Abstract
Mycosporine-like amino acids (MAAs) are valuable molecules that are the basis for important photoprotective constituents. Here we report molecular analysis of mycosporine-like amino acid biosynthetic genes from the halotolerant cyanobacterium Aphanothece halophytica, which can survive at high salinity and alkaline pH. This extremophile was found to have a unique MAA core (4-deoxygadusol)-synthesizing gene separated from three other genes. In vivo analysis showed accumulation of the mycosporine-2-glycine but not shinorine or mycosporine-glycine. Mycosporine-2-glycine accumulation was stimulated more under the stress condition of high salinity than UV-B radiation. The Aphanothece MAA biosynthetic genes also manifested a strong transcript level response to salt stress. Furthermore, the transformed Escherichia coli and Synechococcus strains expressing four putative Aphanothece MAA genes under the control of a native promoter were found to be capable of synthesizing mycosporine-2-glycine. The accumulation level of mycosporine-2-glycine was again higher under the high-salinity condition. In the transformed E. coli cells, its level was approximately 85.2 ± 0.7 μmol/g (dry weight). Successful production of a large amount of mycosporine in these cells provides a new opportunity in the search for an alternative natural sunscreen compound source.
INTRODUCTION
Mycosporine and mycosporine-like amino acids (MAAs) are low-molecular-weight water-soluble molecules containing a substituted cyclohexenone or an imino cyclohexene ring and showing absorption maxima between 310 and 360 nm (1, 2). To date, more than 20 MAAs have been identified that are found in eukaryotic algae, fungi, coral, and cyanobacteria (1, 3). Interestingly, these organisms have evolved the capacity to synthesize and accumulate MAAs upon the adaptation from a detrimental effect of solar UV radiation (1, 4). MAAs are known to be synthesized under osmotic stress or are constitutively present (5, 6). The MAAs protect the cell by absorbing UV radiation and dissipating the energy as heat without generating reactive oxygen species. Due to their high-UV absorption coefficients and their ability to protect skin from UV-mediated damage, MAAs are promising candidates for use in pharmaceutical and cosmetic applications.
The main steps of the MAA biosynthetic pathway and their genetic basis have recently been elucidated (7). A cluster of four genes in Anabaena variabilis ATCC 29413 (Ava_3858 to Ava_3855) was shown to be responsible for MAA biosynthesis from sedoheptulose-7-phosphate (SHP) in the Calvin-Benson cycle. Genes Ava_3858 and Ava_3857 encode demethyl 4-deoxygadusol (DDG) synthase and O-methyltransferase (O-MT), respectively, to synthesize 4-deoxygadusol (4-DG). The product of Ava_3856 catalyzes the addition of glycine to 4-DG to produce mycosporine-glycine. Further condensation of serine onto mycosporine-glycine yields shinorine, which is catalyzed by the product of Ava_3855 encoding a nonribosomal peptide synthetase (NRPS). A similar cluster of four genes in Nostoc punctiforme ATCC 29133 (NpR5600, NpR5599, NpR5598, and NpF5597) has also been shown to catalyze the same reaction, although in this case, NpF5597 encodes d-Ala-d-Ala ligase and the direction of transcription of NpF5597 is opposite that of NpR5600 to NpR5598 (7, 8).
For cyanobacteria, shinorine, porphyra-334, and mycosporine-glycine are often described as the major MAAs (2). It has previously been reported that a community of unicellular cyanobacteria in a hypersaline pond in Israel contains a large amount of MAAs (9). This finding is remarkable because organisms living in the hypersaline environment often accumulate an osmoprotectant, glycine betaine (5). If halophilic cyanobacteria accumulate both glycine betaine and MAAs, they require a large amount of nitrogenous compounds.
A halotolerant cyanobacterium, Aphanothece halophytica, accumulates glycine betaine as an osmoprotectant and can grow under high-salinity conditions up to 3 M NaCl and alkaline pH 11 (10). A. halophytica synthesizes glycine betaine by a three-step methylation of glycine (11). Since both glycine betaine and MAAs are nitrogenous compounds, it was interesting to examine whether A. halophytica can synthesize the MAAs. In this study, we demonstrated that A. halophytica accumulates an MAA, mycosporine-2-glycine. The corresponding putative genes encoding enzymes capable of MAA production in A. halophytica were also identified. Moreover, we found that E. coli and Synechococcus cells transformed with the identified four putative Aphanothece genes under the control of their native promoters were capable of synthesizing mycosporine-2-glycine.
MATERIALS AND METHODS
Strains and growth conditions.
