Abstract
The microbiota inhabiting the mammalian gut is a functional organ that provides a number of services for the host. One factor that may regulate the composition and function of gut microbial communities is dietary toxins. Oxalate is a toxic plant secondary compound (PSC) produced in all major taxa of vascular plants and is consumed by a variety of animals. The mammalian herbivore Neotoma albigula is capable of consuming and degrading large quantities of dietary oxalate. We isolated and characterized oxalate-degrading bacteria from the gut contents of wild-caught animals and used high-throughput sequencing to determine the distribution of potential oxalate-degrading taxa along the gastrointestinal tract. Isolates spanned three genera: Lactobacillus, Clostridium, and Enterococcus. Over half of the isolates exhibited significant oxalate degradation in vitro, and all Lactobacillus isolates contained the oxc gene, one of the genes responsible for oxalate degradation. Although diverse potential oxalate-degrading genera were distributed throughout the gastrointestinal tract, they were most concentrated in the foregut, where dietary oxalate first enters the gastrointestinal tract. We hypothesize that unique environmental conditions present in each gut region provide diverse niches that select for particular functional taxa and communities.
INTRODUCTION
The gut microbiota of mammalian herbivores plays a crucial role in aiding the host to digest low-quality and, often, toxic diets (1–4). Plants typically produce toxic plant secondary compounds (PSCs) as a means to combat herbivory (1, 5, 6). One mechanism by which mammalian herbivores may overcome PSCs is to host microbes capable of degrading such toxins (7). Dozens of species of bacteria capable of metabolizing dietary PSCs have been isolated from the guts of mammals, suggesting a long-term coevolution between hosts and their microbiotas (8–12). Additionally, the structure of the gut microbiota can be altered significantly by exposure to dietary PSCs and other xenobiotics, which is indicative of short-term ecological interactions between the diet and the microbiota (2, 13).
The widely produced PSC oxalate is an ideal compound for studying the effects of PSCs on the composition and function of the mammalian gut microbiota (14). A variety of mammals, including humans and domestic livestock, regularly consume oxalate in their diet (15–19). The consumption of oxalate can have a number of consequences for mammals, which include direct mortality, corrosion of the mouth and gastrointestinal tract, gastric hemorrhaging, diarrhea, kidney stones, and inflammation (15, 20, 21). Despite its prevalence and toxicity, mammals do not produce the enzymes capable of metabolizing oxalate (22, 23). However, oxalate-degrading bacteria are widely distributed within the guts of mammals (8, 11, 24–29).
Numerous oxalate-degrading bacteria that commonly inhabit the mammalian gut have been characterized (30, 31). One of the best-studied species is Oxalobacter formigenes, which requires oxalate as a carbon and energy source for growth (8). However, other oxalate-degrading bacteria include species from the genera Lactobacillus, Bifidobacterium, Streptococcus, Enterococcus, and others (11, 32–35). The majority of studies investigating oxalate-degrading bacteria have been conducted on humans, lab rats, and agricultural systems. Some mammals, such as woodrats and fat sand rats, consume levels of oxalate that would be lethal to other mammals and degrade >90% of the oxalate consumed (36–38). Our understanding of oxalate degradation in these ecologically relevant systems is much less understood.
We conducted a thorough investigation of the oxalate-degrading bacteria residing in the gut of the oxalate specialist Neotoma albigula. Populations of the white-throated woodrat, N. albigula, consume a diet comprised almost entirely of the oxalate-rich Opuntia cactus (37). Woodrats have a complex, segmented gut, with a large hindgut fermentation chamber (cecum) in addition to a foregut chamber, proximal to the stomach, which also harbors a diverse microbial population (2, 39, 40). The foregut community structure shifts in response to dietary toxins and may function in the degradation of toxins (2).
In this work, we tested the hypothesis that unique functional taxa and communities are segregated among gut regions. We predicted that the foregut, the gut chamber where dietary oxalate first enters the gastrointestinal tract, would house the greatest abundance of oxalate-degrading microbes.
MATERIALS AND METHODS
Location, diet, and collection of animals.
Five white-throated woodrats of the species Neotoma albigula were collected with Sherman live traps in Castle Valley, UT, in October 2012. Woodrats were immediately transported to the University of Utah Department of Biology Animal Facility and housed overnight in individual cages (48 × 27 × 20 cm) under a 12-h–12-h light-dark cycle, with a 28°C ambient temperature and 20% humidity. Animals were fed Opuntia cactus and Juniperus osteosperma collected from the location where the woodrats were trapped. To minimize the effects of captivity, animals were euthanized under CO2 and dissected after one night. All methods were approved by the IACUC, under protocol 12-12010.
Isolation of oxalate-degrading bacteria.
Upon dissection, the gut contents of the foregut, stomach, small intestine, cecum, and large intestines were collected along with the feces and were serially diluted in sterile deionized H2O for microbial isolation. A subset of contents was frozen at −80°C until DNA extraction. To isolate oxalate-tolerant and oxalate-degrading bacteria, serial dilutions of gut contents were plated onto brain heart infusion (BHI) agar supplemented with 20 mM sodium oxalate and 1 g/liter calcium chloride. The addition of sodium oxalate and calcium chloride produces calcium oxalate precipitate, which can be used to determine if bacteria are degrading oxalate (8, 41). The genus Lactobacillus is dominant in other woodrat species and contains numerous toxin-degrading species (2, 42, 43). Therefore, we chose to isolate a broad range of species of this genus by culturing gut contents on MRS medium without oxalate supplementation. This approach provided a sample of lactobacilli without specifically selecting for oxalate-degrading species. All media were first pH adjusted to match the gut region from which bacteria were isolated (foregut pH = 4.3, small intestine pH = 7, cecal pH = 6.3, large intestine pH = 6.5, and fecal pH = 6.4). Cultures were incubated at 37°C for 48 h under anaerobic conditions (Anaeropack system; Mitsubishi Gas Chemical Company), and the colony morphology was described for each unique CFU. Unique CFUs from the MRS plates and those that exhibited a zone of clearance around the CFU in the BHI medium, indicative of oxalate degradation, were chosen for further enrichment. Isolates were grown in either MRS or BHI broth with the pH adjusted according to the gut chamber from which they were isolated. Broth was flushed with CO2 and rubber stoppered to maintain an anaerobic environment.
