Abstract
All cultivated ammonia-oxidizing archaea (AOA) within the Nitrososphaera cluster (former soil group 1.1b) are neutrophilic. Molecular surveys also indicate the existence of Nitrososphaera-like phylotypes in acidic soil, but their ecological roles are poorly understood. In this study, we present molecular evidence for the chemolithoautotrophic growth of Nitrososphaera-like AOA in an acidic soil with pH 4.92 using DNA-based stable isotope probing (SIP). Soil microcosm incubations demonstrated that nitrification was stimulated by urea fertilization and accompanied by a significant increase in the abundance of AOA rather than ammonia-oxidizing bacteria (AOB). Real-time PCR analysis of amoA genes as a function of the buoyant density of the DNA gradient following the ultracentrifugation of the total DNA extracted from SIP microcosms indicated a substantial growth of soil AOA during nitrification. Pyrosequencing of the total 16S rRNA genes in the “heavy” DNA fractions suggested that archaeal communities were labeled to a much greater extent than soil AOB. Acetylene inhibition further showed that 13CO2 assimilation by nitrifying communities depended solely on ammonia oxidation activity, suggesting a chemolithoautotrophic lifestyle. Phylogenetic analysis of both 13C-labeled amoA and 16S rRNA genes revealed that most of the active AOA were phylogenetically closely related to the neutrophilic strains Nitrososphaera viennensis EN76 and JG1 within the Nitrososphaera cluster. Our results provide strong evidence for the adaptive growth of Nitrososphaera-like AOA in acidic soil, suggesting a greater metabolic versatility of soil AOA than previously appreciated.
INTRODUCTION
The discovery of ammonia-oxidizing archaea (AOA) has fundamentally changed our perceptions of the global nitrogen cycle (1), as AOA exist in almost every corner of Earth's biosphere (2) and often overwhelmingly outnumber their bacterial counterparts (3). Numerous studies have suggested the enormous diversity of AOA based on 16S rRNA genes and amoA genes encoding the alpha subunit of ammonia monooxygenase, and four major lineages with robust phylogenetic support have recently been revealed, including the Nitrososphaera cluster (also referred to as soil group 1.1b), Nitrosopumilus cluster (marine group 1.1a), Nitrosotalea cluster (group 1.1a associated), and Nitrosocaldus cluster (4, 5). A growing body of evidence has suggested the pH-based selection of these globally distributed archaeal amoA genes, which can be clustered into acidic, acido-neutral, and alkalinophilic ecotype groups (6). However, the mechanisms underlying the ecological coherence of AOA remain poorly understood.
A recent discovery demonstrated the existence of acidophilic Nitrosotalea devanaterra in acidic soil (7). Increasing lines of evidence appeared to support the predominant role of archaeal nitrification in acidic soil by AOA members within the Nitrosotalea cluster (8–10). However, our recent study provides a strong indication that ammonia oxidation in acidic soils was linked to the Nitrososphaera cluster (11), and the putative active AOA in these acidic soils are phylogenetically closely related to the neutrophilic strains Nitrososphaera viennensis EN76 and JG1 (12, 13). The transcriptional activity of AOA was also demonstrated by a single amoA phylotype within the Nitrososphaera cluster in nitrifying acid forest soil with a pH of 4.1 (14). Nevertheless, all AOA cultures determined within the Nitrososphaera cluster grow optimally at the pH of the neutral environments from which they were isolated (2, 12, 13). These observations indicate that some unknown AOA phylotypes within the Nitrososphaera cluster might possess the metabolic capability to survive acid stress and that archaeal ammonia oxidation in nitrifying acidic soil is far more complicated than previously thought.
Molecular surveys have suggested the predominance of AOA members within the Nitrososphaera cluster in neutral and alkaline soils (4, 6, 15, 16). Interestingly, there is accumulating evidence for the presence of these Nitrososphaera-like sequences in low-pH ecosystems, such as agricultural soil (17), forest soil (18), and tea orchard soil (19). In fact, a recent meta-analysis of archaeal amoA genes at global, regional, and local scales strongly suggested that some AOA ecotypes within the Nitrososphaera cluster could be specifically adapted to growth at low pH (6). However, a direct link between archaeal ammonia oxidation in acidic soil and AOA members within the Nitrososphaera cluster has yet to be found. In the present study, a microcosm-based stable isotope probing (SIP) experiment was employed to assess whether Nitrososphaera-like AOA catalyze ammonia oxidation and use a chemolithoautotrophic metabolic strategy in an acidic agricultural soil.
MATERIALS AND METHODS
Soil sampling.
Soil at a depth of 0 to 15 cm was collected from the Red Soil Ecology Experimental Station of the Chinese Academy of Sciences (28°15′N, 116°55′E), Jiangxi Province, China (20). This region has a typical subtropical monsoon climate with a mean annual precipitation of 1,785 mm and a mean annual temperature of 18°C. The soil was derived from quaternary red clay and classified as Hapludult. Fertilization was performed in triplicate field plots with 120 kg urea-N ha−1, 40 kg P2O5-P ha−1, and 118 kg K2O-K ha−1 annually over the last 2 decades. The soil properties are as follows: soil pH 4.92 (H2O) and 3.70 (1 M KCl), 10.7 g organic matter kg−1, 0.75 g total N kg−1, 5.0 mg NH4+-N kg−1, 12 mg NO3−-N kg−1, 36 mg available P kg−1, and 249 mg available K kg−1. The composite samples from each plot were homogenized, passed through a 2.0-mm sieve, and stored at −20°C.
The soil pH was determined using a Mettler Toledo 320-S pH meter (Mettler-Toledo Instruments Co. Ltd., Shanghai, China). The measurement was performed with a water/soil ratio of 2.5 and a 1 M KCl/soil ratio of 2.5. The soil organic matter content was determined using the dichromate oxidation method. Total N was determined using the Kjeldahl method. Ammonium and nitrate were extracted from soil samples with 2 M KCl, and the levels were determined by a Skalar SAN Plus segmented flow analyzer (Skalar, Inc., Breda, The Netherlands). Available P in the soil was extracted using sodium bicarbonate, and the level was determined using the molybdenum blue method. Available K in the soil was extracted using ammonium acetate, and the level was determined by flame photometry.