Escherichia coli strain DH5α cells were grown in Luria-Bertani (LB) medium at 37°C. A. halophytica cells were grown photoautotrophically (70 μE m−2 s−1) in blue-green 11 (BG11) liquid medium containing 18 mM NaNO3 and Turk Island salt solution at 30°C (11). Synechococcus sp. strain PCC 7942 and E. coli cells expressing putative MAA synthetic genes were grown under the same conditions as the wild-type cells but were supplemented with 50 μg/ml of streptomycin. The growth of E. coli and cyanobacterial cells was monitored by measuring the absorbance at 620 and 730 nm, respectively, with a Shimadzu UV-160A spectrophotometer.
Stress treatment.
A. halophytica cells were grown in the growth medium photoautotrophically (normal growth light, 70 μE m−2 s−1) for 14 days prior to salt upshock and UV-B stress experiments. For the upshock experiment, the concentration of NaCl in growth medium was changed from 0.5 to 2.5 M. For the UV-B stress experiment, cells were adjusted to an optical density at 730 nm (OD730) in the culture medium of approximately 0.8 prior to transfer to quartz bottles and exposed to UV-B radiation at 0.1 W/m−2. UV-B exposure was carried out for 6 h each day for 5 days.
The effects of NaCl and UV-B in E. coli and Synechococcus cells expressing putative MAA synthetic genes were also examined. In E. coli, salt stress was achieved by growing cells overnight in LB medium containing NaCl, followed by MAA extraction. For UV-B treatment, cells were grown in LB medium at 37°C until the OD620 reached 0.8, followed by transfer of the cells to quartz bottles and exposure to UV-B radiation at 0.1 W/m−2. UV-B exposure was carried out for 2 h each day for 5 days. The UV irradiances were measured using a UV-X radiometer (UVP, Inc., Upland, CA). Salt stress for Synechococcus cells was achieved by growing cells in BG11 supplemented with 0.3 M NaCl, whereas the applied UV-B stress was the same as mentioned for E. coli.
MAA extraction and analysis.
Ten milliliters of cyanobacterial or E. coli cell cultures was harvested by centrifugation at 5,000 × g for 10 min at 4°C. Cell pellets were extracted in 500 μl of 100% high-performance liquid chromatography (HPLC)-grade methanol overnight at 4°C. Thereafter, aliquots were centrifuged at 15,000 × g for 10 min at 4°C, and the supernatants were transferred to new Eppendorf microcentrifuge tubes. The supernatants were evaporated to dryness at 45°C in a vacuum evaporator, and the extracts were redissolved in 500 μl of Milli-Q water. To this, 2 drops of chloroform was added and mixed by vigorous shaking. Thereafter, the upper water phase was passed through an Ultracel YM-3 membrane (Millipore, USA) by centrifugation (15,000 × g) at 25°C. The resulting flowthrough was further subjected to HPLC analysis with a ShimPack FC-ODS reverse-phase column (3 μm; 150 by 4.6 mm) connected to a guard column (30 by 4.6 mm) that contained the same packing materials as the main column at 35°C. MAAs were detected by using a UV-visible (UV-VIS) detector (330 nm). The mobile phase was run at a flow rate of 0.4 ml/min using water containing 0.1% (vol/vol) trifluoroacetic acid. For purification of MAAs, the eluted fraction containing MAAs was subjected to a second HPLC analysis with a flow rate at 0.4 ml/min using the following linear gradient conditions: first 100% solvent A (0.1 M ammonium acetate, pH 6.5) and then, after 15 min, 20% solvent B (acetonitrile) in solvent A. After evaporation of solvent in the collected fraction, the MAAs were dissolved in water. Then the MAAs were further analyzed by time-of-flight mass spectroscopy (TOF-MS) (AXIMACFR; Shimadzu/Kratos) using gentisic acid as a matrix. The composition of MAAs was determined by alkaline hydrolysis as described in reference 12. The hydrolyzed amino acids were identified by the amino acid analyzer with a Shim Pack Li column (Shimadzu, Japan).
Expression of Aphanothece sunscreen genes in Escherichia coli and a freshwater cyanobacterium, Synechococcus sp. PCC 7942.
Plasmid pUC303-Bm (13) was used to transform E. coli and Synechococcus sp. PCC 7942 cells to serve as appropriate controls. For the construction of an expression vector in E. coli, two coding regions, that of Ap3858 and that of Ap3857 to Ap3855, containing their promoters (here Ap3858 and others represent the A. halophytica genes corresponding to Ava_3858 and others) were PCR amplified using the genomic DNA of A. halophytica with two primer pairs, Ap3858bam/Ap3858His6bam and Ap3857-55xho/Ap3857-55His6xho, respectively (see Table S1 in the supplemental material). Since the His tag sequence was introduced to the primers Ap3858His6bam and Ap3857-55His6xho, the PCR products for Ap3858 and Ap3857 to Ap55 contained histidine extensions at the C-terminal ends. PCR products were subcloned into pCR2.1 TA cloning vector and sequenced. DNA fragments covering Ap3858 and Ap3857 to Ap3855, including promoter regions, were prepared by digestion with BamHI and XhoI and ligated into the corresponding sites of the shuttle vector pUC303. The generated plasmid, pAp3858-55_303, was then used to transform E. coli DH5α and Synechococcus sp. PCC 7942 cells as previously described (14).