Identification of isolates and the oxc gene.
To identify the isolates and the presence of the oxc gene, genomic DNAs were extracted from enrichment cultures by use of a DNeasy tissue kit (Qiagen). The 16S rRNA gene was amplified with the universal bacterial primers 27F and 1492R (5′-AGAGTTTGATCCT-3′ and 5′-GGTTACCTTGTTACGACTT-3′). Primers for the oxc gene (OxcL and OxcR [5′-TCMAMGTAAACACCACCTGGA-3′ and 5′-ATGTATGGTGTTGTMGGYATT-3′]) were designed by aligning sequences of the oxc genes of O. formigenes (accession no. M77128) and Lactobacillus gasseri (accession no. NC_008530) with Mega 5 software. PCR was performed on a Bio-Rad MyCycler with a thermal profile of 95°C for 2 min, followed by 35 cycles of 94°C for 30 s, 50°C (16S rRNA gene) or 52°C (oxc) for 60 s, and 70°C for 60 s, with a final extension step of 72°C for 5 min. Amplified DNA was gel purified following the manufacturer's protocol (Qiagen Qiaquick gel extraction kit) and sent to the DNA Sequencing Core Facility at the University of Utah for Sanger sequencing using the 27F or OxcL primer. Resulting sequences were used in a BLAST search against GenBank. Sequences with >97% identity of the 16S rRNA gene were considered to belong to the same operational taxonomic unit (OTU) (Table 1.)
TABLE 1.
Species identities and oxalate degradation efficiencies (over 48 h) of microbial isolates from different gut regions of N. albigula as determined by sequencing of the 16S rRNA gene and oxalate degradation assays
| Isolate | Gut region | Closest relative in GenBank (accession no.) | % Similarity (E value) | GenBank accession no. | Oxalate degradation efficiency (% [P value])a |
|---|---|---|---|---|---|
| 1-O | Feces | Clostridium sporogenes (JF836014.1) | 100 (0.0) | KF478820 | 18.4 (0.07)A |
| 8-O | Feces | Clostridium sporogenes (JF836013.1) | 99 (0.0) | KF478821 | 40.3 (0.0007)B |
| 11-O | Feces | Clostridium sporogenes (AB627077.1) | 99 (0.0) | KF478822 | 20.5 (0.03)A |
| 21-O | Feces | Clostridium sporogenes (JF836014.1) | 100 (0.0) | KF478823 | 18.4 (0.04)A |
| 6-O | Feces | Enterococcus gallinarum (KF254553.1) | 98 (2 × 10−126) | KF478824 | 24.8 (0.007)A,B |
| 9-O | Feces | Enterococcus gallinarum (KC510236.1) | 100 (0.0) | KF478825 | 35.45 (0.005)B |
| 25-O | Feces | Enterococcus gallinarum (KF254559.1) | 100 (2 × 10−66) | KF478826 | 11.45 (0.4)A |
| 30-L | Large intestine | Lactobacillus gasseri (GU454855.1) | 98 (0.0) | KF478827 | 0 (0.8)A |
| 33-L | Feces | Lactobacillus gasseri (GU454857.1) | 98 (0.0) | KF478828 | 23.7 (0.02)B,C |
| 34-L | Feces | Lactobacillus gasseri (GU454857.1) | 98 (0.0) | KF478829 | 14.5 (0.09)C |
| 38-L | Small intestine | Lactobacillus gasseri (NR_075051.1) | 97 (0.0) | KF478830 | 24.8 (0.02) B |
| 36-L | Small intestine | Lactobacillus animalis (HQ293063.1) | 99 (0.0) | KF478835 | 27.6 (0.06) |
| 41-L | Foregut | Lactobacillus johnsonii (CP002464.1) | 98 (0.0) | KF478831 | 29.3 (0.008)A |
| 49-L | Cecum | Lactobacillus johnsonii (CP002464.1) | 98 (0.0) | KF478832 | 36 (0.004)A |
| 50-L | Cecum | Lactobacillus reuteri (CP006603.1) | 97 (0.0) | KF478833 | 1.8 (0.9)A |
| 55-L | Small intestine | Lactobacillus reuteri (EU381125.1) | 97 (0.0) | KF478834 | 0 (0.9)B |
Oxalate degradation efficiency was estimated by dividing the amount of oxalate remaining in the medium after a 48-h incubation period by the amount of oxalate remaining in uninoculated control medium. Oxalate degradation efficiencies were compared between isolates of the same species by using one-way ANOVA with a post hoc, Bonferroni-corrected paired t test. Different letters indicate significant differences (P < 0.05 for each test).
Oxalate degradation assay.
To evaluate in vitro oxalate-degrading activity, cultures of isolates grown overnight to an optical density at 630 nm (OD630) of 0.6 were subcultured anaerobically in either BHI or MRS broth supplemented with 20 mM sodium oxalate and incubated for 48 h. The pH of the medium was adjusted to 5.5, regardless of the pH from which the bacteria were isolated, as oxalate-degrading activity is maximized at this pH for Lactobacillus (44). The oxalate concentration of the medium after the incubation period was then determined by titration with 0.01 M potassium permanganate (KMnO4) (37). Oxalate was first precipitated by the addition of 4 g/liter CaCl2, which converts soluble sodium oxalate to insoluble calcium oxalate. Following centrifugation, the supernatant was decanted and the precipitate was resuspended in the same volume of deionized H2O. Calcium oxalate was solubilized by the addition of 6 N H2SO4 and titrated at 70 to 90°C, until a pink color persisted for 30 s. Oxalate concentrations in the media were compared to standard dilutions of 0, 5, 10, 15, and 20 mM and to a sterile medium control to determine oxalate concentrations and degradation efficiencies. Standard curves produced R2 values of 0.98 and 0.99 for BHI and MRS media, respectively. Titrations of known oxalate concentrations were 98 to 102% ± 0.7% and 98 to 102% ± 0.9% of the oxalate added for BHI and MRS media, respectively. For each isolated strain, oxalate-degrading assays were run three times. For each assay, the oxalate remaining in the medium was quantified three times and averaged as a single data point. Oxalate-degrading efficiency was determined by dividing the amount of oxalate remaining in the medium after 48 h for each of the isolates by the amount of oxalate remaining in the sterile medium control. To determine if oxalate degradation was significant, the amount of oxalate remaining in the medium of the isolates was compared to the amount of oxalate remaining in the sterile medium by the t test. Differences in oxalate degradation among isolates of the same species were determined by one-way analysis of variance (ANOVA) followed by a Bonferroni-corrected, paired t test post hoc analysis in the R statistical package (P < 0.05 for all tests).
