Abstract
Vibrio vulnificus naturally inhabits a variety of aquatic organisms, including oysters, and is the leading cause of seafood-related death in the United States. Strains of this bacterium are genetically classified into environmental (E) and clinical (C) genotypes, which correlate with source of isolation. E-genotype strains integrate into marine aggregates more efficiently than do C-genotype strains, leading to a greater uptake of strains of this genotype by oysters feeding on these aggregates. The causes of this increased integration of E-type strains into marine “snow” have not been demonstrated. Here, we further investigate the physiological and genetic causalities for this genotypic heterogeneity by examining the ability of strains of each genotype to attach to chitin, a major constituent of marine snow. We found that E-genotype strains attach to chitin with significantly greater efficiency than do C-genotype strains when incubated at 20°C. Type IV pili were implicated in chitin adherence, and even in the absence of chitin, the expression level of type IV pilin genes (pilA, pilD, and mshA) was found to be inherently higher by E genotypes than by C genotypes. In contrast, the level of expression of N-acetylglucosamine binding protein A (gbpA) was significantly higher in C-genotype strains. Interestingly, incubation at a clinically relevant temperature (37°C) resulted in a significant increase in C-genotype attachment to chitin, which subsequently provided a protective effect against exposure to acid or bile, thus offering a clue into their increased incidence in human infections. This study suggests that C- and E-genotype strains have intrinsically divergent physiological programs, which may help explain the observed differences in the ecology and pathogenic potential between these two genotypes.
INTRODUCTION
Vibrio vulnificus is a highly invasive opportunistic human pathogen indigenous to estuarine and coastal waters worldwide (1). Importantly, this bacterium is the causative agent of frequently fatal septicemia in susceptible persons consuming raw or undercooked oysters and is the leading cause of seafood-related death in the United States (2). V. vulnificus is a free-living bacterium but also exists as part of the normal microflora within bivalves (3). As filter feeders, these molluscs tend to concentrate V. vulnificus within their tissues and therefore represent an important ecological niche for this bacterium (4). The mechanisms as to how this opportunistic human pathogen survives and proliferates in the environment and is capable of disease production in humans have been long-standing questions.
V. vulnificus strains are genetically and phenotypically diverse and are grouped into biotypes and genotypes based on biochemical and genetic traits, respectively. Biotype 1 strains are responsible for the majority of human infections and are the focus of this study (5, 6). Genetic polymorphisms within the virulence-correlated gene (vcg) serve as a primary feature to distinguish strains of clinical (C) genotypes from those of environmental (E) genotypes, the former being more often associated with disease (7). Similarly, polymorphisms within the 16S rRNA gene can be used to distinguish between clinically and environmentally associated genotypes, referred to as types B and A, respectively (8). The use of multilocus sequence typing and phylogenetic analyses of sequenced genomes has further delineated C and E genotypes into two distinct evolutionary lineages (9, 10). Indeed, previous studies have shown that C- and E-genotype strains display different ecologies, in which E-genotype strains seem to have a distinctive advantage in inhabiting oysters, whereas C-genotype strains are more successful in infecting the human host (7, 11–13). Furthermore, genome comparisons have allowed the identification of several putative virulence factors (such as the genomic XII region) that could potentially aid C-genotype strains in disease progression (10, 14, 15).
Froelich et al. recently demonstrated that E-genotype cells integrate into marine aggregates more efficiently than do C-genotype cells, thereby resulting in a greater uptake of E-genotype strains by oysters feeding on these aggregates (16). Additionally, experiments with C- and E-genotype cocultures resulted in a significantly greater uptake of E-genotype cells than C-genotype cells into oysters. This finding offered a possible explanation for the predominance of E-genotype strains in oysters; however, the mechanisms for this increased integration of E-genotype strains remain unknown. Marine aggregates are naturally forming conglomerates of organic and inorganic detritus, of which chitin is a primary constituent (17). Chitinous substrates are considered to play a vital role in the ecology of vibrios, serving as a critical reservoir for the survival and persistence of pathogens such as Vibrio cholerae (18). Importantly, these associations with chitin are thought to influence the overall metabolism and physiology of Vibrio spp. (18, 19). The primary goal of the present study was to comparatively assess the efficiencies of chitin attachment by V. vulnificus C- and E-genotype strains in order to further investigate the specific mechanisms responsible for the genotypic disparity observed within marine aggregates and oysters.
Considering the ecological relevance of vibrio associations with chitinous substrates, surface-associated proteins aiding in adherence to chitin have been studied extensively in a variety of Vibrio spp. (19–26). N-Acetylglucosamine (GlcNAc) binding protein A (GbpA) binds to GlcNAc-containing carbohydrates such as chitin. In V. cholerae, GbpA has been demonstrated to bind chitin and the mucin of intestinal epithelial cells, thus linking a single colonization factor critical for both environmental and host survival (19, 27). GbpA also contributes to the persistence of V. cholerae within bivalve tissues, particularly through colonization of mussel hepatopancreas cells (28). Two type IV pili have been identified in V. vulnificus, the mannose-sensitive hemagglutinin pilus (MSHA) and the PilA pilus (also referred to as the chitin-regulated pilus ChiRP in some Vibrio species), both of which are processed by a unique prepilin peptidase (PilD) to form a mature pilus structure that extends from the surface of the cell and interacts with the environment (29). In V. vulnificus, the type IV pilin subunit PilA has been shown to play a role in biofilm formation, adherence to human epithelial cells, and oyster colonization (30, 31). Additionally, pilA and mshA mutants of both Vibrio parahaemolyticus and V. cholerae show a lower fitness for adherence to chitinous substrates (20, 22, 23, 26). In V. cholerae, MSHA is considered to be important for environmental persistence, aiding in attachment to zooplankton exoskeletons, but unlike GbpA, it has been shown to interact with bivalve hemocytes and act as an anticolonization factor in human disease (4, 20, 22, 32). In the current study, the role of these genes in facilitating chitin attachment was assessed for both C- and E-genotype strains.
Stauder et al. demonstrated that increasing temperatures positively affect expression of both mshA and gbpA and consequently enhance V. cholerae attachment to chitin (33). Additionally, due to its ability to resist digestion by acid, chitin has been shown to provide V. cholerae with protection from lethal acid stress, offering an effective means for gastric transit inside the human host (34, 35). V. vulnificus C-genotype strains have been shown to resist the bactericidal effects of human serum compared to their E-genotype counterparts and also to exhibit a more robust cross-protective response in the presence of multiple stressors (12, 13). To our knowledge, the effect of V. vulnificus-chitin interactions on stress resistance has not yet been investigated. Thus, we examined the differences in attachment efficiency between C- and E-genotype cells at the human physiological temperature (37°C) compared to an environmental temperature of 20°C and also examined the ability of chitin-bound cells to resist exposure to acid and bile stress.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
V. vulnificus strains used in this study are listed in Table 1. Bacterial cultures were stored at −80°C in Bacto Luria broth (LB) (BD Difco, NJ) containing 20% glycerol. Clinical isolates selected for this study were characterized as having the vcgC and 16S rRNA type B alleles and have been classified as lineage II strains, possessing genomic region XII. Conversely, environmental isolates were characterized as having vcgE and 16S rRNA type A alleles and have been classified as lineage I strains, lacking genomic region XII (14). All strains were grown in Bacto heart infusion (HI) broth (BD Difco, NJ) for 24 h at 30°C with shaking. PilA and PilD type IV pilus mutants were grown with 25 μg/ml and 50 μg/ml of streptomycin and spectinomycin, respectively.