DNA-SIP microcosms.
Soil DNA-SIP microcosms were constructed to investigate the active soil nitrifying community as previously described (9). Three sets of treatments were performed, including 13CO2-labeled microcosms, 12CO2 control microcosms, and 13CO2-C2H2 (100-Pa) control microcosms. Pairwise comparison between the 13CO2-labeled and12CO2 control treatments was used to assess whether ammonia oxidizers assimilated 13CO2 for autotrophic growth, whereas the 13CO2-C2H2 treatment was employed to assess whether the 13CO2 assimilation by ammonia oxidizers is dependent on the energy generation from ammonia oxidation that could be completely abolished by acetylene (21). For each treatment, 10 g of the sieved fresh soil (equivalent to 8.0 g dry weight g soil [dws]) was placed in a 120-ml serum bottle tightly capped with a black butyl stopper, and the headspace of the bottle contained 5% (vol/vol) 13CO2 or 12CO2. Microcosms were incubated at 60% of the soil's maximum water-holding capacity at 28°C in the dark. The 13CO2 and 13CO2-C2H2 treatments received 100 μg [13C]urea-N weekly, and the 12CO2 treatments received 100 μg [12C]urea-N g−1 dws weekly throughout the 8-week incubation period. The headspace of the each bottle was flushed weekly with pressurized synthetic air (20% O2, 80% N2) for 1 min to maintain oxic conditions. Water loss was replaced by adding sterilized water, and the 13CO2, 12CO2, and C2H2 were also renewed immediately after the headspace air exchange.
The [13C]urea and [12C]urea (99 atom% carbon) were purchased from the Shanghai Engineering Research Center of Stable Isotopes (Shanghai, China), and 13CO2 (99 atom% carbon) was purchased from Sigma-Aldrich Co. (St. Louis, MO).12CO2 was produced by acidifying sodium carbonate. Destructive sampling was performed in triplicate from each treatment during the incubation period, and the soil samples were transferred immediately to a −80°C freezer for subsequent molecular analysis. The rest of the soil replicate was used for the determination of the soil inorganic nitrogen concentrations of NH4+-N, NO2−-N, and NO3−-N.
SIP gradient fractionation.
Soil DNA was extracted using a FastDNA spin kit for soil (MP Biomedicals, Cleveland, OH) according to the manufacturer's instructions. The concentration of extracted DNA was determined with a NanoDrop ND-1000 UV-visible light spectrophotometer (NanoDrop Technologies, Wilmington, DE).
The total DNA extract was subjected to isopycnic centrifugation to separate [13C]DNA from native [12C]DNA in the labeled microcosms as previously described (22). Three micrograms of total DNA extract with an initial CsCl buoyant density of 1.725 g ml−1 was placed in a 5.1-ml Beckman polyallomer ultracentrifuge tube and subjected to 177,000 × g for 44 h at 20°C in a Vti65.2 vertical rotor (Beckman Coulter, Palo Alto, CA). Fifteen DNA fractions (∼380 μl) were obtained by displacing the gradient medium from the top of the ultracentrifuge tube with sterile water using a syringe pump (New Era Pump Systems, Inc., Farmingdale, NY) with a precisely controlled flow rate of 0.38 ml min−1. The buoyant density of each DNA fraction was measured by determining the refractive index of a 60-μl aliquot of each fraction using an AR200 digital hand-held refractometer (Reichert, Inc., Buffalo, NY). The fractionated DNA was precipitated from the CsCl solution by adding 2 volumes of polyethylene glycol 6000 (PEG 6000) in 1.6 M NaCl at 37°C for 1 h followed by centrifugation at 13,000 × g for 30 min. The precipitated DNA sample was purified with 70% ethanol and dissolved in 30 μl Tris-EDTA (TE) buffer.
Real-time quantitative PCR.
To determine the growth of AOA and ammonia-oxidizing bacteria (AOB) during the 8-week incubation and to determine the 13CO2 labeling of amoA-carrying ammonia oxidizers, the abundance of amoA genes in the total DNA from the soil microcosms and in each CsCl fraction from DNA-SIP microcosms was quantified on a CFX96 optical real-time detection system (Bio-Rad Laboratories, Inc., Hercules, CA). The primer pairs Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′) were used for real-time PCR of archaeal amoA genes (23), and the primer pairs amoA1F (5′-GGGGTTTCTACTGGTGGT-3′) and amoA2R (5′-CCCCTCKGSAAAGCCTTCTTC-3′) were used for real-time PCR of bacterial amoA genes (24). The reaction was performed in a 25-μl mixture containing 12.5 μl SYBR Premix Ex Taq (TaKaRa, Dalian, China), 0.5 μM each primer, and 2 μl of DNA template (1 to 10 ng). The real-time PCR conditions were as follows: 95°C for 1 min and 38 cycles of 95°C for 10 s, 55°C for 30 s, and 72°C for 45 s, followed by plate reads at 83°C. The real-time PCR standard was generated using plasmid DNA from one representative clone containing archaeal or bacterial amoA genes. A standard template dilution series from 5.0 × 101 to ∼6.0 × 101 to 5.0 × 108 to ∼6.0 × 108 copies per assay was used. A dilution series of the DNA template was also used to assess whether PCR inhibition occurred during amplification. Real-time PCR was performed in biological triplicates, and each involved three technical replicates. Amplification efficiencies of 96 to 105% were obtained with R2 values of 0.996 to 0.999. In addition to standard agarose gel electrophoresis, a melting curve analysis was performed at the end of each real-time PCR run to check the specificity of the amplification products.
Pyrosequencing.
