Abstract
Sulfur oxidation by chemolithotrophic bacteria is well known; however, sulfur oxidation by heterotrophic bacteria is often ignored. Sulfur dioxygenases (SDOs) (EC 1.13.11.18) were originally found in the cell extracts of some chemolithotrophic bacteria as glutathione (GSH)-dependent sulfur dioxygenases. GSH spontaneously reacts with elemental sulfur to generate glutathione persulfide (GSSH), and SDOs oxidize GSSH to sulfite and GSH. However, SDOs have not been characterized for bacteria, including chemolithotrophs. The gene coding for human SDO (human ETHE1 [hETHE1]) in mitochondria was discovered because its mutations lead to a hereditary human disease, ethylmalonic encephalopathy. Using sequence analysis and activity assays, we discovered three subgroups of bacterial SDOs in the proteobacteria and cyanobacteria. Ten selected SDO genes were cloned and expressed in Escherichia coli, and the recombinant proteins were purified. The SDOs used Fe2+ for catalysis and displayed considerable variations in specific activities. The wide distribution of SDO genes reveals the likely source of the hETHE1 gene and highlights the potential of sulfur oxidation by heterotrophic bacteria.
INTRODUCTION
Biological oxidation of inorganic sulfur compounds has been studied for more than 120 years, ever since Winogradsky's discovery that Beggiatoa oxidizes hydrogen sulfide as an energy source for growth (1). Extensive research has been done with chemolithotrophs that use sulfur oxidation for energy or with some phototrophic bacteria that extract electrons from reduced sulfur for photosynthesis (2–4). Recently, sulfur oxidation has been found in mitochondria from clams (5), worms (6), fish (7), humans (8), and plants (9). In mitochondria, three enzymes are likely involved in oxidizing H2S to sulfite. Sulfide:quinone oxidoreductase (SQR) oxidizes sulfide to sulfane sulfur, as in disulfide (HS-SH) or thiosulfate (10), and two electrons are transferred through the mitochondrial electron transport chain to O2. Then, rhodanese (RHOD), also known as thiosulfate sulfurtransferase (EC 2.8.1.1), is proposed to transfer the sulfane sulfur to GSH to produce glutathione persulfide (GSSH) (10). Finally, sulfur dioxygenase (SDO) (EC 1.13.11.18) oxidizes the sulfane sulfur in GSSH to sulfite (11). Although sulfide oxidation is generally considered a detoxification process in animals, it can generate ATP via oxidative phosphorylation (8, 12).
Since sulfide oxidation is common in mitochondria, it is likely to have important physiological functions. SDO activities were first identified for chemolithotrophs, but the enzymes have not been purified, and the genes are unknown. SDOs are known as GSH-dependent sulfur dioxygenases because GSH spontaneously reacts with sulfur to produce GSSH, which is oxidized by the enzymes to sulfite and GSH (13). The human ETHE1 (hETHE1) (ethylmalonic encephalopathy 1) protein is the human SDO in mitochondria. Mutations in the hETHE1 gene are the cause of a rare recessive hereditary human disease, ethylmalonic encephalopathy (14). Ethylmalonic encephalopathy patients have increased organic acids, such as lactic acid and ethylmalonic acid, in blood and urine, progressive encephalopathy, and a short life-span. The mutant hETHE1 proteins have reduced SDO activities, likely causing elevated H2S concentrations inside cells (11). H2S at low concentrations is a signaling molecule in the brain, and its accumulation may interfere with proper signaling and can inhibit cytochrome c oxidase (15). Inactivation of the ethe1 gene in the plant Arabidopsis thaliana leads to embryo arrest by the early heart stage (9). Thus, SDOs play important physiological roles in plants and animals.
The conservation of ETHE1 in mitochondria implies that it is inherited from the prokaryotic ancestor of mitochondria, which was supposed to be a heterotrophic bacterium instead of a chemolithotrophic bacterium (16). It has been predicted by sequence analysis that ETHE1 homologues are present in Burkholderia vietnamiensis G4 and Myxococcus xanthus DK 1622 (9). Recently, blh has been reported to potentially code for a sulfide dioxygenase (Blh) in Agrobacterium tumefaciens (AtBlh), and its inactivation renders the mutant more sensitive to H2S (17). However, AtBlh has low sequence identity with hETHE1, and its function has not been biochemically characterized. In addition, the identification of ETHE1 homologues by sequence analysis is not straightforward, because ETHE1 belongs to the metallo-β-lactamase superfamily, including β-lactamases, glyoxylase II enzymes, and enzymes that hydrolyze phosphodiester and sulfuric ester bonds (18). Therefore, the distribution of ETHE1 homologues in bacteria has not been reported, and the function of bacterial ETHE1 homologues has not been demonstrated. In this work, we analyzed ETHE1 homologues in bacteria and identified three subgroups of SDOs in the proteobacteria and cyanobacteria. We cloned and expressed putative SDO genes in Escherichia coli, purified the proteins, and characterized 10 SDO enzymes from heterotrophic bacteria.
MATERIALS AND METHODS
Chemicals and enzymes.
All chemicals were purchased from Sigma-Aldrich (Shanghai, China). Restriction enzymes and Phusion DNA polymerase were from Thermo Scientific (Shanghai, China). PCR primers were from Sangon Biotech (Shanghai, China).
Bacterial strains and plasmids.