TABLE 1.
Strains used in this study
| Strain | Source | Genotypesa | Lineageb | Presence of genomic region XIIc |
|---|---|---|---|---|
| JY1701 | Environmental | A, E | II | − |
| JY1305 | Environmental | A, E | II | − |
| SS108-A3A | Environmental | A, E | II | − |
| Env1 | Environmental | A, E | II | − |
| CMCP6 | Clinical | B, C | I | + |
| YJ016 | Clinical | B, C | I | + |
| M06-24 | Clinical | B, C | I | + |
| C7184d | Clinical | B, C | I | + |
| C7184ΩAe | pilA mutant | B, C | I | + |
| C7184ΩDe | pilD mutant | B, C | I | + |
Genotype groupings according to polymorphisms within the 16S rRNA (8) and virulence-correlated (7) genes, respectively.
Lineage groupings based on multilocus sequence typing analysis of six housekeeping genes (9).
+ or − indicates the respective presence or absence of the 33-kb genomic island (region XII) (9).
Parental wild-type strain of type IV pilus mutants.
Type IV pilin mutants were kindly provided by Rohinee Paranjpye of the NOAA Northwest Fisheries Science Center (Seattle, WA).
For coculture competition experiments, in which V. vulnificus C- and E-genotype cells were allowed to attach to chitin simultaneously, 1% mannitol agar containing 1.6% BBL phenol red broth base (BD Difco, NJ) and 1% d-mannitol (Sigma-Aldrich), autoclaved for 5 min at 121°C, was used to differentiate between the two genotypes. C- and E-genotype strains used in this study form yellow and pink colonies on mannitol agar, respectively (36).
Chitin attachment assay.
A chitin attachment assay was performed as previously described, with slight modifications (37). Chitin magnetic beads (New England BioLabs) were washed twice by vortexing in phosphate-buffered saline (PBS). Bacterial cultures were washed twice in PBS by centrifugation and diluted 10-fold into fresh PBS (or 15 ppt artificial seawater [ASW] for temperature experiments). In a 1.5-ml microcentrifuge tube, 900 μl of washed culture was added to 100 μl of washed chitin magnetic beads at a concentration of 5 × 107 cells/ml. The mixture was allowed to incubate at 20°C or 37°C for 1 h on a rotisserie at 8 rpm. The supernatant, containing the unattached cells, was removed by placing the tube onto a 1.5-ml microcentrifuge magnetic stand (Life Technologies) and gently washing the beads three times with PBS. Chitin beads, along with the remaining attached bacteria, were suspended in 1 ml of PBS and prepared as described below for cell quantification using either culture-based methods or fluorescence microscopy.
Culture-based quantification.
A total of 0.2 g of 0.5-mm ZR BashingBead was added to the washed chitin beads suspended in 1 ml of PBS, and the tube was vortexed vigorously for 60 s to detach the bound bacteria. Chitin beads were separated from the supernatant by using a magnetic stand, and the cell suspension containing the detached cells was serially diluted and plated onto HI agar for individual strain experiments or spread plated onto 1% mannitol agar for competition experiments.
Fluorescence microscopy-based quantification.
A total of 10 μl of beads harboring attached V. vulnificus cells was dropped onto a clean microscope slide and allowed to dry. The beads were then fixed with 95% ethanol, stained by using acridine orange, and visualized under a fluorescence microscope. To compare chitin attachment efficiencies, average numbers of cells per bead were quantified for each strain.
Acid/bile stress assay.
For acid stress exposure, washed chitin beads with attached V. vulnificus cells were exposed for 5 min at 20°C to 1 ml of PBS adjusted to pH 3 by using hydrochloric acid. For bile exposure, washed chitin beads with attached cells were exposed to 1 ml of 1% bile (Ox gall powder; Sigma-Aldrich) in PBS for 30 min at 20°C. After incubation, the stress was immediately removed by washing the beads with PBS through gentle inversion of the tube five times. Culture-based quantification was performed as outlined above. The amount of chitin-attached cells that survived exposure to stress was compared to the number of chitin-attached cells that were not exposed to stress and expressed as percent survival. For comparison, percent survival of planktonic cells incubated without chitin but exposed to the same stress conditions was also measured.
Gene expression. (i) RNA harvesting.
V. vulnificus strains C7184 (C genotype) and JY1305 (E genotype), grown in HI broth for 24 h, were washed twice and then resuspended into 15 ppt ASW to a final concentration of ca. 1 × 108 CFU/ml. Each strain was incubated on a rotisserie for 1 h and then treated with RNAprotect (Qiagen) according to the manufacturer's instructions. Cell pellets were resuspended in Tris-EDTA (TE) buffer at pH 8.0 with 10 mg/ml lysozyme and then vortexed at medium speed for 30 min. RNA from lysed cells was extracted by using the RNeasy Minikit (Qiagen) according to the manufacturer's protocol, with optional on-column DNase I treatment. In the final step, RNA was eluted from the column by using nuclease-free water, and a second postextraction DNase treatment was performed by using Turbo DNA-free (Ambion) according to the “rigorous DNase treatment” protocol. RNA quality and quantity were assessed by using a NanoDrop spectrophotometer (Thermo), and all RNA samples were found to have a 260/280-nm absorbance ratio of ≥1.7. RNA samples were stored at −80°C.
(ii) PCR to confirm removal of DNA contamination.
Endpoint PCR was performed on RNA samples, targeting the species-specific vvhA gene (7), to confirm the complete removal of DNA. This was done by using Promega Go-Taq DNA polymerase, 5× Green GoTaq reaction buffer, 10 mM deoxynucleoside triphosphate (dNTP) mix, and primers for vvhA. Cycling parameters were performed according to the manufacturer's recommendations, with an annealing temperature of 53.1°C and 40 cycles of amplification. Any amplification of the vvhA gene was indicative of DNA contamination, in which case the RNA was not used for downstream processes.
(iii) Primer design for qRT-PCR.
