Abstract
Clostridium perfringens causes histotoxic infections and diseases originating in animal or human intestines. A prolific toxin producer, this bacterium also produces numerous enzymes, including sialidases, that may facilitate infection. C. perfringens type D strain CN3718 carries genes encoding three sialidases, including two large secreted sialidases (named NanI and NanJ) and one small sialidase (named NanH) that has an intracellular location in log-phase cultures but is present in supernatants of death phase cultures. Using isogenic mutants of CN3718 that are capable of expressing only NanJ, NanI, or NanH, the current study characterized the properties and activities of each sialidase. The optimal temperature determined for NanJ or NanH enzymatic activity was 37°C or 43°C, respectively, while NanI activity increased until temperature reached 48°C. NanI activity was also the most resistant against higher temperatures. All three sialidases showed optimal activities at pH 5.5. Compared to NanJ or NanH, NanI contributed most to the sialidase activity in CN3718 culture supernatants, regardless of the substrate sialic acid linkage; NanI also released the most sialic acid from Caco-2 cells. Only NanI activity was enhanced by trypsin pretreatment and then only for substrates with an α-2,3- or α-2,6-sialic acid linkage. NanJ and NanI activities were more sensitive than NanH activity to two sialidase inhibitors (N-acetyl-2,3-dehydro-2-deoxyneuraminic acid and siastatin B). The activities of the three sialidases were affected differently by several metal ions. These results indicated that each C. perfringens sialidase has distinct properties, which may allow these enzymes to play different roles depending upon environmental conditions.
INTRODUCTION
Clostridium perfringens is a Gram-positive, spore-forming, rod-shaped anaerobic bacterium that encounters many ecologic niches due to its widespread distribution in the environment, including a presence in sewage, soil, foods, normal intestinal biota, and feces (1). This bacterium is also pathogenic because of its ability to produce more than 16 different toxins (1–3). C. perfringens is classified into five different types (A to E) based upon production of four (α, β, ε, and ι) toxins (1, 4). Besides making one or more of the typing toxins, C. perfringens strains may produce additional toxins such as the enterotoxin perfringolysin O, TpeL, NetB, or beta2 toxin (1, 5–7). All C. perfringens types cause diseases, which differ according to the toxin-producing ability of the infecting strain. Overall, C. perfringens illnesses range from histotoxic infections, such as traumatic gas gangrene, to infections originating in the intestines, such as enteritis or enterotoxemia (4, 8).
Sialidases, also referred to as neuraminidases, are key enzymes for the catabolism of sialic acid-containing oligosaccharides (9, 10). These enzymes are found in higher animals and a variety of microorganisms, including viruses, bacteria, and protozoa (11). Sialidases cleave terminal sialic acid residues that are linked, in the alpha configuration, to oligosaccharide chains present on proteins and lipids (12). Bacterial sialidases can have a nutritional function, enabling a bacterium to procure sialic acids from the host for use as carbon and energy sources (13). Sialidases can also function as virulence factors during bacterial pathogenesis, whereby they can contribute to colonization, increase toxin binding, and cause immunomodulatory effects (14–17).
Individual C. perfringens strains can produce up to three different sialidases, including two large secreted sialidases, named NanI (77 kDa) and NanJ (129 kDa), and one small sialidase named NanH (43 kDa) that has an intracellular location in log-phase cultures but can be found extracellularly in overnight cultures (17–21). These three sialidases are believed to share related, although not identical, catalytic domains (22). In addition, NanI and NanJ possess one and five (respectively) accessory modules, some of which are thought to mediate carbohydrate-binding interactions (22). C. perfringens NanI and NanH have been purified and partially characterized (12). The function of NanH is thought to involve cleavage of short oligosaccharides for nutritional purposes (23). NanI was shown to be the predominant exosialidase produced by C. perfringens type A strain 13 and type D strain CN3718, and this sialidase may also play a nutritional role by releasing sialic acid from higher-order gangliosides (17, 24). In addition, NanI was recently reported to facilitate the adherence of CN3718 to enterocyte-like Caco-2 cells and to increase epsilon toxin binding and cytotoxicity for host MDCK cells (17).
Until now, there has been no characterization of NanJ properties, nor have the properties of the three sialidases yet been compared in a single study. Using preparations of a purified C. perfringens sialidase for such studies entails the risk of effects due to contamination of the preparation with the other two sialidases. Furthermore, there has been only limited analysis of the contributions of each sialidase, when present, to total sialidase activity in C. perfringens culture supernatants. Therefore, in the current study, we inactivated two of the three sialidase genes in type D strain CN3718 to produce a series of mutant strains expressing, at their native levels, only NanJ, NanI, or NanH. These mutant strains were then used to characterize the properties of each C. perfringens sialidase in a background free from the other two sialidases and to analyze the contributions of each of these enzymes to total culture sialidase activity at different points in the growth curve.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
The C. perfringens parent isolate used in this study was type D strain CN3718 (17). BMC205 is a nanJ nanI nanH triple null mutant of CN3718 prepared in a previous study (17). Stock cultures of all isolates were stored in cooked-meat medium (Oxoid) at −20°C. For experiments, all cultures were grown for 3, 6, or 16 h at 37°C in, as specified, fluid thioglycolate (FTG) broth (Difco Laboratories) or Bacto Todd-Hewitt (TH) broth (Becton, Dickinson) with 0.1% sodium thioglycolate (Sigma Chemical). Thioglycolate, a reducing agent, was added to TH broth to allow growth of C. perfringens, an anaerobe. Mutagenesis plasmids used in this study included pJIR750nanJi, pJIR750nanIi, and pJIR750nanHi, which were prepared in a previous study and mediate the insertional inactivation of nanJ, nanI, and nanH, respectively (17). All antibiotics used in this study were purchased from the Sigma-Aldrich Chemical Company or Fisher Scientific Company.
Construction of sialidase null mutants of C. perfringens isolate CN3718.
Using the Sigma Clostridium-modified TargeTron system (25), the nanJ, nanI, or nanH gene of CN3718 was inactivated by the insertion of a target group II intron from mutagenesis plasmid pJIR750NanJi, pJIR750NanIi, or pJIR750NanHi (17). This mutagenesis work resulted in insertional inactivation of the nanI and nanH genes in CN3718 to produce a mutant strain (named ENanJ) expressing only NanJ. Similarly, mutant strains of CN3718 that express only NanI or NanH were constructed, creating strains named ENanI or ENanH, respectively.
The null mutant strains were identified by a colony PCR assay using the following amplification conditions: cycle 1, 95°C for 5 min; cycles 2 through 35, 95°C for 30 s, 55°C for 40 s, and 68°C for 80 s; and a final extension for 8 min at 68°C. An aliquot (20 μl) of each PCR sample was electrophoresed on a 1.5% agarose gel and then visualized by staining the gel with ethidium bromide. This PCR amplifies a 306-bp product from the wild-type nanJ gene using nanJKOF and nanJKOR primers (17), a 467-bp product from the wild-type nanI gene using the primers nanIKOF and nanIKOR (17), and a 285-bp product from wild-type nanH gene using the primers nanHKOF and nanHKOR (17). However, the same PCR will amplify larger products after the 900-bp intron has inserted into these wild-type genes.
DNA isolation and Southern blot analysis of the mutant strains.