To determine the 13CO2 labeling of the 16S rRNA genes of nitrifying communities, pyrosequencing was carried out on a Roche 454 GS FLX Titanium sequencer (Roche Diagnostics Corporation, Branford, CT) by analyzing the V4 regions of the 16S rRNA genes in each CsCl gradient from DNA-SIP microcosms, as previously described (9). Briefly, the DNA sample was amplified using the universal primers 515F (5′-GTGCCAGCMGCCGCGG-3′) and 907R (5′-CCGTCAATTCMTTTRAGTTT-3′) (25), which were fused with A or B adapters, the key sequence, and a unique tag sequence for each DNA sample. The PCR was performed in a 50-μl mixture containing 45 μl Platinum PCR SuperMix (Invitrogen, Shanghai, China), a 200 nM final concentration of each primer, and 2 μl template DNA. The amplification conditions were as follows: 94°C for 5 min, followed by 28 cycles of 94°C for 45 s, 55°C for 30 s, and 72°C for 45 s, followed by extension at 72°C for 5 min. The PCR products were purified using a 2.0% agarose gel and quantified using Picogreen (Invitrogen, Shanghai, China). The purified PCR products from different samples were combined in equimolar ratios into a single tube for pyrosequencing analysis.
Pyrosequencing data were processed using a combination of the RDP pyrosequencing pipeline (26) and the mothur software package (27). Sequence reads from multiple samples were sorted using sample-specific tag sequences, and only sequences of >300 bp in length with an average quality score of >30 and without ambiguous base calls were included for subsequent analyses. The taxonomy of the high-quality reads was further classified using the RDP classifier with a minimum support threshold of 80% and the RDP taxonomic nomenclature (26). The 16S rRNA genes affiliated with the putative nitrifying communities in the 13C-labeled “heavy” fractions from the labeled microcosm were extracted and clustered into an operational taxonomic unit (OTU) at a 0.03 cutoff using mothur. A representative sequence for each OTU was used for subsequent phylogenetic tree analysis.
Cloning and phylogenetic analysis.
A clone library of archaeal amoA genes in the 13C-labeled “heavy” fractions was constructed to infer the phylogenetic relationship of active AOA in this study to those deposited in GenBank. The archaeal amoA genes retrieved from the 13C-labeled DNA from the labeled microcosms were amplified by the CrenamoA 23f (5′-ATGGTCTGGCTWAGACG-3′) and CrenamoA 616f (5′-GCCATACABCKRTANGTCCA-3′) primer pairs, as previously described (28). The PCR products were purified using a 2.0% agarose gel for clone library construction. Escherichia coli JM109 competent cells were used for transformation. Sequencing of clones containing the correct insert was performed by the TaKaRa Sequencing Department (TaKaRa, Shanghai, China). Phylogenetic analysis was conducted with MEGA version 4.0 through a neighbor-joining tree using Kimura 2-parameter distance with 1,000 replicates to produce bootstrap values.
Statistical analysis.
A linear regression analysis was performed for the calculation of the net nitrification rate. One-way analysis of variance (ANOVA) with Tukey's post hoc test was performed for multiple comparisons. All analyses were conducted using the SPSS 13.0 package for Windows (SPSS, Inc.), and P < 0.05 was considered to be statistically significant.
Nucleotide sequence accession numbers.
The nucleotide sequences from this study have been deposited in GenBank under accession no. KF797777 to KF797797 for the archaeal amoA genes from the DNA-SIP experiment. The pyrosequencing reads have been deposited in the DNA Data Bank of Japan (DDBJ) under accession no. DRA001204.
RESULTS
Soil nitrification activity.
Nitrification activity was assessed as the rate of increase in soil nitrate concentrations in SIP microcosms throughout the 8-week incubation period. In the absence of C2H2, a stepwise production of soil nitrate in the labeled microcosms was observed with 85 and 178 μg NO3−-N g−1 dws after incubation for 28 and 56 days, respectively (Fig. 1a). Fertilizing soil microcosms on a weekly basis led to a significant accumulation of soil ammonium, preventing ammonia oxidizers from substrate constraints (Fig. 1b). Similar results were obtained from the 12CO2 control microcosms, demonstrating no significant bias associated with 13CO2 incubation, as previously described (29). The linear regression analysis showed that net nitrification activity was 3.15 μg NO3-N g−1 dws day−1 (R2 = 0.985) over the course of incubation for 56 days. The soil pH value decreased from 4.92 at day 0 to 4.65 and 4.68 at day 56 (P < 0.001) in the nitrifying soil microcosms with the 13CO2 and12CO2 treatments, respectively. However, the addition of acetylene completely abolished soil nitrate production, suggesting that microbial ammonia oxidation was blocked during the 8-week incubation period (Fig. 1a). The urea-N was indeed recovered in almost stoichiometric amounts in the form of ammonium in the SIP control microcosms amended with 13CO2 plus C2H2 (Fig. 1b).
FIG 1.
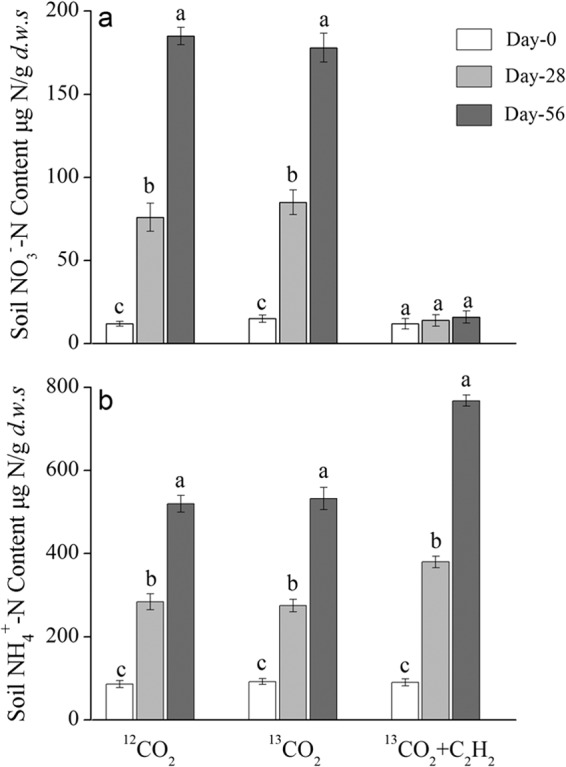
Changes in nitrate (a) and ammonium (b) concentrations in soil microcosms incubated with 13CO2 or 12CO2 or 13CO2 plus C2H2 for 28 and 56 days. The soil microcosms were fertilized with 100 μg urea-N g−1 dws on a weekly basis. Error bars represent standard errors of triplicate microcosms. The same letter above different bars indicates no significant difference (P > 0.05).
