all single vps mutants are viable in S. cerevisiae, but vps32 mutants are lethal in Y. lipolytica, and all vps mutants are lethal in A. nidulans (8–10, 16, 17, 29, 32, 39–42).
One of the major differences between PacC and Rim101 pathways is that only one proteolytic event, corresponding to the step generating PacC53 in A. nidulans, has been demonstrated in S. cerevisiae and is thought to occur in other hemiascomycetous yeasts at alkaline pH. The processed form of Rim101 is thus analogous to the PacC53 form (12, 16, 36) (Fig. 1).
Another difference concerns the PalH pH sensor, which exists in S. cerevisiae as two paralogues, Dfg16 and Rim21, thought to form a heterodimeric receptor (Table 1). Both have been shown to be required for signal transduction (43, 44). Recent results demonstrated that the PalI homologue Rim9 forms a complex with Dfg16p and Rim21p that is localized at the plasma membrane and that these three factors are mutually dependent, with Rim21 the most important and Rim9 and Dfg16 playing secondary roles by maintaining Rim21 levels and guiding the membrane localization of this protein (45).
It was long thought that yeasts had no PALC homologue. YGR122w (called RIM23) has recently been identified as a putative PALC homologue in S. cerevisiae, suggesting that the entire pathway may be conserved in hemiascomycetous yeasts (44). Like PalC in A. nidulans, Rim23 has been shown to be required for Rim101 activation in S. cerevisiae and Y. lipolytica (Fig. 1 and Table 1) (8, 10, 29). In Y. lipolytica, Rim23p also assembles the early ESCRT components Vps27 and Vps23, and vps23 mutants have been shown to have a Rim phenotype (8). Identical domains of Vps23 bind Rim8 (the PalF orthologue) in the pH-sensing Rim pathway and Vps27 in the endocytosis-specific ESCRT pathway, suggesting that Vps23 acts at the crossroads of pH signaling and endocytosis (8).
In S. cerevisiae, interaction between Vps23 and Rim8 is favored by the ubiquitination of Rim8, but these two events have been shown to occur independently of ambient pH and are not critical for Rim101 processing (46). The differential pH dependence of the ubiquitination of Rim8/PalF and its interaction with Vps23 is another distinction between S. cerevisiae and A. nidulans (Table 1). According to the current model (Fig. 1), the ubiquitinated Rim8-Vps23 complex binds to Rim21, leading to the recruitment of Rim23. Rim23 then associates the ESCRT machinery with the plasma membrane complex (27, 46). Furthermore, coimmunoprecipitation studies with S. cerevisiae have indicated that the Vps20/Vps32 heterodimer forms a complex with Vps24, Rim13 (the PalB orthologue), and Rim20 (the PalA orthologue) and that the binding of Rim13 and Rim20 is not affected by vps32 mutations that affect Vps24p binding (47–50). Thus, the recruitment of Rim13 to the endosomal processing complex may differ between yeasts and A. nidulans.
SPECIFIC FEATURES OF Rim101 PROCESSING IN CANDIDA ALBICANS
In C. albicans, the single proteolytic event that mediates Rim101 activation at alkaline pH requires the same protein signaling cascade as in S. cerevisiae. External pH sensing involves Rim21, Dfg16, Rim9, and Rim8, and we and others have shown that the ESCRT-I, -II, and -III endosomal complexes play conserved roles in pH signal transduction in C. albicans (17, 39–42). However, C. albicans differs considerably from A. nidulans and S. cerevisiae in terms of the signaling step, the role of the cortical membrane structure, and its link to the Rim101 proteolytic event (Table 1).
In C. albicans, it has been shown that Rim8 is sequentially posttranslationally phosphorylated rather than being ubiquitinated, and hyperphosphorylation is linked to Rim101 processing, occurring at the same time or after this processing. The ESCRT factors required for Rim101 activation are also required for Rim8 phosphorylation. It has also recently been shown that, under alkaline conditions, Rim8 associates with both Rim21 and Rim101, suggesting that Rim8 may be involved in the link between the cortical signaling complex and the Rim101 proteolysis step (Fig. 1) (51).
Another important key feature specific to the C. albicans pathway is the presence of a negative feedback regulatory loop involving both Rim101-mediated repression and the degradation of Rim8 in the vacuole, leading to a decrease in protein levels at high pH (51–53) (Table 1). The ESCRT factors involved in Rim101 activation are also required for Rim8 phosphorylation, which leads to the association of Vps32 and other Vps factors with the endosomal membrane complex. Vps32 is then able to recruit Rim20, Rim101, and Rim13, leading to the cleavage of Rim101 (9, 17, 32, 51, 54) (Fig. 1). As reported for S. cerevisiae, in which the deletion of RIM9, the PalI homologue, prevents Rim101 cleavage, we have shown that Rim9 is fully required for pH signal transduction in C. albicans, whereas rim9 mutants have a leaky phenotype in A. nidulans and Y. lipolytica (36, 55). Our results also suggest that other factors may be required to bridge the plasma membrane and the endosomal membrane complexes of the Rim pathway and that Rim23 may also be involved in this link in C. albicans (55). A likely candidate for RIM23 in C. albicans is orf19.2914, which encodes a protein carrying a Bro1 domain, like Ygr122w in S. cerevisiae and other PalC orthologues (29).
By contrast to what has been reported for A. nidulans and other yeasts, some authors have observed the Rim13-dependent C-terminal cleavage of Rim101 under acidic conditions in C. albicans, which yields a 65-kDa truncated form. It has been suggested that this form regulates expression of pH-independent genes, involved, for example, in lithium or hygromycin resistance, but which are nevertheless affected by rim101 mutations (56).
SPECIFIC FEATURES OF PacC PROCESSING IN ASPERGILLUS FUMIGATUS
In the major mold pathogen of humans, A. fumigatus, PacC null mutants are abnormally sensitive to alkaline culture media in vitro and display aberrant regulation of zinc homeostasis, an essential component of A. fumigatus virulence in murine hosts (57, 58). Electromobility shift analyses have shown that PacC proteolysis resembles the proteolysis observed in A. nidulans in terms of pH sensitivity and proteolytic cleavage sites (M. Bertuzzi et al., personal communication).
The PacC/Rim101 pathway has also been studied in other Aspergillus species that are usually considered saprophytic. Aspergillus niger is an exceptionally efficient producer of organic acids, resulting in the rapid acidification of its environment. This secretion of acids is thought to help this saprophytic fungus to degrade plant cell walls to obtain nutrients. This species can grow at pH values from 2 to above 8, and this has led to pH regulation and functional genomics studies, resulting in the identification of genes for all steps of the PacC/Rim101 pathway (19, 59, 60). Similarly, in Aspergillus oryzae, a homologue of the A. nidulans PalH has been characterized and shown to be required for pH sensing (61).
