Abstract
Entamoeba histolytica is a protozoan parasite that causes amoebic dysentery and liver abscess. E. histolytica relies on motility, phagocytosis, host cell adhesion, and proteolysis of extracellular matrix for virulence. In eukaryotic cells, these processes are mediated in part by phosphatidylinositol 3-kinase (PI3K) signaling. Thus, PI3K may be critical for virulence. We utilized a functional genomics approach to identify genes whose products may operate in the PI3K pathway in E. histolytica. We treated a population of trophozoites that were overexpressing genes from a cDNA library with a near-lethal dose of the PI3K inhibitor wortmannin. This screen was based on the rationale that survivors would be overexpressing gene products that directly or indirectly function in the PI3K pathway. We sequenced the overexpressed genes in survivors and identified a cDNA encoding a Rap GTPase, a protein previously shown to participate in the PI3K pathway. This supports the validity of our approach. Genes encoding a coactosin-like protein, EhCoactosin, and a serine-rich E. histolytica protein (SREHP) were also identified. Cells overexpressing EhCoactosin or SREHP were also less sensitive to a second PI3K inhibitor, LY294002. This corroborates the link between these proteins and PI3K. Finally, a mutant cell line with an increased level of phosphatidylinositol (3,4,5)-triphosphate, the product of PI3K activity, exhibited increased expression of SREHP and EhCoactosin. This further supports the functional connection between these proteins and PI3K in E. histolytica. To our knowledge, this is the first forward-genetics screen adapted to reveal genes participating in a signal transduction pathway in this pathogen.
INTRODUCTION
Entamoeba histolytica is an enteric protozoan parasite that causes amoebiasis and amoebic liver abscess in humans (1). It is prevalent in developing countries that cannot prevent its fecal-oral spread. E. histolytica enters the human host upon ingestion of water or food contaminated with environmentally stable cysts. After passing through the stomach, excystation leads to the release of trophozoites, which migrate to the bowel lumen for colonization. In 10% of infected individuals, infection can progress from a noninvasive stage to an invasive stage (2), during which the parasite binds to and destroys colonic epithelium. From here, the parasites enter the circulatory system and translocate to other organs. The most common site of extraintestinal infection is the liver, characterized by the formation of amebic liver abscess (ALA).
E. histolytica relies on cell motility, phagocytosis, proteolysis of host extracellular matrix, and host cell adhesion for virulence (3). In other eukaryotic cells, these processes are mediated in part by phosphatidylinositol 3-kinase (PI3K) signaling (4). PI3Ks phosphorylate phosphatidylinositol (PI) and its derivatives to generate signaling lipids such as phosphatidylinositol 3-phosphate (PI3P), phosphatidylinositol 3,4-bisphosphate [PI(3,4)P2], phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2], and phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3] (reviewed in reference 5). These lipids propagate a signal by interacting with proteins harboring specific domains, such as the FYVE finger domains, which bind PI3P, and pleckstrin homology (PH) domains, which bind PI(3,4,5)P3 and PI(3,4)P2 (reviewed in reference 6). The activity of PI3K can be countered by the action of phosphatases, such as phosphatase and tensin homolog (PTEN), which dephosphorylate phosphoinositides (reviewed in reference 7).
In E. histolytica, the products of PI3Ks have been studied using glutathione S-transferase (GST)- and green fluorescent protein (GFP)-tagged versions of FYVE and PH domains (8–10). These studies have shown that PI3P localizes to E. histolytica phagosomes (8, 9) and PI(3,4,5)P3 localizes to both pseudopods and phagosomes (10). Studies using small-molecule inhibitors of PI3K, such as LY294002 and wortmannin, have also been carried out. Treatment of E. histolytica trophozoites with wortmannin inhibits directional cell polarization (11), motility, actin cytoskeletal rearrangements, proteolytic activity, and the development of ALA in an animal model of disease (12). Exposure to either LY294002 (13) or wortmannin (14) inhibits pinocytosis of a fluorescent fluid-phase marker, fluorescein isothiocyanate (FITC)-dextran, and disrupts phagocytosis (8, 10, 14) and adhesion to host cells in a dose-dependent manner (8).
Several unique aspects of PI3K activity in E. histolytica make it worthy of study. First, expression of PI3K is higher in virulent E. histolytica than in nonvirulent E. dispar (15). Second, compared to mammalian cells, E. histolytica has above average levels of PI(3,4,5)P3 in the plasma membrane (10). Third, unlike in mammalian cells (16), serum withdrawal does not affect the steady-state level of PI(3,4,5)P3 in E. histolytica (10). Fourth, the products of PI3K, PI3P and PI(3,4,5)P3, localize to early-forming or newly sealed phagosomes (8–10). In contrast, localization of PI3P to phagosomes in mammalian cells is observed only after their closure (17). Finally, although not much is known about encystation in E. histolytica, wortmannin inhibits encystation in a related reptilian pathogen, E. invadens (18). Therefore, it is possible that E. histolytica encystation also requires PI3K activity. Thus, understanding the unique role of phosphoinositides and PI3K signaling in E. histolytica may provide insight into infection.
In other systems, genomewide overexpression has been used to identify targets of small-molecule drugs. For example, Butcher et al. (19) used overexpression to identify genes regulating rapamycin sensitivity and, hence, TOR kinase signaling. Similarly, genomewide overexpression was used to define targets of a kinase inhibitor, phenylaminopyrimidine (20), and two antifungal drugs, tunicamycin and soraphen (21). Sequencing and annotation of the E. histolytica genome (22, 23) have enabled the development of whole-genome approaches to assign functions to genes. However, to date, the only forward genetics approach that has been applied to E. histolytica is a recent overexpression screen that identified genes that negatively regulate phagocytosis (24).
In the current study, we have adapted an overexpression-based chemical genomic approach (25) to uncover genes that may directly or indirectly participate in PI3K signaling. Specifically, we applied a near-lethal dose of wortmannin to a population of cells that had been transfected with an E. histolytica cDNA library to select for cells that were less sensitive to the drug. We then identified the genes that were overexpressed in the survivors. The screen was based on the hypothesis that cells in which wortmannin toxicity was genetically suppressed may be overexpressing genes that directly or indirectly play a role in PI3K signaling.
MATERIALS AND METHODS
Strains and culture conditions.
Entamoeba histolytica trophozoites (strain HM-1:1MSS) were cultured axenically in TYI-S-33 medium (26) in 15-ml glass screw-cap tubes at 37°C. The methods used to generate E. histolytica cell lines that overexpress a GFP-tagged pleckstrin homology (PH) domain from mammalian Bruton's tyrosine kinase (GFP-PHBtk), EhLimA, or luciferase (here referred to as Eh209) are described elsewhere (10, 24, 27). The GFP-PHBtk and Eh209 transgenic cell lines were maintained in TYI-S-33 medium supplemented with 6 μg/ml G418 and 15 μg/ml hygromycin. Expression of GFP-PHBtk or luciferase was induced by the addition of 5 μg/ml tetracycline to the culture medium 24 h prior to performing assays. The EhLimA-expressing cell line was maintained in a medium supplemented with 23 μg/ml hygromycin, and the drug concentration was increased to 46 μg/ml for 1 week prior to conducting experiments.
The generation of a population of E. histolytica cells transfected with a cDNA library in the E. histolytica expression vector pAH-DEST is described elsewhere (24). Briefly, the cDNA library was constructed using Invitrogen Custom Services (Invitrogen, Carlsbad, CA). RNA was isolated from approximately 2.4 × 107 trophozoites using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Poly(A)+ RNA was selected from 2 mg total RNA and used to generate a primary, uncut, 7-fold normalized cDNA library directionally cloned into the Gateway-compatible entry vector pENTR222(f1−). Proprietary 5′ cap-binding technology was used to select and enrich for full-length clones. The final titer of the amplified library was 3.9 × 106 CFU/ml. Twenty-four random clones, with a minimum of 20 different genes, were analyzed for the presence of inserts. All 24 clones had inserts with an average size of 1.1 kb.
The LR Clonase system (Invitrogen) was used to transfer cDNA inserts from pENTR222 to pAH-DEST (28), a Gateway-compatible E. histolytica expression vector (kind gift from C. A. Gilchrist and W. A. Petri, Jr., Department of Medicine, Division of Infectious Diseases and International Health, University of Virginia, Charlottesville, VA). This plasmid is episomal in nature and confers hygromycin resistance to transfectants. Expression from this plasmid is driven by upstream ferredoxin regulatory sequences. The cDNA library in pAH-DEST was transfected into HM1:IMSS trophozoites using standard protocols (10, 29). Transfectants were maintained by the addition of 23 μg/ml hygromycin to the medium.
E. histolytica trophozoites were transfected with expression plasmids encoding SREHP or EhCoactosin using standard transfection protocols (10, 29). These 2nd-generation SREHP and EhCoactosin overexpressors were maintained in a medium supplemented with 23 μg/ml hygromycin, and the drug concentration was increased to 46 μg/ml for 1 week prior to conducting experiments.
Measurement of wortmannin toxicity and chemical genomics screen.
Log-phase trophozoites (6 × 104 cells) were seeded into 39 ml of TYI-S-33 medium containing a range of concentrations of wortmannin or its diluent, dimethyl sulfoxide (DMSO), and grown for 3 days in 15-ml glass screw-cap tubes at 37°C. Fresh TYI-S-33 medium containing the drug or the diluent was supplied to the cells every 24 h over the course of treatment. Cell viability was measured by microscopy with Trypan blue exclusion. A near-lethal concentration of wortmannin (2 mM) was applied to the library of overexpressing trophozoites for 3 days with a medium change every 24 h.