E. coli BL21(DE3) was cultured in lysogeny broth (LB) or on LB agar plates at 37°C or as specified. Kanamycin (50 μg ml−1) was added to LB when required. Other bacteria and their sources are listed in Table 1, and they were cultured in LB at 30°C. Genomic DNA was extracted by using a genomic DNA isolation kit (Omega Bio-Tek, Shanghai, China). The genomic DNA of M. xanthus DK 1622 was a gift from Yuezhong Li's lab at Shandong University.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmid | Characteristic(s) | Culturing | Source or reference |
|---|---|---|---|
| Strains | |||
| Escherichia coli BL21(DE3) | Cloning strain | LB at 37°C | Invitrogen |
| E. coli MG1655 | Wild type | LB at 37°C | ATCC 47076 |
| Cupriavidus necator JMP134 | Wild type | LB at 30°C | Ron L. Crawford |
| Pseudomonas putida F1 | Wild type | LB at 30°C | ATCC 700007 |
| P. aeruginosa PAO1 | Wild type | LB at 30°C | ATCC 15692 |
| Burkholderia xenovorans LB400 | Wild type | LB at 30°C | William W. Mohn |
| Agrobacterium tumefaciens strain C58 | Wild type | LB at 30°C | Minqin Wang |
| Sinorhizobium meliloti 1021 | Wild type | LB at 30°C | Michael Kahn |
| Haemophilus influenzae R2846 | Wild type | LB at 30°C | Dongqing Yu |
| Plasmids | |||
| pET-30 Ec/Lic | Kmr, expression vector | Invitrogen | |
| pETAtBlh | pET30 Ec/Lic containing Atblh from A. tumefaciens strain C58 | This study | |
| pETSmBlh | pET30 Ec/Lic containing Smblh from S. meliloti 1021 | This study | |
| pETCnSdoA | pET30 Ec/Lic containing CnsdoA from C. necator JMP134 | This study | |
| pETPpSdoA | pET30 Ec/Lic containing PpsdoA from P. putida F1 | This study | |
| pETPaSdoA | pET30 Ec/Lic containing PasdoA from P. aeruginosa PAO1 | This study | |
| pETBxSdoA | pET30 Ec/Lic containing BxsdoA from B. xenovorans LB400 | This study | |
| pETCnETHE1 | pET30 Ec/Lic containing Cnethe1 from C. necator JMP134 | This study | |
| pETMxETHE1a | pET30 Ec/Lic containing Mxethe1a from M. xanthus DK 1622 | This study | |
| pETEcGloB1 | pET30 Ec/Lic containing EcgloB1 from E. coli MG1655 | This study | |
| pETHiGloB1 | pET30 Ec/Lic containing HigloB1 from H. influenzae | This study | |
| pETEcGloB2 | pET30 Ec/Lic containing EcgloB2 from E. coli MG1655 | This study | |
| pETPaGloB2 | pET30 Ec/Lic containing PagloB2 from P. aeruginosa PAO1 | This study |
LB, Luria broth; Kmr, kanamycin resistance.
Cloning, site-directed mutagenesis, expression, and protein purification.
Each selected gene was PCR amplified by using Phusion DNA polymerase from genomic DNA with appropriate primers (see Table S1 in the supplemental material), and the PCR product was cloned into pET30 Ek/LIC previously digested with NdeI and XhoI by using the In-Fusion HD cloning kit (Clontech, Beijing, China). The recombinant plasmid was transformed into E. coli strain BL21(DE3), and the correct clones were identified by PCR and confirmed by sequencing. Genes with site-directed mutations were constructed using a two-step PCR strategy (19) and transformed into E. coli strain BL21(DE3). The transformants were cultivated in LB with 50 μg ml−1 of kanamycin at 37°C with shaking to a turbidity of 0.6 at 600 nm. Then, the cultures were cooled to room temperature, and solid isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a 0.2 mM concentration to induce gene expression. The induced cultures were incubated at 20°C with shaking overnight. Cells were collected via centrifugation, washed twice with ice-cold 20 mM sodium phosphate buffer (pH 7.4) containing 20 mM imidazole and 300 mM NaCl, and broken via sonication at 4°C. The supernatants were collected after centrifugation at 15,000 × g for 15 min at 4°C, and the His-tagged proteins were purified by using nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Shanghai, China) according to the supplier's recommendations. The buffer was exchanged to 20 mM sodium phosphate buffer (pH 7) containing 1 mM dithiothreitol, and 50% glycerol was added to a final concentration of 10% before storage at −80°C. The enzymes were stable for several weeks under the storage conditions.
SDO activity assay and product identification.
SDO activity was analyzed using the method of Suzuki with some modifications (20). The assay was routinely performed at 25°C. GSSH was produced by mixing equal volumes of 17 mM glutathione in distilled water and a saturated sulfur solution, containing about 17 mM elemental sulfur (21). The reaction was carried out in 3 ml of 100 mM potassium phosphate (KPi) buffer (pH 7.4) containing 0.9 mM GSSH, and an SDO protein was added to initiate the reaction. Oxygen consumption was directly monitored using an Orion RDO meter (Thermo Scientific Inc.) and was used to calculate SDO activities. The RDO meter was calibrated with air-saturated water according to the manufacturer's instruction. The consumption of GSSH and the production of sulfite were also determined. Further, SDO activities were determined at various pH values, from 5.8 to 7.8, in 100 mM KPi buffers at 25°C and at different temperatures, from 20°C to 40°C, in 100 mM KPi buffer at pH 7.4.
Kinetics analysis.
The reaction was carried out in 100 mM KPi buffer (pH 7.4) at 25°C with SDO and various concentrations of GSSH. Km and Vmax values were determined by nonlinear regression analysis of the plots of the reaction rates against GSSH concentrations using the GraFit 5.0 software program (Erithacus Software).
Analytical methods.
Protein concentrations were determined according to the method of Bradford (22), with bovine serum albumin as the standard. The purified protein was checked by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The native molecular masses of SDOs were determined via size exclusion chromatography (SEC). The proteins were separated through a TSKgel G3000SWxl column (7.8 mm by 30 cm, 5 mm packing; TOSOH, Japan) at 35°C with a flow rate of 0.5 ml min−1 by using a Shimadzu LC-20AT high-performance liquid chromatography (HPLC) system with a photo diode array detector. The running buffer was 100 mM sodium phosphate buffer (pH 6.7) containing 100 mM sodium sulfate and 0.05% sodium azide. The native protein standards (product number H2899) from Sigma-Aldrich and the YdiV protein (about 26 kDa) (23) were used as the standards. Sulfite was determined by ion chromatography (ICS-1100 system; Dionex) with a 10-ml linear gradient of KOH from 15 mM to 35 mM at a flow rate 1 ml per min. The retention time of sulfite was 8.6 min. GSSH was determined using a colorimetric method (24). Glutathione was detected by the method of Ellman (25). The metals bound to the purified proteins were analyzed by using inductively coupled plasma-mass spectrometry (ICP-MS) at the Shandong Analysis and Test Center, Jinan, Shandong, China.