DNA extraction and microbial inventories.
High-throughput Illumina sequencing and flow cytometry were used to determine the relative abundances and densities of microbial taxa, respectively. DNAs were extracted from the gut contents of each individual by use of a QIAamp DNA Stool minikit (Qiagen). Microbial inventories were generated using primers 515F and 806R for paired-end sequencing on an Illumina MiSeq platform (Argonne National Laboratory, University of Chicago). Sequences were analyzed in the QIIME software package (45) and underwent standard quality control using the default parameters. The Ribosomal Database Project (RDP) was used to classify genera (46). Chloroplast and mitochondrial sequences were removed. Only a subset of the microbial inventories are presented in this study. The complete data set appears elsewhere (47). From microbial inventories, potential oxalate degraders were classified based upon published research. Potential oxalate-degrading bacteria included Oxalobacter (8, 41), Bifidobacterium (32, 35), Ralstonia (48), Janthinobacterium (33), Clostridium (49, 50), Pseudomonas (50) (based on the presence of the frc gene; accession no. NC_021237.1 and NC_019670.1), Lactobacillus (29, 32, 34, 51), Streptococcus (32), Lactococcus (29, 51), and Enterococcus (11). The relative abundance of each genus was determined as a percentage of the whole community. The combined relative abundances of potential oxalate-degrading taxa provided an estimate of the cumulative potential oxalate-degrading capacity of each gut region.
Microbial density was determined by flow cytometry. After dissection, gut contents were diluted in sterile saline and sieved through a 50-mm-mesh cell strainer. Cells were further diluted and stained with a Live/Dead BacLight bacterial viability and counting kit (Life Technologies, Grand Island, NY). Cell counts were determined using a high-speed FACSVantage SE Turbo cell sorter located at the University of Utah's flow cytometry core facility. The density of each microbial taxon was determined by multiplying the relative abundance of the taxon generated by Illumina sequencing by the total microbial density generated by flow cytometry. Illumina data were analyzed in QIIME as described by Kohl et al. (47), and significant differences (P < 0.05) in relative abundance or microbial density between regions were determined using the nonparametric Friedman rank sum test.
Nucleotide sequence accession numbers.
Sequences obtained in this study were deposited in GenBank under accession no. KF478820 to KF478835.
RESULTS
Neotoma albigula diet.
The oxalate concentration of the Opuntia cactus collected from the same location as the woodrats was 1.4% ± 0.05% of dry weight (n = 10), as determined using a modified oxalate assay (37). The oxalate concentration of juniper was 1% ± 0.01% oxalate (n = 10). Stable isotope analysis of hair revealed that this woodrat population had fed almost exclusively on Opuntia cactus over the last 3 months. This amount of cactus corresponds to the consumption of over 900 mg oxalate/kg body weight per day (data not shown).
Identification of isolates and the presence of the oxc gene.
A total of 16 isolates from six OTUs were obtained with two selective and differential media: BHI supplemented with 20 mM oxalate and MRS medium without oxalate. No isolates were obtained from the stomach. From the BHI medium, four isolates of Clostridium sporogenes and three isolates of Enterococcus gallinarum were obtained. All exhibited >97% identity with the closest relatives from GenBank. From the MRS medium without oxalate, four isolates of L. gasseri, one isolate of Lactobacillus animalis, two isolates of Lactobacillus johnsonii, and two isolates of Lactobacillus reuteri were obtained. All exhibited >97% identity to database sequences (Table 1). All nine Lactobacillus isolates exhibited the presence of the oxc gene, while no bands were amplified from the Clostridium or Enterococcus isolates (Fig. 1). Bands from one isolate of L. gasseri (38-L) and one isolate of L. johnsonii (49-L) were excised and gel purified. These isolates were chosen based upon the disparity in in vitro oxalate degradation and intensities of the bands on the gel. Sequence analysis of the L. gasseri band (38-L) revealed 97% sequence identity with the oxc genes from L. gasseri (accession no. CP000413.1), Lactobacillus amylovorus (accession no. CP002338.1), and Lactobacillus acidophilus (accession no. CP002559.1). Analysis of the L. johnsonii band (49-L) revealed 89% sequence identity with the oxc genes from the same OTUs as those described above.
FIG 1.
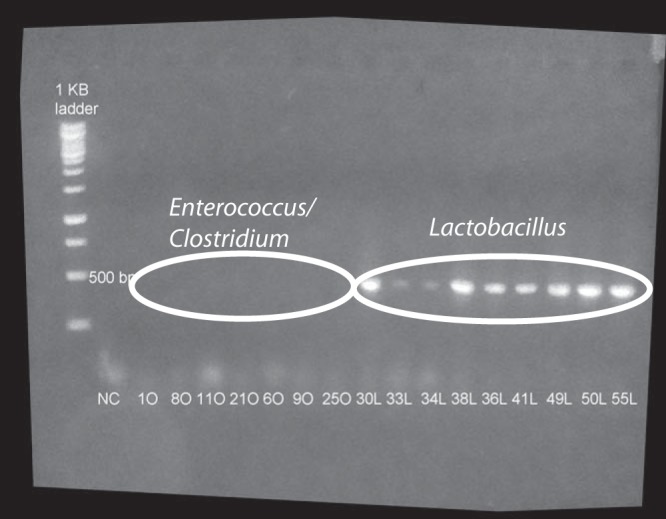
Amplification of the oxc gene. All Lactobacillus isolates exhibited a band of the expected size (400 bp), while none of the Enterococcus or Clostridium isolates exhibited such a band.