Isolation of episomes from survivors and sequencing.
Total DNA was isolated from the population of cells that survived wortmannin selection or from the parental population of cells not exposed to wortmannin (control) using the Wizard genomic DNA purification kit (Promega, Madison, WI). The isolated DNA was transformed into Escherichia coli (XL10 Gold; Stratagene, La Jolla, CA) by standard transformation protocols. The resulting ampicillin-resistant bacterial colonies were randomly chosen from each condition (unselected control or wortmannin selection), transferred to the wells of 96-well plates containing LB medium supplemented with ampicillin (100 μg/ml) and glycerol (50%, vol/vol), and stored at −80°C. Frozen samples were transferred to the Arizona State University BioDesign Institute (Tempe, AZ) for automated plasmid isolation and sequencing.
Analysis of expression by quantitative real-time PCR (RT-qPCR).
Cell lines were grown to mid-log phase (3 × 105 cells/ml). Total RNA was isolated from these cell lines with TRIzol reagent (Invitrogen, Carlsbad, CA), and 2 μg of RNA was treated with DNase I for 30 min at 37°C. The cDNAs were synthesized with oligo(dT) and Superscript III reverse transcriptase (Invitrogen) at 50°C for 2 h.
Expression of SREHP or EhCoactosin transcripts was measured by RT-qPCR using the RT2 SYBR green fluor qPCR mastermix (SABiosciences, Frederick, MD) and the appropriate primers for SREHP (forward primer, 5′-GCTGCATCAAGTCCATTCATTG-3′; reverse primer, 5′-GATAGAAATATTTATCCCAACTAAAGAC-3′) and EhCoactosin (forward primer, 5′-TTAGTGAAGTTGCTGGCCCAGTTG-3′; reverse primer, 5′-GCATCTTCCTTAAGGGCAGCTTTG-3′). Normalization of the levels of cDNA was carried out by comparing expression to that of a housekeeping gene, the small subunit rRNA (ssRNA) (accession no. X61116) (30) (forward primer, 5′-AGGCGCGTAAATTACCCACTTTCG-3′; reverse primer, 5′-CACCAGACTTGCCCTCCAATTGAT-3′). Normalization ratios were calculated using the Pfaffl method (31). At least two independent biological replicates were evaluated. For each cDNA, three technical replicates were performed and the values averaged. The efficiency of each primer pair was assessed by use of a dilution series. In all cases, efficiency values were ≥88% and R2 values were ≥0.935.
LY294002 and cycloheximide sensitivity assays.
To test for LY294002 sensitivity, log-phase trophozoites (2 × 104 cells) were seeded into 13 ml of TYI-S-33 medium containing 50 or 25 μM LY294002 or an equivalent volume of DMSO (diluent control) and grown for 3 days in 15-ml glass screw-cap tubes at 37°C. Fresh TYI-S-33 medium containing the drug or the diluent was supplied to the cells every 24 h over the course of treatment. Cell viability was measured by microscopy with Trypan blue exclusion.
The cycloheximide sensitivity assay was employed as described previously (24). Briefly, trophozoites (3 × 104 cells) were seeded into 13 ml of TYI-S-33 medium supplemented with 100 nM cycloheximide or an equivalent volume of PBS (diluent control) and incubated at 37°C for 48 h. Cell viability was assessed by microscopy with Trypan blue exclusion.
Whole-cell lipid extraction and PI(3,4,5)P3 dot blots.
Total lipid was extracted from trophozoites according to the methods of Gray et al. (32). Briefly, 1 × 106 cells were washed twice with phosphate-buffered saline (PBS). Lipids were precipitated with 5 ml of 0.5 M trichloroacetic acid (TCA) and centrifuged at 500 × g for 5 min at 4°C. The pellets were washed with 3 ml of 5% (wt/vol) TCA–1 mM EDTA and centrifuged at 500 × g for 5 min. To facilitate neutral lipid extraction, 3 ml of methanol-chloroform (2:1) was added to the pellets, and the mixture was vortexed 3 times over a period of 10 min at room temperature. The extracted lipids were centrifuged at 500 × g for 5 min at 4°C. To facilitate acidic lipid extraction, 2.25 ml methanol-chloroform-12.1N HCl (80:40:1) was added, and the mixture was vortexed 4 times during 15 min at room temperature and centrifuged at 500 × g. The resulting supernatant was subjected to a phase split by the addition of 750 μl chloroform and 1.35 ml 0.1N HCl. The solution was centrifuged at 500 × g for 5 min at 4°C. After centrifugation, the organic phase was collected and vacuum dried, and the pellets were used for lipid dot blot analysis.
Lipids were spotted onto Hybond-C nitrocellulose membrane. The membrane was blocked in 1.5% (wt/vol) fatty acid-free bovine serum albumin (BSA) at room temperature for 1 h and probed with mouse anti-PI(3,4,5)P3 antibody (1:1,000 dilution) (Echelon Biosciences, Salt Lake City, UT) followed by incubation with secondary antibody, i.e., peroxidase-conjugated goat anti-mouse IgG (1:2,000 dilution) (Cappel; ICN Pharmaceuticals, Costa Mesa, CA). Immunoblots were visualized using the enhanced chemiluminescence Western blotting detection system (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions. Semiquantitative densitometric analyses of immunoblots were performed using ImageJ software (version 1.42q; U.S. National Institutes of Health, Bethesda, MD).
Measurement of motility.
Motility assays were carried out according to the methods of Zaki et al. (33). Briefly, 8 ml of complete TYI-S-33 medium, supplemented with 0.75% (wt/vol) agarose, was poured into 60-mm petri dishes and allowed to solidify at room temperature. A trough (2 by 30 mm) was cut into the solidified medium to serve as the motility chamber. Trophozoites were incubated in serum-free media for 1 h at 37°C. Following this incubation, 5 × 105 cells were placed in the trough of the motility chamber. A coverslip (22 by 40 mm) was placed over the trough, and the plate was incubated at 37°C for 3 h in 5% CO2. After 3 h, images were captured using an LSM510 confocal microscope (Carl Zeiss MicroImaging, Thornwood, NY). For each trial, at least 3 measurements of the distance between the trough and the leading edge of the population of cells were taken using Zeiss LSM510 image analysis software.
Statistical analyses.
All values are given as means ± standard deviations (SD) from at least 3 trials. To compare means, statistical analyses were performed using GraphPad Instat V.3 with a one-way analysis of variance (ANOVA) and a Tukey-Kramer multiple-comparison test. To compare the composition of the gene set obtained before and after selection, we used a statistical Z-test. In all cases, P values less than 0.001 (***) were considered highly statistically significant, while P values less than 0.01 (**) or 0.05 (*) were considered statistically significant.
In silico analyses.
Structural and functional amino acid residues and domains in the EhCoactosin protein were identified using ExPASy ScanProsite (34). Cartoon renderings of the EhCoactosin protein were generated using the MyDomains-Image Creator in ExPASy PROSITE. The Clustal W algorithm was used for comparison of coactosin protein sequences (MacVector 9.0; MacVector, Cary, NC).
RESULTS
Development of chemical genomic screen to enrich for mutants hyposensitive to wortmannin.
Wortmannin is a cell-permeant, small-molecule metabolite isolated from the fungus Talaromyces wortmanni (35). In mammalian cells, wortmannin can irreversibly inhibit PI3K at low nanomolar concentrations (7). To evaluate wortmannin toxicity in E. histolytica, we exposed trophozoites to a range of concentrations of wortmannin over 3 days. In E. histolytica, PI3P and PI(3,4,5)P3 staining were eliminated by 60 nM (8) and 500 nM (10) wortmannin, respectively. Since we were interested in completely disabling PI3K, we tested a range of concentrations from 500 nM (data not shown) to 4 mM wortmannin. To ensure the presence of active inhibitor, the spent medium was exchanged with fresh wortmannin-containing medium daily. The viability of the trophozoites was assessed and reported as a percentage of untreated control cells. Near 100% death of the trophozoite population was achieved with 2 mM (∼1.5% viable after 3 days), 3 mM (∼0.9% viable after 3 days), and 4 mM (∼0.5% viable after 3 days) wortmannin (Fig. 1). Furthermore, there was no statistically significant difference when 2 mM wortmannin toxicity was compared to that of higher doses. Cell viability was not significantly affected by the diluent, DMSO (Fig. 1). This suggests that cell death was specific to the presence of wortmannin. To minimize off-target effects of wortmannin and to maximize the chances of identifying genes authentically connected to PI3K signaling, 2 mM wortmannin treatment was chosen for the chemical genomics screen.
FIG 1.
