Abstract
A commercially available product containing three probiotic bacterial strains (Lactobacillus helveticus R0052, Bifidobacterium longum subsp. infantis R0033, and Bifidobacterium bifidum R0071) was previously shown in animal trials to modulate both TH1 and TH2 immune responses. Clinical studies on this combination of bacteria have also shown positive health effects against seasonal winter diseases and rotavirus infection. The goal of this study was to use a well-established in vitro intestinal epithelial (HT-29) cell model that has been shown to constitutively express double-stranded RNA (dsRNA) sensors (Toll-like receptor 3 [TLR3], retinoic acid-inducible gene I, melanoma differentiation-associated gene 5, and dsRNA-activated protein kinase). By using the HT-29 cell model, we wanted to evaluate whether or not this combination of three bacteria had the capacity to immune modulate the host cell response to a dsRNA ligand, poly(I·C). Using a custom-designed, two-color expression microarray targeting genes of the human immune system, we investigated the response of HT-29 cells challenged with poly(I·C) both in the presence and in the absence of the three probiotic bacteria. We observed that the combination of the three bacteria had a major impact on attenuating the expression of genes connected to proinflammatory TH1 and antiviral innate immune responses compared to that obtained by the poly(I·C)-only challenge. Major pathways through which the multistrain combination may be eliciting its immune-modulatory effect include the TLR3 domain-containing adapter-inducing beta interferon (TRIF), mitogen-activated protein kinase, and NF-κB signaling pathways. Such a model may be useful for selecting potential biomarkers for the design of future clinical trials.
INTRODUCTION
One of the key protective health effects of probiotics on intestinal epithelial cells (IECs) is the capacity for modulation of innate and adaptive immune responses (1, 2). Several mechanisms of action through which probiotics direct the abrogation of infection have been postulated, such as the modulation of the JAK-STAT, mitogen-activated protein kinase (MAPK), NF-κB, and peroxisome proliferator-activated receptor γ immune-signaling pathways. However, the exact mechanism of action by which specific probiotic bacteria exert their beneficial impact remains to be determined (3, 4). In systems cochallenged with pathogenic microbes, probiotics interact with the host immune system to provide protection, in part, by immune regulation of T cells, including TH1 and TH2 immune responses (1). In systems challenged with proinflammatory stimuli (i.e., pathogenic microbes, lipopolysaccharide, and tumor necrosis factor alpha [TNF-α]), probiotics have the capacity to provide protection by modulating the host immune system, in part, by influencing the activity of T cells, including TH1 and TH2 subsets (1). Proinflammatory responses are characterized by expression of TNF-α, interleukin-1β (IL-1β), and IL-8, which are induced by TH1 cytokines (gamma interferon [IFN-γ]), while cytokines produced by TH2 cells (IL-4, IL-10) are associated with anti-inflammatory activity (1, 5).
A commercial probiotic containing three bacteria (Lactobacillus helveticus R0052, Bifidobacterium longum subsp. infantis R0033, and Bifidobacterium bifidum R0071) is available under the trade names ProbioKid and Biostime. A previous preclinical assessment of this combination using two distinct rat models (TH1 and TH2) of infection reported that it has a synergistic, anti-inflammatory impact and benefited both TH1 and TH2 immune responses, depending on the challenge (5). A clinical trial was performed with this product in combination with intravenous antivirus therapy (ribavirin) to evaluate the potential impact in 78 children (1 to 5 years of age) with rotaviral diarrhea. It was concluded that the product was effective in alleviating the duration and severity of diarrhea (6). Using the same preparation, a randomized, double-blind, placebo-controlled trial was performed in France with 135 school children between the ages of 3 and 7 years. A 3-month supplementation with the combination decreased the risk of occurrence of common seasonal infectious diseases and decreased the incidence of absence from school (7). Overall, there was an indication that the combination of the three bacteria may modulate the immune system, specifically, those pathways associated with antiviral infections.
IECs are one of the cell types in the intestinal epithelial barrier, specifically, the upper layer known as the intestinal mucosa (epithelium), and are an initial point of contact between the host and intestinal microbes (8). These cells are able to discriminate between an assortment of antigens using pathogen recognition receptors (PRRs), such as Toll-like receptors (TLRs) (2, 9).
One such PRR, Toll-like receptor 3 (TLR3), is a receptor for double-stranded RNA (dsRNA). Localized in the endosomal membrane of IECs, TLR3 triggers the activation of the innate antiviral immune response against dsRNA viruses (10, 11). However, new evidence has emerged showing that two cytoplasmic PRRs of the retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) family, RIG-I and melanoma differentiation-associated gene 5 (MDA5), are major sensors for dsRNA viruses, such as rotavirus, and for cell signaling of type I IFN (12, 13). Recent work by Broquet et al. (14) using two colon epithelial cell lines, HCA-7 and HT-29, showed that rotavirus sensing through RIG-I and MDA5, in which signaling occurred via a common adaptor molecule, mitochondrial antiviral signaling protein (MAVS), and not via the TLR3 domain-containing adapter-inducing beta interferon (TRIF) pathway, resulted in the upregulation of a type I IFN-β response in IECs. Overall, there are redundant overlapping sensors in host IECs for the detection of an assortment of dsRNA ligands or RNA viruses that have to be considered.
Poly(I·C), a synthetic double-stranded RNA ligand, was chosen to be a model to stimulate antiviral immune responses in IECs, as it has been extensively studied and established as an accepted model to investigate antiviral immune responses (15). Furthermore, the human colonic adenocarcinoma cell line HT-29 has been extensively used to investigate poly(I·C) induction and has the added benefit of constitutively expressing the dsRNA receptors of TLR3, RIG-I, MDA5, and dsRNA-activated protein kinase (PKR), as reported by Broquet et al. (14). Considering these facts, HT-29 cells are an appropriate in vitro cell model system with which to investigate antiviral immune responses. Previous studies of HT-29 cells challenged with poly(I·C) have been reported, but probiotic cochallenge was not used (16, 17).
In the present study, the rationale was based, in part, on the positive beneficial effects that this combination of bacteria had in the previously reported preclinical and clinical work. The intention was to evaluate at the transcriptional level whether the combination of these three bacteria, here, the probiotic combination (PC), has the capacity to immune modulate an in vitro model of IECs cochallenged with a dsRNA ligand, poly(I·C). To this end, we used a custom-designed, two-color expression microarray targeting genes of the human immune system, here referred to as the immune array (18). The aim of this study was to evaluate by which pathways this combination of bacteria could modulate the host cell response to a dsRNA ligand, poly(I·C), which could potentially help in the design of future clinical trials based on potential biomarkers confirmed in this study.
MATERIALS AND METHODS
Intestinal epithelial cell culture.