Chelating agent treatment.
SDOs were diluted to 0.1 mM with 100 mM KPi buffer (pH 7.4), incubated with 1 mM EDTA for 2 h on ice and then dialyzed in 100 mM KPi (pH 7.4) at 4°C for 2 h. The activities were then assayed. Controls for activity assays were untreated but dialyzed SDOs and reconstituted SDOs, which were the EDTA-treated SDOs further incubated with 10 mM Fe2+ for 10 min.
Bioinformatics.
TBLASTN on the NCBI website was used to find SDO homologues. Maximum identities were from TBLASTN for most homologous regions between the query and target proteins. Sequence identities were obtained by using the ALIGN tool (optimal global alignment of two protein sequences) at the Biology Workbench website. A phylogenetic tree was constructed from the alignment of multiple proteins using the program ClustalX version 2.1, followed by a neighbor-joining analysis using MEGA version 5.10, with a pairwise deletion, p-distance distribution, and bootstrap analysis of 1,000 repeats. The distribution of SDOs in bacteria was vigorously checked from the whole bacteria genome database in NCBI. The amino acid sequences of hETHE1, C. necator SdoA (CnSdoA), and AtBlh were used to analyze by TBLASTN every genus of the available genomes. Individual genomes showing the query coverage of more than 90%, an E value lower than e−30, and a maximum identity level higher than 50% with the query protein were selected and further evaluated. If the homologue had a sequence identity level higher than 40% by using ALIGN at the Biology Workbench website, the genome was considered to harbor a specific type of SDO gene.
RESULTS
TBLASTN search of hETHE1 homologues.
TBLASTN search of bacterial genomes with the hETHE1 protein sequence as the query revealed that hETHE1 homologues were mainly present in the proteobacteria (190 hits of a total of 285 hits) and cyanobacteria (82 hits of total 285 hits), and the top 100 hits showed maximum identities from 43% to 59%. A query using the A. thaliana ETHE1 (AtETHE1) sequence obtained results similar to those for hETHE1, with most homologues present in the proteobacteria (173 hits of a total of 227 hits) and cyanobacteria (53 hits of a total of 227 hits). The top 100 hits showed maximum identities from 44% to 55%.
To further identify potential SDOs with low homology, the hETHE1 sequence was used to analyze individual bacterial genomes by using TBLASTN. Table 2 shows the most similar proteins from E. coli strain MG1655, Burkholderia xenovorans strain LB400, Pseudomonas aeruginosa strain PAO1, Pseudomonas putida strain F1, Cupriavidus necator strain JMP134, A. tumefaciens strain C58, and M. xanthus strain DK 1622. All of them were hypothetical proteins with the metallo-β-lactamase domain. Among the bacterial proteins, AtBlh is the only protein that has been proposed to be a sulfide dioxygenase on the basis of genetic analysis (17). AtBlh had low sequence identity with hETHE1 (Table 2), and a TBLASTN search identified AtBlh homologues mainly in the order Rhizobiales of the Alphaproteobacteria and the order Xanthomonadales of the Gammaproteobacteria, including Blh of Afipia felis (AfBlh), Blh of Oligotropha carboxidovorans (OcBlh), Blh of Sinorhizobium meliloti (SmBlh), and Blh of Xylella fastidiosa (XfBlh).
TABLE 2.
Proteins with the highest identities to hETHE1 in seven bacteria
| Strain | Protein name | GenBank accession no. | Identity (%)a |
|---|---|---|---|
| E. coli MG1655 | EcGloB1 | NP_415447 | 23.4 |
| EcGloB2 | NP_414748 | 20.6 | |
| B. xenovorans LB400 | BxSdoA | YP_554628 | 27.9 |
| BxGloB2 | ABE28683 | 24.7 | |
| P. aeruginosa PAO1 | PaSdoA | NP_251605 | 26.9 |
| PaGloB2 | NP_249523 | 23.2 | |
| P. putida F1 | PpSdoA | ABQ76243 | 26 |
| PpGloB2 | YP_001266145 | 24.3 | |
| C. necator JMP134 | CnSdoA | YP_297791 | 28.9 |
| A. tumefaciens strain C58 | CnETHE1 | YP_297536 | 39.7 |
| AtBlh | Q8UAA9 | 21.2 | |
| AtGloB1 | NP_356997 | 24.8 | |
| M. xanthus DK 1622 | MxETHE1a | YP_633997 | 47.5 |
| MxETHE1b | YP_632494 | 44.8 | |
| MxETHE1c | YP_628462 | 41.3 |
The percentage of identity was the result from the TBLASTN search of a bacterial genome.
Phylogenetic analysis.