Changes in the abundance of soil ammonia oxidizers.
The changes in the population sizes of soil AOA and AOB communities were determined by real-time quantitative PCR of amoA genes during the incubation of the SIP microcosms for 56 days (Fig. 2). The copy number of archaeal amoA genes increased significantly from 1.0 × 108 at the start of the incubation to ∼3.1 × 108 g−1 dws (P < 0.001) in the nitrifying soil microcosms of 13CO2 and12CO2 treatments at day 56 (Fig. 2a), whereas no significant change was observed in the C2H2-treated microcosms (9.3 × 107 g−1 dws; P > 0.05), in which nitrification activity was completely blocked (Fig. 2a). However, the bacterial amoA gene showed no significant changes in abundance over the course of 56 days, ranging from 2.85 to 3.20 × 107 copies g−1 dws (P > 0.05) in all treatments irrespective of acetylene inhibition (Fig. 2b).
FIG 2.
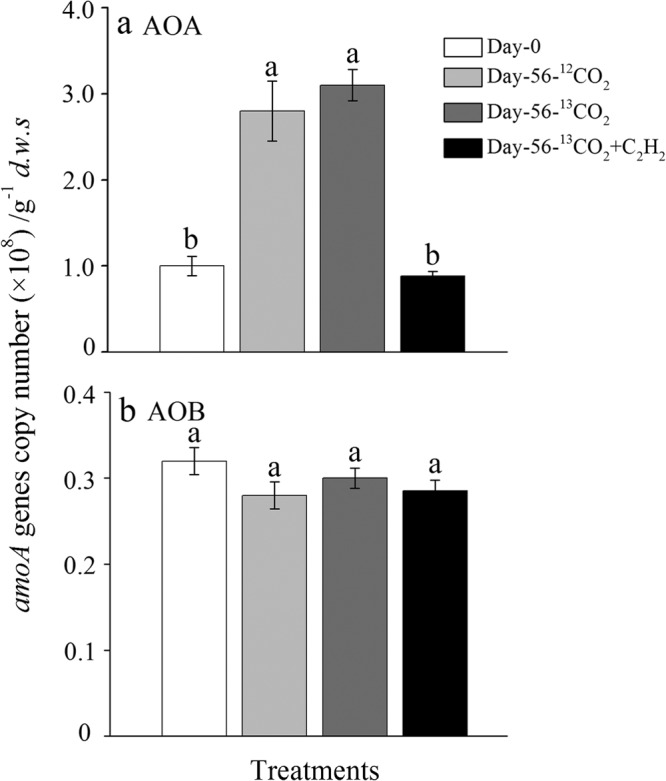
Changes in the copy numbers of archaeal (a) and bacterial (b) amoA genes in soil microcosms over an incubation period of 56 days. Error bars represent the standard errors of triplicate microcosms. The same letter above different bars indicates no significant difference (P > 0.05).
Stable isotope probing of soil nitrifying communities.
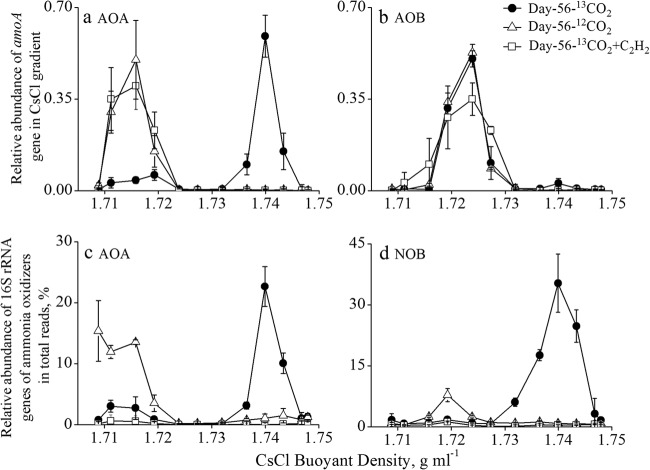
Isopycnic ultracentrifugation was conducted with the total genomic DNA extracted from each SIP microcosm to separate [13C]DNA in the “heavy” DNA fractions from native [12C]DNA in the “light” DNA fractions. The quantitative analysis of amoA genes as a function of the buoyant densities of the DNA gradient provides a powerful means for the interpretation of the labeling of amoA-carrying AOA and AOB communities (28). In the labeled microcosms, the highest copy number of archaeal amoA genes was observed in the “heavy” DNA fractions, with a typical [13C]DNA buoyant density of approximately 1.74 g ml−1, whereas for control microcosms, archaeal amoA genes peaked in the “light” fractions representative of native [12C]DNA (Fig. 3a). Furthermore, bacterial amoA genes were predominantly detected only in the “light” DNA fractions in both the labeled and control microcosms, although a very small number of bacterial amoA genes appeared to be labeled (Fig. 3b). These results suggest substantial 13CO2 assimilation by AOA in the acidic soil tested (Fig. 3).
FIG 3.
Quantitative distribution of amoA (a and b) and relative abundance of 16S rRNA gene reads (c and d) of ammonia-oxidizing communities in the DNA fractions across the entire buoyant density gradient from the labeled and control microcosms after incubation for 56 days. The ratio of amoA genes in each fraction to the sum of amoA genes across the entire gradient of DNA fractions is plotted for AOA (a) and AOB (b). The relative abundance is expressed as the percentage of the targeted archaeal (c) and NOB 16S rRNA gene reads (d) to the total 16S rRNA gene reads in each DNA fraction. The error bars represent the standard errors of the triplicate microcosms.