Human colon adenocarcinoma (HT-29) cells were purchased from the American Type Culture Collection (ATCC; HTB-38) and cultured in a suspension of RPMI 1640 medium (HyClone, Logan, UT) supplemented with 5% bovine calf serum, 5% fetal bovine serum, and 2 mM l-glutamine (Invitrogen, Life Technologies). Cell cultures were grown in standard 75-cm2 tissue culture flasks (Corning Life Sciences, Acton, MA) at 37°C in a humidified, 5% CO2 incubator (Innova CO-170; New Brunswick Scientific, Edison, NJ). Cultures were routinely passaged when they reached a confluence of ∼75 to 90% and used for subsequent microarray experiments between passages 8 and 22. For all microarray experiments, HT-29 cells were seeded at 2 × 106 cells and grown for 48 h in standard tissue culture 25-cm2 flasks (Corning Life Sciences, Acton, MA) to reach a final concentration of 4 × 106 cells. Cells were washed with Dulbecco's phosphate-buffered saline (DPBS; HyClone, Logan, UT) and incubated for 30 min in serum-free RPMI prior to challenge assays.
Bacteria and culture conditions.
A laboratory blend of the multistrain bacterial product ProbioKid was prepared using industrially prepared lyophilized bacterial powders (Lallemand Health Solutions Inc., Montreal, QC, Canada) of Bifidobacterium longum subsp. infantis R0033, B. breve R0071, and Lactobacillus helveticus R0052 in a ratio of 20:20:60, respectively (here referred to as the probiotic combination [PC]). To rehydrate the multistrain blend of lyophilized bacteria, 1 g was mixed for 15 min at room temperature in 99 ml of phosphate-buffered saline (PBS; 0.1% [wt/vol] soy peptone, 0.121% [wt/vol] K2HPO4, 0.034% [wt/vol] KH2PO4). The bacterial pellet from 1 ml of this bacterial suspension was washed in PBS after centrifugation (12,800 × g, 10 min at room temperature) and then resuspended in serum-free RPMI 1640 medium. The bacterial suspension (PC) was added to the culture flask (25-cm2 flasks containing HT-29 cells) to have a multiplicity of infection (MOI) of 100:1 for the ratio of bacteria to HT-29 cells, as described by Audy et al. (18) (see Table S1 in the supplemental material). Viable counts of the bacterial suspension were performed using reinforced clostridial agar, and the suspension was incubated anaerobically for 48 h at 37°C to confirm the calculated ratio.
Cell challenges and RNA extraction.
Cell challenges were performed as explained previously by Audy et al. (18). Briefly, HT-29 cells were incubated for 3 h with poly(I·C) or PC alone or PC in combination with poly(I·C) at 10 μg/ml. RNA was isolated using a phenol-based extraction method (TRIzol reagent; Invitrogen, Burlington, ON, Canada) and Phase Lock Gel-Heavy tubes for phase separation of RNA, per the manufacturer's instructions (5 Prime Inc., Gaithersburg, MD). Total RNA was purified using an RNeasy minikit (Qiagen, Germantown, MD) and eluted in 50 μl of RNase-free water, per the manufacturer's instructions. RNA concentration and purity were determined by use of a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, DE), and quality was determined by use of an Agilent 2100 bioanalyzer and RNA 6000 Nano kits per the manufacturer's instructions (Agilent Technologies, Mississauga, ON, Canada). RNA samples with an A260/A280 ratio of between 1.8 and 2.2 (NanoDrop) and an RNA integrity number (RIN; Agilent 2100 bioanalyzer) of 9 or higher were processed further for reverse transcription (RT) and dye labeling.
RT and direct method for dye labeling.
Fifteen micrograms of control and treated total RNA was used for RT and the direct method of dye labeling as previously described by Audy et al. (18). Briefly, RNA was aliquoted and concentrated with a SpeedVac apparatus for about 45 min at medium temperature. After RNA samples were concentrated with the SpeedVac apparatus, 3 μg/μl of oligo(dT)23 primers (Invitrogen) was added to each sample and RNase-free water was added to obtain a final volume of 10 μl. Samples were incubated at 70°C for 10 min in a dry bath to reduce secondary structure formation and chilled on ice for 5 min. RT of RNA into cDNA was performed by using a SuperScript III reverse transcriptase kit (Invitrogen) by mixing 5× strand buffer, 0.1 M dithiothreitol, 2 mM dCTP, 6.67 mM dATP, 6.67 mM dGTP, 6.67 mM dTTP, 200 units of SuperScript III reverse transcriptase, and 1 mM Cy3-dCTP or Cy5-dCTP dye (GE Healthcare Amersham Biosciences) in a total volume of 20 μl. Samples were incubated at 42°C for 3 h (dry bath) to allow RT, and samples were kept in the dark to avoid photobleaching of the dyes. On completion, RNA was hydrolyzed at 37°C for 30 min with a mix of RNase A (0.05 mg/ml) and RNase H (0.05 U/μl). Reverse-transcribed and dye-labeled samples of cDNA were column purified using a QIAquick PCR purification kit per the manufacturer's instructions (Qiagen). Lastly, samples were concentrated with the SpeedVac apparatus at low temperature for about 1 h and stored in the dark at 4°C until hybridization to a cDNA microarray.
Immune array construction and printing.
As previously described by Audy et al. (18), the immune array was a custom-designed two-color, long oligonucleotide (70-mer) DNA microarray detecting 1,354 human genes belonging to 17 pathways connected to innate immunity and barrier defense. Genes were selected from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and belonged to the following pathways: cell adhesion molecules (CAMs), defensins, mucus production, gap junction, tight junction, apoptosis, adherence, cytokine-cytokine receptor interaction, chemokine signaling, B cell receptor signaling, JAK-STAT signaling, transforming growth factor beta (TGF-β) signaling, T cell receptor signaling, MAPK signaling, TLR signaling, NOD-like receptor (NLR) signaling, and natural killer cell-mediated cytotoxicity (19). Additionally, normalizing genes for quantitative RT-PCR (qRT-PCR) were added to confirm gene expression analysis. These included genes for B2M, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), HPRT1, MAN1B1, MTR, MYC, POLR2A, RPLP0, RPS14, and SI (20). Gene-specific long oligonucleotide sequences (70-mers) were recovered from the Human Exonic Evidence-Based Oligonucleotide (HEEBO) database (21) and were synthesized (IDT Integrated Technology Inc., IA) at a concentration of 25 mM. Oligonucleotides were spotted onto Corning UltraGAPS coated slides using a printing robot from Virtek Vision International Inc. The immune array analysis was repeated twice per microarray slide in six subgrids, and each gene was spotted in duplicate. Immune array slides were printed by the microarray laboratory facility at the National Research Council (NRC), located in Montreal, QC, Canada.
Hybridization and scanning.