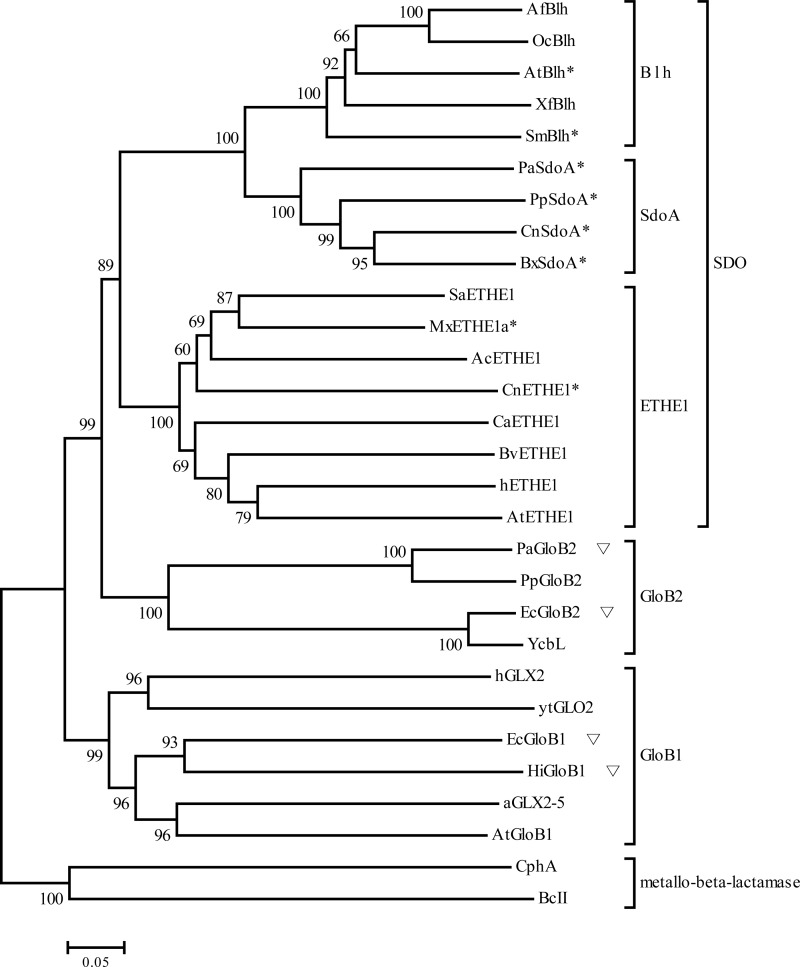
In order to identify potential SDOs from the TBLASTN-identified proteins, phylogenetic analysis was used to group the proteins with hETHE1, Blh, glyoxylase II enzymes, and metallo-β-lactamases (Fig. 1). The metallo-β-lactamases were more distantly related to glyoxylase II enzymes and SDOs. There were two glyoxylase II groups: the GloB1 group contained the well-characterized human glyoxylase II (hGlx2) and plant glyoxylase II (aGlx2-5), and the GloB2 group had a recently characterized bacterial glyoxylase II (YcbL) (26). There were three potential SDO subgroups: Blh, the SdoA group, and the ETHE1 group.
FIG 1.
Phylogenetic analysis of SDOs. The tree was generated with a neighbor-joining method by using MEGA software program, version 5.10. Proteins are listed below, with the organism origin and accession number: AfBlh, A. felis (ZP_11421028); OcBlh, O. carboxidovorans (YP_002287251); AtBlh, A. tumefaciens (Q8UAA9); XfBlh, X. fastidiosa (NP_298058); SmBlh, S. meliloti (NP_435818); PaSdoA, P. aeruginosa (NP_251605); PpSdoA, P. putida (ABQ76243); CnSdoA, C. necator (YP_297791); BxSdoA, B. xenovorans (YP_554628); SaETHE1, Stigmatella aurantiaca (YP_003957083); MxETHE1a, M. xanthus (YP_633997); AcETHE1, A. caldus SM-1 (AEK59246); CnETHE1, C. necator (YP_297536); CaETHE1, Cyanobacterium aponinum (YP_007162862); BvETHE1, B. vietnamiensis (ZP_00420127); hETHE1, Homo sapiens (NP_055112); AtETHE1, A. thaliana (NP_974018); PaGloB2, P. aeruginosa (NP_249523); PpGloB2, P. putida (ABQ76961); EcGloB2, E. coli (NP_415447); YcbL, Salmonella enterica (CAD05397); EcGloB1, E. coli (NP_414748); HiGloB1, Haemophilus influenzae (ADO96205); AtGloB1, A. tumefaciens (NP_356997); aGLX2-5, A. thaliana (NP_850166); ytGLO2, Saccharomyces cerevisiae (CAA71335); hGLX2, Homo sapiens (CAA62483); BcII, Bacillus cereus (M11189); CphA, Aeromonas hydrophila (CAA40386). An asterisk (∗) marks the proteins that tested as having SDO activities, and an inverted triangle (▽) indicates that the proteins tested have no SDO activities in this study.
Detection of SDO activities and native molecular weights.
Selected members from the GloB1, GloB2, ETHE1, Blh, and SdoA groups were produced in E. coli, purified, and tested for using GSSH as a substrate. M. xanthus ETHE1a (MxETHE1a), MxETHE1b, MxETHE1c, C. necator ETHE1 (CnETHE1), CnSdoA, P. aeruginosa SdoA (PaSdoA), P. putida SdoA (PpSdoA), B. xenovorans SdoA (BxSdoA), AtBlh, and SmBlh had activities for GSSH oxidation, while E. coli GloB1 (EcGloB1), EcGloB2, P. aeruginosa GloB2 (PaGloB2), and Haemophilus influenzae GloB1 (HiGloB1) did not. The SDOs oxidized GSSH. The SDOs did not use sulfide, elemental sulfur, thiosulfate, or glutathione as a substrate. The SDOs were monomers as revealed by size exclusion chromatography analysis.
Reaction balance.
The reaction balance was achieved with 30 μg of CnSdoA in a 3-ml reaction volume. After the reaction, 215 ± 15 μM GSSH was decreased, and 220 ± 10 μM oxygen was consumed; 222 ± 21 μM GSH and 212 ± 9 μM sulfite were produced (see Fig. S1 in the supplemental material). Thus, equal molar amounts of GSSH and oxygen were consumed, and the same molar equivalents of GSH and sulfite were produced: GSSH + O2 + H2O → H2SO3 + GSH.
Metal analysis.
When purified SDOs were analyzed using inductively coupled plasma mass spectrometry (ICP-MS), iron was the dominant species (Table 3). In CnSdoA, it contained about 0.55 equivalents of iron, 0.1 equivalents of manganese, and trace amount of cooper and zinc. If one protein bound one metal, there were about 33% of CnSdoA proteins without metal. When 1 μl of 100 mM FeSO4 in distilled water was added to 10 μl of the CnSdoA protein and incubated on ice for 30 min before assaying, the enzyme specific activity was increased by about 30%. Purified CnETHE1 had almost no metal, and its activity was initially undetectable. Its activity was detectable only after CnETHE1 was preincubated with Fe2+. Other SDOs also had iron as the dominant metal species, except PpSdoA and BxSdoA, which had more Mn2+ than Fe2+. Most SDO activities were stimulated by incubating with Fe2+. Mn2+ was as effective as Fe2+ in stimulating the activities for AtBlh and SmBlh. Cu2+ and Zn2+ did not stimulate any SDO activities.