DNA was isolated from wild-type CN3718 or the ENanJ, ENanI, ENanH, and BMC205 mutants, using the MasterPure Gram Positive DNA Purification kit (Epicenter). A 3-μg aliquot of each isolated DNA in Tris-EDTA (TE) buffer (Epicenter) was digested overnight with BsrGI at 37°C according to the manufacturer's instructions (New England BioLabs). The digested DNA samples were then electrophoresed on a 1% agarose gel and transferred onto a positively charged nylon membrane (Roche) for hybridization with the digoxigenin (DIG)-labeled intron sequence-specific probe, which was prepared using the Roche DIG-labeled kit as earlier described (26).
Comparison of sialidase contributions to the intracellular versus supernatant activity of CN3718 log-phase, stationary-phase, and death phase cultures.
After 3 h, 6 h, or 16 h of growth in TH medium at 37°C, equal volumes of each culture supernatant were assayed for extracellular sialidase activity, as described below. At the same time, bacterial cells in these cultures were sonicated and centrifuged. Equal volumes of supernatant from each sonicated culture lysate were then also used to measure the intracellular sialidase activity. In this experiment, sialidase activity was measured by a previously described protocol (17). Briefly, a 20-μl aliquot sample, prepared as described above, was added to 60 μl of 0.05 M Tris-HCl buffer (pH 7.2) in a microtiter plate. A 20-μl aliquot of 4 mM 5-bromo-4-chloro-3-indolyl-α-d-N-acetylneuraminic acid (Sigma) substrate was added, and the mixture was incubated at 37°C for 60 min. The absorbance at 595 nm was then determined using a microplate reader (Bio-Rad). Pilot studies showed that under these assay conditions, sialidase activity in the samples was in the linear range and not saturated.
Western blotting of sialidase production.
For Western blot analysis of sialidase production, a 0.1-ml aliquot of an FTG broth overnight culture of wild-type CN3718 or the ENanJ, ENanI, ENanH, and BMC205 mutants was inoculated into 10 ml of TH medium. After ∼16 h of growth at 37°C, equal volumes of each culture supernatant were mixed with 5× SDS-PAGE loading buffer and boiled for 10 min. A 30-μl aliquot of each SDS sample was electrophoresed on an 8% SDS-polyacrylamide gel and then transferred onto a nitrocellulose membrane. The membrane was then blocked in Tris-buffered saline (TBS) buffer with 5% (wt/vol) nonfat dry milk for 1 h at room temperature. After blocking, the membrane was probed overnight at 4°C with a sialidase rabbit polyclonal antibody (at a 1:1,000 dilution), which was purchased from LifeSpan BioSciences, Inc. After this incubation, the membrane was treated for 1 h at room temperature with TBS buffer containing 5% (wt/vol) nonfat dry milk and a horseradish peroxidase-conjugated secondary anti-rabbit antibody (1:10,000 dilution) (Sigma). Immunoreactivity on the membrane was then detected using the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Effects of temperature and pH on the activity of each C. perfringens sialidase.
To assay the effects of temperature and pH on the activity of each C. perfringens sialidase, the protocol was as described above. Briefly, CN3718, ENanJ, ENanI, ENanH, and BMC205 were grown for 16 h at 37°C in TH broth. To measure temperature effects on sialidase activity, a 20-μl aliquot of supernatant from each of those overnight cultures was added to 60 μl of 0.05 M Tris-HCl buffer (pH 7.2) in a microtiter plate. A 20-μl aliquot of 4 mM 5-bromo-4-chloro-3-indolyl-α-d-N-acetylneuraminic acid (Sigma) substrate was then added, and the different trays were incubated at 25, 37, 43, or 48°C for 60 min. The absorbance at 595 nm was then determined using a microplate reader (Bio-Rad).
The effects of pH on sialidase activity were similarly measured, except that each supernatant aliquot was added to 60 μl of 0.05 M sodium acetate buffer (pH 5.5), 0.05 M Tris-HCl buffer (pH 7.2), or 0.05 M Tris-HCl buffer (pH 9.0) containing 4 mM 5-bromo-4-chloro-3-indolyl-α-d-N-acetylneuraminic acid substrate. After incubation for 60 min at 37°C, the pH of the samples was adjusted back to pH 7.2 and reaction volumes were equalized. The enzymatic activity was then determined with a microplate reader.
Heat inactivation of sialidase activity.
CN3718, ENanJ, ENanI, ENanH, or BMC205 was grown overnight (16 h) at 37°C in TH broth. Supernatants were removed from those cultures and heated at 50°C or 60°C for 0, 5, or 10 min. Sialidase activity was then measured using the methods described above.
Effects of various metal ions and chemicals on sialidase activity.
CN3718, ENanJ, ENanI, ENanH, or BMC205 strains were each grown for ∼16 h at 37°C in TH broth. To avoid the high background caused by metal ions present in TH broth, each TH culture supernatant was desalted and buffer exchanged with phosphate-buffered saline (PBS) buffer (Corning) using a Millipore Ultrafiltration centrifuge tube (10,000 nominal molecular weight limit [NMWL]) (Millipore). A 20-μl aliquot of each desalted supernatant was then added to microtiter wells containing 60 μl of 0.05 M Tris-HCl buffer (pH 7.2) supplemented with one of the following: a 10 mM final concentration of CaCl2, CoCl2, NiCl2, MnCl2, MgCl2, ZnCl2, FeCl2, FeCl3, p-chloromercuribenzoate, or EDTA in a microtiter tray. A 20-μl aliquot of 4 mM 5-bromo-4-chloro-3-indolyl-α-d-N-acetylneuraminic acid was added, and the microtiter tray was incubated at 37°C for 60 min. The absorbance at 595 nm was then determined using a microplate reader.
Substrate specificity assays for C. perfringens sialidases.
To evaluate C. perfringens sialidase substrate specificity, sialidase activity was determined using the EnzyChrom neuraminidase assay kit (BioAssay Systems), which measures sialic acid released by neuraminidase activity. After 20 min or 50 min of incubation at 37°C, a microplate reader (Bio-Rad) was used to measure the sample absorbance at 570 nm, and the optical density (OD) difference was used to calculate the neuraminidase activity according to the kit's instructions. To avoid the background caused by sialic acid present in the TH medium, 16-h TH culture supernatant was desalted and buffer exchanged with PBS buffer using the Millipore Ultrafiltration centrifuge tube as described earlier. Substrates used in this sialidase assay include 100 μg/ml of 3′-sialyl-d-lactose (α-2,3-specific linkage), 6′-sialyl-d-lactose (α-2,6-specific linkage), and colominic acid sodium (α-2,8-specific linkage) (all purchased from Sigma-Aldrich). Pilot studies demonstrated that under these conditions, sialidase activity was in the linear range and not saturated.
A similar experiment was performed using samples that had been trypsin pretreated as described before (17) for 1 h at 37°C. After this incubation with trypsin (Sigma, T4799), trypsin inhibitor (Sigma) (1:1, vol/vol) was added to remove trypsin activity from those samples (17). PBS buffer (Fisher), pH 7.4, trypsin with PBS buffer, trypsin inhibitor with PBS buffer, and trypsin and trypsin inhibitor with PBS buffer were used as negative controls.
If significant levels of sialylate lyase, which degrades free sialic acid, were present in the 16-h TH culture supernatants, this could interfere with the accuracy of sialic acid measurements using the EnzyChrom neuraminidase assay kit, since this kit measures free sialic acid. However, when free sialic acid was added to the supernatants in the absence of substrate, no significant sialic acid degradation was observed after 4 h of incubation at 37°C (data not shown), indicating the absence of significant sialylate lyase activity in the supernatants.