The immune array, as previously described, was dipped in 0.2% sodium dodecyl sulfate (SDS) solution four times, followed by rinsing in water four times, and was immediately immersed in prehybridization solution containing 5× saline sodium citrate (SSC; 20× SSC is 3 M sodium chloride plus 0.3 M sodium citrate, pH 7.0), 0.1% SDS, and 0.1 mg/ml of bovine serum albumin (BSA), and the mixture was incubated for 1 h at 42°C. After 1 h of incubation, the slides were transferred to Coplin jars containing 0.1× SSC and incubated at room temperature for 5 min with gentle agitation, which was repeated twice. Slides were then transferred to a centrifuge box containing high-pressure liquid chromatography-grade water, incubated at room temperature for 30 s with gentle agitation, and dried under centrifugation for 2 min at 600 × g. Dried Cy-labeled cDNA samples were resuspended in 9 μl of hybridization buffer containing DIG Easy Hyb solution (Roche), 0.46 mg/ml Saccharomyces cerevisiae yeast tRNA (Invitrogen), and 0.46 mg/ml sonicated salmon sperm DNA (Invitrogen). Samples were incubated at room temperature for 5 min. Subsequently, samples of Cy3 and Cy5 were mixed together, and the mixture was incubated at 95°C for 5 min, incubated on ice for 1 min, and stored at 42°C until it was ready for hybridization to the immune array. Samples of Cy-labeled cDNA were pipetted (18 μl) onto the immune array under the lifter slips, allowing even dispersion by capillary action, and allowed to hybridize at 50°C for 18 h. Following hybridization, the slides were washed twice for 5 min each time at 42°C in a centrifuge box containing prewarmed 1× SSC–0.1% SDS, then once in 1× SSC for 2 min, and then once in 0.1× SSC for 1 min, with gentle agitation and dipping four times performed between transfers. Lastly, the slides were dried by centrifugation for 2 min at 600 × g. Slides were scanned using a ScanArray 5000 instrument (Perkin-Elmer, Waltham, MA), and the intensities of the individual spots on the slides (from 16-bit TIFF images) were quantified using the QuantArray software package (Perkin-Elmer).
Immune array statistical data analysis.
Assessment of slide quality, LOWESS normalization within the slide, Aquantile normalization between slides, and statistical analysis were conducted with the Limma package from BioConductor in R software (version 2.8.1), freely available web-based software (http://cran.r-project.org/bin/windows/base/). The analysis consisted of comparison of a minimum of four dye-swap hybridizations of HT-29 cells treated with poly(I·C) and PC alone or in combination with unchallenged HT-29 cells. For each treatment, a minimum of 4 biological replicates was performed. Each replicate consisted of challenges that were performed on different days. Genes with significant changes in transcript abundance were selected on the basis of two criteria: (i) a t-test P value of less than 0.05, which was considered statistically significant, and (ii) a cutoff in transcript abundance of least 2-fold. Lastly, two-dimensional clustering analysis was performed with MultiExperiment Viewer, part of TM4 microarray software from the J. Craig Venter Institute (22).
Comparative qRT-PCR.
Measurement of the transcript abundance of differentially expressed and reference genes was confirmed by comparative qRT-PCR of cDNA as described previously by Audy et al. (18) according to MIQE guidelines (23). Briefly, 4 μg of total RNA was treated with RQ1 DNase (Promega, Madison, WI) per the manufacturer's instructions. DNase-treated RNA (1 μg) was reverse transcribed using SuperScript III reverse transcriptase (Invitrogen) per the manufacturer's instructions. Reverse-transcribed cDNA was diluted 1:4 prior to amplification, and 2.5 μl of diluted cDNA was used as the template in qRT-PCR using 300 nmol of gene-specific primers (see Table S1 in the supplemental material) in a 25-μl reaction volume using Perfecta SYBR green SuperMix (Quanta Biosciences, Gaithersburg, MD) per the manufacturer's instructions. An initial incubation of 5 min at 95°C was performed, followed by 40 cycles consisting of template denaturation (15 s at 95°C) and one-step annealing and elongation (30 s at 60°C), with an ABI 7300 real-time PCR system (Applied Biosystems, Carlsbad, CA). As with the immune array, four biological replicates were analyzed for each gene tested, and fold change expression levels were normalized to the expression levels of three reference genes: genes for ACTB, B2M, and RPLP0 (see Table S1 in the supplemental material). Melting curve analysis was used to determine amplification specificity. Reaction efficiency was determined with a LinReg PCR (24), the genes used for normalization were evaluated with the BestKeeper program (25), and relative expression and statistical analyses were conducted with the REST (relative expression software tool) 2009 program (26). The sequences of the primers specific for genes selected for qRT-PCR analysis can be found on Table S1 in the supplemental material.
Microarray data accession number.
Additional information regarding the microarray platform can be located at the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) database under GEO accession no. GPL13933 series 2.
RESULTS
Immune array cluster and Venn diagram analysis.
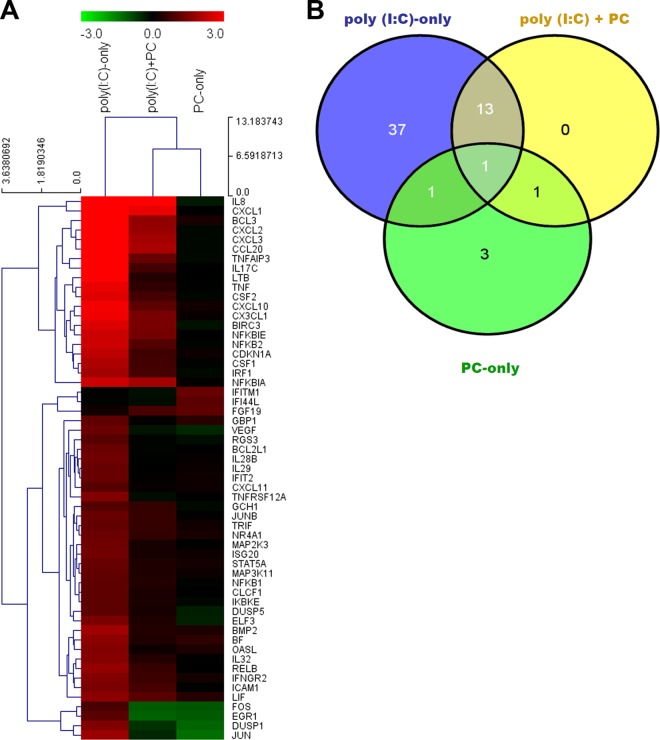
The cluster analysis in Fig. 1A depicts the differentially modulated genes in intestinal epithelial (HT-29) cells challenged for 3 h with poly(I·C) only, PC only, or poly(I·C) plus PC. Genes included in the cluster analysis had a cutoff 2-fold change in differential gene expression and a statistically significant difference in expression with a P value of <0.05 (Fig. 1A). The cochallenge with poly(I·C) plus PC clustered closer to the PC-only challenge than to the poly(I·C)-only challenge.
FIG 1.
(A) Cluster analysis of intestinal epithelial (HT-29) cells challenged with poly(I·C) only, PC (strains R0052, R0033, and R0071) only, and poly(I·C) plus PC. (B) Venn diagram analysis of microarray data from HT-29 cells treated separately with poly(I·C) only, PC only, and poly(I·C) plus PC. Results for all genes were statistically significant with a P value of <0.05 and a cutoff transcript abundance of 2-fold.