SPECIFIC FEATURES OF Rim101 PROCESSING IN CRYPTOCOCCUS NEOFORMANS
As a saprophytic soil fungus, C. neoformans can sense and respond to multiple stresses and changes in its environment. Adaptation to pH is critical for C. neoformans pathogenesis in humans, as this basidiomycete can grow well at physiological pH in the blood and cerebrospinal fluid (pH 7.4) but also at the acidic pH found in the vesicles of phagocytic cells, such as macrophages. Experiments both in vitro and in vivo have demonstrated that phagosomes containing C. neoformans undergo acidification soon after phagocytosis and that the fungus can thrive and multiply for subsequent extrusion in a viable form or exocytosis (62–64). Furthermore, capsule production, a key virulence factor in this fungus, is optimal at physiological neutral-alkaline pH (23). The Rim101 orthologue in C. neoformans has recently been identified and shown to have similar physiological functions to the Rim101/PacC proteins of other fungal species, including roles in pH adaptation, salt tolerance, and iron homeostasis/import.
Rim101 processing and cleavage at alkaline pH occur in two steps, as in A. nidulans, and they are not only triggered by the Rim pathway, but also dependent on the cyclic AMP (cAMP)/protein kinase A (PKA) pathway, in marked contrast to the situation in ascomycetous fungi or yeasts (Table 1). Unlike its ascomycete homologues, Rim101 actually harbors a functional PKA consensus phosphorylation site. Rim101 is located in the nucleus regardless of pH, whereas in other fungi, PacC/Rim101 is mostly cytosolic under acidic conditions, with nuclear localization requiring both Rim13- and Rim20-dependent cleavage of Rim101 and an active PKA pathway (23).
INVOLVEMENT OF pH SIGNALING IN FUNGAL PATHOGENICITY
PacC/Rim101 effectors and downstream targets involved in pathogenesis.
The genes and activities controlled by PacC/Rim101 in the model organisms A. nidulans, S. cerevisiae, and Y. lipolytica can be classified into several functional categories, including siderophore biosynthesis and iron or copper metabolism, ion homeostasis, the production of extracellular alkaline or acidic proteases or permeases, the production of enzymes involved in the synthesis of exported metabolites, membrane or cell wall biosynthesis and remodeling, sporulation, mating, dimorphism, invasive growth, and biofilm formation (12–15, 65–69). In pathogenic fungi, most of these functions are related to pathogenesis and virulence.
pH signaling and Candida albicans virulence.
A comparison of the multiple steps of PacC/Rim101 processing between model and human pathogenic fungi (see above and Table 1) shows that the pathway is more refined in C. albicans than in the model organisms with a specific and accurate negative feedback control system (51). No such negative feedback has yet been described in other species, and its relevance to pathogenicity has not been demonstrated, but this regulatory mechanism may be associated with the higher levels of pH stress with which this commensal organism must deal to survive and to invade various cells or niches of the human body.
The main contribution of the Rim pathway to C. albicans virulence relates to its role in adaptation to neutral-alkaline conditions, through growth, iron transport and metabolism, pH-dependent morphogenesis, cell wall structure, adhesion, and the ability to produce biofilms (7, 68, 70–73). Impaired fungal growth under alkaline conditions results partly from the lower solubility of the ferric ions (Fe3+), leading to iron starvation. One of the principal Rim101-dependent responses to alkaline conditions involves adaptation to iron starvation. Tolerance of iron deprivation at physiological pH is one of the most important virulence determinants of fungi, as freely available iron is strongly limited in human hosts, protecting the host against microorganisms unable to increase iron uptake (7, 52, 68, 74–78). Transcriptional analyses have identified several genes involved in iron metabolism and transport, including some Rim101-dependent targets, such as the siderophore iron transporter genes ARN1 and FET3 and the ferric reductase and ion permease genes ENA1, RBT2, FRE2, FRE5, FRE8, FRP1, FRP2, CTR1, and ZRT1 (52, 74, 79). ALS3 also is dependent on Rim101. In addition to its role in adhesion, Als3 binds to human ferritin, facilitating iron acquisition by C. albicans under host conditions (79, 80).
Both yeasts and hyphae can invade tissues, but the ability to shift from the yeast form to the hyphal form has been clearly linked to virulence (73, 81–83). Mutants unable to undergo morphogenic switching display attenuated virulence (39, 84). According to the pathophysiological model of invasive candidiasis in humans, hyphal forms are better able to adhere and to maintain the colonization of mucosal niches, to disrupt the epithelia of both mucosal and endothelial surfaces and, thus, to disseminate into the bloodstream and to invade deep-seated tissues (73, 82, 83). The yeast-to-hypha transition is also involved in interactions between fungi and bacteria and in the construction of biofilms, which play an important role in the pathogenicity of C. albicans (85–88). Rim101 upregulates filamentation, and many of the genes strongly regulated by Rim101 are hypha-specific genes (ECE1, CSA1, CSA2, SAP5, HYR1, HWP1, RBT1, and IHD1) (52, 70, 73, 79). Genes involved in cell wall structure and remodeling (ALS3, PGA7/RBT6, CHT2, SKN1, PHR1, and PHR2) (18, 68, 79, 89) are also Rim regulated and make a major contribution to Rim-dependent pathogenesis, as many determinant factors for virulence are dependent on the cell wall (reviewed in references 81 and 90). Phr1 and Phr2 are paralogues that are differentially expressed under alkaline-neutral and acidic conditions, respectively. They are β(1-3)glucanosyltranferases involved in glucan remodeling of the cell wall (91, 92). Als3, Pga7/Rbt6, Hwp1, and Sap5 are adhesins that play a key role in invasion and biofilm formation (79, 81, 83, 93). Thus, the main contribution of the PacC/Rim101 pathway to C. albicans virulence is related to its role in adaptation to neutral-alkaline conditions, through iron homeostasis, pH-dependent morphogenesis, cell wall synthesis, and remodeling (7, 18, 68, 71, 94).
Experimental studies in both murine and porcine models have established the key role of the Rim101 pathway in C. albicans pathogenesis in disseminated candidiasis, keratitis, and oropharyngeal and intra-abdominal candidiasis (71, 79, 95, 96). The importance of pH regulation during tissue invasion through hypha formation and pathogen survival has been demonstrated in in vivo and ex vivo genome-wide transcriptional profiling comparisons for liver invasion. ALS3, PHR1, ECE1, HWP1, and DFG16 were shown to be upregulated during liver invasion (93). Also, in a model of intra-abdominal candidiasis, RIM101 was identified as one of the most strongly expressed genes, and a rim101 mutant was found to cause a less-pronounced infiltration of the peritoneal fluid with neutrophils (97).
pH SIGNALING AND ASPERGILLUS FUMIGATUS VIRULENCE
After initial studies demonstrated the role of Rim-mediated pH signaling in C. albicans virulence, further analyses made use of the multiple A. nidulans pH mutants available, together with a neutropenic model of pulmonary aspergillosis to dissect the roles of PacC, PacC processing, and Pal-mediated pH signaling in Aspergillus spp. virulence (98). PacC action was found to be both required for and able to enhance virulence. The A. nidulans pH-responsive transcription factor PacC thus plays a key role in pulmonary pathogenesis. The PalH homologue was recently shown to be required for murine infection (99).