Primers were designed for each gene of interest by using all three sequenced C-genotype strains of V. vulnificus (CMCP6, YJ016, and M06-24) reported in the NCBI database. Primers were designed to also target the three sequenced E-genotype strains of V. vulnificus (JY1701, JY1305, and E64MW) by using whole-genome shotgun contigs deposited in the NCBI database. ClustalW gene alignments were performed to identify conserved nucleotide regions, and primers were designed by using NCBI Primer-BLAST software. Optimal primer quality and fidelity were assessed by using IDT OligoAnalyzer 3.1 software. Primer specificity was analyzed by using in silico PCR (38), and the PCR efficiency of each primer pair was evaluated by using an in silico PCR efficiency estimation tool (39). Primers were purchased from Sigma-Aldrich and validated by using endpoint PCR to ensure specific amplification of the DNA targets of interest for each strain under investigation. Primers used in this study are listed in Table 2.
TABLE 2.
Primers designed for this study
| Gene | Strain(s) amplified | Primer targeta | Sequence (5′–3′) | Expected product size (bp) |
|---|---|---|---|---|
| Glyceraldehyde phosphate dehydrogenase | C7184 and JY1305 | gapdh F | TGAAGGCGGTAACCTAATCG | 97 |
| gapdh R | TACGTCAACACCGATTGCAT | |||
| Type IV pilin protein subunit | JY1305 only | pilA F | TCATTGGTGTGTTAGCCGCA | 73 |
| pilA R | GCTGAGGCAGCTTCTGACTT | |||
| Type IV pilin protein subunit | C7184 only | pilA F | GCACAGCTCCAACCAGTAGT | 57 |
| pilA R | TTGGCGGCACTTCAACAATG | |||
| Type IV pilin protein prepilin peptidase | JY1305 only | pilD F | GGCTTACTGGTAGGCAGCTT | 126 |
| pilD R | GGTTTCTGTCGGCGGTGATA | |||
| Type IV pilin protein prepilin peptidase | C7184 only | pilD F | TTGGCTTACTGGTAGGCAGC | 128 |
| pilD R | GGTTTCTGTCGGTGGTGTGA | |||
| N-Acetylglucosamine binding protein | C7184 and JY1305 | gpbA F | TTGAGTGGACCTTTACCGCC | 91 |
| gpbA R | CGGGCAAGTGGTTGATTTGG | |||
| Mannitol-sensitive hemagglutinin | C7184 and JY1305 | mshA F | CAAGGCGGTTTCACCCTGAT | 90 |
| mshA R | CAGATTTAGAAAACGCGGAGCC |
F, forward; R, reverse.
(iv) Relative qRT-PCR.
The PCR amplification efficiency of each primer set was analyzed by generating a standard curve and evaluating the slope. Standard curves were performed for all genes and all strains under investigation by using five 1:5 serial dilutions of cDNA. To measure relative expression, 1 μg of total RNA was reverse transcribed by using qScript cDNA SuperMix (Quanta Biosciences). cDNA was then diluted, and 50 ng of cDNA template was carried over for quantitative PCR (qPCR). Relative quantitative reverse transcription-PCR (qRT-PCR) was performed on three technical and three biological replicates for each sample by using PerfeCTa SYBR green FastMix, Low ROX (Quanta Biosciences), thus generating nine threshold cycle (CT) values per target. Negative controls consisting of water (in place of cDNA) were employed to rule out the influence of DNA contaminants or other artifacts (e.g., primer-dimers). “No-RT” controls consisting of an equivalent quantity of RNA were added to control wells to rule out the presence of residual genomic DNA. Expression levels of each gene were normalized by using an endogenous control gene (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) to correct for sampling errors. Fold changes in expression levels were measured for E-genotype strains relative to C-genotype strains by using the Pfaffl equation (40), taking into account the differences in PCR efficiencies between primer sets.
Statistical analysis.
For culture-based quantification, attachment was expressed as the percentage of the total number of cells attached to the chitin beads (output) divided by the total number of cells added to the system (input), multiplied by 100. Attachment was also measured by using fluorescence microscopy, with the number of cells attached to 10 beads being averaged. Data were analyzed by using unpaired Student's t test or one-way analysis of variance (ANOVA) followed by Tukey's post hoc test for multiple comparisons. Significance was determined by using a 95% confidence interval. All data were analyzed by using GraphPad Prism (version 5.0; GraphPad Software Inc.). Relative fitness (w) of chitin attachment between genotypes (20, 23) was calculated as w(C,E) = sqrt(CI/CO)/sqrt(EI/EO), where CI and EI represent the initial CFU input and CO and EO represent the output CFU at the end of the experiment. The relative fitness for each type IV pilus mutant was also calculated relative to the fitness of the parental wild-type strain. Gene expression levels of E-genotype strains relative to C-genotype strains were analyzed by using the Mann-Whitney (nonparametric) rank-sum test.
RESULTS AND DISCUSSION
Attachment of V. vulnificus strains of clinical and environmental genotypes to chitin.
The strains selected for this study have several distinct genetic features which have been used to demarcate the evolutionary lineages of clinical and environmental strains (7, 8, 13, 14, 41). We investigated the abilities of four C- and four E-genotype strains to attach to chitin magnetic beads, and while all isolates tested were able to adhere to chitin within 1 h of incubation, E-genotype strains were significantly more capable of chitin binding than were C-genotype strains (Fig. 1A). This finding was further substantiated by visually quantifying the number of cells attached to individual chitin particles using fluorescence microscopy (Fig. 1B and 2). To more accurately reflect the natural environment, C- and E-genotype cells were incubated with chitin in cocultures to examine competitive attachment. Again, E-genotype cells were significantly more efficient in adhering to chitin than were their C-genotype counterparts (Fig. 1C). Differences in attachment efficiency between C- and E-genotype strains were assessed by calculating relative fitness values, in which values of <1 indicated a defect in the competitiveness of the designated strain (see Materials and Methods). In monoculture, the overall fitness of C-genotype strains relative to E-genotype strains was 0.68. This relative fitness was reduced to 0.56 when C- and E-genotype strains were incubated in cocultures with chitin. This indicates that E-genotype strains have a competitive advantage in chitin colonization, which offers an explanation for the enhanced integration of E-genotype strains into marine aggregates along with the subsequent persistence of E-genotype strains within oyster tissues that we have previously reported (16).
FIG 1.