TABLE 3.
Metal analysis of purified SDOsa
| SDO | Protein concn (μM) | Metal concn (μM) |
|||
|---|---|---|---|---|---|
| Iron | Cooper | Manganese | Zinc | ||
| CnSdoA | 460 | 253.6 | 4.4 | 45 | 4.1 |
| PpSdoA | 51.4 | 13.4 | 1.9 | 18.6 | 4.6 |
| PaSdoA | 40.4 | 29.4 | 1.7 | 6.3 | 1.6 |
| BxSdoA | 180 | 38.8 | 1.9 | 68.6 | 26.4 |
| AtBlh | 8.3 | 2.7 | 2.4 | 2 | |
| SmBlh | 39 | 16.6 | 0.9 | 4.7 | 2.2 |
| MxETHE1a | 65 | 25 | 12.3 | 2 | 11.6 |
| CnETHE1 | 125 | 11.2 | 6.9 | 3.8 | 33.3 |
The proteins were eluted from an Ni-NTA agarose column with the elution buffer and directly used for ICP-MS analysis. The data were corrected by subtracting the trace metals present in the elution buffer.
Three SDOs, AtBlh, CnSdoA, and MxETHE1a, were treated with EDTA. The activities for SmBlh and MxETHE1a were unchanged in comparison to those of untreated controls, but the activity for CnSdoA was reduced by 26%. The activity of EDTA-treated CnSdoA was recovered when it was further incubated with Fe2+. The results, together with the result of CnETHE1 that required Fe2+ reconstitution before the activity assay, suggest that individual SDOs have different affinities for Fe2+.
pH and temperature optima.
The tested SDOs were mesophilic enzymes with pH optima at the physiological pH of the cytoplasm, ranging from 7.4 to 7.8 (see Fig. S2A in the supplemental material). The optimal temperature for most SDOs in 100 mM KPi (pH 7.4) was from 35 to 40°C (see Fig. S2B).
Kinetics analysis.
Kinetic parameters of the 10 bacterial SDOs were determined in 100 mM KPi buffer (pH 7.4) at 25°C. The Km values varied from 108 μM to 342 μM GSSH, and Vmax ranged from 4.6 to 90.3 μmol O2 min−1 mg−1 of protein. According to the kcat/Km ratio, MxETHE1a had the best catalytic efficiency (Table 4).
TABLE 4.
Kinetic parameters of SDOsa
| SDO | Vmax (μmol O2 min−1 mg−1) | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| PaSdoA | 10.9 ± 2.1 | 5.8 | 342 ± 192 | 17.0 |
| PpSdoA | 11.9 ± 0.3 | 6.5 | 150 ± 72 | 43.3 |
| BxSdoA | 7.2 ± 1.2 | 3.8 | 204 ± 102 | 18.6 |
| AtBlh | 4. 6 ± 1.0 | 3.6 | 108 ± 84 | 33.3 |
| SmBlh | 5.7 ± 1.0 | 4.5 | 168 ± 90 | 26.8 |
| MxETHE1a | 90.3 ± 13.3 | 38.5 | 132 ± 72 | 291.7 |
| MxETHE1b | 38.6 ± 2.5 | 16.5 | 138 ± 32 | 119.6 |
| MxETHE1c | 63.1 ± 6.0 | 26.6 | 248 ± 60 | 107.3 |
| CnETHE1 | 6.7 ± 0.4 | 3.0 | 108 ± 24 | 27.8 |
| CnSdoA | 23.2 ± 3.1 | 12.8 | 336 ± 132 | 38.1 |
| CnSdoA D217G | 0.25 ± 0.01 | 0.14 | 78 ± 2 | 1.8 |
| CnSdoA D217N | 1.5 ± 0.2 | 0.8 | 258 ± 114 | 3.1 |
Kinetic analysis was done using 100 mM KPi buffer (pH 7.4) at 25°C with varying concentrations of GSSH. The kinetic parameters with standard errors were obtained by using the GraFit 5 software program.
Two conserved amino acid residues specific to SDOs.
AtETHE1 and hETHE1 have conserved amino acid residues for metal binding and for possible substrate binding (9). However, most of the residues are also conserved in other members of the metallo-β-lactamase superfamily (Fig. 2). Two amino acid residues, Asp196 and Asn244 of hETHE1, were identified as conserved in SDOs but not in related proteins (Fig. 2). When the Asp196 equivalent (Asp217) of CnSdoA was mutated to Gly or Asn, the mutant proteins CnSdoA D217G and CnSdoA D217N had significantly reduced activities (Table 4). The decreased kcat/Km values mainly were caused by the decreased kcat values. When the Asn244 equivalent (Asn274) of CnSdoA was mutated to Val or Lys, the mutant proteins CnSdoA N274V and CnSdoA N274K were produced as inclusion bodies in E. coli.
FIG 2.
Multiple sequence alignment of selected SDOs with two known glyoxylase II enzymes. The protein origins and accession numbers are given in the legend for Fig. 1. YcbL, bacterial glyoxylase II; hGLX2, human glyoxylase II. Identical and similar residues are highlighted in black and gray, respectively. Arrow (↓), metal binding sites of all SDOs and glyoxylase II enzymes; asterisk (∗), the amino acid residues whose mutations affect the function of the hETHE1 protein; inverted triangle (▼), two converse and specific residues in SDOs.
Distribution of SDOs in bacteria.
A relatively strict standard was used to identify the distribution of ETHE1, SdoA, and Blh group members in bacteria. When hETHE1 was used as the query protein, the total number of genomes with homologues was 78, including 26 from the cyanobacteria and 52 from the proteobacteria. When searched with CnSdoA, the total number of genomes was 266, with all being in the proteobacteria, including 78 members of the Alphaproteobacteria, 82 of the Betaproteobacteria, 101 of the Gammaproteobacteria, and 5 of the Deltaproteobacteria. When the query protein was AtBlh, only 24 proteobacterial genomes were found to contain its homologues, including 15 of the Rhizobiales order, 5 of the Enterobacteriales order, and 4 of the Xanthomonadales order. Of all the 1,866 completed genomes of the proteobacteria and cyanobacteria (NCBI genome database, June 2013), the SDO-containing bacteria accounted for about 20%. Thus, the bacterial ETHE1 genes in the proteobacteria are likely the source of mitochondrial ETHE1 genes.