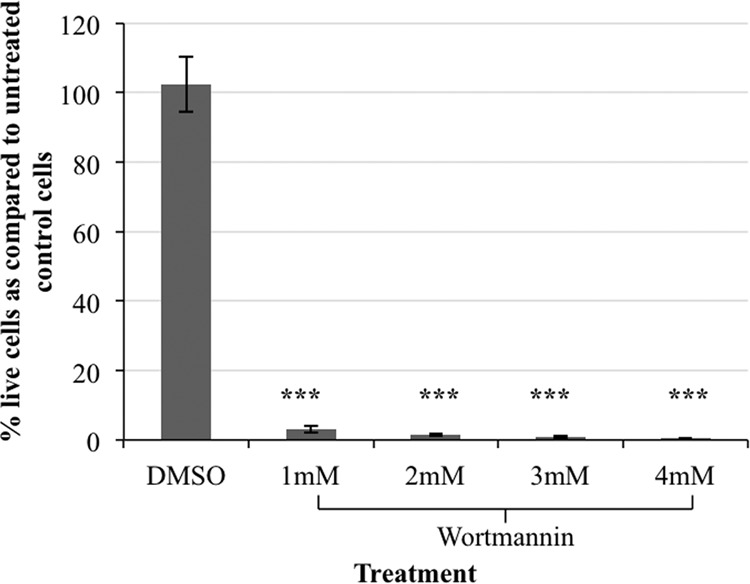
Wortmannin is toxic to E. histolytica. Trophozoites (2 × 104 cells) were exposed to a range of concentrations of wortmannin or its diluent, dimethyl sulfoxide (DMSO), or an equivalent volume of media (control) and grown for 3 days. Cell viability was assessed and expressed as a percentage of untreated control cells. The data represent the means ± SD from 3 trials (***, P < 0.001). Treatment with 2 mM or higher concentrations of wortmannin resulted in the death of nearly 100% of the cells.
Screening with wortmannin identifies genes that may function in PI3K signaling.
E. histolytica cells were transfected with a cDNA overexpression library constructed in the episomal, constitutive expression vector pAH-DEST (28) as described previously (24). The transgenic population of cells was exposed to 2 mM wortmannin over a 3-day period. The surviving cells were allowed to recover for a 48-h period. Episomes from the surviving population were purified and amplified in E. coli. The cDNAs were identified by sequencing. As a control, episomes were also isolated and sequenced from the transgenic population of cells that was not subjected to this selection. Table 1 shows the identity and frequency of cDNAs isolated from trophozoites that survived selection compared to those recovered randomly from a population of overexpressing cells not subjected to selection. To determine if the set of genes enriched after selection was significantly different from that in the control population, we used a statistical Z-test. The statistical difference between genes isolated by selection and those isolated randomly from a control population approached significance (P = 0.05262). This analysis suggests a nonfortuitous enrichment of specific genes by wortmannin treatment.
TABLE 1.
Isolation of cDNAs from E. histolytica overexpressers with and without selection with 2 mM wortmannin
| GI no. | Gene name | No. of isolatesa |
|
|---|---|---|---|
| Control | Selected | ||
| XM_645834.2 | Actin binding protein, EhCoactosin | 2 | 13 |
| AY141199.1 | Ehapt2 retrotransposon | 1 | 12 |
| XM_648457.2 | 40S ribosomal protein S14, putative | 4 | 9 |
| XM_646071.2 | Ras family GTPase, EhRap2 | 1 | 8 |
| XM_647007.2 | 60S ribosomal protein L10, putative | 1 | 7 |
| XM_645248.2 | 60S ribosomal protein L7, putative | 5 | 7 |
| XM_647452.2 | 60S ribosomal protein L17, putative | 3 | 5 |
| M80910.1 | SREHP | 1 | 4 |
| XM_650080.1 | Alcohol dehydrogenase, putative | 7 | 3 |
| XM_650825.1 | 40S ribosomal protein S25, putative | 4 | 3 |
| XM_644144.2 | Hypothetical protein, H644 | 6 | 2 |
| AY141200.1 | Ehapt2 retrotransposon | 2 | 2 |
| XM_646441.2 | 40S ribosomal protein S12, putative | 0 | 2 |
| XM_652064.2 | 40S ribosomal protein S7, putative | 0 | 1 |
| XM_647340.1 | 60S ribosomal protein L7a, putative | 0 | 1 |
| X90911.1 | Profilin | 0 | 1 |
| XM_651795.2 | 60S ribosomal protein L7, putative | 0 | 1 |
| XM_650280.2 | Hypothetical protein, BAR and SH3 domain-containing protein | 0 | 1 |
| XM_644485.1 | 40S ribosomal protein S25, putative | 1 | 1 |
| XM_651024.2 | 60S ribosomal protein L4, putative | 1 | 1 |
| XM_645170.2 | Serine protease inhibitor, putative | 2 | 1 |
| XM_644082.2 | 60S ribosomal protein L14, putative | 2 | 1 |
| XM_645695.2 | 60S ribosomal protein L13, putative | 4 | 1 |
| XM_644069.2 | Enolase, putative | 4 | 1 |
| XM_643252.2 | 60S ribosomal protein L15, putative | 5 | 1 |
| XM_647370.2 | 60S ribosomal protein L9 | 6 | 1 |
| Z48752.1 | Alcohol dehydrogenase 3 | 6 | 0 |
| XM_645775.2 | Hypothetical protein | 9 | 0 |
| XM_646205.2 | 60S ribosomal protein L23, putative | 1 | 0 |
| XM_651193.2 | 40S ribosomal protein S9, putative | 1 | 0 |
| XM_650207.2 | C2 domain containing protein | 3 | 0 |
| XM_651611.2 | Vacuolar ATP synthase subunit E, putative | 1 | 0 |
| XM_643162.2 | Serine-rich protein | 1 | 0 |
| XM_651764.2 | 60S ribosomal protein L11, putative | 1 | 0 |
| XM_648038.2 | Hypothetical protein, mRNA | 1 | 0 |
| XM_001913740.1 | 40S ribosomal protein S7, putative | 1 | 0 |
| XM_645636.1 | Rho GTPase activating protein, putative | 1 | 0 |
| XM_652029.2 | Hypothetical protein | 1 | 0 |
| Total | 89 | 90 | |
E. histolytica overexpressors were left untreated (control) or were subjected to selection with 2 mM wortmannin (selected).
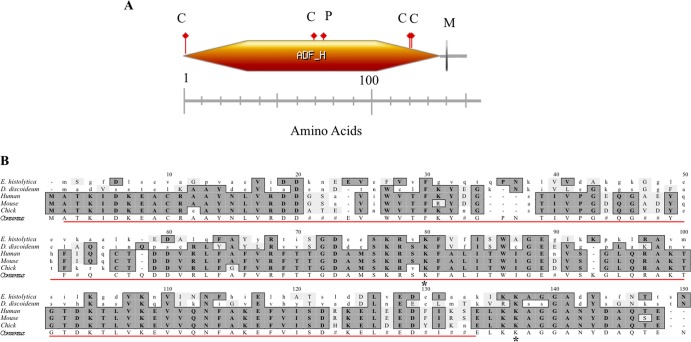
The cDNA encoding an actin-binding protein with an actin depolymerization factor homology (ADF-H) domain (EhCoactosin; XM_645834.2) was the most highly enriched of the cDNAs. It was present in 14.4% of the bacterial clones that had been transformed with episomes isolated from trophozoite survivors. The corresponding protein is predicted to be 148 amino acid long (16.2 kDa) and was previously identified in a proteomic analysis of purified phagosomes from E. histolytica (36). Like other coactosins, EhCoactosin belongs to the ADF/cofilin family of proteins and contains a putative F-actin binding domain. It is also predicted to possess a putative myristoylation, several casein kinase II sites, and protein kinase C phosphorylation sites (Fig. 2A). These phosphorylation sites indicate a possible role for coactosin in cell signaling in E. histolytica. Cofilin, one of the founding members of the ADF/cofilin family, has been shown to be a downstream effector of PI3K activity (37). However, to our knowledge, this is the first study to reveal a functional connection between a coactosin-like protein and PI3K in any system.
FIG 2.
Domains found in EhCoactosin and sequence comparison of coactosin from E. histolytica to those of other species. (A) EhCoactosin is an F-actin binding protein with a postulated molecular mass of 16.2 kDa (148 amino acids). It is predicted to have an actin depolymerization factor homology (ADF-H) domain (orange hexagon), an N-myristoylation site (gray vertical line labeled M), 4 casein kinase II phosphorylation sites (red flags labeled C), and a protein kinase C phosphorylation site (red flag labeled P). (B) The predicted amino acid sequence of EhCoactosin was aligned with coactosin from other organisms using ClustalW multiple sequence alignment, v1.4 (MacVector v9.0). Conserved amino acids are shaded, and the consensus sequence is indicated below the aligned sequence (#, conservation but no consensus). The E. histolytica ADF-H domain is indicated by the red line. Asterisks represent the lysine residues Lys75 and Lys131 of human coactosin that are essential for binding to F-actin and 5-lipoxygenase, respectively.
A sequence alignment revealed that EhCoactosin shares high homology with coactosins from other systems (Fig. 2B). Importantly, Lys75 of human coactosin, which is required for interaction with F-actin, is conserved in EhCoactosin, suggesting that the E. histolytica protein is an authentic actin-binding protein. The 129LKKAGG134 sequence motif, specifically Lys131, on the C-terminal end of human coactosin is necessary for interaction with 5-lipoxygenase (5-LO), an enzyme that is involved in leukotriene biosynthesis (38). Although E. histolytica does not appear to possess 5-LO-like proteins, EhCoactosin harbors a similar motif at its C terminus which may facilitate its interaction with other proteins.
The cDNA encoding the serine-rich E. histolytica protein (SREHP; M80910.0) was present in 4.4% of the bacterial clones that had been transfected with the episomes isolated from trophozoite survivors. It is proposed to be a cell surface receptor that facilitates host-parasite or parasite-parasite interaction through chemoattraction (39). Its potential as a vaccine target has been established (40). SREHP is a cell surface protein, a potential chemoattractant, and is itself phosphorylated. These are all hallmarks of signaling proteins. Thus, it was intriguing to isolate it in a screen for protein partners of PI3K signaling.