Detection of sialic acid release from Caco-2 cells.
Caco-2 cells were cultured in 6-well tissue culture dishes as described previously (17). These Caco-2 cell monolayers were washed three times with PBS buffer and then harvested by gentle scraping in 100 μl of PBS buffer. To avoid background sialic acid present in TH broth, 16-h supernatants from TH cultures of the wild-type, ENanJ, ENanI, or ENanH strain were desalted and buffer exchanged with PBS using Millipore Ultrafiltration centrifuge tubes as described earlier. A 100-μl aliquot of each desalted supernatant was then incubated overnight at 37°C with a 20-μl suspension of Caco-2 cells. The next day, sialic acid released from the Caco-2 cells was measured using the EnzyChrom neuraminidase assay kit according to the kit's instructions.
Sialidase inhibitors and IC50 determination.
Sialidase inhibitors N-acetyl-2,3-dehydro-2-deoxyneuraminic acid (NADNA) and siastatin B (SB) were purchased from Sigma-Aldrich. CN3718, ENanJ, ENanI, ENanH, and BMC205 were each grown for 16 h at 37°C in TH broth. A 20-μl aliquot of each TH culture supernatant was then mixed with different amounts of NADNA (0, 8, 16, or 24 μM) or SB (0, 27, 55, or 110 μM). A 20-μl aliquot of 4 mM 5-bromo-4-chloro-3-indolyl-α-d-N-acetylneuraminic acid substrate was added, and the mixture was incubated at 37°C for 60 min. The absorbance of these samples at 595 nm was then determined using a microplate reader. Based on sample absorbance at 595 nm, the 50% inhibitory concentrations (IC50s) for each supernatant were then estimated using the Excel linear regression method.
RESULTS
Construction and characterization of isogenic CN3718 mutants expressing a single sialidase.
Strain CN3718 naturally produces three sialidases (17). Previous studies have indicated that for this strain, NanH has an intracellular location until 5-h late-log-phase TH cultures but can be found in supernatants of 16-h TH cultures (17, 21), which are in death phase (data not shown). Since CN3718 16-h TH culture supernatants also contain secreted NanI and NanJ sialidases, these samples would be useful sources of sialidases to study, at natural levels, the properties of all three C. perfringens sialidases.
However, to study the properties of each sialidase free from contamination by the other two sialidases, a series of CN3718 mutants were prepared using the Clostridium-modified TargeTron insertional mutagenesis method (25). These efforts resulted in inactivation of two sialidase genes in each mutant, creating null mutant strains (named ENanJ, ENanI, or ENanH) that could express, respectively, only NanJ, NanI, or NanH.
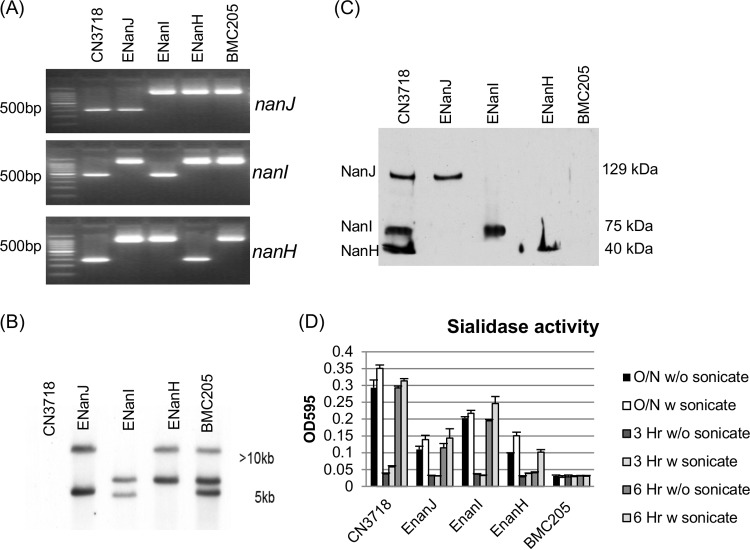
PCR analyses were performed to confirm the identity of each mutant (Fig. 1A). Using DNA from wild-type strain CN3718, a PCR specific for nanJ, nanI, or nanH sequences amplified the expected PCR products of 306 bp for nanJ, 467 bp for nanI, and 285 bp for nanH. The same primers amplified PCR products of ∼1,200 bp, ∼1,400 bp, and ∼1,200 bp using DNA from mutants whose nanJ, nanI, or nanH gene had been targeted for the insertion of an ∼900-bp intron. Combined, the results of these PCR assays (Fig. 1A) confirmed that in the ENanJ mutant both the nanI and nanH genes carried an intron insertion, that in the ENanI mutant, both the nanJ and nanH genes carried an intron insertion, and that in the ENanH mutant, both the nanJ and nanI genes carried an intron insertion. Using DNA from BMC205, a previously prepared CN3718 derivative with all three sialidase genes disrupted by intron insertions (17), the same PCR amplified larger bands for all three sialidase genes (Fig. 1A), consistent with this being a triple sialidase null mutant strain.
FIG 1.
Intron-based insertional mutagenesis to create CN3718 mutants that express only NanJ, NanI, or NanH sialidase. (A) Internal nanJ-, nanI-, and nanH-specific PCR results for wild-type CN3718, ENanJ (expressing only NanJ), ENanI (expressing only NanI), and ENanH (expressing only NanH). Also shown is BMC205, a nanJ nanI nanH triple null mutant strain that had been constructed previously (17). Using DNA from wild-type strain CN3718, a PCR specific for nanJ, nanI, or nanH sequences amplified the expected PCR products of 306 bp for nanJ, 467 bp for nanI, and 285 bp for nanH. The same primers amplified PCR products of ∼1,200 bp, ∼1,400 bp, and ∼1,200 bp using DNA from mutants whose nanJ, nanI, or nanH gene had been targeted for the insertion of an ∼900-bp intron. The migration of 100-bp DNA markers is shown on the left. (B) Southern blot analysis of wild-type CN3718 or the ENanJ, ENanI, ENanH, and BMC205 mutant strains using an intron-specific DIG-labeled probe. DNA size markers are shown on the right. (C) Sialidase Western blot analysis of sialidase expression of wild-type CN3718, ENanJ, ENanI, ENanH, and BMC205 null mutant strains in TH 16-h culture supernatants. Protein size markers are shown on the right. Wild-type CN3718 expresses all three sialidases, including NanJ (129 kDa), NanI (77 kDa), and NanH (43 kDa). (D) Sialidase enzyme activity present after wild-type CN3718 or the ENanJ, ENanI, ENanH, and BMC205 mutant strains were grown for 3 h, 6 h, or 16 h with or without sonication. All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
After curing the intron delivery plasmid from each null mutant strain, DNA from the wild-type or sialidase null mutant strains was subjected to Southern blot analysis using an intron-specific probe (Fig. 1B). Results from these analyses detected no intron probe hybridization to DNA from the wild-type strain, as expected. In contrast, two intron insertions were detected using DNA from the ENanJ, ENanI, or ENanH strains and three intron insertions were noted using DNA from the BMC205 nanJ nanI nanH triple null mutant. These Southern blot results confirmed the expected presence of intron insertions in each mutant and also showed that no undesired extraneous intron insertions had occurred.
Western blot analyses of sialidase production by the ENanH, ENanI, and ENanJ mutants.