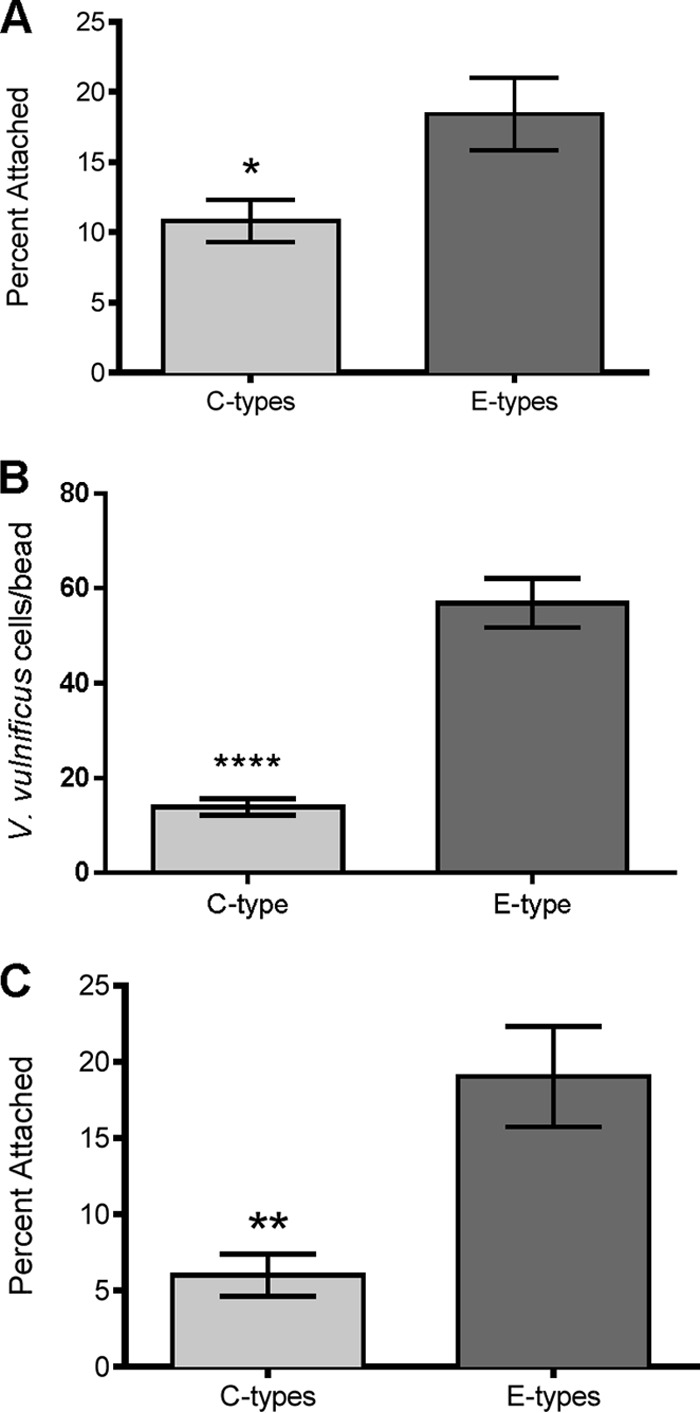
(A) Attachment of V. vulnificus C-genotype strains (C7184, CMCP6, M06-24, and YJ016) and E-genotype strains (JY1305, JY1701, ENV1, and SS108-A3A) to chitin magnetic beads. Error bars represent the standard errors of the means for three replicates of the four strains. E-genotype attachment to chitin was significantly greater than C-genotype attachment (P = 0.0182 by an unpaired Student t test). (B) Microscopic quantification of chitin attachment by V. vulnificus C-genotype (C7184) and E-genotype (JY1305) strains. Error bars represent the standard errors of the means for 10 replicate beads. JY1305 attachment to chitin was significantly greater than that of C7184 (P < 0.0001 by an unpaired Student t test with Welch's correction). (C) Competitive chitin attachment of V. vulnificus C- and E-genotype strains incubated in coculture. Two competitions were performed (M06-24 versus Env1 and C7184 versus JY1305). Error bars represent the standard errors of the means for the two competition studies with triplicate replicates. In coculture, E-genotype cell attachment to chitin was significantly greater than C-genotype cell attachment (P = 0.0045 by a Student t test).
FIG 2.
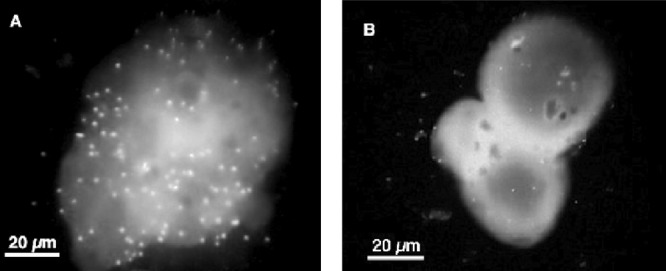
Representative fluorescence microscopy images of V. vulnificus E-genotype (A) and C-genotype (B) cells attached to a single chitin magnetic bead. Image color, contrast, and brightness were applied to each image by using the Macintosh Preview application.
Role of surface proteins in chitin attachment.
We suspected that type IV pili are important for attachment to chitin; thus, we investigated the attachment efficiencies of two V. vulnificus mutants deficient in type IV pilin expression. Both pilA and pilD mutants displayed significantly decreased attachment compared to the parent strain (Fig. 3), with relative fitness values of 0.64 and 0.13, respectively. The much lower fitness value of pilD suggests a crucial role in chitin attachment. It has been demonstrated that a V. vulnificus pilA mutant still expresses pili, whereas the pilD mutant results in a complete loss of surface pili, suggesting that PilD processes both MSHA and PilA type IV pili of V. vulnificus (31, 42). Because chitin is composed of N-acetylglucosamine (GlcNAc) monomers, cells were treated with this sugar to ascertain the specificity of surface proteins in chitin attachment. Incubation with GlcNAc in the presence of chitin resulted in a significant reduction in attachment efficiency for both C- and E-genotype strains (Fig. 4).
FIG 3.
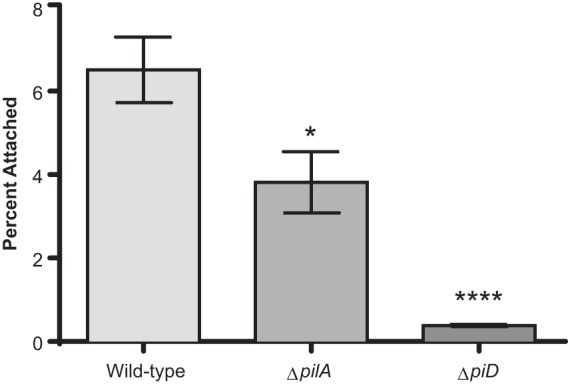
Chitin attachment of V. vulnificus type IV pilin mutants. Depicted are the differences in chitin attachment between parent strain C7184 and strains with mutations in type IV pilin protein A (pilA) (C7184ΩA) and prepilin peptidase (pilD) (C7184ΩD). Error bars represent the standard errors of the means for seven replicates. Asterisks indicate significant differences relative to the parental strain. One-way ANOVA indicated that the loss of type IV pilin genes significantly reduces the ability of C7184 to attach to chitin.
FIG 4.