Oxalate degradation among isolates.
Significant variability in in vitro oxalate-degrading activity was present both between and within species isolated from the woodrat gut (Table 1). Of the 16 isolates examined, over half (53%) exhibited significant oxalate degradation, ranging from 18.4% to 40.3% of the oxalate present. Clostridium sporogenes exhibited the highest degradation activity, while one species, L. reuteri, exhibited no oxalate degradation. Lactobacillus animalis exhibited considerable degradation, but it was not significant due to variability between trials.
Distribution of oxalate-degrading bacteria.
In addition to bacterial isolates, diverse potential oxalate-degrading bacteria were distributed along the length of the gastrointestinal tract, as determined by sequencing of the V4 region of the 16S rRNA gene on the Illumina MiSeq platform. Sequencing of DNAs extracted from the gut contents resulted in 619,625 high-quality sequences (average of 20,654 ± 715 sequences per sample). Genera were grouped as either potential oxalate-degrading or non-oxalate-degrading bacteria, based on published data (see Materials and Methods).
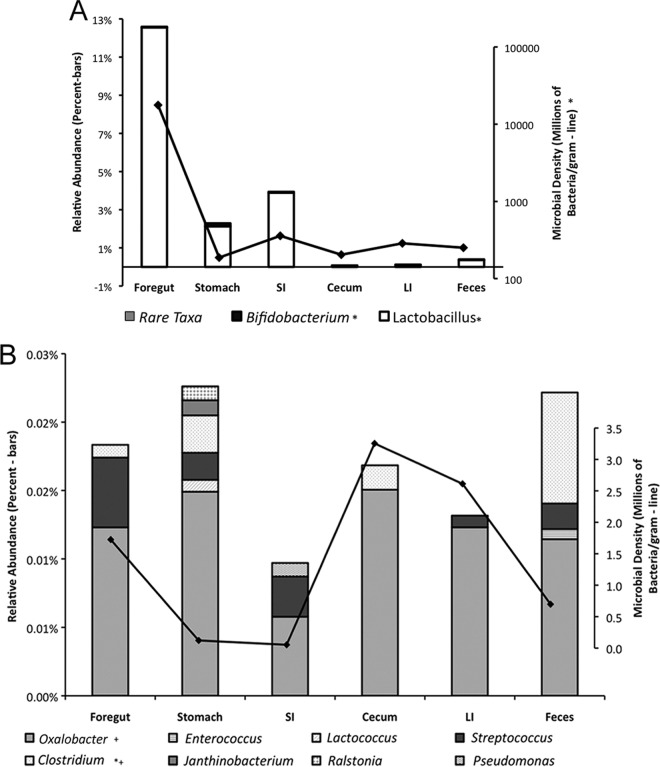
The foregut contained the highest density of potential oxalate-degrading bacteria, dominated by Lactobacillus, and the total density of all potential oxalate-degrading genera combined dropped off precipitously throughout the length of the gastrointestinal tract (Fig. 2). The relative abundances and densities of all potential oxalate-degrading bacteria differed significantly between gut regions (P = 0.03 and 0.03, respectively). Additionally, the relative abundances of Lactobacillus, Bifidobacterium, and Clostridium differed significantly across gut regions (P = 0.001, 0.006, and 0.01, respectively) (Fig. 2A and B). When density was considered, only Clostridium and Oxalobacter varied significantly (P = 0.04 and 0.005, respectively) (Fig. 2B). Oxalobacter, a genus that uniquely requires oxalate as a carbon and energy source, exhibited its highest abundances in the cecum and large intestines. The remaining genera did not vary significantly among gut regions.
FIG 2.
(A) Distribution of potential oxalate-degrading bacteria. The graph shows the microbial densities of all potential oxalate-degrading bacteria throughout the gastrointestinal tract (line) and the relative abundances of the more abundant Lactobacillus and Bifidobacterium genera as a percentage of the whole community (bars). *, significant differences among gut regions as assessed by the Friedman rank sum test (P = 0.006, 0.001, and 0.03 for the relative abundances of Bifidobacterium and Lactobacillus and the microbial density of all oxalate degraders, respectively). SI, small intestine; LI, large intestine. (B) Distribution of rare potential oxalate-degrading taxa. The graph shows the relative abundances of rare potential oxalate-degrading bacteria throughout the gastrointestinal tract as a percentage of the whole community (bars), as well as the density of the Oxalobacter genus (line). *, significant differences in relative abundance; +, significant differences in microbial density (assessed by the Friedman rank sum test; P = 0.01, 0.04, and 0.005 for the relative abundance of Clostridium and the densities of Oxalobacter and Clostridium, respectively).
DISCUSSION
This study identified several microbial taxa that degrade oxalate in the gut of a mammalian herbivore and documented that potential oxalate-degrading communities shift along the gastrointestinal tract. The potential oxalate-degrading microbes were dominated by Lactobacillus in each of the gut regions and comprised several rare taxa, including Oxalobacter. While the highest density of potential oxalate-degrading taxa was present in the foregut, these taxa were distributed throughout the gastrointestinal tract. This distribution may be driven by dietary oxalate. The oxalate content of N. albigula's natural diet of Opuntia is within the range of those of commonly consumed foods, such as rhubarb and spinach, but considerably less than that of Halogeton, which can be as high as 17% (15). However, given that N. albigula consumes ∼100% Opuntia cactus in their diet, they must consume larger amounts of oxalate than many other mammals.