Western blot analysis was then performed to confirm that the mutants produced only their expected sialidase. Results of this experiment demonstrated that 16-h TH culture supernatant of wild-type CN3718 contained the 129-kDa NanJ, the 77-kDa NanI, and the 43-kDa NanH. However, 16-h culture supernatants of the ENanJ strain contained only the 129-kDa NanJ; those of the ENanI strain contained only the 77-kDa NanI, and the 16-h supernatant of the ENanH strain contained only the 43-kDa NanH. All single-sialidase-producing mutants made their sialidase at levels similar to those expressed by CN3718. As expected, the BMC205 did not produce any sialidase, as indicated by the sialidase Western blot results (Fig. 1C).
Sialidase activity assays of the wild-type, EnanJ, EnanI, EnanH, and BMC205 strains were conducted to compare the relative intracellular versus extracellular distribution of NanH, NanJ, and NanI activity after 3, 6, or 12 h of growth, where cultures are in, respectively, the log phase, stationary phase, or death phase (data not shown). Collectively, the results (Fig. 1D) indicated that in these samples, the supernatant NanI sialidase activity of CN3718 was greater than its NanJ or NanH supernatant activity. These assays also confirmed that >80% of NanI and NanJ activities are extracellular at all growth phases, i.e., these are secreted sialidases. These analyses also revealed that substantial NanH activity is found only intracellularly until the death phase. Thus, the NanH activity present in 16-h supernatants is associated with bacterial cell death. BMC205 supernatants were devoid of any sialidase activity; this result also supported production of only three sialidases by CN3718.
Temperature effects on C. perfringens sialidase activity assessed using 16-h TH culture supernatants from null mutant strains producing only NanI, NanJ, or NanH.
To compare temperature effects on the activity of each C. perfringens sialidase, supernatants were removed from 16-h TH cultures of the wild-type strain CN3718 or the ENanJ, ENanI, ENanH, or BMC205 mutants grown at 37°C. Those supernatants were then incubated at temperatures between 25 and 48°C for 60 min in the presence of sialidase substrate. When sialidase activity was measured in supernatants from these cultures (Fig. 2A), 37°C and 43°C were found to be the optimal temperatures for both NanJ and NanH activity. Both of those sialidases exhibited lower activity at 48°C. In contrast, NanI sialidase activity steadily increased with temperature until 48°C. At 25°C, all three sialidases exhibited relatively low activity. The sialidase activity measured in supernatants from 16-h TH cultures of the wild-type strain, ENanJ, ENanI, or ENanH was specifically due to the presence of the known C. perfringens sialidases, since no sialidase activity was detected using supernatants removed from 16-h TH cultures of BMC205, which lacks any known functional sialidase genes. Therefore, these results (Fig. 2A) indicated that NanI activity has a broader temperature optimum than the other two C. perfringens sialidases.
FIG 2.
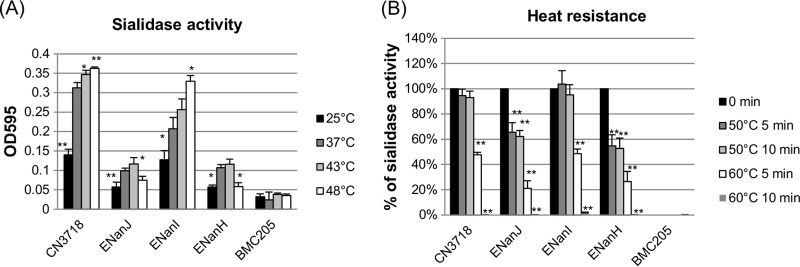
Determination of sialidase optimal temperatures and heat resistance properties using culture supernatants containing NanJ, NanI, or NanH. Wild-type CN3718 expresses all three sialidase genes; BMC205 is a triple sialidase null mutant (17); ENanJ expresses only the nanJ gene, ENanI expresses only the nanI gene, and ENanH expresses only the nanH gene. Supernatants were removed from three independent cultures of each strain grown for 16 h at 37°C in TH broth. (A) The supernatants were incubated for 60 min at 25, 37, 43, or 48°C with sialidase substrate, and sialidase activity was then measured. *, P < 0.05; **, P < 0.001 (values compared to the 37°C value for each supernatant using the Student t test). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations. (B) The supernatants were first treated for 5 min or 10 min at 50°C or 60°C and then incubated for 60 min at 37°C with sialidase substrate to measure the sialidase activity. *, P < 0.05; **, P < 0.001 (values compared to the 0 min value for each supernatant using the Student t test). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
Heat resistance properties of C. perfringens NanI, NanJ, and NanH sialidases.
Supernatants removed from TH cultures of ENanI, which produces only the NanI sialidase, after growth for 16 h at 37°C showed no loss of sialidase activity after a 10-min incubation at 50°C in Tris-HCl buffer, pH 7.2 (Fig. 2B). However, supernatants removed from similar 16-h cultures of ENanJ or ENanH, which contained only NanJ or NanH (respectively), lost about 40% of their sialidase activity after a similar 10-min incubation at 50°C. Following a 5-min incubation in the same buffer, supernatants from the 16-h ENanI TH cultures lost about 50% of their sialidase activity at 60°C, while the supernatants removed from 16-h ENanJ or ENanH TH cultures lost more than 80% of their sialidase activity at 60°C. After a 10-min incubation at 60°C, all 16-h TH culture supernatants lost most of their sialidase activity. These results revealed that compared to the NanJ and NanH sialidases, the NanI sialidase exhibits greater heat resistance.
Effects of pH on C. perfringens sialidase enzymatic activity.
The effects of pH on C. perfringens sialidase activity were assessed by adjusting the pH of the supernatants removed from 16-h TH cultures that had been grown at 37°C. After the substrate 5-bromo-4-chloro-3-indolyl-α-d-N-acetylneuraminic acid was added to those pH-adjusted supernatants, the mixtures were incubated at 37°C for 1 h. At the end of the incubation, the mixtures were adjusted back to pH 7.2 before sialidase activity was measured. Results of this experiment showed that the strongest activity for all three sialidases was detected at pH 5.5 (Fig. 3). At pH 9.0, the activity of all three sialidases had decreased by about 50%. As a negative control, supernatants removed from a culture of the BMC205 triple null mutant grown for 16 h at 37°C in TH broth did not show sialidase activity at any tested pH.
FIG 3.
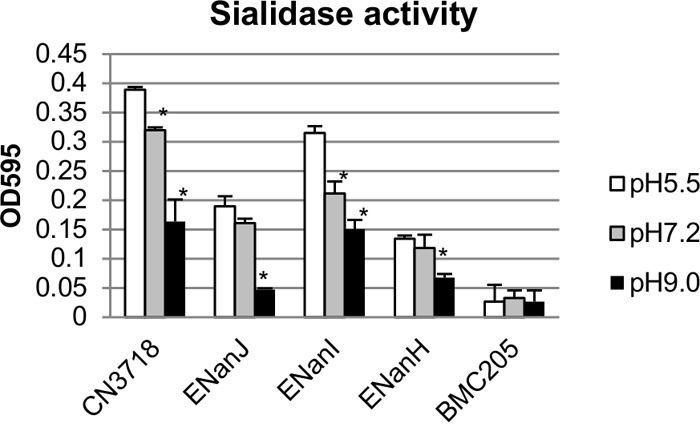
Effect of pH on sialidase activity of 16-h TH culture supernatants containing NanI, NanJ, or NanH. Wild-type CN3718 expresses all three sialidase genes; BMC205 has null mutations in all three sialidase genes (17); ENanJ expresses only the nanJ gene, ENanI expresses only the nanI gene, and ENanH expresses only the nanH gene. Supernatants were removed from three independent cultures of each strain grown 16 h at 37°C in TH broth. The pH of each supernatant was adjusted to pH 5.5, 7.2, or 9.0. Those supernatants were then incubated with substrate at 37°C for 60 min, and sialidase activity was measured after the pH of the samples was adjusted back to 7.2. At each pH value tested, the sialidase activity was determined using supernatants from three independent cultures. *, P < 0.05 (values compared to the corresponding pH 5.5 value for each supernatant, using the Student t test). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
Substrate specificity of C. perfringens NanJ, NanI, and NanH sialidases.