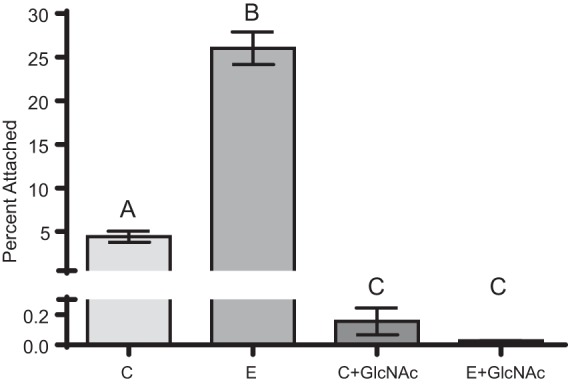
Effect of N-acetylglucosamine (GlcNAc) on V. vulnificus attachment to chitin. Depicted is the attachment efficiencies of two C-genotype (CMCP6 and M06-24) and two E-genotype (ENV1 and JY1305) strains incubated with chitin in the presence or absence of 125 mg/ml GlcNAc. Error bars represent the standard errors of the means for two replicates of two strains. Incubation with GlcNAc resulted in a significant decrease in attachment for strains of both genotypes. Different letters indicate statistically significant differences determined by using one-way ANOVA.
Our study confirmed the relevance of type IV pili in chitin attachment; however, it is important to note that the parent for these mutants was a C-genotype strain. Thus, to further characterize the role of these genes in chitin attachment, we examined relative gene expression levels of C- and E-genotype strains in the presence and absence of chitin. Interestingly, neither of these genes was induced (in strains of either genotype) when cells were incubated with chitin for 1 h (data not shown). This led us to the hypothesis that there may be basal differences in gene expression levels between these two genotypes. Hence, we examined relative gene expression levels between the genotypes when incubated in ASW without chitin addition. We discovered that E-genotype strains inherently express more pilA and pilD, by 23-fold and 9-fold, respectively, than do C-genotype strains (Fig. 5). Additionally, mshA, encoding the principal subunit of the type IV MSHA pilus, was expressed at 2-fold-higher levels in E-genotype strains (Fig. 5).
FIG 5.
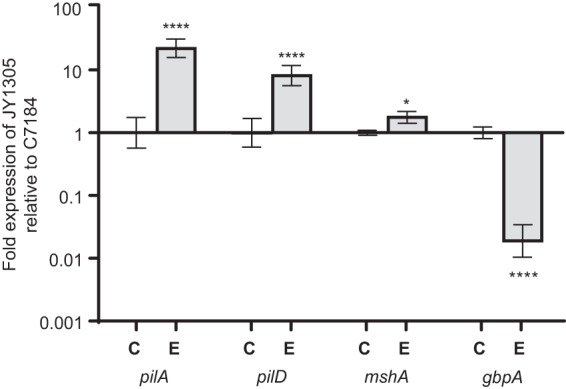
Expression of attachment genes by E-genotype (JY1305) relative to C-genotype (C7184) strains incubated in ASW for 1 h. Error bars represent the standard deviations of three biological and three technical replicates. Asterisks represent statistically significant differences between C- and E-genotype strains for each gene of interest by using one-way ANOVA. JY1305 intrinsically expresses significantly higher levels of pilA, pilD, and mshA than does C7184, whereas C7184 expresses higher levels of gbpA than does JY1305.
The role of N-acetylglucosamine binding protein (gbpA) was also investigated, and we discovered that C-genotype strains expressed significantly (46-fold) more gbpA than did E-genotype strains (Fig. 5). In V. cholerae, GbpA plays an essential role in chitin binding as well as in colonization of the human intestine; thus, the differential expression of gbpA by clinical strains of V. vulnificus likely has very important implications regarding the enhanced pathogenicity of these strains. It is plausible that increased expression levels of gbpA may provide enhanced attachment to epithelial cell surfaces in the human gastrointestinal tract; however, further studies are needed to validate this hypothesis.
Effect of temperature on chitin attachment.
To investigate the effect of temperature on chitin attachment, C- and E-genotype strains were incubated with chitin beads at 20°C and 37°C, in parallel. As has been documented for V. cholerae (33, 43, 44), increasing the incubation temperature resulted in a significant increase in attachment for C-genotype strains (Fig. 6). As a result, the fitness of C-genotype strains relative to E-genotype strains in attachment to chitin increased from 0.450 at 20°C to 0.997 at 37°C. In contrast, this increase in temperature caused a significant decrease in attachment for E-genotype strains (Fig. 6). This inverse effect of temperature on chitin attachment likely has important clinical and ecological relevance and warrants further investigation.
FIG 6.
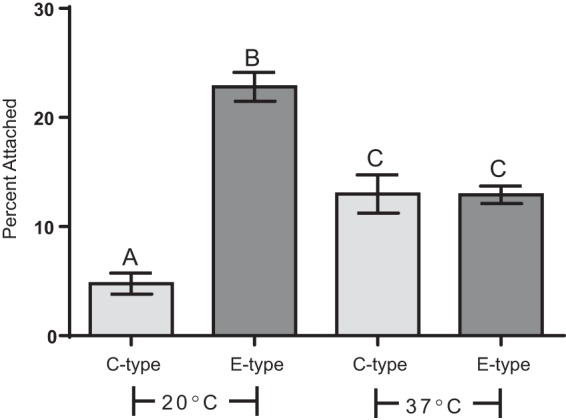
Effect of incubation temperature on attachment of C- and E-genotype strains to chitin. Depicted is attachment of two C-genotype strains (CMCP6 and M06-24) and two E-genotype strains (ENV1 and JY1305) incubated with chitin at room temperature (20°C) or the human physiological temperature (37°C). Error bars represent the standard errors of the means for two replicates of two strains. Different letters indicate statistically significant differences determined by using one-way ANOVA. Increasing the incubation temperature from 20°C to 37°C resulted in a significant increase in attachment for C-genotype strains and a significant decrease in attachment for E-genotype strains.
Protective effect of chitin.
We examined whether adherence to chitin allowed V. vulnificus C- and E-genotype strains to withstand stressors that would be encountered by this bacterium upon entry into the human host. To this end, cells attached to chitin beads were incubated in acidified PBS. Chitin particles were then washed to remove the acid stress, and the percentage of culturable cells remaining on chitin particles was quantified. Despite the rapid lethality (>99%) of pH 3 to planktonic (control) cultures of V. vulnificus, C-genotype cells significantly survived acid exposure when attached to chitin particles (Fig. 7A). Conversely, E-genotype cells were not significantly protected by chitin in the face of acid stress, emphasizing the difference in genetic programming between these two genotypes. Along with a temperature upshift, the acidic environment of the human stomach is likely to be one of the first physiological stresses encountered by the bacterium when ingested. V. vulnificus is known to possess several mechanisms for acid resistance and adaptation (45, 46), such as the cadBA operon, which is induced under conditions of low pH and which neutralizes the external pH through simple acid/base chemistry (47). Nonetheless, the genetic mechanisms responsible for combating acid stress in C-genotype strains may be superior to those in E-genotype strains, and it is tempting to speculate that attachment to chitin may grant C-genotype strains with temporary protection upon entry into the human host, thus providing them with a competitive advantage over E-genotype strains. Notably, although there is no clear role for chitin in oyster-associated infections, it is well known that bacterial cells in an attached state exhibit observable physiological changes relative to their planktonic counterparts (48); thus, attachment to some other more relevant oyster-based substratum may offer similar protection during the transition into the human host.