Transcriptome analyses comparing in vitro responses to alkaline stress and gene expression during the initiation of murine infection recently identified A. fumigatus adaptation to alkaline stress as a key component in the early stages of mouse infection (100). Consistent with these findings, the virulence of A. fumigatus PacC null mutants is attenuated in murine models of pulmonary aspergillosis. In addition to zinc homeostasis, many other virulence-enabling functions are also under the control of PacC regulation during A. fumigatus infection. This suggests that in A. fumigatus, PacC acts as a master regulator of multiple virulence factors and that the abolition of PacC-mediated pH signaling can greatly decrease virulence (M. Bertuzzi et al., personal communication).
pH SIGNALING AND CRYPTOCOCCUS NEOFORMANS VIRULENCE
Several virulence factors have been identified in C. neoformans, but the capsule is probably the major one (101). This complex structure is composed mainly of polysaccharides that are linked to the cell wall (102). Mutant strains unable to synthesize a capsule are avirulent, and the capsular polysaccharides exert profound depressive effects on both innate and adaptive immune responses (103, 104). Different factors, including pH, have been shown to influence the capsule size and structure (105). Rim101 plays a role in capsule formation in association with the cAMP/PKA pathway, probably by facilitating capsule anchorage to the cell wall rather than regulating capsule biosynthesis (23). Transcriptional profiling has identified several Rim101-dependent genes, including those encoding a UDP-glucose dehydrogenase, a manosyltranferase, and a phosphomannomutase, which are essential for capsule synthesis. However, many of the genes involved in capsule production are not dependent on Rim101. Other downstream targets of Rim101, such as ENA1, which is directly regulated by Rim101, are associated with the alkaline response or with iron or metal homeostasis (CFT1, FET3, and SIT1). As in other fungi, rim101 mutants display impaired growth under alkaline or iron starvation conditions. Interestingly, Vps23 has been shown to be involved in iron acquisition, capsule formation, and virulence. However, these functions may be explained by the role of Vps23 in endocytosis, and its contribution, like other ESCRT factors, in Rim101 processing in C. neoformans has not been demonstrated yet (106). Many of the genes involved in cell wall biosynthesis and remodeling are regulated by Rim101 in C. neoformans (CHS4, CHS5, CHS6, CHS8, SKN1, FKS1, KRE6, CDA3, CHI22, and AGS1) (107). Rim101 is also required for the formation of titan cells, a type of enlarged yeast cell. Titan cells are more resistant than normal yeast cells to engulfment by immune cells, and they are also more resistant to oxidative and nitrosative stresses, providing normal cryptococcal cells with cross-protection (107–109). The role of the Rim pathway in this pathogenesis-related morphological switch in C. neoformans is reminiscent of the Rim-dependent yeast-to-hypha transition displayed by C. albicans in the host.
Despite these defects in the capsule, alkaline pH adaptation, iron homeostasis, and morphological switching, the rim101-deleted strain was found to be hypervirulent in a mouse model of disseminated cryptococcosis with inhalation as the route of inoculation (23, 110). These rather unexpected results were initially attributed to greater survival of the rim101 mutant within macrophages due to the derepression of acid response genes, leading to increased survival under acidic conditions (23). However, it now also appears likely that the capsule attachment defect has a profound effect on the host immune response. First, enhanced diffusion of the capsule polysaccharides into the surrounding tissues may increase their immunomodulatory effects. Second, as the cell wall of the rim101 mutant cells contained higher than normal levels of antigenic mannoproteins (MP88 and MP98), inappropriate recognition and responses may result in excessive host inflammation and, eventually, in cell death. Recent in vivo experiments have confirmed that there is an exacerbation of the neutrophil and cytokine defenses that results in a deleterious and ineffective inflammatory response (107). The capsule defect itself worsens this disproportionate immune response by unmasking the hyperimmunogenic cell wall. The hypervirulent phenotype displayed by the rim101 mutant and the role of the pathway in altering the antigenic properties of the cell wall have not been demonstrated in other species, further highlighting the profound differences in the PacC/Rim101 pathway between ascomycetes and basidiomycetes. Further studies focusing on the implication of pH signaling in alterations to host-pathogen interactions through cell wall remodeling are warranted (20, 22, 23, 110).
THE POSSIBLE USE OF pH SIGNALING AS A DRUG TARGET FOR ANTIFUNGAL STRATEGIES
Recent studies have highlighted the potential of the Pac/Rim pathway as a source of targets for improving antifungal strategies. The results detailed above highlight the role of this pathway in iron homeostasis, morphological switching under host conditions, cell wall synthesis, and remodeling. Together, they provide a basis for this hypothesis: targeting iron uptake by blocking siderophore biosynthesis, or through the use of anti-Als3 antibodies or chelators, has been shown to reduce Aspergillus, Candida, Fusarium, and Zygomycetes infections in animal models and in clinical trials (80, 111–115). Given its unique composition and its role in the host-pathogen interface in modulating both the immune response and morphogenetic switching, the cell wall remains an ideal target. However, its dynamic structure makes adaptation possible, and fungi are thus able to respond to drugs targeting the cell wall (116, 117). We summarize here additional studies from our group and others whose findings implicate the Pac/Rim pathway in the cellular responses underlying antifungal tolerance.
Tolerance to azoles and caspofungin was first described by Sanglard and coworkers, who established the role of the calcineurin pathway in such tolerance in C. albicans (118–120). The compensatory mechanisms involved in antifungal tolerance are mediated by a complex interplay between the calcineurin, Tor, HOG, and PKC-cAMP-PKA signaling pathways, resulting in an increase in chitin synthesis (120–127). Compromise of PKC signaling and the chaperone Hsp90 leads to similar killing effects as calcineurin inhibitors on ergosterol biosynthesis inhibitors and echinocandins (3, 124, 128–132). The mechanisms of antifungal drug tolerance in A. fumigatus are similar to those described for C. albicans, although they have been explored less thoroughly. The calcineurin blockade has been shown to enhance the inhibition of 1,3-β-d-glucan by caspofungin in A. fumigatus and to convert caspofungin into a fungicidal drug (133, 134). Hsp90 is also involved in the echinocandin tolerance of A. fumigatus and Aspergillus terreus (129, 135). In C. neoformans too, the combination of azoles or caspofungin with calcineurin inhibitors was found to be synergistic, but this may reflect the known ability of FK506 to block the pumps involved in multidrug resistance (136). As with C. albicans, mutants of the PKC pathway display enhanced susceptibility to caspofungin (137).
A role of pH regulation in antifungal activity was suggested by the observation that the azole compound D0870 had higher levels of fungicidal activity against C. neoformans at low pH (138). Similar results were subsequently obtained in susceptibility tests under acidic conditions, which were found to inhibit the trailing effect of the azoles in Candida spp. (139). This trait, which hampers interpretation of the activity of azoles and echinocandins against yeasts, is not fully understood, but it may relate to the lack of fungicidal activity of these drugs (which are instead fungistatic) (139, 140).