The Venn diagram analysis in Fig. 1B was used to find genes that were uniquely and commonly modulated between the three challenges. As with the cluster analysis, only those genes with a 2-fold change in differential expression and a P value of <0.05 were considered. Of the 56 differentially expressed genes in the Venn diagram analysis, 37 were unique to the poly(I·C)-only challenge, 3 were unique to the PC-only challenge, 13 were common to the poly(I·C)-only challenge and poly(I·C) plus PC challenge, 2 were common to the poly(I·C)-only challenge and the PC-only challenge, 2 were common to the PC-only challenge and the poly(I·C) plus PC challenge, and only 1 gene was common to all three challenges (Fig. 1B; Tables 1 to 3).
TABLE 1.
Unique genes from Venn diagram analysis of HT-29 cells challenged with poly(I·C) only and PC only in descending order of fold change in expressiona
| Challenge (no. of unique genes), immune gene protein | Function | Immune pathway(s) | Fold change in expressionb |
|---|---|---|---|
| Poly(I·C) only (n = 37) | |||
| IL-17C (cytokine) | T cell-derived cytokine that stimulates the release of TNF-α and IL-1β | Inflammatory cytokine and receptors | 12.41 |
| LTB (cytokine) | Cytokine with a specific role in immune response regulation | Cytokine-cytokine receptor interaction | 8.20 |
| TNF-α (cytokine) | Potent proinflammatory cytokine, involved in inflammatory responses | MAPK signaling pathway, TGF-β signaling pathway, cytokine-cytokine receptor interaction | 6.77 |
| CSF2 (cytokine) | Cytokine that stimulates growth and differentiation | JAK-STAT signaling, cytokine-cytokine receptor interaction, T cell receptor signaling | 6.31 |
| NF-κB2 | Pleiotropic transcription factor involved in inflammation, immunity, differentiation, cell growth, tumorigenesis, and apoptosis | MAPK signaling pathway, NF-κB signaling pathway | 5.32 |
| CDKN1A | Intermediate inhibitor of cellular proliferation in response to DNA damage | p53 signaling, cell cycle | 4.72 |
| CSF1 (cytokine) | Granulocyte-macrophage colony-stimulating factors that have a role in immune defenses | Cytokine-cytokine receptor interaction | 4.14 |
| IRF1 | Transcriptional activator for IFN-α-, IFN-β-, and IFN-γ-stimulated genes, a modulator of virus-induced signaling | NF-κB signaling pathway, TLR signaling pathway | 3.86 |
| BMP2 | Member of the TGF-β superfamily | TGF-β signaling pathway, cytokine-cytokine receptor interaction | 3.77 |
| RELB | Component of the NF-κB RelB-p52 complex | MAPK signaling pathway, NF-κB signaling pathway | 3.38 |
| BF | Not in Gene Cards database | No pathway | 3.17 |
| IL-32 (cytokine) | Inducer of TNF-α and IL-8, activating cytokine signaling pathways of NF-κB and p38 MAPK | Inflammatory response and autoimmunity | 3.04 |
| OASL | Binder to double-stranded RNA and DNA | No KEGG pathway | 2.95 |
| IFNGR2 | Accessory factor of the IFN-γ signal transduction pathway | Cytokine-cytokine receptor interaction | 2.93 |
| TNFRSF12A | Inducer of apoptosis in some cell types | Cytokine-cytokine receptor interaction, NF-κB signaling pathway, apoptosis | 2.89 |
| ICAM1 | Adhesion molecule, intercellular 1, upregulated by cytokines | CAMs | 2.80 |
| ELF3 | Transcriptional activator that binds and transactivates external transcribed spacer (ETS) sequences | Selected targets of ESR1 | 2.74 |
| ISG20 | Exonuclease involved in the antiviral function of IFN against RNA viruses | IFN-α and IFN-β response | 2.59 |
| IL-28B (cytokine) | Protein with immunomodulatory activity that displays potent antiviral activity | Cytokine-cytokine receptor interaction, JAK-STAT signaling pathway | 2.52 |
| MAP2K3 | Mitogen-activated protein kinase 3, critical component of TNF signaling | MAPK signaling pathway, TLR signaling pathway | 2.48 |
| NR4A1 | Regulator of expression of delayed-early genes during liver regeneration | MAPK signaling pathway | 2.43 |
| VEGF | Signaling protein involved in angiogenesis | Cytokine-cytokine receptor interaction | 2.39 |
| IFIT2 | IFN-α-inducible protein induced by viral infections | Interferons and receptors | 2.38 |
| JUNB | Transcription factor involved in regulating primary growth factor response | MAPK signaling pathway, JAK-STAT signaling pathway | 2.36 |
| BCL2L1 | Potent inhibitor of cell death, regulating cell death by blocking the voltage-dependent anion channel (VDAC) | Apoptosis, JAK-STAT signaling pathway | 2.36 |
| IL-29 (cytokine) | Protein involved in antiviral immunity and induced by viral infections or dsRNA | Cytokine-cytokine receptor interaction, JAK-STAT signaling pathway | 2.35 |
| STAT5A | Protein involved in signal transduction and activation of transcription in response to cytokines | JAK-STAT signaling pathway | 2.35 |
| TRIF | Adaptor used by TLR3 and TL4R to mediate NF-κB and interferon-regulatory factor (IRF) activation | NF-κB signaling, TLR signaling pathway | 2.26 |
| MAP3K11 | Kinase that activates MAPK8/JNK kinase and functions as a positive regulator of the Jun N-terminal protein kinase signaling pathway | MAPK signaling pathway | 2.25 |
| NFKB1 | Rapidly acting primary transcription factor that plays a key role in regulating the immune response to infection | MAPK signaling pathway, NF-κB signaling pathway | 2.20 |
| CLCF1 (cytokine) | Cytokine with a B cell-stimulating capability | Cytokine-cytokine receptor interaction, JAK-STAT signaling pathway | 2.20 |
| GBP1 | Guanylate binding protein mediating an antiviral response | JAK-STAT signaling pathway | 2.20 |
| DUSP5 | Phosphatase that inactivates members of the MAPK superfamily | MAPK signaling pathway | 2.15 |
| IKBKE | Noncanonical IκB kinase (IKK) that is essential for regulating antiviral signaling pathways | NF-κB signaling pathway, TLR signaling pathway | 2.14 |
| RGS3 | Downregulator of G-protein-mediated release of inositol phosphates and activation of MAPKs | G-protein signaling, G-protein alpha-q signaling cascades | 2.05 |
| CXCL11 (chemokine) | Chemotactic protein for interleukin-activated T cells | Cytokine-cytokine receptor interaction, chemokine signaling pathway, TLR signaling pathway | 2.04 |
| GCH1 | Member of the GTP cyclohydrolase family in BH4 biosynthesis | Nitric oxide signaling pathway, neurotransmitter receptors/regulators, IFN-α and IFN-β response | 2.03 |
| PC only (n = 3) | |||
| IFITM1 | IFN-induced antiviral protein that mediates cellular innate immunity | Interferon signaling pathway, B cell receptor signaling pathway | 2.36 |
| FGF19 | Protein involved in the suppression of bile acid biosynthesis | MAPK signaling pathway | 2.20 |
| IFI44L | Member of the IFI44 family | Interferons and receptors pathway | 2.10 |
Immune gene function data are from GeneCards (http://www.genecards.org), and immune pathway data are from KEGG (http://www.genome.jp/kegg/pathway.html).