FIG 7.
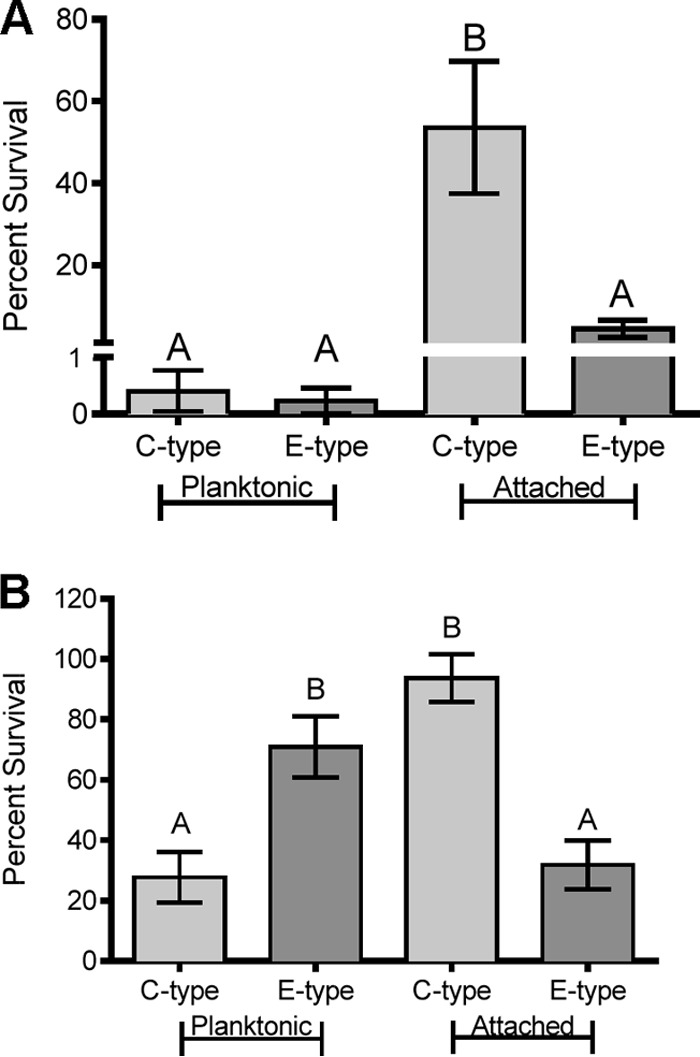
(A) Protective effect of chitin attachment upon exposure to acid stress. Depicted is the survival of two C-genotype strains (CMCP6 and M06-24) and two E-genotype strains (ENV1 and JY1305) when exposed to acid stress (pH 3) for 5 min in the planktonic state or when attached to chitin. Error bars represent the standard errors of the means for four replicates of two strains. Different letters indicate significant differences determined by using one-way ANOVA. Attachment to chitin provided a significant protective effect against acid stress for C-genotype strains only. (B) Effect of chitin attachment on exposure to bile stress. Depicted is the survival of two C-genotype strains (CMCP6 and M06-24) and two E-genotype strains (ENV1 and JY1305) when exposed to 1% bile for 30 min in the planktonic state or when attached to chitin. Error bars represent the standard errors of the means for two (planktonic) or four (attached) replicates of two strains. Different letters indicate significant differences determined by using one-way ANOVA. E-genotype strains survived bile stress significantly better than did C-genotype strains when in the planktonic state, while C-genotype strains survived bile stress significantly better than did E-genotype strains when attached to chitin.
A bacterial pathogen attempting to successfully traverse the human gastrointestinal tract will inevitably encounter the antimicrobial properties of bile, an emulsifying fluid produced by the liver and excreted into the small intestine. A variety of enteric bacteria have been shown to respond to and subsequently develop resistance to bile exposure (49). Furthermore, pathogenic Vibrio spp., such as V. cholerae and V. parahaemolyticus, have been shown to possess mechanisms for bile resistance resulting in modulated expression of virulence factors (50–52). We exposed planktonic and chitin-attached cells to bile stress for 30 min and observed that while planktonic E-genotype cells survived bile stress significantly better than planktonic C-genotype cells, chitin-attached C-genotype cells survived bile stress significantly better than chitin-attached E-genotype cells (Fig. 7B). This opposing trend highlights the divergent physiological responses of strains of these two genotypes when exposed to clinically relevant stress. Further investigation into the acid and bile stress resistance of chitin-attached C-genotype cells would likely provide important insights into the virulent nature of strains of this genotype. Our recent comparative analysis of C- and E-genotype genomes revealed a number of genes unique to C-genotype strains, many of which could bestow the cell with enhanced stress resistance (10). We are currently investigating genes unique to C-genotype strains that may equip them with this heightened stress response.
Conclusion.
V. vulnificus is widely recognized as a human pathogen, although genetic analyses have revealed that strains of this species are not equally pathogenic. Genotyping has revealed differences not only in pathogenic potential but also in the ecological niches in which these strains reside (11, 12, 53). Genetic dimorphisms among V. vulnificus strains have been documented on a genome-wide scale; thus, it has been proposed that C- and E-genotype strains may reflect distinct ecotypes (10, 13). Chitin is considered to play a pivotal role in the ecology of vibrios, and adherence to this substrate has been demonstrated for a number of species, including V. vulnificus, V. cholerae, V. parahaemolyticus, and V. alginolyticus (18, 25, 34, 54). The ability to associate with chitin in the aquatic environment has a profound influence on the life-style of Vibrio spp., providing them with a number of advantages, including food availability, adaptation to environmental nutrient gradients, tolerance to stress, and protection from predators (33, 55).
The ecologically distinct C- and E-genotype strains examined here displayed different degrees of chitin attachment, providing deeper insight into the natural ecology of this organism. Investigation of a larger set of C- and E-genotype strains should be performed to further substantiate this phenomenon. In this study, type IV pili were found to be important for chitin adherence and appear to be a key player in attachment by E-genotype strains. Conversely, gbpA expression was enhanced in C-genotype strains compared to E-genotype strains, which we suspect has significant implications in human intestinal colonization and survival. Attachment to substrates such as chitin is likely to shift the physiological regimen of the bacterium, resulting in different adaptive capabilities in the face of stress. As demonstrated here, chitin-associated cells exhibited substantially different stress responses compared to those of free-living planktonic cells, particularly for C-genotype strains.
Sequence analyses of type IV pili among several Vibrio spp. have revealed interesting clues into the immense genetic diversity observed within this genus. For many bacterial species, including numerous pathogens, type IV pili have been implicated in environmental survival as well as host colonization. Aagesen and Hase proposed that the heterogeneities among pilA and mshA sequences are likely a result of positive selective pressures, which would be expected to have ecological implications for the bacterium (29). Sequence alignment of V. vulnificus type IV pilus genes (pilA, pilD, and mshA) as well as gbpA revealed a considerable number of genetic polymorphisms, many of which correlate with genotype. Indeed, our study reveals that C- and E-genotype strains differentially express these four genes within the same environment, even in the absence of chitin. Thus, the observed differences in chitin attachment do not appear to be a result of induction of gene expression but rather an inherent predisposition for adherence to chitin. We propose that this variance can help explain the observed genotypic differences in oyster colonization and possibly pathogenic potential. The environmental and molecular mechanisms facilitating chitin attachment deserve further examination and may provide a better understanding of the ecological and clinical divergence of these two genotypes.