The role of the PacC/Rim101 pathway in azole tolerance has been explored in S. cerevisiae. In a chemical-genetic and genetic interactions study, all mutants of the pathway were found to be hypersensitive to fluconazole, and the double rim20-erg11 mutant was not viable, with Erg11p the enzyme targeted by azoles (141). We previously showed that a rim101-deleted strain of C. albicans also displayed enhanced azole susceptibility. Deletions of VPS28 and VPS32, encoding endocytic components of the endosomal membrane complex of the PacC/Rim101 pathway, resulted in the same phenotype of azole hypersusceptibility. The constitutively active truncated RIM101SL gene restores azole susceptibility in all rim101 and vps mutant strains, demonstrating the involvement of the PacC/Rim101 pathway in C. albicans tolerance to azoles (142). The azole hypersensitivity phenotype was subsequently confirmed by others as part of a large phenotypic profiling experiment on a collection of strains with deletions of genes encoding transcriptional regulators (143). Furthermore, we recently confirmed that all C. albicans rim mutants, not just the rim101 mutant, were hypersensitive to all the triazoles used to treat humans (fluconazole, voriconazole, and posaconazole) and that Rim disruption rendered these compounds fungicidal rather than fungistatic (144). In A. fumigatus, iron homeostasis has been linked to ergosterol synthesis and azole resistance (112). A recent study demonstrated a role for the PacC/Rim101 pathway in C. neoformans tolerance to azoles and amphotericin B through regulation of the cation transporters Ena1 and Nha1. As these transporters are involved in membrane stability and cation homeostasis, their inhibition enhances azole and polyene activity (145).
CONCLUSIONS
Many of the features described above support the hypothesis that the pH signaling pathways could be targeted in a novel approach to decrease fungal development and virulence in the infected host. We assume that blockade of the PacC/Rim101 pathway is likely to have beneficial and synergistic effects, by reducing the virulence and growth of pathogenic fungi and by decreasing the tolerance and resistance of these fungi to existing antifungal drugs. Furthermore, the specificity of the PacC/Rim101 pathway and its conservation throughout the fungal kingdom suggest that this pathway has considerable potential as a therapeutic target.
Defining the proper target(s) is of course an essential step. While multiple targets can be envisioned to inhibit the Pac/Rim pathway, targets that are both well conserved in pathogenic fungi and are fungus specific should be prioritized (25, 28, 99, 146). Fungus-specific components of the Pac/Rim pathways in pathogenic yeasts and fungi need to be fully characterized, both from a genetic and a biochemical point of view, in terms of mutant phenotype, protein structure, subcellular localization, and interactions. The molecular mechanism of pH sensing remains itself mysterious, and definite proof of the role of the alleged sensors is still missing. Interactions between the pH-sensing pathway and mechanisms of fungal tolerance represent another area poorly explored up to now. Genetic screens for chemical-genetic interactions (141) of Pac/Rim mutants with antifungal drugs may shed light on this aspect. These experiments will identify targets for adjunctive therapies to existing drugs in order to obtain fungicidal rather than fungistatic activity or to lower fungal tolerance to existing drugs, thus decreasing emergence of resistance.
Finally, it has become apparent that the fungal pH signaling pathways, while being highly conserved in terms of components, are entangled in highly diversified genetic networks in different organisms. These remain to be precisely deciphered to appreciate fully the range of variations that can be expected. Further studies of the pH signaling pathway in different pathogenic fungi may thus answer some of these questions and hopefully help to design new drugs badly needed in both medicine and agriculture.
ACKNOWLEDGMENTS
We are very grateful to Guilhem Janbon and Elaine Bignell and to two anonymous referees for their insightful comments on the manuscript.
Footnotes
Published ahead of print 17 January 2014
REFERENCES
- 1.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv13. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- 2.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53. 10.3109/1048410903241444 [DOI] [PubMed] [Google Scholar]
- 3.Cowen LE. 2008. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol. 6:187–198. 10.1038/nrmicro1835 [DOI] [PubMed] [Google Scholar]
- 4.Sanglard D, Coste A, Ferrari S. 2009. Antifungal drug resistance mechanisms in fungal pathogens from the perspective of transcriptional gene regulation. FEMS Yeast Res. 9:1029–1050. 10.1111/j.1567-1364.2009.00578.x [DOI] [PubMed] [Google Scholar]
- 5.Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J, Cardenas ME. 2007. Sensing the environment: lessons from fungi. Nat. Rev. Microbiol. 5:57–69. 10.1038/nrmicro1578 [DOI] [PubMed] [Google Scholar]
- 6.Bastidas RJ, Reedy JL, Morales-Johansson H, Heitman J, Cardenas ME. 2008. Signaling cascades as drug targets in model and pathogenic fungi. Curr. Opin. Investig. Drugs 9:856–864 [PMC free article] [PubMed] [Google Scholar]
- 7.Davis DA. 2009. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr. Opin. Microbiol. 12:365–370. 10.1016/j.mib.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 8.Blanchin-Roland S. 2011. Identical domains of Yarrowia lipolytica Vps23 are required for both ESCRT and Rim pathways, but the latter needs an interaction between the Vps23 UEV domain and Rim8/PalF. FEMS Yeast Res. 11:473–486. 10.1111/j.1567-1364.20113.00735.x [DOI] [PubMed] [Google Scholar]
- 9.Blanchin-Roland S, Costa GD, Gaillardin C. 2005. ESCRT-I components of the endocytic machinery are required for Rim101-dependent ambient pH regulation in the yeast Yarrowia lipolytica. Microbiology 151:3627–3637. 10.1009/mic.0.28196-0 [DOI] [PubMed] [Google Scholar]
- 10.Blanchin-Roland S, Da Costa G, Gaillardin C. 2008. Ambient pH signalling in the yeast Yarrowia lipolytica involves YlRim23p/PalC, which interacts with Snf7p/Vps32p, but does not require the long C terminus of YlRim9p/PalI. Microbiology 154:1668–1676. 10.1099/mic.0.2008/017046-0 [DOI] [PubMed] [Google Scholar]
- 11.Gonzalez-Lopez CI, Szabo R, Blanchin-Roland S, Gaillardin C. 2002. Genetic control of extracellular protease synthesis in the yeast Yarrowia lipolytica. Genetics 160:417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamb TM, Xu W, Diamond A, Mitchell AP. 2001. Alkaline response genes of Saccharomyces cerevisiae and their relationship to the RIM101 pathway. J. Biol. Chem. 276:1850–1856. 10.1074/jbc.M008381200 [DOI] [PubMed] [Google Scholar]
- 13.Penalva MA, Arst HNJ. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 66:426–446. 10.1128/MMBR.66.3.426-446.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Penalva MA, Arst HN. 2004. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Annu. Rev. Microbiol. 58:425–451. 10.1146/annurev.micro.58.030603.123715 [DOI] [PubMed] [Google Scholar]
- 15.Penalva MA, Tilburn J, Bignell E, Arst HN. 2008. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 16:291–300. 10.1016/j.tim.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 16.Xu W, Mitchell AP. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917–6923. 10.1128/JB.183.23.6917-6923.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Smith FJJ, Subaran R, Mitchell AP. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528–5537. 10.1091/mbc.E04-08-0666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr. Genet. 44:1–7. 10.1007/s00296-003-0415-2 [DOI] [PubMed] [Google Scholar]