All genes were upregulated. Fold change or expression ratio = 2log2ratio. A 2-fold change in differentially expressed genes and a P value of <0.05 were considered statistically significant.
TABLE 3.
Common gene from Venn diagram analysis of HT-29 cells cochallenged with PC only versus poly(I·C) only, PC only versus poly(I·C) plus PC, and PC only versus poly(I·C) only versus poly(I·C) plus PCa
| Challenges compared, common immune gene protein | Function | Immune pathway | Fold changeb |
||
|---|---|---|---|---|---|
| PC | Poly(I·C) | Poly (I·C) + PC | |||
| PC only vs poly(I·C) only | |||||
| JUN | Early response gene interacting with MYC to regulate AP1 genes | B cell receptor signaling pathway | 2.50 ↓ | 3.60 ↑ | |
| DUSP1 | Dual-specificity phosphatase 1 that reverses the activation of MAPK | MAPK signaling pathway | 2.27 ↓ | 2.87 ↑ | |
| PC only vs poly(I·C) + PC, FOS | Nuclear phosphoprotein that forms a complex with the JUN/AP-1 complex | MAPK signaling pathway Toll-like receptor signaling | 2.00 ↓ | 2.08 ↓ | |
| PC only vs poly(I·C) only vs poly(I·C) + PC, EGR1 | Activator of the transcription of target genes required for mitogenesis and differentiation | MAPK signaling pathway | 2.08 ↓ | 2.10 ↑ | 2.22 ↓ |
Immune gene function data are from GeneCards (http://www.genecards.org), and immune pathway data are from KEGG (http://www.genome.jp/kegg/pathway.html).
Fold change or expression ratio = 2log2ratio. ↑, upregulation; ↓, downregulation. A 2-fold change in differentially expressed genes and a P value of <0.05 were considered statistically significant.
Closer examination of the results of the cluster and Venn diagram analyses revealed a number of observations regarding the poly(I·C)-only challenge. First, it was evident that the poly(I·C)-only challenge induced the upregulation of various proinflammatory genes associated with the TLR3-TRIF, MAPK, NF-κB, and cytokine-cytokine receptor signaling pathways (Fig. 1 and Table 1). The genes associated with the upregulation of the MAPK signaling pathway included mitogen-activated protein kinase 3 (MAP2K3), which is a critical component of the TNF-stimulated signaling pathway that causes increased expression of inflammatory cytokines, and MAP3K11, a kinase that activates MAPK8-JNK kinase, which is involved in the transcriptional activity of NF-κB (Fig. 1 and Table 1). Unique genes connected with upregulation of the NF-κB signaling pathway induced by poly(I·C) only included the genes for TRIF, IRF3, NF-κB1, NF-κB2, RELB, and IKBKE, to name a few (Fig. 1A and Table 1). Other unique genes associated with the upregulation of cytokines and chemokines included the genes for TNF-α, IL-17C, IL-28B, IL-29, IL-32, LTB, CLCF1, CSF1, and CXCL11 (Fig. 1B and Table 1).
The second observation from the 37 uniquely modulated genes of the poly(I·C)-only treatment was the number of genes connected to antiviral functions in HT-29 cells, such as the genes for IRF1, ICAM1, TRIF, ISG20, GBP1, IL-28B, IL-29, IFIT2, and IKBKE (Fig. 1A and B; Table 1). In addition, the gene for OASL, which binds double-stranded RNA and DNA, was also upregulated uniquely in HT-29 cells challenged with poly(I·C) only (Fig. 1A and B; Table 1). Other uniquely upregulated genes associated with cellular proliferation and differentiation in apoptosis, JAK-STAT, MAPK, and cytokine-cytokine receptor signaling pathways included those for STAT5A, IFNGR2, BCL2L1, DUSP5, CDKN1A, CSF1, BMP2, CSF2, NR4A1, TNFRSF12A, JUNB, and vascular endothelial growth factor (VEGF) (Fig. 1 and Table 1).
Lastly, with respect to the poly(I·C)-only challenge, we report the upregulation of a number of known negative regulators of the proinflammatory innate immune response, such as TNF-α-induced protein 3 (TNFAIP3 or A20), NFKBIA, and NFKBIE.
It was also apparent from the cluster and Venn diagram analysis that the cochallenge with poly(I·C) plus PC did not lead to upregulation of the aforementioned proinflammatory genes which were uniquely induced by poly(I·C) only, such as genes for MAP2K3, MAP3K11, TNF-α, LTB, NF-κB1, NF-κB2, RELB, IL-17, CSF1, IL-28B, IL-29, IL-32, LTB, and CLCF1 (Fig. 1 and Table 1). In addition, the genes connected to antiviral functions in HT-29 cells (genes for IRF1, ICAM1, TRIF, ISG20, GBP1, IL-28B, IL-29, IFIT2, and IKBKE) and the gene for OASL, which were previously upregulated in the poly(I·C)-only challenge, were not significantly modulated by the poly(I·C) plus PC cochallenge (Fig. 1 and Table 1).
Considering the 13 common genes that were upregulated in both the poly(I·C)-only challenge and the poly(I·C) plus PC cochallenge, there were a number of proinflammatory genes associated with NF-κB and cytokine-cytokine receptor signaling pathways, such as the gens for IL-8, CXCL1, CXCL2, CXCL3, CXCL10, BCL3, TNFAIP3, NFKBIA, and NFKBIE (Fig. 1 and Table 2). All 13 genes that were upregulated in response to poly(I·C) only were attenuated by the cochallenge with poly(I·C) plus PC (Fig. 1 and 1B; Table 2); i.e., the presence of the probiotic combination reduced the ability of poly(I·C) only to upregulate these genes. Moreover, all 13 common upregulated genes, the majority of which were connected to the proinflammatory response, had a change in the fold change of expression (Δ fold change attenuation) ranging from 1.33 to 4.17 in the upregulation of genes in HT-29 cells when cells were cochallenged with the multistrain PC and poly(I·C) (Fig. 1A and B; Table 2).
TABLE 2.