Pyrosequencing analysis of 16S rRNA genes at the community level was further performed in each DNA fraction across the entire density of CsCl gradients from triplicate soil microcosms after incubation for 56 days. Approximately 685,000 high-quality sequence reads were obtained with an average length of 430 bp in the V4 region of the 16S rRNA gene (see Table S1 in the supplemental material). Across the entire DNA gradient, archaeal 16S rRNA genes were highly enriched in the 13C-labeled DNA fractions, accounting for up to 22.6% of the total microbial communities for the labeled treatment (Fig. 3c). In stark contrast, a very low background of archaeal 16S rRNA genes was detected in the “heavy” DNA fractions, with <1.06% of the total microbial communities for the control microcosms of 12CO2 and 13CO2-C2H2 treatments (see Table S1). It is noteworthy that a small number of AOB 16S rRNA gene reads were enriched up to 1.63% in the “heavy” DNA fractions from the labeled microcosm, whereas the level remained constantly low for the control microcosms, ranging from 0.01 to 0.03% (see Table S1). Interestingly, up to 35.3% of the total 16S rRNA genes in the “heavy” DNA fractions were affiliated with nitrite-oxidizing bacteria (NOB) in the labeled microcosms (Fig. 3d), similar to our previous findings (22).
Phylogenetic analysis of active nitrifying communities.
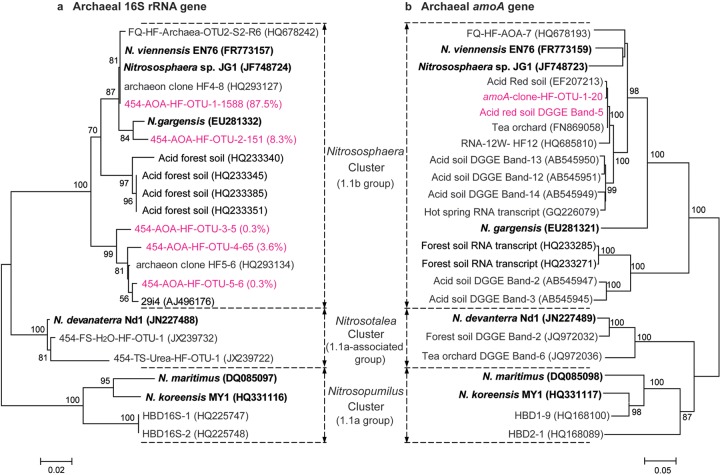
Phylogenetic analysis showed that the 13C-labeled archaeal 16S rRNA genes retrieved from the labeled microcosms fell exclusively within the Nitrososphaera cluster (Fig. 4a), and up to 87.5% of these reads were phylogenetically most closely related to the neutrophilic AOA strains JG1 and N. viennensis EN76 (Fig. 4a). Intriguingly, 4.1% and 8.3% of the 13C-labeled 16S rRNA genes were affiliated with the fosmid 29i4 lineage and moderately thermophilic Nitrososphaera gargensis, respectively (Fig. 4a). The phylogeny of the 13C-labeled archaeal amoA genes remains largely congruent with that of 16S rRNA genes (Fig. 4b). All clones of the 13C-labeled archaeal amoA sequences formed a single OTU (at 97% sequence identity) and were phylogenetically closely related to strain JG1 and N. viennensis EN76 (Fig. 4b). These putatively active amoA-carrying AOA were closely related to the 13C-labeled amoA genes (22) and transcripts (30) in alkaline soils and amoA transcripts in hot springs (31) within the Nitrososphaera cluster (Fig. 4b).
FIG 4.
Phylogenetic analysis of archaeal 16S rRNA (a) and amoA genes (b) in the [13C]DNA from the labeled microcosms after incubation for 56 days. The 16S rRNA reads of archaeal 16S rRNA genes in “heavy” DNA fractions 5, 6, and 7 were pooled for analysis with a total of 1,815 reads. The notation “454-AOA-HF-OTU-1-1588-(87.5%)” indicates that OTU-1 contains 1,588 sequences with >97% identity, accounting for 87.5% of the total archaeal 16S rRNA gene reads retrieved from the [13C]DNA. The notation “amoA-clone-OTU-1-20” indicates that all 20 cloned archaeal amoA genes in the [13C]DNA showed sequence identity of >97% and could be taxonomically classified into a single phylotype. One representative sequence was extracted using the mothur software for the tree construction of the archaeal amoA and 16S rRNA genes. The notation “Acid red soil DGGE band-5” represents the sequence of the newly emerging DGGE bands in the acidic soil studied after incubation for 56 days (data not shown). The putatively active amoA genes within the Nitrososphaera cluster were retrieved from various publications for analysis, including FQ-HF-AOA-7 (22) and RNA-12W-HF-12 (30) in alkaline soil, hot spring RNA transcripts (31), putatively active AOA DGGE bands 12, 13, and 14 in acidic soils (11), and acidic forest soil RNA transcripts (14).
It is interesting to note that the 13C-labeled 16S rRNA genes of AOB were closely related to the strains of Nitrosospira AF and L115, which were isolated from acidic soil with a pH value of 4.0 (32) and placed within the Nitrosospira cluster 3 (see Fig. S1 in the supplemental material). Sequence analysis further demonstrated that the labeled NOB were predominantly composed of Nitrospira-like 16S rRNA genes, whereas less than 0.4% of 16S rRNA genes were related to Nitrobacter-like NOB (see Table S2 in the supplemental material).