DISCUSSION
Conserved amino acid residues of SDOs.
All the metal binding amino acid residues are conserved in glyoxylase II enzymes (27, 28). The conserved amino acid residues are also involved in metal binding in AtETHE1 (9) and in bacterial SDOs (Fig. 2). Despite the conserved amino acid residues, glyoxylase II enzymes bind two metal ions, whereas AtETHE1 binds one metal ion. The structural comparison of AtETHE1 and human glyoxylase II (hGlx2) has revealed minor structure variations that allow the binding of one metal or two metals (29). The results from our metal analysis and metal reconstitution suggest that the bacterial SDOs bind one Fe2+ for catalysis and some SDOs in the Blh group may also use Mn2+ for catalysis. AtETHE1 and hETHE1 are known to use Fe2+ for catalysis (11, 29). The conserved amino acid residues for metal binding among SDOs and glyoxylase II enzymes indicate the close relationship between the two types of enzymes.
The glyoxylase II enzymes have conserved amino acid residues for the binding of the glutathionyl moiety of their substrate, S-d-lactoylglutathione, but they are not the same in SDOs despite the fact that SDOs also use a substrate with the glutathionyl moiety (9). The specific amino acid residues involved in substrate binding in SDOs have not been confirmed by structural analysis. However, residues Tyr38, Thr136, Thr152, Cys161, Arg163, Leu185, and Asp196 are important for the function of hETHE1 (14, 30), and their mutations are associated with ethylmalonic encephalopathy in humans (14). Since most of these residues are also conserved in other members of the metallo-β-lactamase superfamily (Fig. 2), their specific functions in SDOs are unknown. Among them, the only amino acid residue specific to SDOs is Asp196 of hETHE1. In the AtETHE1 structure (2GCU), the Asp184 (the hETHE1 Asp196 equivalent) side chain carboxylate stabilizes a loop between two β-sheets via interaction with the backbone amide nitrogen and carbonyl oxygen of other amino acid residues within the loop, and a detailed account has been given (11). The mutant hETHE1 D196N has a reduced catalytic efficiency (11), which is in agreement with our observation that the mutants CnSdoA D217G and CnSdoA D217N had reduced catalytic efficiencies (Table 4). The mutant CnSdoA D217N still preserves some of the interactions, and the mutant CnSdoA D217G does not maintain any of the interactions. Consequently, the loop should become more disordered. For CnSdoA, the mutations resulted in reduced Km values and kcat values with a net loss of catalytic efficiencies for both mutant proteins (Table 4). Thus, the loop may play a role in substrate binding, and the disordered loop may increase its affinity for the substrate, but a tight binding may decrease the catalytic turnover. Further, we identified that hETHE1 Asn244 was conserved and specific to SDOs. In the AtETHE1 structure (2GCU), Asn232 the (hETHE1 Asn244 equivalent) is in the last α-helix at the C terminus, and the carbonyl oxygen and amine group of its carboxamide interact with the backbone amide nitrogen of Thr68 and the backbone carbonyl oxygen of His66, respectively. Since His66 is next to three metal-binding residues (His65, His63, and Asp61), the interactions may be important for metal binding or the stabilization of the C terminus. When CnSdoA Asn274 (hETHE1 Asn244 equivalent) was mutated to Val or Lys, the mutant proteins could not be produced as soluble proteins in E. coli.
SDO genes are often associated with other sulfur-related genes in bacterial genomes.
We characterized 10 bacterial SDOs from the proteobacteria. The SDO genes are often present as a single copy per bacterial genome, and they are often linked to other sulfur-related genes, such as the genes coding for the putative SQR, rhodanese, a sulfite exporter. Some genomes harbor more than one SDO gene. C. necator JMP134 has two SDO genes, encoding CnSdoA and CnETHE1, and M. xanthus DK1622 contains three ETHE1 genes, coding for MxETHE1a, MxETHE1b, and MxETHE1c. When a genome contains more than one SDO gene, not all of them are physically linked to other sulfur-related genes.
The SDO gene and its associated genes reveal the sulfur-metabolizing potential. For example, in C. necator JMP134, the CnSdoA gene (Reut_B3589) is linked to Reut_B3588, Reut_B3590, and Reut_B3591. Reut_B3590 encodes a Fis family transcriptional regulator, and Reut_B3591 codes for a potential sulfite exporter (TauE). Reut_B3588 encodes a hypothetical protein (YP_297790) containing two domains: putative phosphatase (DUF442) and uncharacterized NAD (FAD)-dependent dehydrogenase (HcaD). Further analysis suggests that DUF442 codes for a rhodanese domain with a catalytic Cys residue, similar to E. coli rhodanese YnjE (17), and HcaD is likely an SQR because its homologue (PP_0053) in P. putida KT2440 has recently been reported to be an FAD-containing SQR (31). SQR oxidizes sulfide to sulfane sulfur, and rhodanese may transfer the sulfane sulfur to glutathione to produce GSSH, which is the substrate for SDO (10). Thus, the bacteria with these genes have the potential to oxidize sulfide to sulfite before exporting the latter into the medium.
Rhodanese is present in various forms in bacteria. Individual rhodanese genes are often associated with SDO genes in bacteria, such as in M. xanthus DK1622. It can also be fused with SQR, such as in C. necator JMP134, or with SDO, such as in Blh proteins, which contain a rhodanese domain at the N terminus. In addition, some ETHE1 proteins, such as the B. vietnamiensis G4 protein (YP_001116099), have a rhodanese domain at the C terminus.
SDO may be involved in sulfur oxidation in chemolithotrophs.