The cDNA encoding a Ras family GTPase (EhRap2; XM_646071.2) was isolated from 8.9% of bacterial clones that had been transfected with episomes from trophozoite survivors. Rap GTPases are known to participate in PI3K signaling in other systems (41, 42), supporting the validity of our screen. Because of its known connection to PI3K signaling, we did not conduct additional studies with EhRap2. The cDNA encoding the 40S ribosomal protein S14 (XM_648457.2), the 60S ribosomal protein L10 (XM_647007.2), and the ehapt2 retrotransposable element (AY141199.1) were isolated from 10%, 7.8%, and 13.3%, respectively, of bacterial clones that had been transfected with episomes from trophozoite survivors. These genes also were not chosen for further study, because overexpression of ribosomal proteins or retrotransposons may have profound nonspecific effects on protein synthesis and gene expression. Furthermore, several of the cDNAs encoding ribosomal proteins also were identified in the unselected control population (Table 1).
Authentication of EhCoactosin and SREHP.
EhCoactosin was selected for additional analysis, because it was the most abundant cDNA isolated in the screen and because it may be the first coactosin-like protein to exhibit a functional connection to PI3K. SREHP was selected because of its potential as a signaling molecule and its importance as a vaccine target. Expression vectors encoding EhCoactosin and SREHP, isolated in the original screen, were transfected into wild-type trophozoites to construct 2nd-generation cell populations overexpressing these proteins. Overexpression of SREHP and EhCoactosin transcripts was confirmed by RT-qPCR (Fig. 3A and B). SREHP and EhCoactosin transcripts exhibited 3.6- and 7.1-fold higher abundance, respectively, in the 2nd-generation transgenic cell lines than in the parental controls (Fig. 3A and B).
FIG 3.
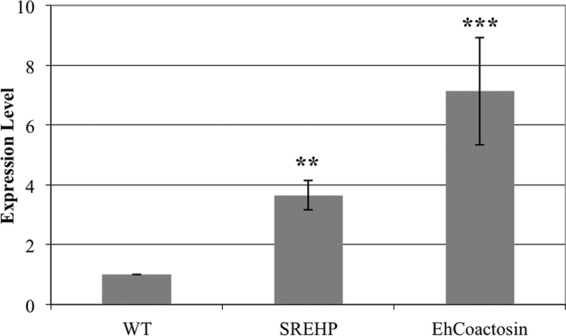
RT-qPCR confirms overexpression of EhCoactosin and SREHP. RNA from untransfected (wild type; WT) log-phase E. histolytica trophozoites or from trophozoites transfected with an expression vector encoding SREHP or EhCoactosin was used for RT-qPCR analysis of expression. The ssRNA gene was used as a loading control. SREHP and EhCoactosin were expressed at approximately 3.6 (±0.5)-fold and 7.1 (±1.8)-fold, respectively, above wild-type levels. The data represent the means ± SD from 2 biological replicates and 3 technical replicates per biological replicate (**, P < 0.01; ***, P < 0.001).
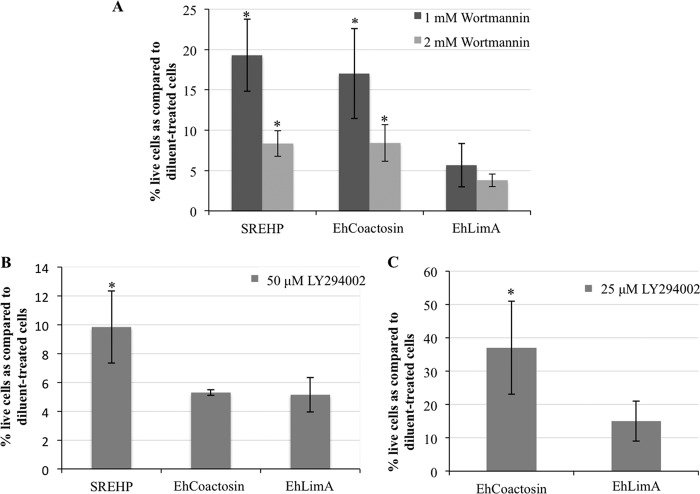
To validate the isolation of EhCoactosin and SREHP in the screen, we exposed the 2nd-generation transgenic cell lines to selection with 1 mM and 2 mM wortmannin for 3 days and reassessed toxicity. These cell lines exhibited significantly higher survival after exposure to wortmannin than control diluent-treated cells (Fig. 4A). To test if the decreased susceptibility to the drug was due simply to the presence of episomal DNA, we tested wortmannin toxicity in another cell line overexpressing an irrelevant protein, EhLimA, from the same nonintegrating expression vector (24). There was no significant increase in the number of survivors after wortmannin treatment in the EhLimA-expressing population (Fig. 4A). This suggests that the genetic suppression of wortmannin toxicity by EhCoactosin or SREHP overexpression is genuine.
FIG 4.
SREHP and EhCoactosin overexpressors are less susceptible to wortmannin and LY294002 toxicity. Transgenic trophozoites were exposed to 1 mM or 2 mM wortmannin (A), 50 μM LY294002 (B), or 25 μM LY294002 (C) for 3 days, after which viability was assessed. The data are reported as a percentage of diluent-treated control amoebae and represent the means ± SD from 3 trials (*, P < 0.05). (A) SREHP- and EhCoactosin-overexpressing cells exhibited increased survival in the presence of wortmannin compared to untransfected wild-type (WT) cells or another transgenic cell line overexpressing an irrelevant gene, EhLimA, from the same expression vector. (B) SREHP-overexpressing cells exhibited enhanced survival in the presence of 50 μM LY294002. (C) EhCoactosin-overexpressing cells exhibited enhanced survival in the presence of 25 μM LY204002.
If overexpression of EhCoactosin or SREHP authentically rescues E. histolytica cells from wortmannin toxicity, it is conceivable that overexpression should also confer resistance to other PI3K inhibitors. To test this, we quantified viability in the 2nd-generation cell lines exposed to 50 μM LY294002, a concentration known to strongly inhibit staining with a PI3P biosensor in E. histolytica (8). Cells overexpressing SREHP exhibited reduced sensitivity to LY294002 toxicity (Fig. 4B), supporting a functional connection between SREHP and PI3K signaling. However, overexpression of EhCoactosin did not protect cells from exposure to LY204002 at this concentration. One possibility was that our test with LY294002 was too stringent. Therefore, we assessed the viability of the EhCoactosin-expressing cell line after exposure to a lower concentration (25 μM) of LY294002. In this case, we observed increased survival of the EhCoactosin-expressing cell line (Fig. 4C) compared to diluent-treated control cells. This corroborates the isolation of EhCoactosin in the original screen and indicates that expression of SREHP is more efficient at protecting cells from LY294002 toxicity than expression of EhCoactosin.
EhCoactosin or SREHP could represent false positives if their overexpression leads to a nonspecific multidrug-resistant phenotype. To address this possibility, we examined the specificity of the response by testing the susceptibility of EhCoactosin- and SREHP-overexpressing cells to an unrelated small molecule, cycloheximide, as previously described (24). There was no significant difference in survival of the 2nd-generation transgenic and parental control cell lines (Fig. 5). This indicated that the isolation of EhCoactosin and SREHP from our original screen was not due to a multidrug resistance phenotype characterized by nonspecific extrusion of small molecules.
FIG 5.
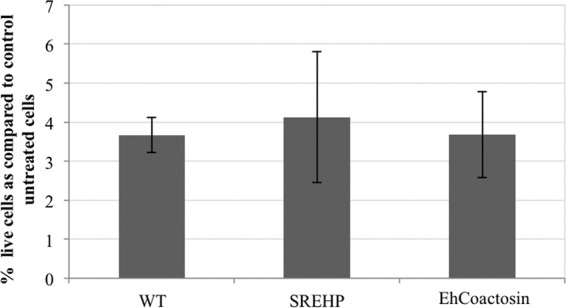
Cycloheximide is toxic to E. histolytica cells overexpressing SREHP and EhCoactosin. Transgenic trophozoites were exposed to 100 nM cycloheximide for 48 h, and viability was assessed. The data are presented as a percentage of untreated control cells and represent the means ± SD from 3 trials. Cycloheximide was toxic to both wild-type (WT) and transgenic amoebae. Therefore, cells overexpressing SREHP and EhCoactosin may not have a multidrug resistance phenotype.
Although SREHP has been characterized (39, 40, 43, 44), little is known about EhCoactosin, except that it is associated with E. histolytica phagosomes (36). In other systems, increased expression of coactosin correlates with enhanced cell motility (45, 46). Therefore, to verify EhCoactosin as an authentic coactosin family member, we performed a previously published under-agar motility assay (47) on the 2nd-generation EhCoactosin overexpressors. EhCoactosin-overexpressing cells traveled a significantly longer distance in a 3-h time frame than untransfected control trophozoites (Fig. 6), suggesting that this protein, like other coactosins, has a cell migration function in E. histolytica.
FIG 6.
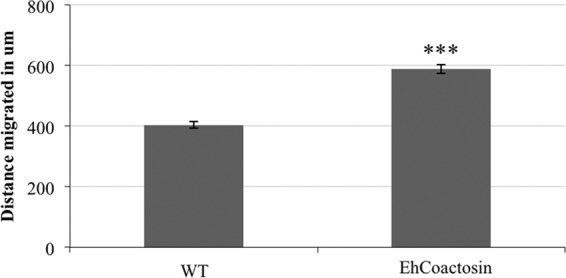
Cells overexpressing EhCoactosin display increased cell motility. A motility assay was performed on EhCoactosin overexpressors or untransfected wild-type (WT) cells. Images of cells moving under agar were captured using confocal microscopy. The maximum distance migrated by lead trophozoites was measured. The data represent the means ± SD from 3 trials (***, P < 0.001).