RT-PCR analysis.
Total RNA was extracted from A. halophytica cells using the RNeasy kit (Qiagen, Hilden, Germany). Five micrograms of total RNA was reverse transcribed using the Superscript II RT kit (Invitrogen, CA) as per the manufacturer's instructions. The PCR amplification was performed with oligonucleotides specific for targeted genes (Ap3858, Ap3857, Ap3856, and Ap3855) and AprnpB as a positive control (see Table S1 in the supplemental material). The PCR-amplified samples were electrophoresed on 1.2% (wt/vol) agarose gels and stained with 0.1 μg/ml of ethidium bromide. All reverse transcription-PCR (RT-PCR) experiments were repeated at least three times.
Other methods.
SDS-PAGE and Western blot analyses were performed as previously described (11). Protein concentration was determined by the Bradford method. Protein bands on SDS-PAGE gels were detected with Coomassie brilliant blue (CBB R-250) stain. For Western blot analysis, SDS-PAGE-separated protein bands were transferred to a polyvinylidene difluoride (PVDF) membrane. The anti-6-His (6×His tag) antibody was purchased from R & D Systems (Minneapolis, MN).
Nucleotide sequence accession numbers.
Nucleotide sequence data for Ap3858 and Ap3857 to Ap3855 are available in the DDBJ database under accession numbers AB854643 and AB854644, respectively.
RESULTS
UV-absorbing compound in A. halophytica.
To examine whether A. halophytica accumulates MAAs, A. halophytica cells were grown in the BG11 liquid medium as described in Materials and Methods. The cells were then extracted with methanol. The absorption spectra of the methanol-extracted fraction showed a shoulder at around 330 nm (see Fig. S1A in the supplemental material). After evaporation of methanol, the aliquots were dissolved in water and vigorously shaken together with chloroform, and the upper water phase was filtered and centrifuged. The absorption spectra of the resulting phase exhibited a single peak at 331 nm (see Fig. S1B). HPLC analysis revealed a main single peak at a retention time of about 9.1 min (Fig. 1A). A few smaller, unknown peaks were also detected at 14.7, 15.1, and 15.8 min.
FIG 1.
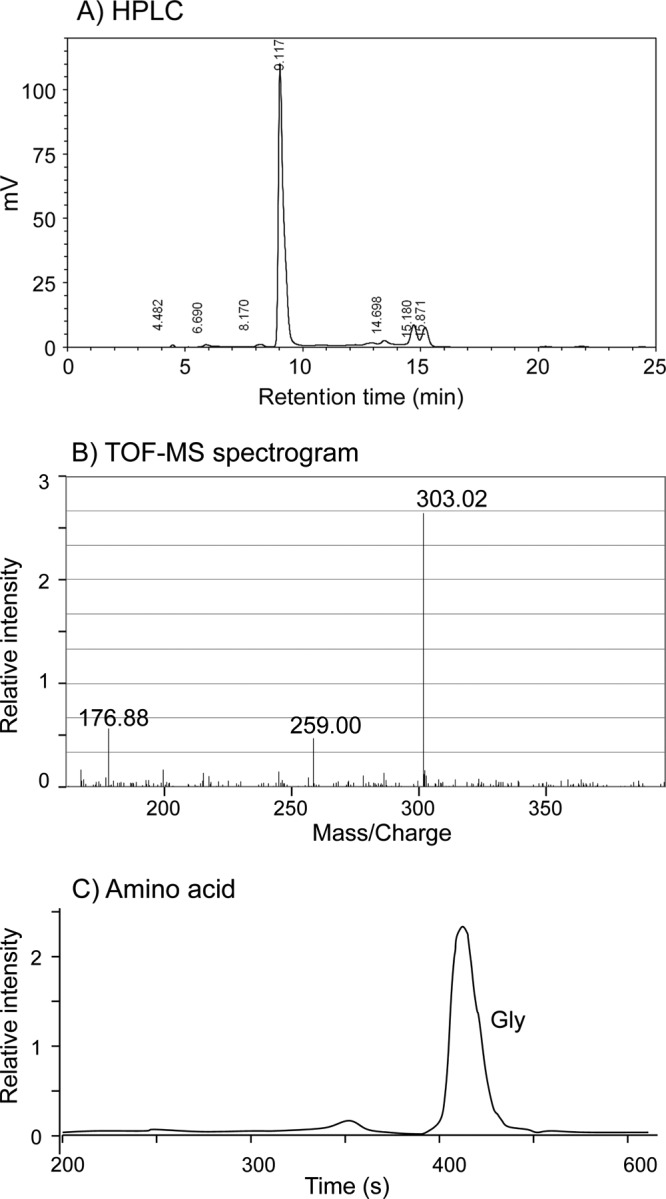
HPLC (A), TOF-MS (B), and amino acid (C) analyses of Ap-MAA. (A) HPLC chromatogram of UV-absorbing compound produced by A. halophytica. A Shim Pack FC-ODS reverse-phase column connected to a guard column that contained the same packing materials as the main column was used for analysis. UV-absorbing compound was detected by using a UV-VIS detector at 330 nm. The mobile phase was run at a flow rate of 0.4 ml/min using water containing 0.1% (vol/vol) trifluoroacetic acid. (B) TOF-MS spectrogram of purified Ap-MAA measured with gentisic acid as the matrix. (C) Amino acid composition of purified Ap-MAA after alkaline hydrolysis. Aliquots of purified fraction were hydrolyzed with alkaline solution as described in Materials and Methods. The hydrolyzed amino acids were identified by the amino acid analyzer with a Shim Pack Amino-Li column.