This study next examined the ability of 16-h TH culture supernatants containing a single sialidase to release sialic acid from various natural substrates with a defined sialic acid linkage. The results obtained, shown in Fig. 4A, indicated that wild-type supernatant possesses activity against the three sialic acid linkages tested, i.e., an α-2,3-specific linkage, an α-2,6-specific linkage, and an α-2,8-specific linkage. The observed activity was in the following order: α-2,3 > α-2,6 > α-2,8 sialic acid linkages.
FIG 4.
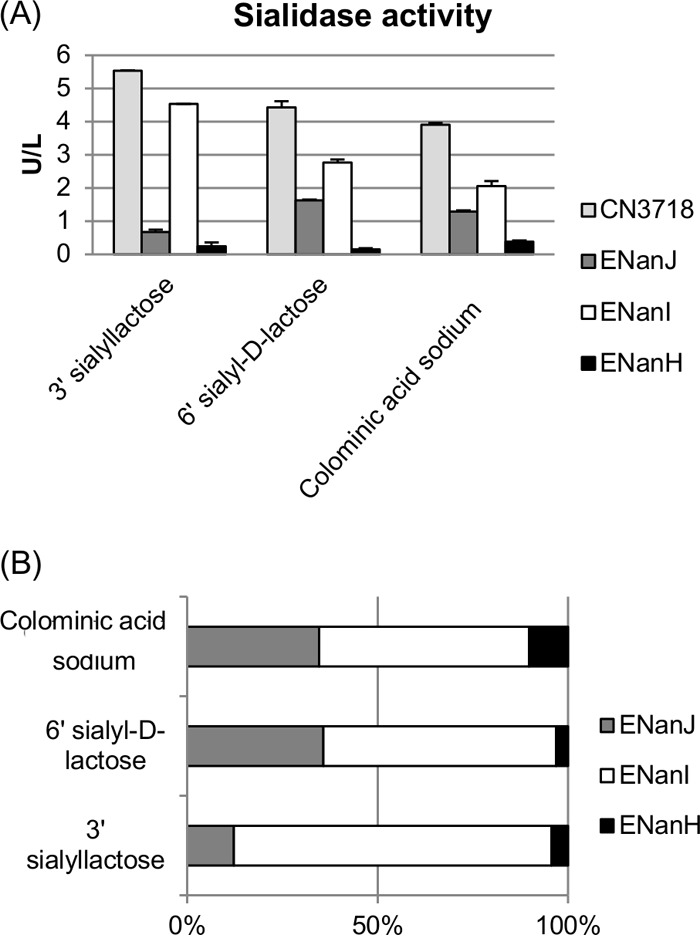
Sialidase activity of culture supernatants containing NanI, NanJ, or NanH using different substrates. Sialidase activity assays were performed using the EnzyChrom neuraminidase assay kit. For each substrate, the sialidase activity was determined using supernatants removed from five independent cultures (wild-type CN3718 expresses all three sialidase genes; BMC205 has null mutations in all three sialidase genes [17]; ENanJ expresses only the nanJ gene, ENanI expresses only the nanI gene, and ENanH expresses only the nanH gene) grown for 16 h at 37°C in TH broth. Substrates assayed included 3′ sialyllactose (2,3-linkage), 6′ sialyl-d-lactose (2,6-linkage), and colominic acid sodium (2,8-linkage). (A) Sialidase activity (after subtraction of the background using BMC205 results) was determined using three sialic acid linkage substrates. (B) Percent contribution of each sialidase to wild-type supernatant sialic acid release from each substrate (wild-type value equals 100%). All experiments were repeated at least three times.
Furthermore, these studies revealed that in 16-h TH supernatants, NanI is responsible for most of the total supernatant sialidase activity against each of the tested substrates, regardless of their sialic acid linkage (Fig. 4A and B). NanI showed preferential activity in the order of α-2,3 > α-2,6 > α-2,8 linkages, similar to the pattern of the wild-type supernatant. NanJ activities showed a preference for α-2,6 > α-2,8 > α-2,3 sialic acid linkages for the tested substrates. NanH activities showed a preference for α-2,8 > α-2,3 > α-2,6 linkages using the tested substrates. Supernatants from 16-h TH cultures of the wild-type strain, which contained all three C. perfringens sialidases, showed strong activity for all three tested substrates, consistent with the three sialidases working together in combination (Fig. 4B).
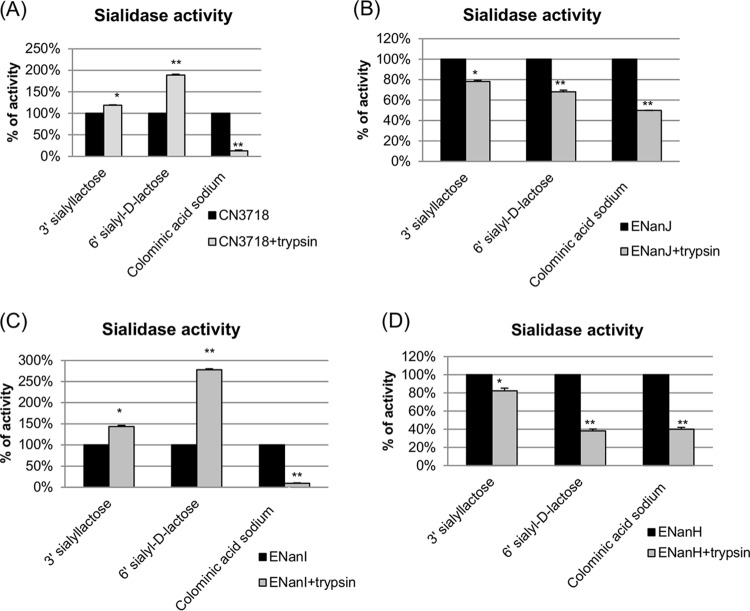
We previously reported that the sialidase activity of NanI (but not NanJ or NanH) could be enhanced by trypsin pretreatment (17), which may have relevance for C. perfringens type D diseases originating in the intestines since this could increase bacterial adherence and toxin binding (17). Therefore, in the current study, different substrates were preincubated with trypsin-pretreated NanJ, NanI, or NanH supernatants. The results obtained (Fig. 5) indicated that trypsin pretreatment enhanced NanI activity, but only for substrates with an α-2,6-sialic acid linkage or, to a lesser extent, an α-2,3-sialic acid linkage. The sialidase activity of either NanJ or NanH was decreased by trypsin pretreatment, and this pretreatment did not affect their substrate preference. As expected with NanI being the predominant sialidase made by this type D strain, the wild-type supernatant showed the same substrate preference patterns as NanI supernatant after trypsin pretreatment. Sialidase Western blot analyses (not shown) determined that NanI was processed to ∼65 kDa, as described previously (17), while both NanJ and NanH were substantially degraded after trypsin treatment.