Alteration in the level of phosphatidylinositol (3,4,5)-triphosphate affects expression of EhCoactosin and SREHP.
We used an independent genetic test to affirm the connections between PI3K and EhCoactosin or SREHP. Previously, it was reported that a transgenic cell line overexpressing a GFP-tagged pleckstrin homology (PH) domain, from mammalian Bruton's tyrosine kinase (GFP-PHBtk), exhibited reduced phagocytosis (10), increased motility (10), and decreased levels of PI(4,5)P2 (48). Since the levels of PI(4,5)P2 and PI(3,4,5)P3 are tightly coordinated, we predicted that PI(3,4,5)P3 abundance also was altered in this cell line. Thus, we measured the level of PI(3,4,5)P3 using a lipid dot blot analysis of whole-cell extracts from untransfected parent cells and the GFP-PHBtk cell line. Compared to untransfected trophozoites, GFP-PHBtk-expressing cells exhibited a 2.6-fold increase in PI(3,4,5)P3 (Fig. 7A and B). To determine if the increase in PI(3,4,5)P3 was due to the presence of episomal expression plasmids, we also measured PI(3,4,5)P3 in a control cell line (Eh209) expressing an irrelevant protein, luciferase, from the same pGIR209 (27) expression vector. The level of PI(3,4,5)P3 was not significantly higher in the control transgenic cell line, suggesting that the quantity of this signaling lipid is authentically higher in GFP-PHBtk-expressing cells. Given that PI(3,4,5)P3 is the product of PI3K, increased levels of this lipid may be the genetic equivalent of enhanced PI3K activity.
FIG 7.
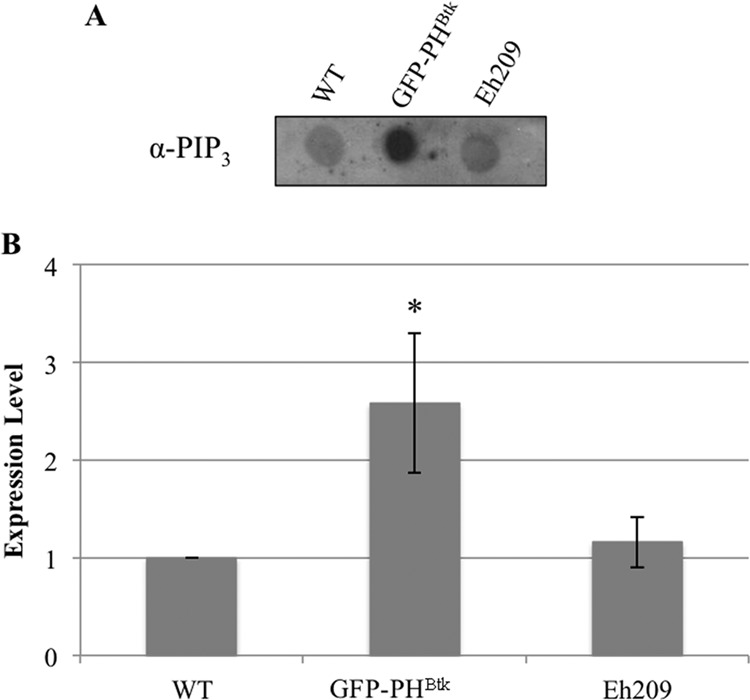
GFP-PHBtk overexpressors display altered PI(3,4,5)P3 levels. Phosphoinositides were extracted from whole-cell lysates, and PI(3,4,5)P3 levels were measured using dot blots with antibodies specific to PI(3,4,5)P3. (A) A typical dot blot is shown. (B) Blot densities were analyzed by ImageJ software (version 1.42q). The density for the untransfected wild type (WT) was arbitrarily set to 1, and all other data are reported as a ratio to the WT. The data represent the means ± SD from 3 trials. PI(3,4,5)P3 levels were higher in GFP-PHBtk-expressing cells (*, P < 0.05) than in WT cells or in a control cell line expressing an irrelevant protein, luciferase (Eh209).
Products of PI3K may propagate a signal by interacting with and phosphorylating downstream effector proteins (reviewed in reference 7). One outcome of signaling is changes in gene expression (49–51). Therefore, we measured the level of EhCoactosin and SREHP transcript in the GFP-PHBtk cell line to assess the impact of altered levels of PI(4,5)P2 (48) and PI(3,4,5)P3 (this study). Compared to wild-type cells, there was a 1.65-fold increase in the abundance of SREHP transcript (Fig. 8A) and a 3.4-fold increase in the abundance of EhCoactosin transcript (Fig. 8B) in cells expressing GFP-PHBtk. SREHP and EhCoactosin transcript levels were not significantly higher in a control transgenic cell line expressing an irrelevant protein, luciferase (Eh209) (Fig. 8A and B). This indicated that the increase in SREHP and EhCoactosin transcripts in the GFP-PHBtk-expressing cells was not due to the presence of episomal DNA or the application of selection. These expression data support the connection between PI3K signaling and EhCoactosin and SREHP.
FIG 8.
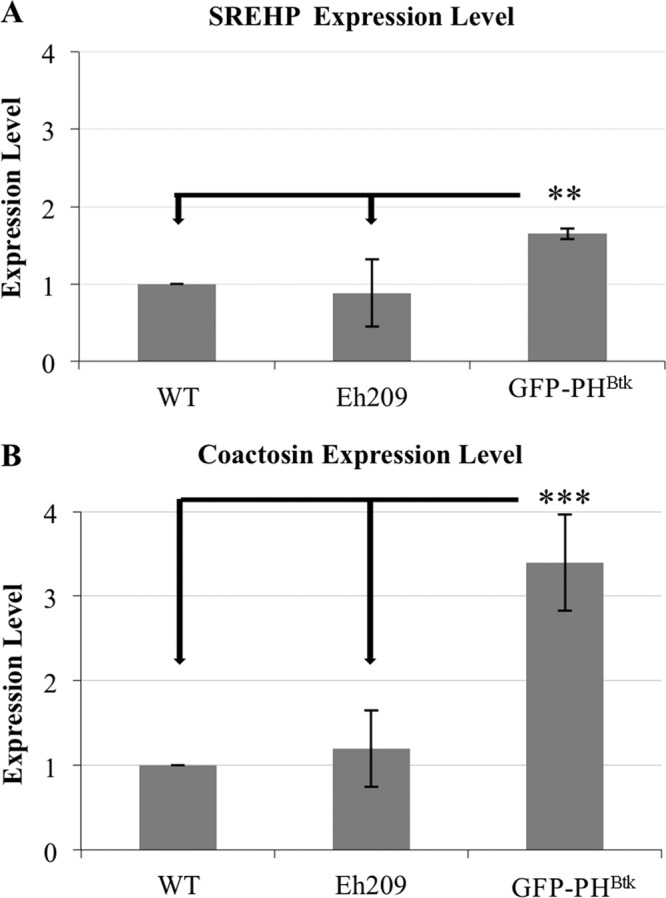
GFP-PHBtk overexpressors possess increased levels of EhCoactosin and SREHP transcripts. RNA from untransfected (wild type; WT) log-phase E. histolytica trophozoites, from a cell line expressing a GFP-tagged PH domain (GFP-PHBtk), and from a cell line expressing an irrelevant protein, luciferase (Eh209), was used for qPCR analysis of expression. The ssRNA gene was used as a loading control. (A) SREHP was expressed in the GFP-PHBtk cell line at a level that was approximately 1.65 (±0.07)-fold (**, P < 0.01) higher than that in WT cells. (B) EhCoactosin was expressed in the GFP-PHBtk cell line at a level that was approximately 3.4 (±0.57)-fold (***, P < 0.001) higher than that in WT levels cells. Neither SREHP nor EhCoactosin was expressed at a higher level in the control cell line (Eh209), suggesting that the increase seen in the GFP-PHBtk cell line was authentic. The data represent the means ± SD from ≥3 biological replicates.
DISCUSSION
In this study, we utilized a functional genomics approach to identify genes that may directly or indirectly function in the PI3K pathway in E. histolytica. We uncovered at least one gene, a Rap GTPase, that participates in PI3K signaling in other systems. Our study also identified several genes not previously identified as partners in PI3K signaling, including SREHP and a coactosin-like protein, EhCoactosin. A mutant cell line with an increased level of PI(3,4,5)P3, the product of PI3K activity, exhibited increased expression of SREHP and EhCoactosin. This supports the functional connection between these proteins and PI3K in E. histolytica. The assignment of SREHP and EhCoactosin as partners operating with PI3K signaling is novel.
Although not examined in detail, the enrichment of the EhRap2-coding gene in our screen was intriguing. E. histolytica possesses two Rap isoforms, EhRap1 and EhRap2, which share over 91% identity (52). EhRap2 shares at least 60% identity with Rap1 proteins from other systems and at least 50% identity with Rap2 proteins from other systems. Like other Rap1 proteins (41, 53), EhRap2 localizes to phagosomes (54). In platelets, PI3K can activate Rap1 (54, 55). In Dictyostelium discoideum, Rap1 interacts with the Ras-binding domain of PI3K to modulate cell polarity and pseudopod formation (42). EhRap2 possesses all of the effector binding domains and switch I and switch II regions (data not shown) that are necessary for interaction with PI3K (reviewed in references 56 and 57). Since, in E. histolytica, EhRap2 (54), PI3K (36), and the products of PI3K (8–10) localize to phagosomes, and since EhRap2 was highly enriched among the wortmannin-selected population (this study), it is reasonable to hypothesize that similar local EhRap2/PI3K associations exist.