Common genes from Venn diagram analysis of HT-29 cells cochallenged with poly(I·C) only versus poly(I·C) plus PC in descending order of Δ fold change of attenuationa
| Immune gene protein | Function | Immune pathway(s) | Fold change on challenge withb,c: |
Δ fold change of attenuationc,d | |
|---|---|---|---|---|---|
| Poly(I·C) only | Poly(I·C) + PC | ||||
| TNFAIP3 | Component of ubiquitin-editing protein complex that inhibits NF-κB activation | NF-κB signaling, apoptosis | 9.80 ↑ | 2.35 ↑ | 4.17 ↓ |
| CXCL10 (chemokine) | Protein with pleiotropic effects, including stimulation of monocytes and natural killer cells and T cell migration | Cytokine-cytokine receptor interaction, chemokine signaling pathway, TLR signaling pathway | 7.41 ↑ | 2.17 ↑ | 3.42 ↓ |
| CXCL2 (chemokine) | Protein expressed at sites of inflammation | Cytokine-cytokine receptor interaction, chemokine signaling pathway | 11.02 ↑ | 3.46 ↑ | 3.18 ↓ |
| CXCL3 (chemokine) | Protein with chemotactic activity for neutrophils | Cytokine-cytokine receptor interaction, chemokine signaling pathway | 12.36 ↑ | 3.90 ↑ | 3.17 ↓ |
| BCL3 | Contributor to the regulation of transcriptional activation of NF-κB pathway | NF-κB signaling, apoptosis | 10.29 ↑ | 3.33 ↑ | 3.09 ↓ |
| CX3CL1 (chemokine) | Protein with chemotactic activity for T cells and monocytes | Cytokine-cytokine receptor interaction, chemokine signaling pathway | 7.20 ↑ | 2.68 ↑ | 2.68 ↓ |
| CCL20 (chemokine) | Chemotactic factor that attracts lymphocytes and neutrophils | Cytokine-cytokine receptor interaction, chemokine signaling pathway | 10.55 ↑ | 4.07 ↑ | 2.59 ↓ |
| CXCL1 (chemokine) | Protein with chemotactic activity for neutrophils | Cytokine-cytokine receptor interaction, chemokine signaling pathway | 16.31 ↑ | 6.77 ↑ | 2.41 ↓ |
| BIRC3 | Inhibitor of apoptosis by binding to TNF receptor-associated factors TRAF1 and TRAF2 (apoptotic suppressor) | Apoptosis | 6.08 ↑ | 2.72 ↑ | 2.24 ↓ |
| NFKBIE | Inhibitor of NF-κB by complexing with and trapping it in the cytoplasm | NF-κB signaling | 5.28 ↑ | 2.63 ↑ | 2.00 ↓ |
| LIF (cytokine) | Inducer of terminal differentiation in leukemic cells | Cytokine-cytokine receptor interaction, JAK-STAT signaling pathway | 3.17 ↑ | 2.09 ↑ | 1.52 ↓ |
| NFKBIA | Inhibitor of NF-κB/REL complexes by trapping REL dimers in the cytoplasm | NF-κB signaling | 5.35 ↑ | 3.90 ↑ | 1.37 ↓ |
| IL-8 (chemokine) | Potent chemotactic factor that attracts neutrophils, basophils, and T cells | Cytokine-cytokine receptor interaction, chemokine signaling pathway, TLR signaling pathway | 12.98 ↑ | 9.79 ↑ | 1.33 ↓ |
Immune gene function data are from GeneCards (http://www.genecards.org), and immune pathway data are from KEGG (http://www.genome.jp/kegg/pathway.html).
Fold change or expression ratio = 2log2ratio. ↑, upregulation.
A 2-fold change in differentially expressed genes and a P value of <0.05 were considered statistically significant.
Δ fold change = expression with poly(I·C)/[expression with poly(I·C) + expression with PC]. ↓, a reduction in the Δ fold change of HT-29 cells challenged with poly(I·C).
Finally, in the HT-29 cells challenged with PC only, the expression of only six genes was significantly modulated, of which the expression of only three was unique to the PC-only challenge (Fig. 1; Tables 1 and 3). More importantly, none of the genes whose expression was modulated by the PC-only challenge were connected to proinflammatory responses (Fig. 1; Tables 1 and 3). However, the gene for IFITM1, which plays a key role in the antiproliferative action of IFN-γ and which mediates innate immunity, was uniquely upregulated in just the PC-only challenge (Fig. 1; Tables 1 and 3).
Comparative qRT-PCR.
qRT-PCR analysis was performed to validate the immune array results and to confirm the attenuation of upregulated genes associated with a proinflammatory response in the HT-29 cells cochallenged with poly(I·C) plus PC (Table 4). By comparing HT-29 cells challenged with poly(I·C) only versus poly(I·C) plus PC, we could evaluate the reduction in expression of selected genes that were modulated by the multistrain probiotic (PC). The expression ratios were calculated using the relative expression software tool REST 2009 (v2.0.13). The expression ratio for the upregulation of genes is equal to the given value, whereas the values for the downregulated genes are the reciprocal expression ratio values shown in Table 4 (26).
TABLE 4.
Relative expression ratio by RT-qPCR of genes of HT-29 cells challenged with poly(I·C) only versus poly(I·C) plus PCa
| Immune gene protein | Expression ratio | 1/expression ratio | P value |
|---|---|---|---|
| TNF | 0.153 | 6.54 | 0.022 |
| FOS | 0.199 | 5.03 | 0.027 |
| CXCL10 | 0.225 | 4.44 | 0.013 |
| IL-17C | 0.228 | 4.39 | 0.024 |
| IL-29 | 0.265 | 3.77 | 0.019 |
| IL-28B | 0.292 | 3.42 | 0.042 |
| IL-8 | 0.335 | 2.99 | 0.000 |
| IRF1 | 0.352 | 2.84 | 0.007 |
| NFKB1 | 0.545 | 1.83 | 0.007 |
| NFKBIA | 0.564 | 1.77 | 0.031 |
Relative expression ratios were determined by RT-qPCR using REST 2009 (v2.0.13) analysis software. All genes selected were statistically significantly downregulated with a P value of <0.05.
Ten genes were selected on the basis of the immune array results and included the genes for IL-8, CXCL10, IL-29, NFKBIA, NFKB1, TNF, IL-28B, FOS, IRF1, and IL-17C, as shown in Table 4. The data collected from qRT-PCR and REST analysis software of the gene expression of HT-29 cells challenged with poly(I·C) relative to that of cells cochallenged with poly(I·C) plus PC showed that the challenge with PC resulted in the statistically significant (P < 0.05) downregulation of all the aforementioned genes (Table 4). The fold change reported by the microarrays is an underestimate, and the two methods cannot be directly compared quantitatively because qRT-PCR is more sensitive (27). The results presented in Table 4 show that the expression patterns obtained by qRT-PCR were relatively similar to the results of the microarray analysis, thus confirming the attenuation of proinflammatory genes by PC and validating the immune array results.
DISCUSSION
The aim of this study was to evaluate at the cellular level whether the combination of three bacteria in PC had the capacity to immune modulate the host cell response to a dsRNA ligand, poly(I·C), using a well-established IEC line (HT-29).
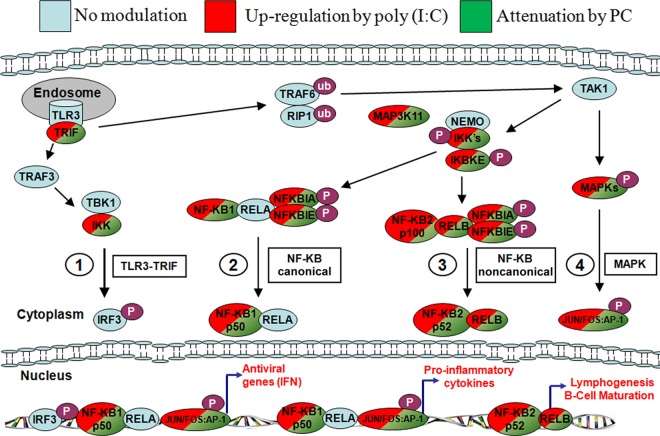
The binding of dsRNA ligand poly(I·C) to TLR3, RIG-I, or MDA5 has been reported to activate the downstream signaling cascade that ends with the induction of type I interferons (IFNs) and the production of cytokines/chemokines (a TH1 response) (10, 11, 14). The results of the poly(I·C)-only challenge in HT-29 cells of our study are in accordance with previously published data on the upregulation of a number of proinflammatory and antiviral genes connected to the TLR3-TRIF, NF-κB, and MAPK signaling pathways (Fig. 2).