FIG 5.
Sialidase activity of trypsin-pretreated 16-h TH culture supernatants containing NanI, NanJ, or NanH using different substrates (3′ sialyllactose, 6′ sialyl-d-lactose, and colominic acid). (A) Wild-type CN3718 supernatant sialidase activity after trypsin pretreatment (wild-type supernatant sialidase activity without trypsin treatment was considered 100%). (B) ENanJ strain supernatant sialidase activity after trypsin pretreatment (ENanJ supernatant sialidase activity without trypsin treatment was considered 100%). (C) ENanI strain supernatant sialidase activity after trypsin pretreatment (ENanI supernatant sialidase activity without trypsin treatment was considered 100%). (D) ENanH strain supernatant sialidase activity after trypsin pretreatment (ENanH supernatant sialidase activity without trypsin treatment was considered 100%). *, P < 0.05; **, P < 0.001 (values compared to corresponding nontreated samples of each supernatant, using the Student t test). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
Sialic acid release from Caco-2 cells after treatment with 16-h TH culture supernatants containing only NanJ, NanI, or NanH.
To further explore the potential pathogenic contributions of each sialidase, Caco-2 cells were incubated in suspension with 16-h TH culture supernatants containing only NanJ, NanI, or NanH. The results showed that at their natural levels present in these culture supernatants, both NanJ and NanI released substantial amounts of sialic acid from Caco-2 cells (Fig. 6). In contract, at its native levels present in 16-h TH culture supernatants, NanH caused only limited release of sialic acid from Caco-2 cells. The negative-control BM205 did not release any sialic acid from Caco-2 cells, as expected since this strain cannot produce any sialidases.
FIG 6.
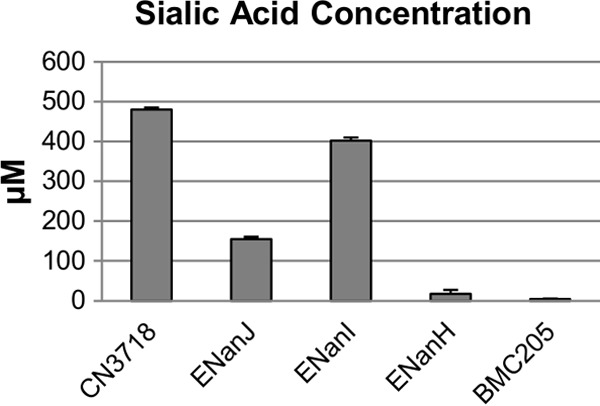
Sialic acid release from Caco-2 cells after treatment with 16-h TH culture supernatants containing NanI, NanJ, or NanH. Supernatants were removed from five independent cultures (wild-type CN3718 expresses all three sialidase genes; BMC205 has null mutations in all three sialidase genes [17]; ENanJ expresses only the nanJ gene, ENanI expresses only the nanI gene, and ENanH expresses only the nanH gene) that had been grown for 16 h at 37°C in TH broth. The supernatants were then incubated with Caco-2 cells (see Materials and Methods) for 16 h at 37°C, and the concentration of sialic acid released from the Caco-2 cells was then detected using the EnzyChrom neuraminidase assay kit. All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
Effects of various metal ions and compounds on the activity of C. perfringens sialidases.
The effects of various metal ions on the enzymatic activity of the three C. perfringens sialidases were measured using supernatants removed from each culture grown for 16 h at 37°C in TH broth (Table 1). For these assays, cations were supplied as chloride salts and the pH of the incubation mixture was adjusted to pH 7.2 in 0.05 M Tris-HCl buffer based on the results shown in Fig. 3. By this approach, several metal ions (Fe2+, Mn2+, and Mg2+) were found at these tested concentrations to enhance NanI enzyme activity, while Fe3+ and Zn2+ decreased NanI enzyme activity. Besides Fe2+ and Mn2+, Co2+, Mg2+, Ni2+, and Zn2+ also increased NanJ activity. In contrast, only Fe3+ decreased NanJ activity. For NanH activity, all tested metal ions, except Mg2+, were found to decrease its activity.
TABLE 1.
Effects of various compounds and metal ions on sialidase activity
| Compound or metal ion (10 mM) | % sialidase activity (relative to control) |
|||
|---|---|---|---|---|
| CN3718 | ENanJ | ENanI | ENanH | |
| None (control) | 100 | 100 | 100 | 100 |
| Fe2+ | 101 | 188 | 111 | 65 |
| Fe3+ | 56 | 87 | 33 | 79 |
| Ca2+ | 90 | 100 | 100 | 71 |
| Co2+ | 110 | 200 | 104 | 78 |
| Ni2+ | 94 | 155 | 95 | 90 |
| Mn2+ | 110 | 170 | 115 | 53 |
| Mg2+ | 115 | 117 | 124 | 99 |
| Zn2+ | 61 | 136 | 65 | 88 |
| p-Chloromercuribenzoate | 18 | 54 | 13 | 5 |
| EDTA | 104 | 105 | 86 | 97 |
p-Chloromercuribenzoate (10 mM) substantially inhibited the enzymatic activity of all three sialidases, but NanJ was the most resistant sialidase against this agent. Another experiment showed that EDTA has a minimal inhibitory effect on NanI, while NanH and NanJ are essentially unaffected by this divalent cation chelator. These results further confirm differences between the activities of the C. perfringens sialidases.
Effects of sialidase inhibitors on NanJ, NanI, or NanH activities.
Various inhibitors have been reported to reduce the activity of C. perfringens sialidases (27–29). However, most of those previous inhibitor studies did not specify which C. perfringens sialidase was used and none of them had compared the inhibitor sensitivity of all three C. perfringens sialidases in a side-by-side experiment. In the current research, the sialidase activities of C. perfringens 16-h TH culture supernatants that contained natural levels of only NanJ, NanI, or NanH were tested in the presence of two sialidase inhibitors, i.e., NADNA and SB. Results of these analyses showed that the sialidase activity of NanH was relatively more resistant to both tested sialidase inhibitors, especially NADNA, than the activity of NanI or NanJ (Fig. 7). The sialidase activity of NanJ was equally sensitive to the two inhibitors, while NanI showed more sensitivity to NANDA than to SB. The IC50s of the inhibitors determined for each of the three sialidases are shown in Table 2.
FIG 7.
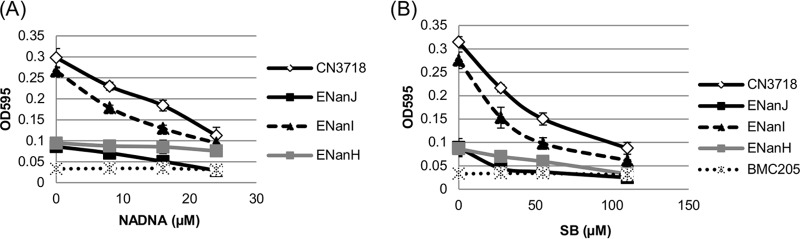
Inhibition of C. perfringens sialidase activity by sialidase inhibitors NADNA and SB. Supernatants were removed from five independent cultures (wild-type CN3718 expresses all three sialidase genes; BMC205 has null mutations in all three sialidase genes [17]; ENanJ expresses only the nanJ gene, ENanI expresses only the nanI gene, and ENanH expresses only the nanH gene) grown for 16 h at 37°C in TH broth. (A) Sialidase activity of wild-type CN3718, ENanJ, ENanI, ENanH, and BMC205 supernatants in the presence of the NADNA sialidase inhibitor (0, 8, 16, and 24 μM). (B) Sialidase activity of wild-type CN3718, ENanJ, ENanI, ENanH, and BMC205 supernatants in the presence of the SB sialidase inhibitor (0, 27.5, 55, 110 μM). All experiments were repeated three times, and mean values are shown. The error bars indicate standard deviations.