In our study, the cDNA encoding ehapt2 was also highly enriched in the wortmannin-selected population. The genome of E. histolytica contains non-long terminal repeat (non-LTR) retrotransposons (58, 59). These include the long interspersed repetitive elements (LINEs) and the short interspersed repetitive elements (SINEs). Ehapt2 is a member of the EhSINE1 family of retrotransposable elements. Ehapt2 is present in the vicinity of protein-coding genes (60) and may influence the expression of such genes. Although Yadav et al. (61) reported that laboratory strains of E. histolytica may be retrotransposition incompatible, discerning the genomic position of ehapt2 in trophozoites that are less sensitive to wortmannin is necessary to fully understand the connection, if any, to PI3K signaling.
Several hypothetical protein-coding genes were also isolated in our screen. One such protein, H644 (XM_644144.2), was previously identified in a screen for negative regulators of phagocytosis (24). Overexpression of H644 strongly reduced phagocytosis of human red blood cells by E. histolytica (24). A second hypothetical protein (XM_650280.2) contained an N-terminal Bin-Amphiphysin-Rvs (BAR) superfamily domain and a C-terminal SH3 domain. BAR domain proteins are evolutionarily conserved and bind to the plasma membrane. Specifically, BAR domains induce, stabilize, or detect membrane curvatures and facilitate the interaction between the plasma membrane and the underlying cytoskeleton (62). Interestingly, H644 possesses a significant number of putative phosphorylation sites (24), and many BAR domain-containing proteins possess SH3 domains (63, 64). Thus, these hypothetical proteins may be authentic signaling molecules, and their identification in our screen may not be fortuitous.
Several ribosomal protein-coding genes were highly enriched in our screen. These may or may not be authentic participants in PI3K signaling. In a screen of yeast deletion mutants for wortmannin sensitivity, 119 of the 1,067 genes, including several ribosomal protein-coding genes, were functionally characterized to be involved in protein synthesis (25). Therefore, it is possible that in our study, overexpression of ribosomal proteins has a direct or indirect connection to PI3K-based signaling.
The most highly enriched cDNA in our screen was EhCoactosin. Coactosin, an F-actin binding protein, was first isolated from the actin-myosin complex of Dictyostelium discoideum (45). Subsequently, coactosin-like proteins (CLPs) have been identified in other systems (46, 65, 66). Coactosin contains an actin depolymerizing factor homology (ADF-H) domain, a structurally conserved motif that allows proteins to interact with actin and to remodel the actin cytoskeleton (reviewed in reference 67). In general, coactosins only bind F-actin (46, 65, 68). With the exception of D. discoideum coactosin, which weakly interferes with barbed-end capping of actin filaments (68), coactosins do not seem to promote actin filament elongation or disassembly. It has been suggested that, in vivo, coactosins regulate actin dynamics as a member of a complex with other proteins (69). EhCoactosin possesses an ADF-H domain; however, it remains to be seen if it binds actin and if it has additional interacting protein partners.
Coactosin has been implicated in cell motility. In D. discoideum, coactosin is concentrated in pseudopods (45). In developing chick embryos, coactosin is expressed in migrating neural crest cells (46). Additionally, the levels of coactosin mRNA are higher in motile and migrating cells. For example, in D. discoideum, coactosin mRNA reaches a peak level in aggregating cells that are responding chemotactically to cyclic AMP (45). There is also evidence for local translation of coactosin mRNAs at cellular extensions in oculomotor nerves (46) and in the pseudopods of human metastatic tumor cells (70). We showed that the level of EhCoactosin transcript is higher in the GFP-PHBtk overexpressors. Interestingly, this cell line also exhibits increased motility (10). Furthermore, we demonstrated that overexpression of EhCoactosin is sufficient to increase motility. Together, these data suggest that EhCoactosin is an authentic member of the coactosin family.
Coactosin may also play a role in phagocytosis in lower eukaryotes, as evidenced by its identification in the proteome of purified phagosomes of D. discoideum (71, 72) and E. histolytica (36). PI3K itself also localizes to E. histolytica phagosomes (36). The mechanism by which increased levels of PI(3,4,5)P3 lead to increased expression of EhCoactosin has not been discerned. However, given that both proteins localize to the same organelle, and given that EhCoactosin has several phosphorylation sites, it is possible that, in addition to regulating expression of EhCoactosin, PI3K also regulates activity of EhCoactosin by phosphorylation. It remains to be seen if EhCoactosin physically interacts with PI3K or its products.
SREHP has also been implicated in E. histolytica phagocytosis. In particular, trophozoites can induce host cell apoptosis via caspase-3 activation (73). This is followed by preferential phagocytosis of apoptotic cells over healthy cells (73). SREHP is thought to be a cell surface receptor facilitating the binding and uptake of apoptotic host cells (44). SREHP also undergoes posttranslational modifications, including phosphorylation, O-linked terminal N-acetylation, and acylation (39). Given its subcellular localization, posttranslational modifications, and function as a receptor for apoptotic cells, it has been hypothesized to be involved in cell signaling (44). In the current study, identification of SREHP supports its role in signal transduction, perhaps through a PI3K-mediated pathway.
Recently, we discovered that SREHP localizes to lipid rafts in E. histolytica (data not shown). Lipid rafts are small cholesterol- and sphingolipid-rich membrane domains that regulate signaling, including PI3K signaling. In E. histolytica the PI3K substrate, PI(4,5)P2, is also enriched in rafts (47). Although it remains to be seen if PI(3,4,5)P3 is localized to rafts in E. histolytica, the connection between SREHP and PI3K is strengthened by the fact that SREHP colocalizes with PI3K-based signaling lipids in rafts.
It is conceivable that overexpression of PI3K itself would have protected the cells from near-lethal doses of a PI3K inhibitor. However, we did not identify cDNAs encoding PI3Ks. There are several possible explanations for our inability to isolate PI3K-encoding genes in this screen. First, the screen was not exhaustive. The cDNAs from only 96 E. coli transformants were sequenced. Therefore, it is possible that additional sequencing will identify cDNAs encoding E. histolytica PI3Ks. The E. histolytica genome possesses at least 8 putative PI3Ks (data not shown), but it remains to be seen which of these are authentic PI3Ks. Second, the screen may have been too stringent. Although the false-positive discovery rate would have increased, screening with lower concentrations of wortmannin may have revealed genes encoding authentic PI3Ks.
This study enhances our knowledge of PI3K signaling in E. histolytica, which, in turn, provides insight into virulence. This study also illustrates the utility of full-genome functional genetic screens in this parasite. Our screen was based on a simple phenotype, cell viability. Predictably, future forward and reverse genetic screens, based on more complex phenotypes, are likely to lead to the functional annotation of many genes in this organism. This will, undoubtedly, reveal new targets for the development of vaccines or therapies against E. histolytica infection.
ACKNOWLEDGMENTS
We thank William A. Petri, Jr., and Carol A. Gilchrist (University of Virginia School of Medicine, Charlottesville, VA) for the E. histolytica expression plasmid, pAH-DEST. We thank Ada V. King for the creation of the E. histolytica overexpression library. We also thank Lisa J. Bain (Department of Biological Sciences, Clemson University), Michael J. Childress (Department of Biological Sciences, Clemson University), and Terri F. Bruce (Clemson Light Imaging Facility and Department of Biological Sciences, Clemson University) for assistance with RT-qPCR, statistical analyses, and microscopy, respectively.
This project was supported by grant no. R21 AI081100-01 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, to L.A.T. Research reported in this publication was also supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant no. P20 GM103444. This material is based upon work supported by NIFA/USDA, under project number SC-1700457.
The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The contents are solely the responsibility of the authors and do not necessarily represent the views of the National Institute of Allergy and Infectious Diseases, the National Institutes of Health, or the USDA.
Footnotes
Published ahead of print 17 January 2014
This article is technical contribution no. 6140 of the Clemson University Experiment Station.