DISCUSSION
Our results provide compelling evidence for the chemolithoautotrophic growth of archaeal ammonia oxidizers in an acidic soil that are phylogenetically closely related to neutrophilic AOA within the Nitrososphaera cluster. Soil nitrification activity was paralleled by an increase in the abundance of archaeal rather than bacterial amoA gene copy numbers (Fig. 2). This observation was substantiated by stable isotope tracing of active ammonia oxidizers, showing considerable 13CO2 assimilation by AOA but slight 13CO2 assimilation by AOB in the acidic soil tested (Fig. 3). All labeled AOA were phylogenetically classified into the Nitrososphaera cluster, and up to 87.5% of 13C-labeled archaeal 16S rRNA genes were phylogenetically closely related to neutrophilic strains JG1 and N. viennensis EN76 (Fig. 4a). These results provide strong evidence for the better adaptation of archaeal ammonia oxidizers to growth at low pH compared to bacterial oxidizers and suggest that the ecological significance of AOA members within the Nitrososphaera cluster is not limited to neutral and alkaline ecosystems.
Nitrification rarely occurs in media at pH values below 6.0 for all known AOA cultures within the Nitrososphaera cluster. A physiological study demonstrated that the neutrophilic strain JG1 grows optimally at pH of 6.5 to 7.0 (12), whereas strain EN76 is also a considered neutrophilic, with an optimal pH slightly higher than 7.5 (Christa Schleper, personal communication) (13). The nitrification activity of these AOA significantly decreased or even ceased at pH values below 5.5 (12, 13). Both the JG1 and EN76 strains appeared to be specifically adapted to growth at the bulk pH of the soil environment from which they were cultivated. For instance, agricultural and garden soils used as inoculums for JG1 and EN76 had pH values of 6.5 and 8.0, respectively. However, the 13C-labeled AOA in this study were phylogenetically closely related to neutrophilic JG1 and N. viennensis EN76 stains (Fig. 4), despite their active growth in the acidic soil tested. These results indicate that phylogenetic relatedness does not necessarily indicate similarity in the physiology of soil ammonia oxidizers and highlights the importance of efforts to cultivate environmentally representative AOA in a wide variety of natural habitats where representative AOA could have evolved distinctly different metabolisms under constant selective pressures.
A global analysis of archaeal amoA genes has suggested the distribution of AOA members within the Nitrososphaera cluster even in strongly acidic soil, although the numerical predominance of these organisms is observed in neutral and alkaline soils (6, 16). Soil ammonia availability decreases exponentially with decreasing pH (8, 11). It thus appears plausible that the ability to capture ammonia at low concentrations could explain the predominant role of archaeal nitrification in acidic soil, as all cultivated AOA have a much higher affinity for ammonia than AOB (5). However, our results provide a strong indication that acid tolerance ability, rather than high substrate affinity, enables archaeal ammonia oxidizers to grow in acidic soil. For instance, the soil ammonia content was estimated to be as high as 28 nM NH3 in the acidic soil used in our study (5.0 μg NH4+-N g−1 dsw) with pH 4.92 under field conditions (NH3 plus H+ ↔ NH4+; pKa of 9.5) (32). This value is high enough to meet the substrate demand of the neutrophilic strain JG1, which has a Km value of 12.7 nM, despite the fact that this strain does not grow at a pH of less than 6.0 (12). In fact, a recent discovery has demonstrated that the growth of Nitrosotalea devanaterra occurs only under pH 6.0, although this species shows the greatest substrate affinity for AOA discovered to date (7). These findings further suggest that acid tolerance plays a more important role than substrate affinity in the AOA activity in ammonia-poor acidic soils. We speculate that acidic soil exerts a selection pressure favoring acid-tolerant AOA, and the labeled active AOA possess a unique metabolism of acidity tolerance, which is apparently absent in the neutrophilic JG1 and EN76 strains. Cultivation of these acid-resistant AOA and comparative genomic analysis will likely shed new light on ammonia uptake and oxidation mechanisms at low pH.
The 13C-labeled active AOB are most closely affiliated with strains AF and L115, which were isolated from acidic soils with pH 4.0 (see Fig. S1 in the supplemental material) (32). Interestingly, culture studies indicate neutrophilic lifestyles of strains AF and L115, with optimal growth at pH 7.0. The ammonia oxidation activity of these two strains was restricted below pH 5.5, and growth ceased at pH 6.5. This pattern represents a common theme for nearly all AOB isolated from acidic soils within the genus Nitrosospira (32, 33). These observations imply the absence of acid-tolerant metabolism in AOB or that the mechanism by which AOB cope with acid stress is unknown. In addition, the existence of acidophilic AOA within the Nitrososphaera cluster remains uncertain. The possibility of ammonia oxidation occurring within the microneutrophilic environment of soil aggregates could not be excluded, despite the pH declines from 4.92 to 4.68 over the 8-week incubation. It is also noteworthy that urea hydrolysis can provide a greater advantage for bacterial ammonia oxidation at lower pH (34, 35) and for archaeal nitrification (36, 37), although the mechanism is still under debate (9).
DNA-SIP indicated that AOA was the leading cause of nitrification activity in the soil tested. Using the abundance of amoA gene biomarkers as a function of DNA buoyant density from labeled and control microcosms, the incorporation of 13C into the genomes of AOA and/or AOB could be demonstrated with great confidence (29). Pyrosequencing of the total 16S rRNA genes in DNA fractions is a new profiling strategy for the interpretation of labeling of organisms actively involved in the metabolism of labeled substrates (22). Although the assimilation of 13C by targeted microorganisms could not be quantitatively analyzed, these approaches facilitate the ecological interpretation of the labeled functional microbial guilds. For instance, up to 84.6% of archaeal amoA was detected in the “heavy” DNA fraction, suggesting that the majority of AOA was labeled and resolved in the [13C]DNA from labeled microcosms (Fig. 4a). The pyrosequencing of SIP DNA fractions demonstrated that the relative frequency of archaeal 16S rRNA reads was enriched up to 22.6% of the total microbial communities in the “heavy” DNA fraction, lending further support to AOA-mediated labeling. Assuming that soil nitrate production resulted solely from archaeal nitrification and that the genome of AOA contains only one amoA copy, the cell-specific ammonia oxidation rate of 13C-labeled AOA was estimated to be 0.036 fmol cell−1 h−1 (see Table S3 in the supplemental material), slightly lower than that of the JG1 strain, which was 0.058 fmol cell−1 h−1 (12). In stark contrast, the observed nitrate production would necessitate a cell-specific rate of ammonia oxidation of up to 35.2 fmol N cell−1 h−1 by labeled AOB (see Table S3), five times higher than that of their closest relative strains L115 and AF (32). Acetylene inhibition further demonstrated that the autotrophic growth of AOA depended solely on ammonia oxidation, suggesting that AOA dominated nitrification activity in SIP microcosms following urea amendment. However, DNA-SIP relies entirely on cell proliferation, and it is likely that bacterial ammonia oxidation occurred without cell division. RNA-SIP would be helpful to better understand bacterial nitrification in acidic soils under field conditions.