TABLE 2.
Effect of sialidase inhibition on sialidase activity
| Sample | IC50 (μM) |
|
|---|---|---|
| NADNA | SB | |
| CN3718 | 18.9 | 42.2 |
| ENanJ | 12.4 | 15.1 |
| ENanI | 13.4 | 27.5 |
| ENanH | 44.6 | 50.9 |
DISCUSSION
Sialic acids can be important molecules during bacterial pathogenesis (28, 30). Some bacterial pathogens have evolved to use these molecules beneficially in several ways: (i) as a nutrient; (ii) to cloak themselves, thus avoiding the host innate immune response by a molecular mimicry effect; and (iii) for biofilm formation (9, 11, 13, 30, 31). The mammalian mucosal surface contains many different kinds of sialic acid, almost all of which are bound to proteins or lipid acceptors, making them unavailable for microbial metabolism (32). Therefore, some bacterial pathogens secrete one or more sialidases, which release sialic acid from a diverse range of host sialoglycoconjugates (28). In addition, sialidases can unmask cryptic host ligands important for bacterial adherence or toxin binding (10, 17, 30) and exert immunomodulatory effects (9).
Previous studies (11, 12) had suggested that differences exist between the activities of C. perfringens NanI and NanH. The current work significantly extends those previous findings by comparing, in 16-h TH culture supernatants, the activities of all three C. perfringens sialidases at natural levels and in a background free of contamination by the other sialidases. With respect to heat tolerance, it was previously reported that NanI activity is more heat resistant than NanH activity (11, 12). The current study confirmed those previous findings and further determined that NanJ resembles NanH by exhibiting much less sialidase activity above 43°C. It had also been previously reported that the pH optima of NanI and NanH were pH 5 and pH 6, respectively (12). The current study confirmed that previous report and then further determined that NanJ also functions best at pH ∼5. The enzymatic activity of each sialidase was also found to vary in sensitivity to various metal ions. Furthermore, while the enzymatic activities of all three C. perfringens sialidases were shown to be sensitive to p-chloromercuribenzoate, which reacts with thiol groups in proteins, NanJ was relatively more resistant to this treatment than the other two C. perfringens sialidases. The sensitivity of NanH to p-chloromercuribenzoate observed in the current study is in agreement with results of a previous study (33). These results also distinguished the three C. perfringens sialidases from Streptomyces sialidase, which is unaffected by treatment with p-chloromercuribenzoate (11). Finally, our results extend previous reports (12) by now indicating that all three C. perfringens sialidases are active against a broad range of sialic acid linkages. NanI was found to be responsible for most of the overnight TH culture supernatant sialidase activity against all substrates, regardless of whether they had an α-2,3, α-2,6, or α-2,8 sialic acid linkage.
The role of sialidases in C. perfringens virulence is still unproven. Studies using NanI and NanJ null mutants of type A strain 13, which does not produce NanH, did not support a requirement for these sialidases in the mouse gas gangrene model (24). However, as acknowledged in that study, the large C. perfringens inoculum necessary for the mouse gas gangrene model could mask virulence contributions by sialidases. In addition, those gas gangrene results do not address possible contributions of sialidases to intestinal infections, as suggested by a recent study (17) showing that NanI is important for adherence of CN3718 to enterocyte-like cells and for enhancing epsilon toxin binding and activity. Given the potential for sialidases to contribute to C. perfringens virulence, our current study used the availability of mutants expressing only natural levels of a single sialidase to investigate the relative contributions of each sialidase to sialic acid release from cultured host cells. Both NanI and NanJ, but not NanH, released sialic acid from Caco-2 cells. These effects may be relevant for pathogenesis since, in vivo, these sialidases could release sialic acid from host cells for nutritional use by C. perfringens. These findings are also consistent with previous studies indicating that NanI can modify the host cell surface to increase CN3718 adherence and binding of epsilon toxin (17, 21). Since the current results indicated that NanJ releases sialic acid from Caco-2 cells, yet previous studies (17) had indicated that NanJ cannot facilitate CN3718 adherence or epsilon toxin, NanI and NanJ likely target different sialic acid-containing molecules on the Caco-2 cell surface. Identification of those targets requires further studies.
The results obtained using two sialidase inhibitors (NADNA and SB) were consistent with each of the C. perfringens sialidases possessing different enzymatic properties. When the effects of NADNA and SB were tested on sialidase activities in the various culture supernatants, the results showed that both inhibitors could inhibit overall CN3718 sialidase activity. However, the major NanI sialidase was the most sensitive sialidase to both NADNA and SB, while NanH was the most insensitive sialidase to these inhibitors. Similar inhibitors have been developed with activity against sialidases of influenza viruses, and those agents have been applied clinically for the treatment of influenza. Since C. perfringens sialidases, especially NanI, can enhance adherence of this bacterium to host cells and promote the activity of some C. perfringens toxins on sensitive cells (17), our current results suggest that these enzymes may also represent potential therapeutic targets.
The distinct properties of each C. perfringens sialidase, as identified in this study, may help this bacterium to grow and survive under the various environmental conditions it encounters, such as soil, sewage, muscle tissue, animal intestines, and feces. It is interesting that our study found that each C. perfringens sialidase, but particularly NanI, functions well at relatively high (>37°C) temperatures. This observation is consistent with the 43°C optimum growth temperature of C. perfringens, which exceeds temperatures found in animal hosts. However, temperatures of 43°C or higher are found in soil surfaces in tropical regions, suggesting that this bacterium may be well adapted for growth in soil. Finally, our results would suggest that, when produced together, these sialidases work effectively in combination, perhaps utilizing their different properties. In this regard, while NanH does not appear to release sialic acid from Caco-2 cells, the release of NanH from dying bacteria could still contribute to exosialidase activity in some environments, possibly helping to generate nutrients for viable bacteria remaining in the local C. perfringens population.
While the current and previous studies (17) have provided some initial insights into why C. perfringens produces three sialidases, many questions remain. For example, during pathogenesis, does expression of C. perfringens sialidases facilitate bacterial survival in mucosal niche environments? Regarding this question, it is interesting that sialic acid can reportedly enhance sialidase gene expression by C. perfringens (23), given that sialic acid also induces the expression of nanA and nanB, but not nanC, in Streptococcus pneumoniae and this sialidase induction then contributes to both biofilm formation and virulence in that bacterium (31). Other questions yet to be addressed include the following. Do these sialidases provide nutrients to C. perfringens growing in the intestines or other environments? Do they promote effective in vivo resistance against host defenses? The relative contributions of NanH, NanI, and NanJ when C. perfringens is present in the intestines, or other environments, will require additional studies.
ACKNOWLEDGMENTS
This research was generously supported by NIAID through R01 AI056177-8 (B. McClane, principal investigator [PI]), a project grant to B. McClane from the master MARCE grant 2U54AI057168-08 (M. Levine, PI), and R03 AI105635-01 (J. Li, PI).