REFERENCES
- 1.World Health Organization. 1997. Amoebiasis. Wkly. Epidemiol. Rec. 72:97–999100475 [Google Scholar]
- 2.Haque R, Duggal P, Ali IM, Hossain MB, Mondal D, Sack RB, Farr BM, Beaty TH, Petri WA., Jr 2002. Innate and acquired resistance to amebiasis in Bangladeshi children. J. Infect. Dis. 186:547–552. 10.1086/341566 [DOI] [PubMed] [Google Scholar]
- 3.Laughlin RC, Temesvari LA. 2005. Cellular and molecular mechanisms that underlie Entamoeba histolytica pathogenesis: prospects for intervention. Expert Rev. Mol. Med. 7:1–19. 10.1017/S1462399405009622 [DOI] [PubMed] [Google Scholar]
- 4.Halet G. 2005. Imaging phosphoinositide dynamics using GFP-tagged protein domains. Biol. Cell 97:501–518. 10.1042/BC20040080 [DOI] [PubMed] [Google Scholar]
- 5.Payrastre B, Missy K, Giuriato S, Bodin S, Plantavid M, Gratacap M. 2001. Phosphoinositides: key players in cell signalling, in time and space. Cell Signal. 13:377–387. 10.1016/S0898-6568(01)00158-9 [DOI] [PubMed] [Google Scholar]
- 6.Lemmon MA. 2008. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9:99–111. 10.1038/nrm2328 [DOI] [PubMed] [Google Scholar]
- 7.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. 2001. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70:535–602. 10.1146/annurev.biochem.70.1.535 [DOI] [PubMed] [Google Scholar]
- 8.Powell RR, Welter BH, Hwu R, Bowersox B, Attaway C, Temesvari LA. 2006. Entamoeba histolytica: FYVE-finger domains, phosphatidylinositol 3-phosphate biosensors, associate with phagosomes but not fluid filled endosomes. Exp. Parasitol. 112:221–231. 10.1016/j.exppara.2005.11.013 [DOI] [PubMed] [Google Scholar]
- 9.Nakada-Tsukui K, Okada H, Mitra BN, Nozaki T. 2009. Phosphatidylinositol-phosphates mediate cytoskeletal reorganization during phagocytosis via a unique modular protein consisting of RhoGEF/DH and FYVE domains in the parasitic protozoon, Entamoeba histolytica. Cell. Microbiol. 11:1471–1491. 10.1111/j.1462-5822.2009.01341.x [DOI] [PubMed] [Google Scholar]
- 10.Byekova YA, Powell RR, Welter BH, Temesvari LA. 2010. Localization of phosphatidylinositol (3,4,5)-trisphosphate to phagosomes in Entamoeba histolytica achieved using glutathione S-transferase- and green fluorescent protein-tagged lipid biosensors. Infect. Immun. 78:125–137. 10.1128/IAI.00719-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivière C, Marion S, Guillén N, Bacri J, Gazeau F, Wilhelm C. 2007. Signaling through the phosphatidylinositol 3-kinase regulates mechanotaxis induced by local low magnetic forces in Entamoeba histolytica. J. Biomech. 40:64–77. 10.1016/j.jbiomech.2005.11.012 [DOI] [PubMed] [Google Scholar]
- 12.López-Contreras L, Hernández-Ramírez V, Flores-García Y, Chávez-Munguía B, Talamás-Rohana P. 2013. Src and PI3 K inhibitors affect the virulence factors of Entamoeba histolytica. Parasitology 140:202–209. 10.1017/S0031182012001540 [DOI] [PubMed] [Google Scholar]
- 13.Meza I, Clarke M. 2004. Dynamics of endocytic traffic of Entamoeba histolytica revealed by confocal microscopy and flow cytometry. Cell Motil. Cytoskel. 59:215–226. 10.1002/cm.20038 [DOI] [PubMed] [Google Scholar]
- 14.Ghosh SK, Samuelson J. 1997. Involvement of p21racA, phosphoinositide 3-kinase, and vacuolar ATPase in phagocytosis of bacteria and erythrocytes by Entamoeba histolytica: suggestive evidence for coincidental evolution of amebic invasiveness. Infect. Immun. 65:4243–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacFarlane RC, Singh U. 2006. Identification of differentially expressed genes in virulent and nonvirulent Entamoeba species: potential implications for amebic pathogenesis. Infect. Immun. 74:340–351. 10.1128/IAI.74.1.340-351.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sephton C, Mousseau D. 2008. Dephosphorylation of Akt in C6 cells grown in serum-free conditions corresponds with redistribution of p85/PI3K to the nucleus. J. Neurosci. Res. 86:675–682. 10.1002/jnr.21516 [DOI] [PubMed] [Google Scholar]
- 17.Vieira OV, Botelho RJ, Rameh L, Brachmann SM, Matsuo T, Davidson HW, Schreiber A, Backer JM, Cantley LC, Grinstein S. 2001. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155:19–26. 10.1083/jcb.200107069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makioka A, Kumagai M, Ohtomo H, Kobayashi S, Takeuchi T. 2001. Inhibition of encystation of Entamoeba invadens by wortmannin. Parasitol. Res. 87:371–375. 10.1007/s004360000339 [DOI] [PubMed] [Google Scholar]
- 19.Butcher RA, Bhullar BS, Perlstein EO, Marsischky G, LaBaer J, Schreiber SL. 2006. Microarray-based method for monitoring yeast overexpression strains reveals small-molecule targets in TOR pathway. Nat. Chem. Biol. 2:103–109. 10.1038/nchembio762 [DOI] [PubMed] [Google Scholar]
- 20.Luesch H, Wu TY, Ren P, Gray NS, Schultz PG, Supek F. 2005. A genome-wide overexpression screen in yeast for small-molecule target identification. Chem. Biol. 12:55–63. 10.1016/j.chembiol.2004.10.015 [DOI] [PubMed] [Google Scholar]
- 21.Rine J, Hansen W, Hardeman E, Davis RW. 1983. Targeted selection of recombinant clones through gene dosage effects. Proc. Natl. Acad. Sci. U. S. A. 80:6750–6754. 10.1073/pnas.80.22.6750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenzi HA, Puiu D, Miller JR, Brinkac LM, Amedeo P, Hall N, Caler EV. 2010. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl. Trop. Dis. 4:e716. 10.1371/journal.pntd.0000716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loftus B, Anderson I, Davies R, Alsmark UCM, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, Nozaki T, Suh B, Pop M, Duchene M, Ackers J, Tannich E, Leippe M, Hofer M, Bruchhaus I, Willhoeft U, Bhattacharya A, Chillingworth T, Churcher C, Hance Z, Harris B, Harris D, Jagels K, Moule S, Mungall K, Ormond D, Squares R, Whitehead S, Quail MA, Rabbinowitsch E, Norbertczak H, Price C, Wang Z, Guillén N, Gilchrist C, Stroup SE, Bhattacharya S, Lohia A, Foster PG, Sicheritz-Ponten T, Weber C, Singh U, Mukherjee C, El-Sayed NM, Petri WA, Jr, Clark CG, Embley TM, Barrell B, Fraser CM, Hall N. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433:865–868. 10.1038/nature03291 [DOI] [PubMed] [Google Scholar]
- 24.King AV, Welter BH, Koushik AB, Gordon LN, Temesvari LA. 2012. A genome-wide over-expression screen identifies genes involved in phagocytosis in the human protozoan parasite, Entamoeba histolytica. PLoS One 7:e43025. 10.1371/journal.pone.0043025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zewail A, Xie MW, Xing Y, Lin L, Zhang PF, Zou W, Saxe JP, Huang J. 2003. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc. Natl. Acad. Sci. U. S. A. 100:3345–3350. 10.1073/pnas.0530118100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diamond LS, Harlow DR, Cunnick CC. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432. 10.1016/0035-9203(78)90144-X [DOI] [PubMed] [Google Scholar]
- 27.Ramakrishnan G, Vines RR, Mann BJ, Petri WA., Jr 1997. A tetracycline-inducible gene expression system in Entamoeba histolytica. Mol. Biochem. Parasitol. 84:93–100. 10.1016/S0166-6851(96)02784-3 [DOI] [PubMed] [Google Scholar]
- 28.Abhyankar MM, Hochreiter AE, Connell SK, Gilchrist CA, Mann BJ, Petri WA., Jr 2009. Development of the Gateway system for cloning and expressing genes in Entamoeba histolytica. Parasitol. Int. 58:95–97. 10.1016/j.parint.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Welter BH, Temesvari LA. 2009. Overexpression of a mutant form of EhRabA, a unique Rab GTPase of Entamoeba histolytica, alters endoplasmic reticulum morphology and localization of the Gal/GalNAc adherence lectin. Eukaryot. Cell 8:1014–1026. 10.1128/EC.00030-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ehrenkaufer GM, Eichinger DJ, Singh U. 2007. Trichostatin A effects on gene expression in the protozoan parasite Entamoeba histolytica. BMC Genomics 8:216. 10.1186/1471-2164-8-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45–e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray A, Olsson H, Batty IH, Priganica L, Peter Downes C. 2003. Nonradioactive methods for the assay of phosphoinositide 3-kinases and phosphoinositide phosphatases and selective detection of signaling lipids in cell and tissue extracts. Anal. Biochem. 313:234–245. 10.1016/S0003-2697(02)00607-3 [DOI] [PubMed] [Google Scholar]
- 33.Zaki M, Andrew N, Insall RH. 2006. Entamoeba histolytica cell movement: a central role for self-generated chemokines and chemorepellents. Proc. Natl. Acad. Sci. U. S. A. 103:18751–18756. 10.1073/pnas.0605437103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34:W362–W365. 10.1093/nar/gkl124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ui M, Okada T, Hazeki K, Hazeki O. 1995. Wortmannin as a unique probe for an intracellular signalling protein, phosphoinositide 3-kinase. Trends Biochem. Sci. 20:303–307. 10.1016/S0968-0004(00)89056-8 [DOI] [PubMed] [Google Scholar]