Chemolithotrophs employ multiple systems for sulfur oxidation (32), including sulfur oxygenase reductase (SOR) and SDO. SOR converts S0 to sulfide, thiosulfate, and sulfite (33, 34). On the other hand, SDOs oxidize sulfur to sulfite (35). Although SDOs were initially identified in the cell extracts of some chemolithotrophic bacteria (13), they have not been characterized (36, 37). We searched the genome of Acidithiobacillus caldus SM-1 and found one SDO gene encoding a hypothetical protein (AEK59246) belonging to the ETHE1 group (Fig. 1). The protein does not has a signal peptide as determined by using SignalP analysis (38), and all of the characterized bacterial SDOs do not have a signal peptide either.
Physiological functions of heterotrophic bacterial SDOs.
The bacterial SDOs may have various functions. The detoxification function of AtBlh has been demonstrated. In A. tumefaciens, the inactivation of Atblh causes the blh mutant to accumulate high levels of H2S in the cell and to grow more slowly under low-oxygen conditions or with externally introduced H2S (17). We tried but failed to identify a gene coding for a potential SQR in the A. tumefaciens genome. Perhaps due to the same reason, Guimaraes et al. proposed that AtBlh oxidized H2S to sulfite (17). Here, we showed that AtBlh oxidized only GSSH. Thus, it is unclear how A. tumefaciens converts H2S to GSSH, which can be oxidized by AtBlh to sulfite. Mitochondria also oxidize H2S to sulfite for detoxification and possibly for the production of extra ATP (8). However, the produced ATP may not significantly contribute to the animal's energy supply, partly due to the toxicity and limited availability of sulfide under aerobic conditions (12). When heterotrophic bacteria possess SQR, rhodanese, and SDO, they may play a detoxification role. It remains to be demonstrated whether these heterotrophic bacteria can use H2S oxidation for energy production and whether they have relatively high activities for removing H2S from gas emissions.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yuezhong Li (Shandong University) for the M. xanthus DK1622 genomic DNA, Minqin Wang (Shandong University) for A. tumefaciens strain C58, William W. Mohn (University of British Columbia) for B. xenovorans LB400, Michael Kahn (Washington State University) for S. meliloti 1021, Ron L. Crawford (University of Idaho) for C. necator JMP134, Dongqing Yu (Shandong Provincial Hospital) for H. influenzae R2846, and Lichuan Gu (Shandong University) for the YdiV protein.
This work was supported by the State Key Laboratory of Microbial Technology at Shandong University. Research was done at Shandong University.
Footnotes
Published ahead of print 3 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03281-13.
REFERENCES
- 1.Winogradsky S. 1887. Über Schwefelbakterien. Bot. Zeitung 45:489–610 [Google Scholar]
- 2.Brune DC. 1995. Sulfur compounds as photosynthetic electron donors. Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 3.Masau RJ, Oh JK, Suzuki I. 2001. Mechanism of oxidation of inorganic sulfur compounds by thiosulfate-grown Thiobacillus thiooxidans. Can. J. Microbiol. 47:348–358. 10.1139/w01-015 [DOI] [PubMed] [Google Scholar]
- 4.Stetter K. 1996. Hyperthermophilic procaryotes. FEMS Microbiol. Rev. 18:149–158. 10.1111/j.1574-6976.1996.tb00233.x [DOI] [Google Scholar]
- 5.Powell MA, Somero GN. 1986. Hydrogen sulfide oxidation is coupled to oxidative phosphorylation in mitochondria of Solemya reidi. Science 233:563–566. 10.1126/science.233.4763.563 [DOI] [PubMed] [Google Scholar]
- 6.Völkel S, Grieshaber M. 1997. Sulphide oxidation and oxidative phosphorylation in the mitochondria of the lugworm. J. Exp. Biol. 200:83–92 [DOI] [PubMed] [Google Scholar]
- 7.Bagarinao T, Vetter RD. 1990. Oxidative detoxification of sulfide by mitochondria of the California killifish Fundulus parvipinnis and the speckled sanddab Citharichthys sitgmaeus. J. Comp. Physiol. B 160:519–527 [Google Scholar]
- 8.Goubern M, Andriamihaja M, Nubel T, Blachier F, Bouillaud F. 2007. Sulfide, the first inorganic substrate for human cells. FASEB J. 21:1699–1706. 10.1096/fj.06-7407com [DOI] [PubMed] [Google Scholar]
- 9.Holdorf MM, Owen HA, Lieber SR, Yuan L, Adams N, Dabney-Smith C, Makaroff CA. 2012. Arabidopsis ETHE1 encodes a sulfur dioxygenase that is essential for embryo and endosperm development. Plant Physiol. 160:226–236. 10.1104/pp.112.201855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jackson MR, Melideo SL, Jorns MS. 2012. Human sulfide:quinone oxidoreductase catalyzes the first step in hydrogen sulfide metabolism and produces a sulfane sulfur metabolite. Biochemistry 51:6804–6815. 10.1021/bi300778t [DOI] [PubMed] [Google Scholar]
- 11.Kabil O, Banerjee R. 2012. Characterization of patient mutations in human persulfide dioxygenase (ETHE1) involved in H2S catabolism. J. Biol. Chem. 287:44561–44567. 10.1074/jbc.M112.407411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grieshaber MK, Volkel S. 1998. Animal adaptations for tolerance and exploitation of poisonous sulfide. Annu. Rev. Physiol. 60:33–53. 10.1146/annurev.physiol.60.1.33 [DOI] [PubMed] [Google Scholar]
- 13.Rohwerder T, Sand W. 2003. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149:1699–1710. 10.1099/mic.0.26212-0 [DOI] [PubMed] [Google Scholar]