These results agree with our previous findings of archaeal predominance in nitrifying acid agricultural soils (11). The sequence analysis further indicated that archaeal nitrification in the acid agricultural soil was most likely dominated by AOA members of denaturing gradient gel electrophoresis (DGGE) bands 12 to 14, which have high sequence similarity to the 13C-labeled AOA in this study (11) (Fig. 4b). In addition to phylogenetically closely related phylotypes to N. viennensis EN76, active AOA communities were also composed of archaeal ammonia oxidizers affiliated with the 29i4 lineage (4.2%) and moderately thermophilic N. gargensis (8.3%) in the [13C]DNA fraction (Fig. 4a). These observations suggest that the metabolic versatility of AOA within the Nitrososphaera cluster is more complicated than previously suggested (38). Furthermore, the high affinity of Nitrospira for nitrite substrate might have resulted in their overwhelming predominance over Nitrobacter among active NOB communities (22, 39). The diversity of nitrifying communities is likely associated with distinct metabolism and/or life strategies, and the cultivation of environmentally relevant AOA would be helpful in understanding the complex interactions and evolution within nitrifying communities in natural environments.
Taken together, our results provide strong evidence for the autotrophic growth of AOA within the Nitrososphaera cluster in an acidic soil, the numerical predominance of which is broadly observed in neutral and alkaline soils. This finding expands the metabolic spectrum of Nitrososphaera cluster AOA and implies that the physiological adaptation of microbial functional guilds to environmental pressures may represent niche specialization of nitrifying communities in natural habitats. The results also demonstrate that the phylogenetic relatedness of AOA does not necessarily indicate functional correlations, highlighting the importance of cultivating AOA representatives across a wide variety of environments on Earth.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant no. 41101227, 41090281, and 31201690) and the Innovation Project of the Institute of Soil Science, Chinese Academy of Sciences (grant no. ISSASIP1104).
We gratefully acknowledge Jianbo Fan for sampling assistance and Jiuyu Li and our lab colleagues for helpful discussion on soil pH determination.
Footnotes
Published ahead of print 27 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03633-13.
REFERENCES
- 1.Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546. 10.1038/nature03911 [DOI] [PubMed] [Google Scholar]
- 2.Hatzenpichler R. 2012. Diversity, physiology, and niche differentiation of ammonia-oxidizing archaea. Appl. Environ. Microbiol. 78:7501–7510. 10.1128/AEM.01960-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, Prosser JI, Schuster SC, Schleper C. 2006. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 442:806–809. 10.1038/nature04983 [DOI] [PubMed] [Google Scholar]
- 4.Pester M, Rattei T, Flechl S, Gröngröft A, Richter A, Overmann J, Reinhold-Hurek B, Loy A, Wagner M. 2012. amoA-based consensus phylogeny of ammonia-oxidizing archaea and deep sequencing of amoA genes from soils of four different geographic regions. Environ. Microbiol. 14:525–539. 10.1111/j.1462-2920.2011.02666.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahl DA, de la Torre JR. 2012. Physiology and diversity of ammonia-oxidizing archaea. Annu. Rev. Microbiol. 66:83–101. 10.1146/annurev-micro-092611-150128 [DOI] [PubMed] [Google Scholar]
- 6.Gubry-Rangin C, Hai B, Quince C, Engel M, Thomson BC, James P, Schloter M, Griffiths RI, Prosser JI, Nicol GW. 2011. Niche specialization of terrestrial archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:21206–21211. 10.1073/pnas.1109000108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. 2011. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc. Natl. Acad. Sci. U. S. A. 108:15892–15897. 10.1073/pnas.1107196108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu L, Han WY, Zhang JB, Wu YC, Wang BZ, Lin XG, Zhu JG, Cai ZC, Jia ZJ. 2012. Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. ISME J. 6:1978–1984. 10.1038/ismej.2012.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu L, Jia ZJ. 2013. Urease gene-containing archaea dominate autotrophic ammonia oxidation in two acid soils. Environ. Microbiol. 15:1795–1809. 10.1111/1462-2920.12071 [DOI] [PubMed] [Google Scholar]
- 10.Zhang LM, Hu HW, Shen JP, He JZ. 2012. Ammonia-oxidizing archaea have more important role than ammonia-oxidizing bacteria in ammonia oxidation of strongly acidic soils. ISME J. 6:1032–1045. 10.1038/ismej.2011.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang R, Wu YC, Zhang JB, Zhong WH, Jia ZJ, Cai ZC. 2012. Nitrification activity and putative ammonia-oxidizing archaea in acidic red soils. J. Soils Sediments 12:420–428. 10.1007/s11368-011-0450-4 [DOI] [Google Scholar]
- 12.Kim JG, Jung MY, Park SJ, Rijpstra WIC, Sinninghe Damsté JS, Madsen EL, Min D, Kim JS, Kim GJ, Rhee SK. 2012. Cultivation of a highly enriched ammonia-oxidizing archaeon of thaumarchaeotal group I. 1b from an agricultural soil. Environ. Microbiol. 14:1528–1543. 10.1111/j.1462-2920.2012.02740.x [DOI] [PubMed] [Google Scholar]