- 36.Marion S, Laurent C, Guillén N. 2005. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: a proteomic approach. Cell. Microbiol. 7:1504–1518. 10.1111/j.1462-5822.2005.00573.x [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Eddy R, Condeelis J. 2007. The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer 7:429–440. 10.1038/nrc2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Provost P, Doucet J, Hammarberg T, Gerisch G, Samuelsson B, Rådmark O. 2001. 5-Lipoxygenase interacts with coactosin-like protein. J. Biol. Chem. 276:16520–16527. 10.1074/jbc.M011205200 [DOI] [PubMed] [Google Scholar]
- 39.Stanley SL, Jr, Tian K, Koester JP, Li E. 1995. The serine-rich Entamoeba histolytica protein is a phosphorylated membrane protein containing O-linked terminal N-acetylglucosamine residues. J. Biol. Chem. 270:4121–4126. 10.1074/jbc.270.8.4121 [DOI] [PubMed] [Google Scholar]
- 40.Zhang T, Li E, Stanley SL., Jr 1995. Oral immunization with the dodecapeptide repeat of the serine-rich Entamoeba histolytica protein (SREHP) fused to the cholera toxin B subunit induces a mucosal and systemic anti-SREHP antibody response. Infect. Immun. 63:1349–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pizon V, Desjardins M, Bucci C, Parton RG, Zerial M. 1994. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J. Cell Sci. 107:1661–1670 [DOI] [PubMed] [Google Scholar]
- 42.Kortholt A, Bolourani P, Rehmann H, Keizer-Gunnink I, Weeks G, Wittinghofer A, Van Haastert PJ. 2010. A Rap/phosphatidylinositol 3-kinase pathway controls pseudopod formation. Mol. Biol. Cell 21:936–945. 10.1091/mbc.E09-03-0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanley SL, Jr, Becker A, Kunz-Jenkins C, Foster L, Li E. 1990. Cloning and expression of a membrane antigen of Entamoeba histolytica possessing multiple tandem repeats. Proc. Natl. Acad. Sci. U. S. A. 87:4976–4980. 10.1073/pnas.87.13.4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teixeira JE, Huston CD. 2008. Participation of the serine-rich Entamoeba histolytica protein in amebic phagocytosis of apoptotic host cells. Infect. Immun. 76:959–966. 10.1128/IAI.01455-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Hostos E, Bradtke B, Lottspeich F, Gerisch G. 1993. Coactosin, a 17 kDa F-actin binding protein from Dictyostelium discoideum. Cell Motil. Cytoskel. 26:181–191. 10.1002/cm.970260302 [DOI] [PubMed] [Google Scholar]
- 46.Hou X, Katahira T, Kimura J, Nakamura H. 2009. Expression of chick Coactosin in cells in morphogenetic movement. Dev. Growth Differ. 51:833–840. 10.1111/j.1440-169X.2009.01146.x [DOI] [PubMed] [Google Scholar]
- 47.Koushik AB, Powell RR, Temesvari LA. 2013. Localization of phosphatidylinositol 4, 5-bisphosphate to lipid rafts and uroids in the human protozoan parasite, Entamoeba histolytica. Infect. Immun. 81:2145–2155. 10.1128/IAI.00040-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goldston AM, Powell RR, Koushik AB, Temesvari LA. 2012. Exposure to host ligands correlates with co-localization of Gal/GalNAc lectin subunits in lipid rafts and PIP2 signaling in Entamoeba histolytica. Eukaryot. Cell 11:743–751. 10.1128/EC.00054-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du K, Montminy M. 1998. CREB is a regulatory target for the protein kinase Akt/PKB. J. Biol. Chem. 273:32377–32379. 10.1074/jbc.273.49.32377 [DOI] [PubMed] [Google Scholar]
- 50.Caravatta L, Sancilio S, di Giacomo V, Rana R, Cataldi A, Di Pietro R. 2008. PI3-K/Akt-dependent activation of cAMP-response element-binding (CREB) protein in Jurkat T leukemia cells treated with TRAIL. J. Cell. Physiol. 214:192–200. 10.1002/jcp.21186 [DOI] [PubMed] [Google Scholar]
- 51.Meier R, Alessi DR, Cron P, Andjelković M, Hemmings BA. 1997. Mitogenic activation, phosphorylation, and nuclear translocation of protein kinase Bβ. J. Biol. Chem. 272:30491–30497. 10.1074/jbc.272.48.30491 [DOI] [PubMed] [Google Scholar]
- 52.Shen P, Lohia A, Samuelson J. 1994. Molecular cloning of ras and rap genes from Entamoeba histolytica. Mol. Biochem. Parasitol. 64:111–120. 10.1016/0166-6851(94)90139-2 [DOI] [PubMed] [Google Scholar]
- 53.Seastone DJ, Zhang L, Buczynski G, Rebstein P, Weeks G, Spiegelman G, Cardelli JA. 1999. The small Mr ras-like GTPase Rap1 and the phospholipase C pathway act to regulate phagocytosis in Dictyostelium discoideum. Mol. Biol. Cell 10:393–406. 10.1091/mbc.10.2.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okada M, Huston CD, Mann BJ, Petri WA, Jr, Kita K, Nozaki T. 2005. Proteomic analysis of phagocytosis in the enteric protozoan parasite Entamoeba histolytica. Eukaryot. Cell 4:827–831. 10.1128/EC.4.4.827-831.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woulfe D, Jiang H, Mortensen R, Yang J, Brass LF. 2002. Activation of Rap1B by Gi family members in platelets. J. Biol. Chem. 277:23382–23390. 10.1074/jbc.M202212200 [DOI] [PubMed] [Google Scholar]
- 56.Castellano E, Downward J. 2011. RAS interaction with PI3K more than just another effector pathway. Genes Cancer 2:261–274. 10.1177/1947601911408079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caron E. 2003. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. J. Cell Sci. 116:435–440. 10.1242/jcs.00238 [DOI] [PubMed] [Google Scholar]
- 58.Sharma R, Bagchi A, Bhattacharya A, Bhattacharya A. 2001. Characterization of a retrotransposon-like element from Entamoeba histolytica. Mol. Biochem. Parasitol. 116:45–53. 10.1016/S0166-6851(01)00300-0 [DOI] [PubMed] [Google Scholar]
- 59.Mandal PK, Bagchi A, Bhattacharya A, Bhattacharya S. 2004. An Entamoeba histolytica LINE/SINE pair inserts at common target sites cleaved by the restriction enzyme-like LINE-encoded endonuclease. Eukaryot. Cell 3:170–179. 10.1128/EC.3.1.170-179.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Willhoeft U, Buß H, Tannich E. 2002. The abundant polyadenylated transcript 2 DNA sequence of the pathogenic protozoan parasite Entamoeba histolytica represents a nonautonomous non-long-terminal-repeat retrotransposon-like element which is absent in the closely related nonpathogenic species Entamoeba dispar. Infect. Immun. 70:6798–6804. 10.1128/IAI.70.12.6798-6804.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yadav VP, Mandal PK, Bhattacharya A, Bhattacharya S. 2012. Recombinant SINEs are formed at high frequency during induced retrotransposition in vivo. Nat. Commun. 3:854. 10.1038/ncomms1855 [DOI] [PubMed] [Google Scholar]
- 62.Frost A, Unger VM, De Camilli P. 2009. The BAR domain superfamily: membrane-molding macromolecules. Cell 137:191–196. 10.1016/j.cell.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Safari F, Suetsugu S. 2012. The BAR domain superfamily proteins from subcellular structures to human diseases. Membranes 2:91–117. 10.3390/membranes2010091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahashi Y, Meyerkord CL, Wang H. 2009. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ. 16:947–955. 10.1038/cdd.2009.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Provost P, Doucet J, Stock A, Gerisch G, Samuelsson B, Rådmark O. 2001. Coactosin-like protein, a human F-actin-binding protein: critical role of lysine-75. Biochem. J. 359:255–263. 10.1042/0264-6021:3590255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Doucet J, Provost P, Samuelsson B, Rådmark O. 2002. Molecular cloning and functional characterization of mouse coactosin-like protein. Biochem. Biophys. Res. Commun. 290:783–789. 10.1006/bbrc.2001.6236 [DOI] [PubMed] [Google Scholar]
- 67.Poukkula M, Kremneva E, Serlachius M, Lappalainen P. 2011. Actin-depolymerizing factor homology domain: a conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton 68:471–490. 10.1002/cm.20530 [DOI] [PubMed] [Google Scholar]
- 68.Röhrig U, Gerisch G, Morozova L, Schleicher M, Wegner A. 1995. Coactosin interferes with the capping of actin filaments. FEBS Lett. 374:284–286. 10.1016/0014-5793(95)01130-7 [DOI] [PubMed] [Google Scholar]
- 69.Hellman M, Paavilainen VO, Naumanen P, Lappalainen P, Annila A, Permi P. 2004. Solution structure of coactosin reveals structural homology to ADF/cofilin family proteins. FEBS Lett. 576:91–96. 10.1016/j.febslet.2004.08.068 [DOI] [PubMed] [Google Scholar]
- 70.Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. 2010. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 70:3780–3790. 10.1158/0008-5472.CAN-09-4439 [DOI] [PubMed] [Google Scholar]
- 71.Gotthardt D, Blancheteau V, Bosserhoff A, Ruppert T, Delorenzi M, Soldati T. 2006. Proteomics fingerprinting of phagosome maturation and evidence for the role of a Gα during uptake. Mol. Cell. Proteomics 5:2228–2243. 10.1074/mcp.M600113-MCP200 [DOI] [PubMed] [Google Scholar]
- 72.Shevchuk O, Batzilla C, Hägele S, Kusch H, Engelmann S, Hecker M, Haas A, Heuner K, Glöckner G, Steinert M. 2009. Proteomic analysis of Legionella-containing phagosomes isolated from Dictyostelium. Int. J. Med. Microbiol. 299:489–508. 10.1016/j.ijmm.2009.03.006 [DOI] [PubMed] [Google Scholar]
- 73.Huston CD, Boettner DR, Miller-Sims V, Petri WA., Jr 2003. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect. Immun. 71:964–972. 10.1128/IAI.71.2.964-972.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]