- 14.Tiranti V, Briem E, Lamantea E, Mineri R, Papaleo E, De Gioia L, Forlani F, Rinaldo P, Dickson P, Abu-Libdeh B, Cindro-Heberle L, Owaidha M, Jack RM, Christensen E, Burlina A, Zeviani M. 2006. ETHE1 mutations are specific to ethylmalonic encephalopathy. J. Med. Genet. 43:340–346. 10.1136/jmg.2005.036210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lloyd D. 2006. Hydrogen sulfide: clandestine microbial messenger? Trends Microbiol. 14:456–462. 10.1016/j.tim.2006.08.003 [DOI] [PubMed] [Google Scholar]
- 16.Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, Robbertse B, Spatafora JW, Rappe MS, Giovannoni SJ. 2011. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci. Rep. 1:13. 10.1038/srep00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guimaraes BG, Barbosa RL, Soprano AS, Campos BM, de Souza TA, Tonoli CC, Leme AF, Murakami MT, Benedetti CE. 2011. Plant pathogenic bacteria utilize biofilm growth-associated repressor (BigR), a novel winged-helix redox switch, to control hydrogen sulfide detoxification under hypoxia. J. Biol. Chem. 286:26148–26157. 10.1074/jbc.M111.234039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bebrone C. 2007. Metallo-beta-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 74:1686–1701. 10.1016/j.bcp.2007.05.021 [DOI] [PubMed] [Google Scholar]
- 19.Landt O, Grunert HP, Hahn U. 1990. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96:125–128. 10.1016/0378-1119(90)90351-Q [DOI] [PubMed] [Google Scholar]
- 20.Suzuki I. 1965. Oxidation of elemental sulfur by an enzyme system of Thiobacillus thiooxidans. Biochim. Biophys. Acta 104:359–371. 10.1016/0304-4165(65)90341-7 [DOI] [PubMed] [Google Scholar]
- 21.Visser JM, Robertson LA, Van Verseveld HW, Kuenen JG. 1997. Sulfur production by obligately chemolithoautotrophic Thiobacillus species. Appl. Environ. Microbiol. 63:2300–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 23.Li B, Li N, Wang F, Guo L, Huang Y, Liu X, Wei T, Zhu D, Liu C, Pan H, Xu S, Wang HW, Gu L. 2012. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res. 40:11073–11085. 10.1093/nar/gks869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flavin M. 1962. Microbial transsulfuration: the mechanism of an enzymatic disulfide elimination reaction. J. Biol. Chem. 237:768–777 [PubMed] [Google Scholar]
- 25.Riener CK, Kada G, Gruber HJ. 2002. Quick measurement of protein sulfhydryls with Ellman's reagent and with 4,4′-dithiodipyridine. Anal. Bioanal. Chem. 373:266–276. 10.1007/s00216-002-1347-2 [DOI] [PubMed] [Google Scholar]
- 26.Stamp AL, Owen P, El Omari K, Nichols CE, Lockyer M, Lamb HK, Charles IG, Hawkins AR, Stammers DK. 2010. Structural and functional characterization of Salmonella enterica serovar Typhimurium YcbL: an unusual type II glyoxalase. Protein Sci. 19:1897–1905. 10.1002/pro.475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marasinghe GP, Sander IM, Bennett B, Periyannan G, Yang KW, Makaroff CA, Crowder MW. 2005. Structural studies on a mitochondrial glyoxalase II. J. Biol. Chem. 280:40668–40675. 10.1074/jbc.M509748200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suttisansanee U, Honek JF. 2011. Bacterial glyoxalase enzymes. Semin. Cell Dev. Biol. 22:285–292. 10.1016/j.semcdb.2011.02.004 [DOI] [PubMed] [Google Scholar]
- 29.McCoy JG, Bingman CA, Bitto E, Holdorf MM, Makaroff CA, Phillips GN., Jr 2006. Structure of an ETHE1-like protein from Arabidopsis thaliana. Acta Crystallogr. D Biol. Crystallogr. 62:964–970. 10.1107/S0907444906020592 [DOI] [PubMed] [Google Scholar]
- 30.Mineri R, Rimoldi M, Burlina AB, Koskull S, Perletti C, Heese B, von Dobeln U, Mereghetti P, Di Meo I, Invernizzi F, Zeviani M, Uziel G, Tiranti V. 2008. Identification of new mutations in the ETHE1 gene in a cohort of 14 patients presenting with ethylmalonic encephalopathy. J. Med. Genet. 45:473–478. 10.1136/jmg.2008.058271 [DOI] [PubMed] [Google Scholar]
- 31.Shibata H, Kobayashi S. 2006. Characterization of a HMT2-like enzyme for sulfide oxidation from Pseudomonas putida. Can. J. Microbiol. 52:724–730. 10.1139/w06-022 [DOI] [PubMed] [Google Scholar]
- 32.Valdés J, Pedroso I, Quatrini R, Holmes DS. 2008. Comparative genome analysis of Acidithiobacillus ferrooxidans, A. thiooxidans and A. caldus: insights into their metabolism and ecophysiology. Hydrometallurgy 94:180–184. 10.1016/j.hydromet.2008.05.039 [DOI] [Google Scholar]
- 33.Urich T, Bandeiras TM, Leal SS, Rachel R, Albrecht T, Zimmermann P, Scholz C, Teixeira M, Gomes CM, Kletzin A. 2004. The sulphur oxygenase reductase from Acidianus ambivalens is a multimeric protein containing a low-potential mononuclear non-haem iron centre. Biochem. J. 381:137–146. 10.1042/BJ20040003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen ZW, Liu YY, Wu JF, She Q, Jiang CY, Liu SJ. 2007. Novel bacterial sulfur oxygenase reductases from bioreactors treating gold-bearing concentrates. Appl. Microbiol. Biotechnol. 74:688–698. 10.1007/s00253-006-0691-0 [DOI] [PubMed] [Google Scholar]
- 35.Suzuki I. 1999. Oxidation of inorganic sulfur compounds: chemical and enzymatic reactions. Can. J. Microbiol. 45:97–105. 10.1139/w98-223 [DOI] [Google Scholar]
- 36.Chen L, Ren Y, Lin J, Liu X, Pang X. 2012. Acidithiobacillus caldus sulfur oxidation model based on transcriptome analysis between the wild type and sulfur oxygenase reductase defective mutant. PLoS One 7:e39470. 10.1371/journal.pone.0039470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mangold S, Valdes J, Holmes DS, Dopson M. 2011. Sulfur metabolism in the extreme acidophile Acidithiobacillus caldus. Front. Microbiol. 2:17. 10.3389/fmicb.2011.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.