- 19.Andersen MR, Lehmann L, Nielsen J. 2009. Systemic analysis of the response of Aspergillus niger to ambient pH. Genome Biol. 10:R47. 10.1186/gb-2009-10-5-r47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cervantes-Chavez JA, Ortiz-Castellanos L, Tejeda-Sartorius M, Gold S, Ruiz-Herrera J. 2010. Functional analysis of the pH responsive pathway Pal/Rim in the phytopathogenic basidiomycete Ustilago maydis. Fungal Genet. Biol. 47:446–457. 10.1016/j.fgb.2010.02.004 [DOI] [PubMed] [Google Scholar]
- 21.Silveira HC, Gras DE, Cazzaniga RA, Sanches PR, Rossi A, Martinez-Rossi NM. 2010. Transcriptional profiling reveals genes in the human pathogen Trichophyton rubrum that are expressed in response to pH signaling. Microb. Pathog. 48:91–96. 10.1016/j.micpath.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 22.Arechiga-Carvajal ET, Ruiz-Herrera J. 2005. The RIM101/pacC homologue from the basidiomycete Ustilago maydis is functional in multiple pH-sensitive phenomena. Eukaryot. Cell 4:999–1008. 10.1128/EC.4.6.999-1008.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. 2010. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 6(2):e1000776. 10.1371/journal.ppat.1000776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arst HN, Penalva MA. 2003. pH regulation in Aspergillus and parallels with higher eukaryotic regulatory systems. Trends Genet. 19:224–231. 10.1016/S0168-9525(03)00052-0 [DOI] [PubMed] [Google Scholar]
- 25.Herranz S, Rodriguez JM, Bussink HJ, Sanchez-Ferrero JC, Arst HNJ, Penalva MA, Vincent O. 2005. Arrestin-related proteins mediate pH signaling in fungi. Proc. Natl. Acad. Sci. U. S. A. 102:12141–12146. 10.1073/pnas.0504776102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calcagno-Pizarelli AM, Negrete-Urtasun S, Denison SH, Rudnicka JD, Bussink HJ, Munera-Huertas T, Stanton L, Hervas-Aguilar A, Espeso EA, Tilburn J, Arst HNJ, Penalva MA. 2007. Establishment of the ambient pH signaling complex in Aspergillus nidulans: PalI assists plasma membrane localization of PalH. Eukaryot. Cell 6:2365–2375. 10.1128/EC.00275-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hervas-Aguilar A, Galindo A, Penalva MA. 2010. Receptor-independent ambient pH signaling by ubiquitin attachment to fungal arrestin-like PalF. J. Biol. Chem. 285:18095–18102. 10.1074/jbc.M110.114371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bignell EM. 2012. Conservation in Aspergillus fumigatus of pH-signaling seven transmembrane domain and arrestin proteins, and implications for drug discovery. Ann. N. Y. Acad. Sci. 1273:35–43. 10.1111/j.1749-6632.2012.06814.x [DOI] [PubMed] [Google Scholar]
- 29.Galindo A, Hervas-Aguilar A, Rodriguez-Galan O, Vincent O, Arst HNJ, Tilburn J, Penalva MA. 2007. PalC, one of two Bro1 domain proteins in the fungal pH signalling pathway, localizes to cortical structures and binds Vps32. Traffic 8:1346–1364. 10.1111/j.1600-0854.2007.00620.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Galindo A, Calcagno-Pizarelli AM, Arst HN, Jr, Peñalva ḾA. 2012. An ordered pathway for the assembly of ESCRT-containing fungal ambient pH signalling complexes at the plasma membrane. J. Cell Sci. 125:1784–1795. 10.1242/jcs.098897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Galan O, Galindo A, Hervas-Aguilar A, Arst HNJ, Penalva MA. 2009. Physiological involvement in pH signaling of Vps24-mediated recruitment of Aspergillus PalB cysteine protease to ESCRT-III. J. Biol. Chem. 284:4404–4412. 10.1074/jbc.M808645200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent O, Rainbow L, Tilburn J, Arst HNJ, Penalva MA. 2003. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol. Cell. Biol. 23:1647–1655. 10.1128/MCB.23.5.1647-1655.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diez E, Alvaro J, Espeso EA, Rainbow L, Suarez T, Tilburn J, Arst HN, Penalva MA. 2002. Activation of the Aspergillus PacC zinc finger transcription factor requires two proteolytic steps. EMBO J. 21:1350–1359. 10.1093/emboj/21.6.1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Penas MM, Hervas-Aguilar A, Munera-Huertas T, Reoyo E, Penalva MA, Arst HNJ, Tilburn J. 2007. Further characterization of the signaling proteolysis step in the Aspergillus nidulans pH signal transduction pathway. Eukaryot. Cell 6:960–970. 10.1128/EC.00047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hervas-Aguilar A, Rodriguez JM, Tilburn J, Arst HNJ, Penalva MA. 2007. Evidence for the direct involvement of the proteasome in the proteolytic processing of the Aspergillus nidulans zinc finger transcription factor PacC. J. Biol. Chem. 282:34735–34747. 10.1074/jbc.M706723200 [DOI] [PubMed] [Google Scholar]
- 36.Li W, Mitchell AP. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics 145:63–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su SS, Mitchell AP. 1993. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics 133:67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Su SS, Mitchell AP. 1993. Molecular characterization of the yeast meiotic regulatory gene RIM1. Nucleic Acids Res. 21:3789–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cornet M, Bidard F, Schwarz P, Da Costa G, Blanchin-Roland S, Dromer F, Gaillardin C. 2005. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect. Immun. 73:7977–7987. 10.1128/IAI.73.12.7977-7987.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kullas AL, Li M, Davis DA. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3:1609–1618. 10.1128/EC.3.6.1609-1618.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolf JM, Johnson DJ, Chmielewski D, Davis DA. 2010. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot. Cell 9:1203–1215. 10.1128/EC.00056-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf JM, Davis DA. 2010. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics 184:673–694. 10.1534/genetics.109.112029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barwell KJ, Boysen JH, Xu W, Mitchell AP. 2005. Relationship of DFG16 to the Rim101p pH response pathway in Saccharomyces cerevisiae and Candida albicans. Eukaryot. Cell 4:890–899. 10.1128/EC.4.5.890-899.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothfels K, Tanny JC, Molnar E, Friesen H, Commisso C, Segall J. 2005. Components of the ESCRT pathway, DFG16, and YGR122w are required for Rim101 to act as a corepressor with Nrg1 at the negative regulatory element of the DIT1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 25:6772–6788. 10.1128/MCB.25.15.6772-3788.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Obara K, Yamamoto H, Kihara A. 2012. Membrane protein Rim21 plays a central role in sensing ambient pH in Saccharomyces cerevisiae. J. Biol. Chem. 287:38473–38481. 10.1074/jbc.M112.394205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Herrador A, Herranz S, Lara D, Vincent O. 2010. Recruitment of the ESCRT machinery to a putative seven-transmembrane-domain receptor is mediated by an arrestin-related protein. Mol. Cell. Biol. 30:897–907. 10.1128/MCB.00132-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss P, Huppert S, Kölling R. 2009. Analysis of the dual function of the ESCRT-III protein Snf7 in endocytic trafficking and in gene expression. Biochem. J. 424:89–97. 10.1042/BJ20090957 [DOI] [PubMed] [Google Scholar]