- 13.Tourna M, Stieglmeier M, Spang A, Könneke M, Schintlmeister A, Urich T, Engel M, Schloter M, Wagner M, Richter A, Schleper C. 2011. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U. S. A. 108:8420–8425. 10.1073/pnas.1013488108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stopnisek N, Gubry-Rangin C, Hofferle S, Nicol GW, Mandic-Mulec I, Prosser JI. 2010. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl. Environ. Microbiol. 76:7626–7634. 10.1128/AEM.00595-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J. 5:908–917. 10.1038/ismej.2010.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu HW, Zhang LM, Dai Y, Di HJ, He JZ. 2013. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J. Soils Sediments 13:1439–1449. 10.1007/s11368-013-0726-y [DOI] [Google Scholar]
- 17.Nicol GW, Leininger S, Schleper C, Prosser JI. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978. 10.1111/j.1462-2920.2008.01701.x [DOI] [PubMed] [Google Scholar]
- 18.Ying JY, Zhang LM, He JZ. 2010. Putative ammonia-oxidizing bacteria and archaea in an acidic red soil with different land utilization patterns. Environ. Microbiol. Rep. 2:304–312. 10.1111/j.1758-2229.2009.00130.x [DOI] [PubMed] [Google Scholar]
- 19.Yao HY, Gao YM, Nicol GW, Campbell CD, Prosser JI, Zhang LM, Han WY, Singh KS. 2011. Links between ammonia oxidizer community structure, abundance, and nitrification potential in acidic soils. Appl. Environ. Microbiol. 77:4618–4625. 10.1128/AEM.00136-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He JZ, Zheng Y, Chen CR, He YQ, Zhang LM. 2008. Microbial composition and diversity of an upland red soil under long-term fertilization treatments as revealed by culture-dependent and culture-independent approaches. J. Soils Sediments 8:349–358. 10.1007/s11368-008-0025-1 [DOI] [Google Scholar]
- 21.Berg P, Klemedtsson L, Rosswall T. 1982. Inhibitory effect of low partial pressures of acetylene on nitrification. Soil Biol. Biochem. 14:301–303. 10.1016/0038-0717(82)90041-4 [DOI] [Google Scholar]
- 22.Xia WW, Zhang CX, Zeng XW, Feng YZ, Weng JH, Lin XG, Zhu JG, Xiong ZQ, Xu J, Cai ZC, Jia ZJ. 2011. Autotrophic growth of nitrifying community in an agricultural soil. ISME J. 5:1226–1236. 10.1038/ismej.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688. 10.1073/pnas.0506625102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rotthauwe J, Witzel K, Liesack W. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704–4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stubner S. 2002. Enumeration of 16S rDNA of Desulfotomaculum lineage 1 in rice field soil by real-time PCR with SybrGreen detection. J. Microbiol. Methods 50:155–164. 10.1016/S0167-7012(02)00024-6 [DOI] [PubMed] [Google Scholar]
- 26.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145. 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tourna M, Freitag TE, Nicol GW, Prosser JI. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357–1364. 10.1111/j.1462-2920.2007.01563.x [DOI] [PubMed] [Google Scholar]
- 29.Jia ZJ, Conrad R. 2009. Bacteria rather than archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658–1671. 10.1111/j.1462-2920.2009.01891.x [DOI] [PubMed] [Google Scholar]
- 30.Pratscher J, Dumont MG, Conrad R. 2011. Ammonia oxidation coupled to CO2 fixation by archaea and bacteria in an agricultural soil. Proc. Natl. Acad. Sci. U. S. A. 108:4170–4175. 10.1073/pnas.1010981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang HC, Huang QY, Dong HL, Wang P, Wang FP, Li WJ, Zhang CL. 2010. RNA-based investigation of ammonia-oxidizing archaea in hot springs of Yunnan Province, China. Appl. Environ. Microbiol. 76:4538–4541. 10.1128/AEM.00143-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang QQ, Bakken LR. 1999. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol. Ecol. 30:171–186. 10.1111/j.1574-6941.1999.tb00646.x [DOI] [PubMed] [Google Scholar]
- 33.De Boer W, Kowalchuk GA. 2001. Nitrification in acid soils: micro-organisms and mechanisms. Soil Biol. Biochem. 33:853–866. 10.1016/S0038-0717(00)00247-9 [DOI] [Google Scholar]
- 34.De Boer W, Duyts H, Laanbroek HJ. 1989. Urea stimulated autotrophic nitrification in suspensions of fertilized, acid heath soil. Soil Biol. Biochem. 21:349–354. 10.1016/0038-0717(89)90142-9 [DOI] [Google Scholar]
- 35.Burton SAQ, Prosser JI. 2001. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl. Environ. Microbiol. 67:2952–2957. 10.1128/AEM.67.7.2952-2957.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay J-É, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedrós-Alió C, Bertilsson S. 2012. Role for urea in nitrification by polar marine Archaea. Proc. Natl. Acad. Sci. U. S. A. 109:17989–17994. 10.1073/pnas.1201914109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spang A, Poehlein A, Offre P, Zumbrägel S, Haider S, Rychlik N, Nowka B, Schmeisser C, Lebedeva EV, Rattei T, Böhm C, Schmid M, Galushko A, Hatzenpichler R, Weinmaier T, Daniel R, Schleper C, Spieck E, Streit W, Wagner M. 2012. The genome of the ammonia-oxidizing Candidatus Nitrososphaera gargensis: insights into metabolic versatility and environmental adaptations. Environ. Microbiol. 14:3122–3145. 10.1111/j.1462-2920.2012.02893.x [DOI] [PubMed] [Google Scholar]
- 38.Alves RJE, Wanek W, Zappe A, Richter A, Svenning MM, Schleper C, Urich T. 2013. Nitrification rates in Arctic soils are associated with functionally distinct populations of ammonia-oxidizing archaea. ISME J. 7:1620–1631. 10.1038/ismej.2013.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Off S, Alawi M, Spieck E. 2010. Enrichment and physiological characterization of a novel Nitrospira-like bacterium obtained from a marine sponge. Appl. Environ. Microbiol. 76:4640–4646. 10.1128/AEM.00320-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.