ACKNOWLEDGMENTS
We thank Rohinee Paranjpye of the NOAA Northwest Fisheries Science Center (Seattle, WA) for kindly providing type IV pilin mutants.
This study is based on work supported by Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, award no. 2009-03571 and by the Duke University Marine Laboratory Mary Derrickson McCurdy Scholar Award granted to James D. Oliver.
Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Department of Agriculture or the Duke University Marine Laboratory.
Footnotes
Published ahead of print 20 December 2013
REFERENCES
- 1.Oliver JD. 2006. Vibrio vulnificus, p 349–366 In Thompson FL, Austin B, Swings J. (ed), The biology of vibrios. ASM Press, Washington, DC [Google Scholar]
- 2.Oliver JD. 2006. Vibrio vulnificus, p 253–276 In Belkin S, Colwell RR. (ed), Oceans and health: pathogens in the marine environment. Springer Science, New York, NY [Google Scholar]
- 3.Froelich B, Oliver JD. 2013. The interactions of Vibrio vulnificus and the oyster Crassostrea virginica. Microb. Ecol. 65:807–816. 10.1007/s00248-012-0162-3 [DOI] [PubMed] [Google Scholar]
- 4.Pruzzo C, Gallo G, Canesi L. 2005. Persistence of vibrios in marine bivalves: the role of interactions with haemolymph components. Environ. Microbiol. 7:761–772. 10.1111/j.1462-2920.2005.00792.x [DOI] [PubMed] [Google Scholar]
- 5.Tison DL, Nishibuchi M, Greenwood JD, Seidler RJ. 1982. Vibrio vulnificus biogroup 2: new biogroup pathogenic for eels. Appl. Environ. Microbiol. 44:640–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bisharat N, Agmon V, Finkelstein R, Raz R, Ben-Dror G, Lerner L, Soboh S, Colodner R, Cameron DN, Wykstra DL, Swerdlow DL, Farmer JJ. 1999. Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421–1424. 10.1016/S0140-6736(99)02471-X [DOI] [PubMed] [Google Scholar]
- 7.Rosche TM, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol. Immunol. 49:381–389. 10.1111/j.1348-0421.2005.tb03731.x [DOI] [PubMed] [Google Scholar]
- 8.Nilsson WB, Paranjype RN, DePaola A, Strom MS. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442–446. 10.1128/JCM.41.1.442-446.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen ALV, Oliver JD, DePaola A, Feil EJ, Fidelma Boyd E. 2007. Emergence of a virulent clade of Vibrio vulnificus and correlation with the presence of a 33-kilobase genomic island. Appl. Environ. Microbiol. 73:5553–5565. 10.1128/AEM.00635-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrison SS, Williams T, Cain A, Froelich B, Taylor C, Baker-Austin C, Verner-Jeffreys D, Hartnell R, Oliver JD, Gibas CJ. 2012. Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One 7:e37553. 10.1371/journal.pone.0037553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner E, Oliver JD. 2008. Population structures of two genotypes of Vibrio vulnificus in oysters (Crassostrea virginica) and seawater. Appl. Environ. Microbiol. 74:80–85. 10.1128/AEM.01434-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bogard RW, Oliver JD. 2007. Role of iron in human serum resistance of the clinical and environmental Vibrio vulnificus genotypes. Appl. Environ. Microbiol. 73:7501–7505. 10.1128/AEM.01551-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosche TM, Binder EA, Oliver JD. 2010. Vibrio vulnificus genome suggests two distinct ecotypes. Environ. Microbiol. Rep. 2:128–132. 10.1111/j.1758-2229.2009.00119.x [DOI] [PubMed] [Google Scholar]
- 14.Sanjuan E, Fouz B, Oliver JD, Amaro C. 2009. Evaluation of genotypic and phenotypic methods to distinguish clinical from environmental Vibrio vulnificus strains. Appl. Environ. Microbiol. 75:1604–1613. 10.1128/AEM.01594-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulig PA, de Crecy-Lagard V, Wright AC, Walts B, Telonis-Scott M, McIntyre LM. 2010. SOLiD sequencing of four Vibrio vulnificus genomes enables comparative genomic analysis and identification of candidate clade-specific virulence genes. BMC Genomics 11:512. 10.1186/1471-2164-11-512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froelich B, Ayrapetyan M, Oliver JD. 2013. Integration of Vibrio vulnificus into marine aggregates and its subsequent uptake by Crassostrea virginica oysters. Appl. Environ. Microbiol. 79:1454–1458. 10.1128/AEM.03095-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchner M. 1995. Microbial colonization of copepod body surfaces and chitin degradation in the sea. Helgolander Meeresunters. 49:201–212. 10.1007/BF02368350 [DOI] [Google Scholar]
- 18.Vezzulli L, Pezzati E, Repetto B, Stauder M, Giusto G, Pruzzo C. 2008. A general role for surface membrane proteins in attachment to chitin particles and copepods of environmental and clinical vibrios. Lett. Appl. Microbiol. 46:119–125. 10.1111/j.1472-765X.2007.02269.x [DOI] [PubMed] [Google Scholar]
- 19.Kirn TJ, Jude BA, Taylor RK. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863–866. 10.1038/nature04249 [DOI] [PubMed] [Google Scholar]
- 20.Meibom KL, Li XB, Nielsen AT, Wu CY, Roseman S, Schoolnik GK. 2004. The Vibrio cholerae chitin utilization program. Proc. Natl. Acad. Sci. U. S. A. 101:2524–2529. 10.1073/pnas.0308707101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svitil AL, Chadhain S, Moore JA, Kirchman DL. 1997. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol. 63:408–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiavelli DA, Marsh JW, Taylor RK. 2001. The mannose-sensitive hemagglutinin of Vibrio cholerae promotes adherence to zooplankton. Appl. Environ. Microbiol. 67:3220–3225. 10.1128/AEM.67.7.3220-3225.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shime-Hattori A, Iida T, Arita M, Park KS, Kodama T, Honda T. 2006. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 264:89–97. 10.1111/j.1574-6968.2006.00438.x [DOI] [PubMed] [Google Scholar]
- 24.Frischkorn KR, Stojanovski A, Paranjpye R. 2013. Vibrio parahaemolyticus type IV pili mediate interactions with diatom-derived chitin and point to an unexplored mechanism of environmental persistence. Environ. Microbiol. 15:1416–1427. 10.1111/1462-2920.12093 [DOI] [PubMed] [Google Scholar]
- 25.Pruzzo C, Crippa A, Bertone S, Pane L, Carli A. 1996. Attachment of Vibrio alginolyticus to chitin mediated by chitin-binding proteins. Microbiology 142(Part 8):2181–2186 [DOI] [PubMed] [Google Scholar]