- 48.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3:271–282. 10.1016/S1534-5807(02)00220-4 [DOI] [PubMed] [Google Scholar]
- 49.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283–289. 10.1016/S1534-5807(02)00219-8 [DOI] [PubMed] [Google Scholar]
- 50.Katzmann DJ, Stefan CJ, Babst M, Emr SD. 2003. Vps27 recruits ESCRT machinery to endosomes during MVB sorting. J. Cell Biol. 162:413–423. 10.1083/jcb.200302136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gomez-Raja J, Davis DA. 2012. The β-arrestin-like protein Rim8 is hyperphosphorylated and complexes with Rim21 and Rim101 to promote adaptation to neutral-alkaline pH. Eukaryot. Cell 11:683–693. 10.1128/EC.05211-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 54:1335–1351. 10.1111/j.1365-2958.2004.04350.x [DOI] [PubMed] [Google Scholar]
- 53.Porta A, Ramon AM, Fonzi WA. 1999. PRR1, a homolog of Aspergillus nidulans palF, controls pH-dependent gene expression and filamentation in Candida albicans. J. Bacteriol. 181:7516–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boysen JH, Mitchell AP. 2006. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol. Biol. Cell 17:1344–1353. 10.1091/mbc.E05-10-0949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cornet M, Richard ML, Gaillardin C. 2009. The homologue of the Saccharomyces cerevisiae RIM9 gene is required for ambient pH signalling in Candida albicans. Res. Microbiol. 160:219–223. 10.1016/j.resmic.2009.02.002 [DOI] [PubMed] [Google Scholar]
- 56.Li M, Martin SJ, Bruno VM, Mitchell AP, Davis DA. 2004. Candida albicans Rim13p, a protease required for Rim101p processing at acidic and alkaline pHs. Eukaryot. Cell 3:741–751. 10.1128/EC.3.3.741-751.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amich J, Vicentefranqueira R, Leal F, Calera JA. 2010. Aspergillus fumigatus survival in alkaline and extreme zinc-limiting environments relies on the induction of a zinc homeostasis system encoded by the zrfC and aspf2 genes. Eukaryot. Cell 9:424–437. 10.1128/EC.00348-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moreno MA, Ibrahim-Granet O, Vicentefranqueira R, Amich J, Ave P, Leal F, Latgé J-P, Calera JA. 2007. The regulation of zinc homeostasis by the ZafA transcriptional activator is essential for Aspergillus fumigatus virulence. Mol. Microbiol. 64:1182–1197. 10.1111/j.1365-2958.2007.05726.x [DOI] [PubMed] [Google Scholar]
- 59.Hesse SJ, Ruijter GJ, Dijkema C, Visser J. 2002. Intracellular pH homeostasis in the filamentous fungus Aspergillus niger. Eur. J. Biochem. 269:3485–3494. 10.1046/j.1432-1033.2002.03042.x [DOI] [PubMed] [Google Scholar]
- 60.Van den Hombergh JP, MacCabe AP, van de Vondervoort PJ, Visser J. 1996. Regulation of acid phosphatases in an Aspergillus niger pacC disruption strain. Mol. Gen. Genet. 251:542–550 [DOI] [PubMed] [Google Scholar]
- 61.Dohmoto M, Inoue Y, Kobayashi A, Ohashi S, Sano M. 2010. Characterization of the palH gene from Aspergillus oryzae. Biosci. Biotechnol. Biochem. 74:188–190 http://dx.doi.org/10.1271/bbb.90575 [DOI] [PubMed] [Google Scholar]
- 62.Nyberg K, Johansson U, Johansson A, Camner P. 1992. Phagolysosomal pH in alveolar macrophages. Environ. Health Perspect. 97:149–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Levitz SM, Nong SH, Seetoo KF, Harrison TS, Speizer RA, Simons ER. 1999. Cryptococcus neoformans resides in an acidic phagolysosome of human macrophages. Infect. Immun. 67:885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nicola AM, Robertson EJ, Albuquerque P, Derengowski Lda S, Casadevall A. 2011. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. mBio 2(4):e00167–11. 10.1128/mBio.00167-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lambert M, Blanchin-Roland S, Le Louedec F, Lepingle A, Gaillardin C. 1997. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol. Cell. Biol. 17:3966–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Treton B, Blanchin-Roland S, Lambert M, Lepingle A, Gaillardin C. 2000. Ambient pH signalling in ascomycetous yeasts involves homologues of the Aspergillus nidulans genes palF and paIH. Mol. Gen. Genet. 263:505–513. 10.1007/s004380051195 [DOI] [PubMed] [Google Scholar]
- 67.Lamb TM, Mitchell AP. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677–686. 10.1128/MCB.23.2.677-686.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Selvig K, Alspaugh JA. 2011. pH response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology 39:249–256. 10.5941/MYCO.2011.39.4.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Voordeckers K, De Maeyer D, van der Zande E, Vinces MD, Meert W, Cloots L, Ryan O, Marchal K, Verstrepen KJ. 2012. Identification of a complex genetic network underlying Saccharomyces cerevisiae colony morphology. Mol. Microbiol. 86:225–239. 10.1111/j.1365-2958.2012.08192.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berman J, Sudbery PE. 2002. CANDIDA ALBICANS: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930. 10.1038/nrg948 [DOI] [PubMed] [Google Scholar]
- 71.Davis D, Edwards JE, Mitchell AP, Ibrahim AS. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect. Immun. 68:5953–5959. 10.1128/IAI.68.10.5953-5959.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mitchell BM, Wu TG, Jackson BE, Wilhelmus KR. 2007. Candida albicans strain-dependent virulence and Rim13p-mediated filamentation in experimental keratomycosis. Invest. Ophthalmol. Vis. Sci. 48:774–780. 10.1167/iovs.06-0793 [DOI] [PubMed] [Google Scholar]
- 73.Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat. Rev. Microbiol. 9:737–748. 10.1038/nrmicro2636 [DOI] [PubMed] [Google Scholar]
- 74.Baek Y-U, Li M, Davis DA. 2008. Candida albicans ferric reductases are differentially regulated in response to distinct forms of iron limitation by the Rim101 and CBF transcription factors. Eukaryot. Cell 7:1168–1179. 10.1128/EC.00108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jung WH, Kronstad JW. 2008. Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cell. Microbiol. 10:277–284. 10.1111/j.1462-5822.2007.01077.x [DOI] [PubMed] [Google Scholar]
- 76.Lan C-Y, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 53:1451–1469. 10.1111/j.1365-2958.2004.04214.x [DOI] [PubMed] [Google Scholar]
- 77.Tangen KL, Jung WH, Sham AP, Lian T, Kronstad JW. 2007. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiology 153:29–41. 10.1099/mic.0.2006/000927-0 [DOI] [PubMed] [Google Scholar]
- 78.Kronstad JW, Hu G, Jung WH. 2013. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends Microbiol. 21:457–465. 10.1016/j.tim.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, Mitchell AP, Filler SG. 2008. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell. Microbiol. 10:2180–2196. 10.1111/j/1462-5822.2008.01198.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot. Cell 10:168–173. 10.1128/EC.00279-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gow NA, Hube B. 2012. Importance of the Candida albicans cell wall during commensalism and infection. Curr. Opin. Microbiol. 5:406–412. 10.1016/j.mib.2012.04.005 [DOI] [PubMed] [Google Scholar]
- 82.Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. 2012. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat. Rev. Microbiol. 10:112–122. 10.1038/nrmicro2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lionakis MS, Netea MG. 2013. Candida and host determinants of susceptibility to invasive candidiasis. PLoS Pathog. 9(1):e1003079. 10.1371/journal.ppat.1003079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lo HJ, Kohler JR, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink GR. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 85.Douglas LJ. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30–36. 10.1016/S0966-842X(02)00002-1 [DOI] [PubMed] [Google Scholar]