Previous studies have implicated the Lactobacillus genus as a potentially important contributor to oxalate degradation in the mammalian gut (34, 51). The results of this study suggest that lactobacilli are the primary oxalate-degrading bacteria present in N. albigula. Lactobacillus isolates were obtained using a genus-specific medium without oxalate, which effectively provided a random sample. Of the nine Lactobacillus isolates, 100% of them harbored the oxc gene, which suggests that members of the Lactobacillus genus within the woodrat gut have a considerable potential to degrade oxalate. However, in vitro analysis of oxalate degradation revealed that only half of the Lactobacillus isolates actually degraded oxalate under the given conditions. Consistent with previous reports, there were significant differences between isolates within some OTUs in regard to oxalate degradation (34, 51). The difference between potential activity indicated by the presence of the oxc gene and actual activity quantified in the assays underscores the role of the environment in mediating oxalate degradation. The complex gut environment could produce numerous microniches suitable for oxalate degradation by different taxa throughout the gut. The disparity between oxalate degradation and the presence of the oxc gene could also be due to mutations or truncation in the oxc genes such that they no longer function to degrade oxalate (52).
There is little work to date on oxalate degradation by the genera Clostridium and Enterococcus. One study found that Enterococcus faecalis is capable of degrading oxalate for use as a sole carbon and energy source (11). Another showed that Enterococcus faecium and E. faecalis could degrade oxalate in the presence of MRS broth, similar to the case for the Lactobacillus isolates in our study (29). Our study is the first to show that E. gallinarum is also capable of degrading oxalate. Similarly, while members of the Clostridium genus exhibit oxalate-degrading activity (49, 50, 53), this is the first report of oxalate degradation in C. sporogenes. It is not known if these genera use the same pathway as Oxalobacter, Lactobacillus, Bifidobacterium, and others. The oxc gene was not amplified from the Enterococcus and Clostridium isolates by using the oxc primers. These results could be due to either primer incompatibility or the lack of the oxc gene. To date, other oxalate-degrading genes that may be responsible for oxalate degradation have been found in different Clostridium and Enterococcus species. These include the oxdD gene, encoding oxalate decarboxylase, in Clostridium botulinum (accession no. NC_009495.1) and a gene encoding oxalate decarboxylase in Enterococcus gilvus (accession no. AJDQ01000012.1) (58; A. Earl et al., unpublished data). Therefore, these genera may use a different pathway for oxalate degradation than that of Oxalobacter, Lactobacillus, and other genera that contain the oxc and frc genes.
The mammalian gut microbiota is best understood in the context of community ecology. There has been a wide range of oxalate-degrading rates reported for oxalate-degrading bacteria that reflect both short-term and long-term environmental influences (Table 2). Some genera, such as Lactobacillus, Clostridium, and Enterococcus, exhibit relatively low rates of oxalate degradation (0 to 7 μmol/ml medium/day), whereas others, such as Providencia, Oxalophagus, and Oxalobacter, have much higher rates (20 to 400 μmol/ml medium/day). Furthermore, the oxalate-degrading rates of whole communities, as well as the proportion of oxalate-degrading bacteria comprising a community, fluctuate with oxalate concentrations in the immediate environment (41, 54). Thus, oxalate in the immediate environment of microbial communities may drive the oxalate-degrading function in the short term to respond to dynamic concentrations of oxalate. In the long term, persistent exposure to oxalate may drive the evolution of the oxalate-degrading function within taxonomic groups. For O. formigenes, this function provides an effective means to extract carbon and energy (8). In other genera, such as Lactobacillus, it may function as a means to detoxify compounds in the environment. A detoxification function is inferred from the fact that increasing oxalate concentrations lead to lower microbial yields and oxalate-degrading efficiencies in Lactobacillus and other genera (32).
TABLE 2.
Oxalate-degrading rates of several species isolated from a range of sources
| Species | Isolation source | Oxalate-degrading rate (μmol oxalate/ml medium/day) | Reference |
|---|---|---|---|
| Oxalobacter formigenes | Sheep rumen | 408 | 17 |
| Oxalobacter vibrioformis | Anaerobic soils | 20.4–39 | 59 |
| Lactobacillus acidophilus | Not determined | 0.227–0.393 | 46 |
| Dairy | 0.35–1 | 48 | |
| Lactobacillus gasseri | Dairy | 0.35–1 | 48 |
| Lactobacillus johnsonii | Dairy | 0 | 48 |
| Lactobacillus murinus | Canine/feline gut | 7 | 28 |
| Lactobacillus animalis | Canine/feline gut | 6.5 | 28 |
| Lactobacillus reuteri | Canine/feline gut | 0 | 28 |
| Lactobacillus gasseri | Woodrat gut | 0–2.8 | This study |
| Lactobacillus reuteri | Woodrat gut | 0–1.8 | This study |
| Lactobacillus johnsonii | Woodrat gut | 2.9–3.6 | This study |
| Lactobacillus animalis | Woodrat gut | 2.5 | This study |
| Providencia rettgeri | Human feces | 28 | 38 |
| Clostridium sporogenes | Woodrat gut | 1.8–4.3 | This study |
| Oxalophagus oxalicus (formerly Clostridium oxalicus) | Anaerobic soils | 20.8–39.8 | 59 |
| Enterococcus gallinarum | Woodrat gut | 2.5–3.5 | This study |
The highest densities of the genera Lactobacillus, Bifidobacterium, and Oxalobacter seem to coincide with their optimal pHs for oxalate degradation within the N. albigula gut. Thus, the pH of each gut region may be another important factor driving in vivo oxalate degradation and the densities of different species of oxalate-degrading bacteria. The genera Bifidobacterium and Lactobacillus degrade oxalate optimally at pH 4.5 and 5.5, respectively (35, 44). However, optimal pH for oxalate-degrading activity within the Oxalobacter genus was reported as 6.4, after examination over the range of pH 5 to 8 (55). The pH of the foregut of N. albigula was measured at 4.3, compared to a pH of 1.4 in the stomach and 6.3 to 7.0 in the hindgut regions. Given the densities of each genus and the pH of each region, Lactobacillus and Bifidobacterium are likely responsible for most of the oxalate degradation in the foregut, whereas Oxalobacter is likely the principal oxalate-degrading bacterium in the hindgut. The distribution of potential oxalate-degrading taxa in the woodrat gut supports the hypothesis that the foregut of woodrats acts as a microbial detoxification chamber (2).