Footnotes
Published ahead of print 27 December 2013
REFERENCES
- 1.McClane BA, Robertson SL, Li J. 2013. Clostridium perfringens, p 465–489 In Doyle MP, Buchanan RL. (ed), Food microbiology: fundamentals and frontiers, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 2.Hatheway C. 1990. Toxigenic clostridia. Clin. Microbiol. Rev. 3:66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonel JL. 1986. Toxins of Clostridium perfringens types A, B, C, D, and E, p 477–517 In Dorner F, Drews H. (ed), Pharmacology of bacterial toxins. Pergamon Press, Oxford, England [Google Scholar]
- 4.Petit L, Gilbert M, Popoff M. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104–110. 10.1016/S0966-842X(98)01430-9 [DOI] [PubMed] [Google Scholar]
- 5.Rood JI. 1998. Virulence genes of Clostridium perfringens. Annu. Rev. Microbiol. 52:333–360. 10.1146/annurev.micro.52.1.333 [DOI] [PubMed] [Google Scholar]
- 6.Gurjar A, Li J, McClane BA. 2010. Characterization of toxin plasmids in Clostridium perfringens type C isolates. Infect. Immun. 78:4860–4869. 10.1128/IAI.00715-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26. 10.1371/journal.ppat.0040026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McClane BA, Uzal FA, Miyakawa MF, Lyerly D, Wilkins TD. 2006. The enterotoxic clostridia, p 688–752 In Dworkin M, Falkow S, Rosenburg E, Schleifer H, Stackebrandt E. (ed), The prokaryotes, 3rd ed. Springer, New York, NY [Google Scholar]
- 9.Lewis AL, Lewis WG. 2012. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell. Microbiol. 14:1174–1182. 10.1111/j.1462-5822.2012.01807.x [DOI] [PubMed] [Google Scholar]
- 10.Vimr ER. 1994. Microbial sialidases: does bigger always mean better? Trends Microbiol. 2:271–277. 10.1016/0966-842X(94)90003-5 [DOI] [PubMed] [Google Scholar]
- 11.Traving C, Schauer R. 1998. Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 54:1330–1349. 10.1007/s000180050258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roggentin P, Kleineidam RG, Schauer R. 1995. Diversity in the properties of two sialidase isoenzymes produced by Clostridium perfringens spp. Biol. Chem. Hoppe-Seyler 376:569–575. 10.1515/bchm3.1995.376.9.569 [DOI] [PubMed] [Google Scholar]
- 13.Vimr ER, Kalivada KA, Deszo EL, Steenbergen SM. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132–153. 10.1128/MMBR.68.1.132-153.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastoriza Gallego M, Hulen C. 2006. Influence of sialic acid and bacterial sialidase on differential adhesion of Pseudomonas aeruginosa to epithelial cells. Colloids Surf. B Biointerfaces 52:154–156. 10.1016/j.colsurfb.2006.04.013 [DOI] [PubMed] [Google Scholar]
- 15.Honma K, Mishima E, Sharma A. 2011. Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect. Immun. 79:393–401. 10.1128/IAI.00629-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King SJ. 2010. Pneumococcal modification of host sugars: a major contributor to colonization of the human airway. Mol. Oral Microbiol. 25:15–24. 10.1111/j.2041-1014.2009.00564.x [DOI] [PubMed] [Google Scholar]
- 17.Li J, Sayeed S, Robertson S, Chen J, McClane BA. 2011. Sialidases affect the host cell adherence and epsilon toxin-induced cytotoxicity of Clostridium perfringens type D strain CN3718. PLoS Pathog. 7:e1002429. 10.1371/journal.ppat.1002429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyristis M, Bryant AE, Sloan J, Awad MM, Nisbet IT, Stevens DL, Rood JI. 1994. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol. Microbiol. 12:761–777. 10.1111/j.1365-2958.1994.tb01063.x [DOI] [PubMed] [Google Scholar]
- 19.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, KH, Dimitrov GI, Watkins KL, Mulligan S, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031–1040. 10.1101/gr.5238106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001. 10.1073/pnas.022493799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Ma M, Sarker MR, McClane BA. 2013. CodY is a global regulator of virulence-associated properties for Clostridium perfringens type D strain CN3718. mBio 4:e00770–13. 10.1128/mBio.00770-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boraston AB, Ficko-Blean E, Healey M. 2007. Carbohydrate recognition by a large sialidase toxin from Clostridium perfringens. Biochemistry 46:11352–11360. 10.1021/bi701317g [DOI] [PubMed] [Google Scholar]
- 23.Walters DM, Stirewalt VL, Melville SB. 1999. Cloning, sequence, and transcriptional regulation of the operon encoding a putative N-acetyl-mannosamine-6-phosphate epimerase (nanE) and sialic acid lyase (anaA) in Clostridium perfringens. J. Bacteriol. 181:4526–4532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiarezza M, Lyras D, Pidot SJ, Flore-Diaz M, Awad MM, Kennedy CL, Cordner LM, Phumoonna T, Poon R, Hughes ML, Emmins JJ, Alape-Giron A, Rood JI. 2009. The NanI and NanJ sialidases of Clostridium perfringens are not essential for virulence. Infect. Immun. 77:4421–4428. 10.1128/IAI.00548-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Y, McClane BA, Fisher DJ, Rood JI, Gupta P. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71:7542–7547. 10.1128/AEM.71.11.7542-7547.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, McClane BA. 2008. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 4:e1000056. 10.1371/journal.ppat.1000056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holzer CT, von Itzstein M, Jin B, Pegg MS, Stewart WP, Wu WY. 1993. Inhibition of sialidases from viral, bacterial and mammalian sources by analogues of 2-deoxy-2,3-didehydro-N-acetylneuraminic acid modified at the C-4 position. Glycoconj. J. 10:40–44. 10.1007/BF00731185 [DOI] [PubMed] [Google Scholar]
- 28.Corfield T. 1992. Bacterial sialidases—roles in pathogenicity and nutrition. Glycobiology 2:509–521. 10.1093/glycob/2.6.509 [DOI] [PubMed] [Google Scholar]
- 29.Streicher H. 2004. Inhibition of microbial sialidases—what has happened beyond the influenza virus? Curr. Med. Chem. Anti-Infect. Agents 3:149–161. 10.2174/1568012043353964 [DOI] [Google Scholar]
- 30.Severi E, Hood DW, Thomas GH. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 153:2817–2822. 10.1099/mic.0.2007/009480-0 [DOI] [PubMed] [Google Scholar]
- 31.Trappetti C, Kadioglu A, Carter M, Hayre J, Iannelli F, Pozzi G, Andrew PW, Oggioni MR. 2009. Sialic acid: a preventable signal for pneumococcal biofilm formation, colonization, and invasion of the host. J. Infect. Dis. 199:1497–1505. 10.1086/598483 [DOI] [PubMed] [Google Scholar]
- 32.Sillanaukee P, Ponnio M, Jaaskelainen IP. 1999. Occurrence of sialic acids in healthy humans and different disorders. Eur. J. Clin. Invest. 29:413–425. 10.1046/j.1365-2362.1999.00485.x [DOI] [PubMed] [Google Scholar]
- 33.Kruse S, Pommerencke J, Kleineidam RG, Roggentin P, Schauer R. 1998. Effect of cysteine modifications on the activity of the ‘small' Clostridium perfringens sialidase. Glycoconj. J. 15:769–775 [DOI] [PubMed] [Google Scholar]