- 86.Giacometti R, Kronberg F, Biondi RM, Passeron S. 2011. Candida albicans Tpk1p and Tpk2p isoforms differentially regulate pseudohyphal development, biofilm structure, cell aggregation and adhesins expression. Yeast 28:293–308. 10.1002/yea.1839 [DOI] [PubMed] [Google Scholar]
- 87.Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Mylonakis E. 2008. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 105:14585–14590. 10.1073/pnas.0805048105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bonhomme J, d'Enfert C. 2013. Candida albicans biofilms: building a heterogeneous, drug-tolerant environment. Curr. Opin. Microbiol. 16:398–403. 10.1016/j.mib.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 89.De Bernardis F, Muhlschlegel FA, Cassone A, Fonzi WA. 1998. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect. Immun. 66:3317–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gow NA, Brown AJ, Odds FC. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366–371 http://dx.doi.org/10.1016/S1369-5274(02)00338-7 [DOI] [PubMed] [Google Scholar]
- 91.Fonzi WA. 1999. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J. Bacteriol. 181:7070–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mouyna I, Fontaine T, Vai M, Monod M, Fonzi WA, Diaquin M, Popolo L, Hartland RP, Latge JP. 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275:14882–14889. 10.1074/jbc.275.20.14882 [DOI] [PubMed] [Google Scholar]
- 93.Thewes S, Kretschmar M, Park H, Schaller M, Filler SG, Hube B. 2007. In vivo and ex vivo comparative transcriptional profiling of invasive and non-invasive Candida albicans isolates identifies genes associated with tissue invasion. Mol. Microbiol. 63:1606–1628. 10.1111/j.1365-2958.2007.05614.x [DOI] [PubMed] [Google Scholar]
- 94.Davis D, Wilson RB, Mitchell AP. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol. Cell. Biol. 20:971–978. 10.1128/MCB.20.3.971-978.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuan X, Mitchell BM, Hua X, Davis DA, Wilhelmus KR. 2010. The RIM101 signal transduction pathway regulates Candida albicans virulence during experimental keratomycosis. Invest. Ophthalmol. Vis. Sci. 51:4668–4676. 10.1167/iovs.09-4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheng S, Clancy CJ, Xu W, Schneider F, Hao B, Mitchell AP, Nguyen MH. 2013. Profiling of Candida albicans gene expression during intra-abdominal candidiasis identifies biologic processes involved in pathogenesis. J. Infect. Dis. 208:1529–1537. 10.1093/infdis/jit335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng S, Clancy C, Mitchell AP, Fanning S, Solis N, Filler SG, Nguyen M. 2012. Nanostring analysis of Candida albicans gene expression during intra-abdominal candidiasis (IAC) provides insights into pathogenesis and reveals rewiring of regulatory pathways, abstr M1056 Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA American Society for Microbiology, Washington, DC [Google Scholar]
- 98.Bignell E, Negrete-Urtasun S, Calcagno AM, Haynes K, Arst HNJ, Rogers T. 2005. The Aspergillus pH-responsive transcription factor PacC regulates virulence. Mol. Microbiol. 55:1072–1084. 10.1111/j.1365-2958.2004.04472.x [DOI] [PubMed] [Google Scholar]
- 99.Grice CM, Bertuzzi M, Bignell EM. 2013. Receptor-mediated signaling in Aspergillus fumigatus. Front. Microbiol. 4:26. 10.3389/fmicb.2013.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McDonagh A, Fedorova ND, Crabtree J, Yu Y, Kim S, Chen D, Loss O, Cairns T, Goldman G, Armstrong-James D, Haynes K, Haas H, Schrettl M, May G, Nierman WC, Bignell E. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 4(9):e1000154. 10.1371/journal.ppat.1000154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zaragoza O, Rodrigues ML, De Jesus M, Frases S, Dadachova E, Casadevall A. 2009. The capsule of the fungal pathogen Cryptococcus neoformans. Adv. Appl. Microbiol. 68:133–216. 10.1016/S0065-2164(09)01204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Reese AJ, Doering TL. 2003. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol. Microbiol. 50:1401–1409. 10.1046/j.1365-2958.2003.03780.x [DOI] [PubMed] [Google Scholar]
- 103.Chang YC, Kwon-Chung KJ. 1994. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol. Cell. Biol. 14:4912–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vecchiarelli A, Pericolini E, Gabrielli E, Kenno S, Perito S, Cenci E, Monari C. 2013. Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol. 8:1107–1116. 10.2217/fmb.13.84 [DOI] [PubMed] [Google Scholar]
- 105.O'Meara TR, Alspaugh JA. 2012. The Cryptococcus neoformans capsule: a sword and a shield. Clin. Microbiol. Rev. 25:387–408. 10.1128/CMR.00001-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hu G, Caza M, Cadieux B, Chan V, Liu V, Kronstad J. 2013. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect. Immun. 81:292–302. 10.1128/IAI.01037-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O'Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. 2013. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. mBio 4(1):e00522-12. 10.1128/mBio.00522-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chrétien F, Heitman J, Dromer F, Nielsen K. 2010. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 6(6):e1000953. 10.1371/journal.ppat.1000953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Okagaki LH, Wang Y, Ballou ER, O'Meara TR, Bahn Y-S, Alspaugh JA, Xue C, Nielsen K. 2011. Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot. Cell 10:1306–1316. 10.1128/EC.05179-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM. 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135:174–188. 10.1016/j.cell.2008.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chung D, Haas H, Cramer RA. 2012. Coordination of hypoxia adaptation and iron homeostasis in human pathogenic fungi. Front. Microbiol. 3:381. 10.3389/fmicb.2012.00381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Haas H. 2012. Iron: a key nexus in the virulence of Aspergillus fumigatus. Front. Microbiol. 3:28. 10.3389/fmicb.2012.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ibrahim AS, Gebremariam T, French SW, Edwards JE, Jr, Spellberg B. 2010. The iron chelator deferasirox enhances liposomal amphotericin B efficacy in treating murine invasive pulmonary aspergillosis. J. Antimicrob. Chemother. 65:289–292. 10.1093/jac/dkp426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leal SM, Jr, Roy S, Vareechon C, Carrion S deJesus Clark H, Lopez-Berges MS, diPietro A, Schrettl M, Beckmann N, Redl B, Haas H, Pearlman E. 2013. Targeting iron acquisition blocks infection with the fungal pathogens Aspergillus fumigatus and Fusarium oxysporum. PLoS Pathog. 9(7):e1003436. 10.1371/journal.ppat.1003436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Spellberg B, Ibrahim AS, Chin-Hong PV, Kontoyiannis DP, Morris MI, Perfect JR, Fredricks D, Brass EP. 2012. The Deferasirox-AmBisome Therapy for Mucormycosis (DEFEAT Mucor) study: a randomized, double-blinded, placebo-controlled trial. J. Antimicrob. Chemother. 67:715–722. 10.1093/jac/dkr375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aimanianda V, Latgé J-P. 2010. Problems and hopes in the development of drugs targeting the fungal cell wall. Expert Rev. Anti Infect. Ther. 8:359–364. 10.1586/eri.10.13 [DOI] [PubMed] [Google Scholar]