Significant populations of Oxalobacter present in the cecum and large intestines indicate that oxalate degradation may occur throughout the gut. The presence of Oxalobacter in the hindgut can induce the secretion of oxalate into this region, where it can be degraded further (56, 57). The density and relative abundance of Oxalobacter in the feces vary between mammals. In the current study, there were ∼1 million Oxalobacter cells/g, or 0.01% of the community. In humans, there are ∼1 million Oxalobacter cells/g (24). Rats harbor ∼10 million Oxalobacter cells/g, or <0.1% of the community (41). In sheep, Oxalobacter makes up ∼0.007% of the community in the rumen (54). In the current study, organisms in fecal samples were among the least representative of Oxalobacter abundances across the gut. This finding is of particular importance given that most studies estimate the presence of oxalate-degrading bacteria in fecal samples (e.g., see references 26 to 29).
The PSC oxalate is a widely produced compound that has a considerable impact on the ecology and evolution of mammalian herbivores, as well as humans. While most studies in the past have focused on fecal communities or a single microbial species capable of degrading large amounts of oxalate, recent research reveals that highly diverse oxalate-degrading bacteria inhabit the mammalian gut (8, 11, 25–29). We have shown that consortia of oxalate-degrading bacteria are present and distributed throughout the gut in mammalian herbivores that specialize on oxalate-rich plants, with fecal communities being among the least representative of potential oxalate-degrading bacteria. The oxalate-degrading function of the gut microbiota may be the result of a responsive community (54). Complex ecological interactions between host, diet, and microbiota can have considerable impacts on the composition and function of the microbiota and, subsequently, the host phenotype. These must be taken into account when we consider how the diet and xenobiotics affect the gut microbiota and, ultimately, the host phenotype.
ACKNOWLEDGMENTS
We declare that we have no conflicts of interest.
We thank Jesse Nelson for help with isolating bacteria and conducting oxalate degradation assays.
We thank the National Science Foundation (NSF grant DEB-1342615) for funding.
Footnotes
Published ahead of print 20 December 2013
REFERENCES
- 1.Karasov WH, Carey HV. 2009. Metabolic teamwork between gut microbes and host. Microbe 4:323–328 [Google Scholar]
- 2.Kohl KK, Dearing MD. 2012. Experience matters: prior exposure to plant toxins enhances diversity of gut microbes in herbivores. Ecol. Lett. 15:1008–1015. 10.1111/j.1461-0248.2012.01822.x [DOI] [PubMed] [Google Scholar]
- 3.O'Hara AM, Shanahan F. 2006. The gut microbiota as a forgotten organ. EMBO Rep. 7:688–693. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barboza PS, Bennet A, Lignot JH, Mackie RI, McWhorter TJ, Secor SM, Skovgaard N, Sundset MA, Wang T. 2010. Digestive challenges for vertebrate animals: microbial diversity, cardiorespiratory coupling, and dietary specialization. Physiol. Biochem. Zool. 83:764–774. 10.1086/650472 [DOI] [PubMed] [Google Scholar]
- 5.Dearing MD, Mangione AM, Karasov WH. 2000. Diet breadth of mammalian herbivores: nutrient versus detoxification constraints. Oecologia 123:397–405. 10.1007/s004420051027 [DOI] [PubMed] [Google Scholar]
- 6.Dearing MD, Foley WJ, McLean S. 2005. The influence of plant secondary metabolites on the nutritional ecology of herbivorous terrestrial vertebrates. Annu. Rev. Ecol. Evol. Syst. 36:169–189. 10.1146/annurev.ecolsys.36.102003.152617 [DOI] [Google Scholar]
- 7.Freeland WJ, Janzen DH. 1974. Strategies in herbivory by mammals: the role of plant secondary compounds. Am. Nat. 108:269–287. 10.1086/282907 [DOI] [Google Scholar]
- 8.Allison MJ, Dawson KA, Mayberry WR, Foss JG. 1985. Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch. Microbiol. 141:1–7. 10.1007/BF00446731 [DOI] [PubMed] [Google Scholar]
- 9.Jones RJ, Megarrity RG. 1986. Successful transfer of DHP-degrading bacteria from Hawaiian goats to Australian ruminants to overcome the toxicity of Leucaena. Aust. Vet. J. 63:259. 10.1111/j.1751-0813.1986.tb02990.x [DOI] [PubMed] [Google Scholar]
- 10.Kageyama A, Benno Y, Nakase T. 1999. Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 49:1725–1732 [DOI] [PubMed] [Google Scholar]
- 11.Hokama S, Honma Y, Toma C, Ogawa Y. 2000. Oxalate-degrading Enterococcus faecalis. Microbiol. Immunol. 44:235–240. 10.1111/j.1348-0421.2000.tb02489.x [DOI] [PubMed] [Google Scholar]
- 12.Sundset MA, Barboza PS, Green TK, Folkow LP, Blix AS, Mathiesen SD. 2010. Microbial degradation of usnic acid in the reindeer rumen. Naturwissenschaften 97:273–278. 10.1007/s00114-009-0639-1 [DOI] [PubMed] [Google Scholar]
- 13.Maurice CF, Haiser HJ, Turnbaugh PJ. 2013. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 152:39–50. 10.1016/j.cell.2012.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franceschi VR, Nakata PA. 2005. Calcium oxalate in plants: formation and function. Annu. Rev. Plant Biol. 56:41–71. 10.1146/annurev.arplant.56.032604.144106 [DOI] [PubMed] [Google Scholar]