FIG 2.
Simplified diagram of proposed immune pathways modulated by poly(I·C) and PC. Red, upregulation by poly(I·C); green, attenuation when PC is added to the challenge.
It has previously been reported that TLR3 mRNA and the TLR3 protein are constitutively expressed in HT-29 cells at high levels to anticipate viral antigens (28). It has also been confirmed by Broquet et al. (14) that TLR3, RIG-I, MDA5, and PKR are all constitutively expressed at the mRNA level to anticipate viral detection. This explains why no specific upregulation of TLR3 or the other dsRNA detection sensors was observed upon poly(I·C) induction.
We also did not observe the upregulation of the complete NF-κB pathway. The expression of TBK1, TRAF3, TRAF6, and RELA, to name a few other important mediators in the NF-κB signaling pathway (29), was not modified in our system. However, this could be explained by the fact that activation of the canonical NF-κB pathway transpires within minutes, whereas the activation of the noncanonical NF-κB pathway occurs several hours later, according to Zarnegar et al. (30). Alternatively, these genes may be constitutively expressed, and the proteins may be activated and deactivated by phosphorylation/dephosphorylation events.
When HT-29 cells were cochallenged with both poly(I·C) and PC, we observed a relative attenuation of the proinflammatory response compared to that with the poly(I·C)-only challenge. The results indicated that PC reduced the proinflammatory TH1 innate immune response of genes induced by the poly(I·C)-only challenge by inhibiting the expression of key genes. Similarly, antiviral genes induced by the poly(I·C)-only challenge, such as the genes for IRF1, ICAM1, TRIF, ISG20, GBP1, IL-28B, IL-29, IFIT2, and IKBKE, were inhibited when the PC plus poly(I·C) cochallenge was used. The expression of all 37 genes (the majority of which were connected to proinflammatory and antiviral innate immune responses) that were upregulated in the poly(I·C)-only challenge, including the genes for IL-17C and OASL, was inhibited when the multistrain probiotic (PC) was added.
The expression of all 13 commonly modulated genes that were upregulated by the poly(I·C)-only challenge and that were connected to the proinflammatory responses of NF-κB and cytokine/chemokine production was attenuated, with the Δ fold change reduction ranging from 1.33 to 4.17 by the poly(I·C) plus PC cochallenge (Fig. 1B). Overall, it was apparent from the results that PC had a major impact on attenuating proinflammatory responses, such as the NF-κB pathway and cytokine/chemokine production (Table 2 and Fig. 2).
The treatment of quiescent (i.e., unchallenged) HT-29 cells by PC only had almost no impact on gene expression (Fig. 1B). This finding agrees with our previously reported findings with HT-29 cells challenged with the individual probiotic strains (18). However, the gene for IFITM1, an IFN-induced antiviral protein that mediates cellular innate immunity, was uniquely upregulated by the PC-only challenge (Table 1), which may explain the positive protective effect of this probiotic combination against a poly(I·C)-only challenge that attenuates the proinflammatory and antiviral response.
Many studies have reported that probiotics can modulate key pathways in IECs and that various probiotics prevent NF-κB activation by inhibiting NFKBIA phosphorylation, ubiquitination, proteasomal degradation, or translocation of NF-κB into the nucleus (2). Here we propose, on the basis of the results obtained in this study, that the multistrain probiotic PC may be acting on the NF-κB pathway specifically to inhibit it and thereby block the induction of type I IFNs and the many cytokine/chemokine genes but that its activity is not limited to the NF-κB pathway. The exact mechanism by which this probiotic blocked the expression of these genes will require further investigation. We showed that the probiotic bacteria did not degrade the dsRNA ligand (data not shown). Therefore, the reduction of the proinflammatory response occurred at the cellular expression level and was induced by bacteria in our in vitro model.
Some clues for the mechanism can be found in previous studies performed by Jandu et al. (4) using L. helveticus R0052, one of the strains in the multistrain formulation used in this study. Their studies have suggested that R0052 could be acting via the JAK-STAT pathway upstream of the NF-κB pathway. It has also been known that L. helveticus R0052 has a lactocepin, prtH4 (protease), and von Schillde and coworkers (31) have shown that the lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Moreover, L. helveticus R0052 has a surface layer protein (SlpA), and Johnson-Henry et al. (32) showed that the S layer from R0052 inhibited enterohemorrhagic Escherichia coli O157:H7 adhesion to IECs. Martinez et al. (33) have reported that treatment with the S layer was able to inhibit viral infection in dendritic cells; however, the inhibition was contingent upon the S layer being used in the early stages of viral infection and not after. Recently, Taverniti et al. (34), using an in vitro model of IECs (Caco-2), reported that the strain L. helveticus MIMLh5 and its S-layer protein induced anti-inflammatory effects by reducing the activation of NF-κB. Additional studies will attempt to discern a possible role of the S-layer protein in the immune response of HT-29 cells to dsRNA ligand poly(I·C) challenge.
Overall, the results obtained in this study clearly show that PC has the capacity to modulate the host cell response to a dsRNA ligand, poly(I·C). Moreover, we were able to show that the key TLR3-TRIF, NF-κB, and MAPK pathways were implicated in the immune modulation by these probiotics. It was also shown that key genes from these pathways, as well as antiviral genes, such as IRF1, TRIF, ISG20, IL-28B, IL-29, IFIT2, IKBKE, and IFITM1, could be useful in the selection of potential biomarkers for the design of future clinical trials of dsRNA viruses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jocelyn Belvis for maintaining the HT-29 cell lines, Andre Nantel for his expertise in data-mining analysis of the immune array, and Pierre Burguiere for his input on the writing of the manuscript. We also thank Philippe Langella, INRA Jouy-en-Josas, and Julie Green-Johnson, UOIT, Canada, for their insightful comments in the preparation of the manuscript.
We all declare that we are or were employees in the Research and Development sector of Lallemand Health Solutions Inc., a company that manufactures and sells probiotic bacteria.
Footnotes
Published ahead of print 27 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03411-13.