- 117.Munro CA. 2013. Chitin and glucan, the yin and yang of the fungal cell wall, implications for antifungal drug discovery and therapy. Adv. Appl. Microbiol. 83:145–172. 10.1016/B978-0-12-407678-5.00004-0 [DOI] [PubMed] [Google Scholar]
- 118.Marchetti O, Entenza JM, Sanglard D, Bille J, Glauser MP, Moreillon P. 2000. Fluconazole plus cyclosporine: a fungicidal combination effective against experimental endocarditis due to Candida albicans. Antimicrob. Agents Chemother. 44:2932–2938. 10.1128/AAC.44.11.2932-2938.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Marchetti O, Moreillon P, Glauser MP, Bille J, Sanglard D. 2000. Potent synergism of the combination of fluconazole and cyclosporine in Candida albicans. Antimicrob. Agents Chemother. 44:2373–2381. 10.1128/AAC.44.9.2373-2381.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48:959–976. 10.1046/j.1365-2958.2003.03495.x [DOI] [PubMed] [Google Scholar]
- 121.Fuchs BB, Mylonakis E. 2009. Our paths might cross: the role of the fungal cell wall integrity pathway in stress response and cross talk with other stress response pathways. Eukaryot. Cell 8:1616–1625. 10.1128/EC.001932-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walker LA, Gow NAR, Munro CA. 2010. Fungal echinocandin resistance. Fungal Genet. Biol. 47:117–126. 10.1016/j.fgb.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Munro CA, Selvaggini S, de Bruijn I, Walker L, Lenardon MD, Gerssen B, Milne S, Brown AJP, Gow NAR. 2007. The PKC, HOG and Ca2+ signalling pathways co-ordinately regulate chitin synthesis in Candida albicans. Mol. Microbiol. 63:1399–1413. 10.1111/j.1365-2958.2007.05588.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.LaFayette SL, Collins C, Zaas AK, Schell WA, Betancourt-Quiroz M, Gunatilaka AA, Perfect JR, Cowen LE. 2010. PKC signaling regulates drug resistance of the fungal pathogen Candida albicans via circuitry comprised of Mkc1, calcineurin, and Hsp90. PLoS Pathog. 6(8):e1001069. 10.1371/journal.ppat.1001069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Reinoso-Martin C, Schuller C, Schuetzer-Muehlbauer M, Kuchler K. 2003. The yeast protein kinase C cell integrity pathway mediates tolerance to the antifungal drug caspofungin through activation of Slt2p mitogen-activated protein kinase signaling. Eukaryot. Cell 2:1200–1210. 10.1128/EC.2.6.1200-1210.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Perlin DS. 2011. Current perspectives on echinocandin class drugs. Future Microbiol. 6:441–457. 10.2217/fmb.11.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Perlin DS. 2007. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10:121–130. 10.1016/j.drup.2007.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cowen LE. 2009. Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog. 5(8):e1000471. 10.1371/journal.ppat.1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309:2185–2189. 10.1126/science.1118370 [DOI] [PubMed] [Google Scholar]
- 130.Cowen LE, Steinbach WJ. 2008. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot. Cell 7:747–764. 10.1128/EC.00041-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE. 2009. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 5(7):e1000532. 10.1371/journal.ppat.1000532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist S. 2006. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot. Cell 5:2184–2188. 10.1128/EC.00274-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Steinbach WJ, Singh N, Miller JL, Benjamin DK, Jr, Schell WA, Heitman J, Perfect JR. 2004. In vitro interactions between antifungals and immunosuppressants against Aspergillus fumigatus isolates from transplant and nontransplant patients. Antimicrob. Agents Chemother. 48:4922–4925. 10.1128/AAC.48.12.4922-4925.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Steinbach WJ, Cramer RA, Jr, Perfect BZ, Henn C, Nielsen K, Heitman J, Perfect JR. 2007. Calcineurin inhibition or mutation enhances cell wall inhibitors against Aspergillus fumigatus. Antimicrob. Agents Chemother. 51:2979–2981. 10.1128/AAC.01394-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cowen LE, Singh SD, Köhler JR, Collins C, Zaas AK, Schell WA, Aziz H, Mylonakis E, Perfect JR, Whitesell L, Lindquist S. 2009. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc. Natl. Acad. Sci. U. S. A. 106:2818–2823. 10.1073/pnas.0813394106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Del Poeta M, Cruz MC, Cardenas ME, Perfect JR, Heitman J. 2000. Synergistic antifungal activities of bafilomycin A(1), fluconazole, and the pneumocandin MK-0991/caspofungin acetate (L-743,873) with calcineurin inhibitors FK506 and L-685,818 against Cryptococcus neoformans. Antimicrob. Agents Chemother. 44:739–746. 10.1128/AAC.44.3.739-746.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kraus PR, Fox DS, Cox GM, Heitman J. 2003. The Cryptococcus neoformans MAP kinase Mpk1 regulates cell integrity in response to antifungal drugs and loss of calcineurin function. Mol. Microbiol. 48:1377–1387. 10.1046/j.1365-2958.2003.03508.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yamada H, Watanabe T, Kato K, Mochizuki H. 1997. Fungicidal mechanism of action of D0870 against Cryptococcus neoformans under acidic conditions. Antimicrob. Agents Chemother. 41:2710–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Marr KA, Rustad TR, Rex JH, White TC. 1999. The trailing end point phenotype in antifungal susceptibility testing is pH dependent. Antimicrob. Agents Chemother. 43:1383–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Khlif M, Bogreau H, Michel-Nguyen A, Ayadi A, Ranque S. 2010. Trailing or paradoxical growth of Candida albicans when exposed to caspofungin is not associated with microsatellite genotypes. Antimicrob. Agents Chemother. 54:1365–1368. 10.1128/AAC.00530-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Parsons AB, Brost RL, Ding H, Li Z, Zhang C, Sheikh B, Brown GW, Kane PM, Hughes TR, Boone C. 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22:62–69. 10.1038/nbt919 [DOI] [PubMed] [Google Scholar]
- 142.Cornet M, Gaillardin C, Richard ML. 2006. Deletions of the endocytic components VPS28 and VPS32 in Candida albicans lead to echinocandin and azole hypersensitivity. Antimicrob. Agents Chemother. 50:3492–3495. 10.1128/AAC.00391-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Homann OR, Dea J, Noble SM, Johnson AD. 2009. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 5(12):e1000783. 10.1371/journal.pgen.1000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sahli S, Boulahfa S, Cornet M. 2012. pH signaling inhibition in Candida albicans leads to enhanced activity and restore fungicidal effect of ergosterol synthesis inhibitors, abstr M983 Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., San Francisco, CA. American Society for Microbiology, Washington, DC [Google Scholar]
- 145.Jung K-W, Strain AK, Nielsen K, Jung K-H, Bahn Y-S. 2012. Two cation transporters Ena1 and Nha1 cooperatively modulate ion homeostasis, antifungal drug resistance, and virulence of Cryptococcus neoformans via the HOG pathway. Fungal Genet. Biol. 49:332–345. 10.1016/j.fgb.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Abadio AKR, Kioshima ES, Teixeira MM, Martins NF, Maigret B, Felipe MSS. 2011. Comparative genomics allowed the identification of drug targets against human fungal pathogens. BMC Genomics 12:75. 10.1186/1471-2164-12-75 [DOI] [PMC free article] [PubMed] [Google Scholar]