- 15.James LF, Butcher JE. 1972. Halogeton poisoning of sheep: effect of high level of oxalate intake. J. Anim. Sci. 35:1233–1238 [DOI] [PubMed] [Google Scholar]
- 16.Lincoln SD, Black B. 1980. Halogeton poisoning in range cattle. J. Am. Vet. Med. Assoc. 176:717–718 [PubMed] [Google Scholar]
- 17.Holmes RP, Goodman HO, Assimos DG. 2001. Contribution of dietary oxalate to urinary oxalate excretion. Kidney Int. 59:270–276. 10.1046/j.1523-1755.2001.00488.x [DOI] [PubMed] [Google Scholar]
- 18.Ruiz N, Ward D, Saltz D. 2002. Calcium oxalate crystals in leaves of Pancratium sickenbergeri: constitutive or induced defense? Funct. Ecol. 16:99–105. 10.1046/j.0269-8463.2001.00594.x [DOI] [Google Scholar]
- 19.Massey LK. 2007. Food oxalate: factors affecting measurement, biological variation, and bioavailability. J. Am. Diet. Assoc. 107:1191–1194. 10.1016/j.jada.2007.04.007 [DOI] [PubMed] [Google Scholar]
- 20.Gontzea I, Sutzescu P. 1968. Natural antinutritive substances in foodstuffs and forages. S. Karger, Basel, Switzerland [Google Scholar]
- 21.Concon JM. 1988. Food toxicology—principles and concepts. Marcel Dekker, New York, NY [Google Scholar]
- 22.Hodgkinson A. 1977. Oxalic acid in biology and medicine. Academic Press, New York, NY [Google Scholar]
- 23.Conyers RAJ, Bals R, Rofe AM. 1990. The relation of clinical catastrophes, endogenous oxalate production, and urolithiasis. Clin. Chem. 36:1717–1730 [PubMed] [Google Scholar]
- 24.Allison MJ, Cook HM, Milne DB, Gallagher S, Clayman RV. 1986. Oxalate degradation by gastrointestinal bacteria from humans. J. Nutr. 116:455–460 [DOI] [PubMed] [Google Scholar]
- 25.Ito H, Miura N, Masai M, Yamamoto K, Hara T. 1996. Reduction of oxalate content of foods by the oxalate degrading bacterium, Eubacterium lentum WYH-1. Int. J. Urol. 3:31–34. 10.1111/j.1442-2042.1996.tb00626.x [DOI] [PubMed] [Google Scholar]
- 26.Sidhu H, Enatska L, Ogden S, Williams WN, Allison MJ, Peck AB. 1997. Evaluating children in the Ukraine for colonization with the intestinal bacterium Oxalobacter formigenes, using a polymerase chain reaction-based detection system. Mol. Diagn. 2:89–97. 10.1016/S1084-8592(97)80015-X [DOI] [PubMed] [Google Scholar]
- 27.Hokama S, Toma C, Iwanaga M, Morozumi M, Sagaya K, Ogawa Y. 2005. Oxalate-degrading Providencia rettgeri isolated from human stools. Int. J. Urol. 12:533–538. 10.1111/j.1442-2042.2005.01083.x [DOI] [PubMed] [Google Scholar]
- 28.Weese JS, Weese HE, Yuricek L, Rousseau J. 2004. Oxalate degradation by intestinal lactic acid bacteria in dogs and cats. Vet. Microbiol. 101:161–166. 10.1016/j.vetmic.2004.03.017 [DOI] [PubMed] [Google Scholar]
- 29.Ren Z, Pan C, Jiang L, Wu C, Liu Y, Zhong Z, Ran L, Ren F, Chen X, Wang Y. 2011. Oxalate-degrading capabilities of lactic acid bacteria in canine feces. Vet. Microbiol. 152:368–373. 10.1016/j.vetmic.2011.05.003 [DOI] [PubMed] [Google Scholar]
- 30.Sahin N. 2003. Oxalotrophic bacteria. Res. Microbiol. 154:399–407. 10.1016/S0923-2508(03)00112-8 [DOI] [PubMed] [Google Scholar]
- 31.Abratt VR, Reid SJ. 2010. Oxalate-degrading bacteria of the human gut as probiotics in the management of kidney stone disease. Adv. Appl. Microbiol. 72:63–87. 10.1016/S0065-2164(10)72003-7 [DOI] [PubMed] [Google Scholar]
- 32.Campieri C, Campieri M, Bertuzzi V, Swennen E, Matteuzzi D, Stefoni S, Pirovano F, Centi C, Ulisse S, Famularo G, De Simone C. 2001. Reduction of oxaluria after an oral course of lactic acid bacteria at high concentration. Kidney Int. 60:1097–1105. 10.1046/j.1523-1755.2001.0600031097.x [DOI] [PubMed] [Google Scholar]
- 33.Audic S, Robert C, Campagna B, Parinello H, Claverie JM, Raoult D, Drancourt M. 2007. Genome analysis of Minibacterium massiliensis highlights the convergent evolution of water-living bacteria. PLoS Gen. 3:e138. 10.1371/journal.pgen.0030138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turroni S, Vitali B, Bendazzoli C, Candela M, Gotti R, Federici F, Pirovano F, Brigidi P. 2007. Oxalate consumption by lactobacilli: evaluation of oxalyl-CoA decarboxylase and formyl-CoA transferase activity in Lactobacillus acidophilus. J. Appl. Microbiol. 103:1600–1607. 10.1111/j.1365-2672.2007.03388.x [DOI] [PubMed] [Google Scholar]
- 35.Turroni S, Bendazzoli C, Dipalo SCF, Candela M, Vitali B, Gotti R, Brigidi P. 2010. Oxalate-degrading activity in Bifidobacterium animalis subsp. lactis: impact of acidic conditions on the transcriptional levels of the oxalyl-CoA decarboxylase and formyl-CoA transferase genes. Appl. Environ. Microbiol. 76:5609–5620. 10.1128/AEM.00844-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirley EK, Schmidt-Nielsen K. 1967. Oxalate metabolism in the pack rat, sand rat, hamster, and white rat. J. Nutr. 91:496–502 [DOI] [PubMed] [Google Scholar]
- 37.Justice KE. 1985. Oxalate digestability in Neotoma albigula and Neotoma mexicana. Oecologia 67:231–234. 10.1007/BF00384290 [DOI] [PubMed] [Google Scholar]