REFERENCES
- 1.Duary RK, Bhausaheb MA, Batish VK, Grover S. 2012. Anti-inflammatory and immunomodulatory efficacy of indigenous probiotic Lactobacillus plantarum Lp91 in colitis mouse model. Mol. Biol. Rep. 39:4765–4775. 10.1007/s11033-011-1269-1 [DOI] [PubMed] [Google Scholar]
- 2.Thomas CM, Versalovic J. 2010. Probiotics-host communication: modulation of signaling pathways in the intestine. Gut Microbes 1:148–163. 10.4161/gmic.1.3.11712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherman PM, Ossa JC, Johnson-Henry K. 2009. Unraveling mechanisms of action of probiotics. Nutr. Clin. Pract. 24:10–14. 10.1177/0884533608329231 [DOI] [PubMed] [Google Scholar]
- 4.Jandu N, Zeng ZJ, Johnson-Henry KC, Sherman PM. 2009. Probiotics prevent enterohaemorrhagic Escherichia coli O157:H7-mediated inhibition of interferon-gamma-induced tyrosine phosphorylation of STAT-1. Microbiology 155:531–540. 10.1099/mic.0.021931-0 [DOI] [PubMed] [Google Scholar]
- 5.Cazzola M, Tompkins TA, Matera MG. 2010. Immunomodulatory impact of a synbiotic in T(h)1 and T(h)2 models of infection. Ther. Adv. Respir. Dis. 4:259–270. 10.1177/1753465810379009 [DOI] [PubMed] [Google Scholar]
- 6.Mei LX, Chen Z. 2008. Evaluation of the efficacy of a synbiotic preparation on rotaviral infection in children. Med. Information 21:893–895 [Google Scholar]
- 7.Cazzola M, Pham-Thi N, Kerihuel JC, Durand H, Bohbot S. 2010. Efficacy of a synbiotic supplementation in the prevention of common winter diseases in children: a randomized, double-blind, placebo-controlled pilot study. Ther. Adv. Respir. Dis. 4:271–278. 10.1177/1753465810379010 [DOI] [PubMed] [Google Scholar]
- 8.Bron PA, van Baarlen P, Kleerebezem M. 2012. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 10:66–78. 10.1038/nrmicro2690 [DOI] [PubMed] [Google Scholar]
- 9.Westendorf AM, Fleissner D, Hansen W, Buer J. 2010. T cells, dendritic cells and epithelial cells in intestinal homeostasis. Int. J. Med. Microbiol. 300:11–18. 10.1016/j.ijmm.2009.08.009 [DOI] [PubMed] [Google Scholar]
- 10.Hosoya S, Villena J, Shimazu T, Tohno M, Fujie H, Chiba E, Shimosato T, Aso H, Suda Y, Kawai Y, Saito T, Alvarez S, Ikegami S, Itoh H, Kitazawa H. 2011. Immunobiotic lactic acid bacteria beneficially regulate immune response triggered by poly(I:C) in porcine intestinal epithelial cells. Vet. Res. 42:111. 10.1186/1297-9716-42-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto M, Seya T. 2008. TLR3: interferon induction by double-stranded RNA including poly(I:C). Adv. Drug Deliv Rev. 60:805–812. 10.1016/j.addr.2007.11.005 [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi O, Akira S. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20:17–22. 10.1016/j.coi.2008.01.002 [DOI] [PubMed] [Google Scholar]
- 13.Wilkins C, Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47. 10.1016/j.coi.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broquet AH, Hirata Y, McAllister CS, Kagnoff MF. 2011. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 186:1618–1626. 10.4049/jimmunol.1002862 [DOI] [PubMed] [Google Scholar]
- 15.Fortier ME, Kent S, Ashdown H, Poole S, Boksa P, Luheshi GN. 2004. The viral mimic, polyinosinic:polycytidylic acid, induces fever in rats via an interleukin-1-dependent mechanism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 287:R759–R766. 10.1152/ajpregu.00293.2004 [DOI] [PubMed] [Google Scholar]
- 16.Furrie E, Macfarlane S, Thomson G, Macfarlane GT. 2005. Toll-like receptors-2, -3 and -4 expression patterns on human colon and their regulation by mucosal-associated bacteria. Immunology 115:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berchtold S, Manncke B, Klenk J, Geisel J, Autenrieth IB, Bohn E. 2008. Forced IFIT-2 expression represses LPS induced TNF-alpha expression at posttranscriptional levels. BMC Immunol. 9:75. 10.1186/1471-2172-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Audy J, Mathieu O, Belvis J, Tompkins TA. 2012. Transcriptomic response of immune signalling pathways in intestinal epithelial cells exposed to lipopolysaccharides, Gram-negative bacteria or potentially probiotic microbes. Benef. Microbes 3:273–286. 10.3920/BM2012.0027 [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28:27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dydensborg AB, Herring E, Auclair J, Tremblay E, Beaulieu JF. 2006. Normalizing genes for quantitative RT-PCR in differentiating human intestinal epithelial cells and adenocarcinomas of the colon. Am. J. Physiol. Gastrointest. Liver Physiol. 290:G1067–G1074. 10.1152/ajpgi.00234.2005 [DOI] [PubMed] [Google Scholar]
- 21.Kim YH, Pollack JR. 2009. Comparative genomic hybridization on spotted oligonucleotide microarrays. Methods Mol. Biol. 556:21–32. 10.1007/978-1-60327-192-9_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. 2003. TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378 [DOI] [PubMed] [Google Scholar]
- 23.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 24.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 339:62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- 25.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26:509–515. 10.1023/B:BILE.0000019559.84305.47 [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ. 2000. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 28:4552–4557. 10.1093/nar/28.22.4552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cario E, Podolsky DK. 2000. Differential alteration in intestinal epithelial cell expression of Toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect. Immun. 68:7010–7017. 10.1128/IAI.68.12.7010-7017.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Witte K, Witte E, Sabat R, Wolk K. 2010. IL-28A, IL-28B, and IL-29: promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 21:237–251. 10.1016/j.cytogfr.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 30.Zarnegar B, Yamazaki S, He JQ, Cheng G. 2008. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc. Natl. Acad. Sci. U. S. A. 105:3503–3508. 10.1073/pnas.0707959105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Schillde MA, Hormannsperger G, Weiher M, Alpert CA, Hahne H, Bauerl C, van Huynegem K, Steidler L, Hrncir T, Perez-Martinez G, Kuster B, Haller D. 2012. Lactocepin secreted by Lactobacillus exerts anti-inflammatory effects by selectively degrading proinflammatory chemokines. Cell Host Microbe 11:387–396. 10.1016/j.chom.2012.02.006 [DOI] [PubMed] [Google Scholar]
- 32.Johnson-Henry KC, Hagen KE, Gordonpour M, Tompkins TA, Sherman PM. 2007. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell. Microbiol. 9:356–367. 10.1111/j.1462-5822.2006.00791.x [DOI] [PubMed] [Google Scholar]
- 33.Martinez MG, Prado Acosta M, Candurra NA, Ruzal SM. 2012. S-layer proteins of Lactobacillus acidophilus inhibits JUNV infection. Biochem. Biophys. Res. Commun. 422:590–595. 10.1016/j.bbrc.2012.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taverniti V, Stuknyte M, Minuzzo M, Arioli S, De Noni I, Scabiosi C, Cordova ZM, Junttila I, Hamalainen S, Turpeinen H, Mora D, Karp M, Pesu M, Guglielmetti S. 2013. S-layer protein mediates the stimulatory effect of Lactobacillus helveticus MIMLh5 on innate immunity. Appl. Environ. Microbiol. 79:1221–1231. 10.1128/AEM.03056-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.