- 26.Watnick PI, Fullner KJ, Kolter R. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhowmick R, Ghosal A, Das B, Koley H, Saha DR, Ganguly S, Nandy RK, Bhadra RK, Chatterjee NS. 2008. Intestinal adherence of Vibrio cholerae involves a coordinated interaction between colonization factor GbpA and mucin. Infect. Immun. 76:4968–4977. 10.1128/IAI.01615-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stauder M, Huq A, Pezzati E, Grim CJ, Ramoino P, Pane L, Colwell RR, Pruzzo C, Vezzulli L. 2012. Role of GbpA protein, an important virulence-related colonization factor, for Vibrio cholerae's survival in the aquatic environment. Environ. Microbiol. Rep. 4:439–445. 10.1111/j.1758-2229.2012.00356.x [DOI] [PubMed] [Google Scholar]
- 29.Aagesen AM, Hase CC. 2012. Sequence analyses of type IV pili from Vibrio cholerae, Vibrio parahaemolyticus, and Vibrio vulnificus. Microb. Ecol. 64:509–524. 10.1007/s00248-012-0021-2 [DOI] [PubMed] [Google Scholar]
- 30.Paranjpye RN, Johnson AB, Baxter AE, Strom MS. 2007. Role of type IV pilins in persistence of Vibrio vulnificus in Crassostrea virginica oysters. Appl. Environ. Microbiol. 73:5041–5044. 10.1128/AEM.00641-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paranjpye RN, Strom MS. 2005. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73:1411–1422. 10.1128/IAI.73.3.1411-1422.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao A, Liu Z, Joelsson A, Zhu J. 2006. Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc. Natl. Acad. Sci. U. S. A. 103:14542–14547. 10.1073/pnas.0604650103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stauder M, Vezzulli L, Pezzati E, Repetto B, Pruzzo C. 2010. Temperature affects Vibrio cholerae O1 El Tor persistence in the aquatic environment via an enhanced expression of GbpA and MSHA adhesins. Environ. Microbiol. Rep. 2:140–144. 10.1111/j.1758-2229.2009.00121.x [DOI] [PubMed] [Google Scholar]
- 34.Nalin DR, Daya V, Reid A, Levine MM, Cisneros L. 1979. Adsorption and growth of Vibrio cholerae on chitin. Infect. Immun. 25:768–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nahar S, Sultana M, Naser MN, Nair GB, Watanabe H, Ohnishi M, Yamamoto S, Endtz H, Cravioto A, Sack RB, Hasan NA, Sadique A, Huq A, Colwell RR, Alam M. 2011. Role of shrimp chitin in the ecology of toxigenic Vibrio cholerae and cholera transmission. Front. Microbiol. 2:260. 10.3389/fmicb.2011.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Froelich BA, Oliver JD. 2011. Orientation of mannitol related genes can further differentiate strains of Vibrio vulnificus possessing the vcgC allele. Adv. Stud. Biol. 3:151–160 http://m-hikari.com/asb/asb2011/asb1-4-2011/froelichASB1-4-2011.pdf [Google Scholar]
- 37.Jude BA, Martinez RM, Skorupski K, Taylor RK. 2009. Levels of the secreted Vibrio cholerae attachment factor GbpA are modulated by quorum-sensing-induced proteolysis. J. Bacteriol. 191:6911–6917. 10.1128/JB.00747-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bikandi J, San Millan R, Rementeria A, Garaizar J. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinformatics 20:798–799. 10.1093/bioinformatics/btg491 [DOI] [PubMed] [Google Scholar]
- 39.Mallona I, Weiss J, Egea-Cortines M. 2011. pcrEfficiency: a Web tool for PCR amplification efficiency prediction. BMC Bioinformatics 12:404. 10.1186/1471-2105-12-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vickery MC, Nilsson WB, Strom MS, Nordstrom JL, DePaola A. 2007. A real-time PCR assay for the rapid determination of 16S rRNA genotype in Vibrio vulnificus. J. Microbiol. Methods 68:376–384. 10.1016/j.mimet.2006.02.018 [DOI] [PubMed] [Google Scholar]
- 42.Paranjpye RN, Lara JC, Pepe JC, Pepe CM, Strom MS. 1998. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect. Immun. 66:5659–5668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hood MA, Winter PA. 1997. Attachment of Vibrio cholerae under various environmental conditions and to selected substrates. FEMS Microbiol. Ecol. 22:215–223. 10.1111/j.1574-6941.1997.tb00373.x [DOI] [Google Scholar]
- 44.Castro-Rosas J, Escartín EF. 2002. Adhesion and colonization of Vibrio cholerae O1 on shrimp and crab carapaces. J. Food Prot. 65:492–498 [DOI] [PubMed] [Google Scholar]
- 45.Kim JS, Sung MH, Kho DH, Lee JK. 2005. Induction of manganese-containing superoxide dismutase is required for acid tolerance in Vibrio vulnificus. J. Bacteriol. 187:5984–5995. 10.1128/JB.187.17.5984-5995.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones MK, Oliver JD. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723–1733. 10.1128/IAI.01046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rhee JE, Rhee JH, Ryu PY, Choi SH. 2002. Identification of the cadBA operon from Vibrio vulnificus and its influence on survival to acid stress. FEMS Microbiol. Lett. 208:245–251. 10.1111/j.1574-6968.2002.tb11089.x [DOI] [PubMed] [Google Scholar]
- 48.Petrova OE, Sauer K. 2012. Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 194:2413–2425. 10.1128/JB.00003-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunn JS. 2000. Mechanisms of bacterial resistance and response to bile. Microbes Infect. 2:907–913. 10.1016/S1286-4579(00)00392-0 [DOI] [PubMed] [Google Scholar]
- 50.Pace JL, Chai TJ, Rossi HA, Jiang X. 1997. Effect of bile on Vibrio parahaemolyticus. Appl. Environ. Microbiol. 63:2372–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta S, Chowdhury R. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 65:1131–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect. Immun. 68:1491–1497. 10.1128/IAI.68.3.1491-1497.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfeffer CS, Hite MF, Oliver JD. 2003. Ecology of Vibrio vulnificus in estuarine waters of eastern North Carolina. Appl. Environ. Microbiol. 69:3526–3531. 10.1128/AEM.69.6.3526-3531.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaneko T, Colwell RR. 1975. Adsorption of Vibrio parahaemolyticus onto chitin and copepods. Appl. Microbiol. 29:269–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pruzzo C, Vezzulli L, Colwell RR. 2008. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol. 10:1400–1410. 10.1111/j.1462-2920.2007.01559.x [DOI] [PubMed] [Google Scholar]