- 38.Palgi N, Ronen Z, Pinshow B. 2008. Oxalate balance in fat sand rats feeding on high and low calcium diets. J. Comp. Physiol. B 178:617–622. 10.1007/s00360-008-0252-1 [DOI] [PubMed] [Google Scholar]
- 39.Carleton MD. 1973. A survey of gross stomach morphology in New World Cricetinae (Rodentia, Muroidea) with comments on functional interpretations. Museum of Zoology, University of Michigan, Ann Arbor, MI [Google Scholar]
- 40.Kohl KD, Weiss RB, Dale C, Dearing MD. 2011. Diversity and novelty of the gut microbial community of an herbivorous rodent (Neotoma bryanti). Symbiosis 54:47–54. 10.1007/s13199-011-0125-3 [DOI] [Google Scholar]
- 41.Daniel SL, Hartman PA, Allison MJ. 1987. Microbial degradation of oxalate in the gastrointestinal tract of rats. Appl. Environ. Microbiol. 53:1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada T, Saitoh T, Sasaki E, Nishitani Y, Osawa R. 2006. Role of tannin-binding salivary proteins and tannase-producing bacteria in the acclimation of the Japanese wood mouse to acorn tannins. J. Chem. Ecol. 32:1165–1180. 10.1007/s10886-006-9078-z [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez H, Landete JM, Rivas BDL, Muñoz R. 2008. Metabolism of food phenolic acids by Lactobacillus plantarum CECT 748T. Food Chem. 107:1393–1398. 10.1016/j.foodchem.2007.09.067 [DOI] [Google Scholar]
- 44.Azcarate-Peril MA, Bruno-Bárcena JM, Hassan HM, Klaenhammer TR. 2006. Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus. Appl. Environ. Microbiol. 72:1891–1899. 10.1128/AEM.72.3.1891-1899.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kohl KD, Miller AW, Marvin J, Mackie R, Dearing MD. 24 December 2013. Herbivorous rodents (Neotoma spp.) harbor an abundant and active foregut microbiota. Environ. Microbiol. 10.1111/1462-2920.12376 [DOI] [PubMed] [Google Scholar]
- 48.Sahin N, Gokler I, Tamer AU. 2002. Isolation, characterization and numerical taxonomy of novel oxalate-oxidizing bacteria. J. Microbiol. Seoul 40:109–118 [Google Scholar]
- 49.Gnanandarajah JS, Johnson TJ, Kim HB, Abrahante JE, Lulich JP, Murtaugh MP. 2012. Comparative faecal microbiota of dogs with and without calcium oxalate stones. J. Appl. Microbiol. 113:745–756. 10.1111/j.1365-2672.2012.05390.x [DOI] [PubMed] [Google Scholar]
- 50.Chandra TS, Shethna YI. 1975. Isolation and characterization of some new oxalate-decomposing bacteria. Antonie Van Leeuwenhoek 41:101–111. 10.1007/BF02565041 [DOI] [PubMed] [Google Scholar]
- 51.Murphy C, Murphy S, O'Brien F, O'Donoghue M, Boileau T, Sunvold G, Reinhart G, Kiely B, Shanahan F, O'Mahony L. 2009. Metabolic activity of probiotics—oxalate degradation. Vet. Microbiol. 136:100–107. 10.1016/j.vetmic.2008.10.005 [DOI] [PubMed] [Google Scholar]
- 52.Balasubramanian S, Zheng D, Liu YJ, Fang G, Frankish A, Carriero N, Robilotto R, Cayting P, Gerstein M. 2009. Comparative analysis of processed ribosomal protein pseudogenes in four mammalian genomes. Genome Biol. 10:R2. 10.1186/gb-2009-10-1-r2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MT, Mitchell WJ, Carter AT, Bentley ST, Mason DR, Crossman L, Paul CJ, Ivens A, Wells-Bennik MHJ, Davis IJ, Cerdeño-Tárraga AM, Churcher C, Quail MA, Chillingworth T, Feltwell T, Fraser A, Goodhead I, Hance Z, Jagels K, Larke N, Maddison M, Moule S, Mungall K, Norbertczak H, Rabbinowitsch E, Sanders M, Simmonds M, White B, Whithead S, Parkhill J. 2007. Genome sequence of a proteolytic (group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 17:1082–1092. 10.1101/gr.6282807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belenguer A, Ben Bati M, Hervás G, Toral PG, Yáñez-Ruiz DR, Frutos P. 2013. Impact of oxalic acid on rumen function and bacterial community in sheep. Animal 7:940–947. 10.1017/S1751731112002455 [DOI] [PubMed] [Google Scholar]
- 55.Kuhner CH, Hartman PA, Allison MJ. 1996. Generation of a proton motive force by the anaerobic oxalate-degrading bacterium, Oxalobacter formigenes. Appl. Environ. Microbiol. 62:2494–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatch M, Cornelius J, Allison M, Sidhu H, Peck A, Freel RW. 2006. Oxalobacter sp. reduces urinary oxalate excretion by promoting enteric oxalate secretion. Kidney Int. 69:691–698. 10.1038/sj.ki.5000162 [DOI] [PubMed] [Google Scholar]
- 57.Hatch M, Gjymishka A, Salido EC, Allison MJ, Freel RW. 2011. Enteric oxalate elimination is induced and oxalate is normalized in a mouse model of primary hyperoxaluria following intestinal colonization with Oxalobacter. Am. J. Physiol. Gastrointest. Liver Physiol. 300:G461–G469. 10.1152/ajpgi.00434.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sidhu H, Schmidt ME, Cornelius JG, Van Thamilsel S, Khan SR, Hesse A, Peck AB. 1999. Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract-dwelling bacterium Oxalobacter formigenes: possible prevention by gut recolonization or enzyme replacement therapy. J. Am. Soc. Nephrol. 10:334–340 [PubMed] [Google Scholar]
- 59.Dehning I, Schink B. 1989. Two new species of anaerobic oxalate-fermenting bacteria, Oxalobacter vibrioformis sp. nov. and Clostridium oxalicum sp. nov., from sediment samples. Arch. Microbiol. 153:79–84. 10.1007/BF00277545 [DOI] [Google Scholar]