The abundant Aphanothece UV-absorbing compound is mycosporine-2-glycine.
Identification of the UV-absorbing compound from A. halophytica (hereafter referred to as Ap-MAA) was performed by two methodologies: (i) TOF-MS to identify the molecular mass and fragmentation pattern and (ii) amino acid analysis after alkaline hydrolysis to identify the amino acid composition of the Ap-MAA. As shown in Fig. 1B, a TOF-MS spectrogram of the purified Ap-MAA showed 3 signal peaks at m/z 176.88, 259.00, and 303.02. The mass of this observation at peak 303.02 was in agreement with the mass of the protonated form of mycosporine-2-glycine, implying that the detected MAA is likely mycosporine-2-glycine. The signal peak at 259 was in agreement with the protonated form of decarboxylated product of mycosporine-2-glycine, and the signal peak 176.88 might be sodium salt of gentisic acid.
Furthermore, amino acid composition analysis revealed the presence of only a single amino acid corresponding to glycine (Fig. 1C). Thus, glycine can be considered the sole amino acid in the structure of the Ap-MAA molecule. Hence, we concluded that the purified Ap-MAA is mycosporine-2-glycine.
Stress-induced accumulation of Ap-MAA.
Stress treatments were carried out to determine the accumulation level of Ap-MAA. The salt upshock condition (0.5 M to 2.5 M NaCl) caused a significant increase in the Ap-MAA level, from 0.35 ± 0.02 to 31.97 ± 2.5 μmol/g (dry weight). The accumulation of Ap-MAA reached a maximum after 48 h of salt upshock (Fig. 2A). The UV-B exposure of A. halophytica cells grown at 0.5 M NaCl showed an increase in the Ap-MAA level to approximately 16.64 ± 0.6 μmol/g (dry weight) at day 5 (Fig. 2B, white bars). When A. halophytica cells were grown at 2.0 M NaCl prior to UV-B exposure, the MAA level was increased to 28.38 ± 1.2 μmol/g (dry weight) (Fig. 2B, gray bars).
FIG 2.
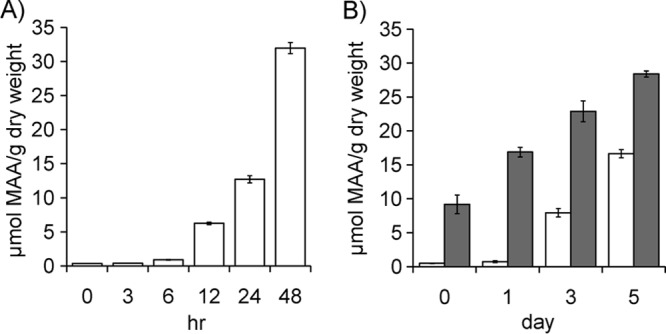
Time course of Ap-MAA accumulation as stressed by salt upshock and UV-B radiation. (A) Ap-MAA accumulation under salt upshock (0.5 M to 2.5 M NaCl). (B) Ap-MAA accumulation under UV-B radiation. White bars represent Aphanothece cells grown in 0.5 M NaCl and subjected to UV-B stress. Gray bars represent Aphanothece cells grown in 2.0 M NaCl and subjected to UV-B stress. Data are the means ± standard deviations of three independent experiments.
Identification of the MAA biosynthetic genes.
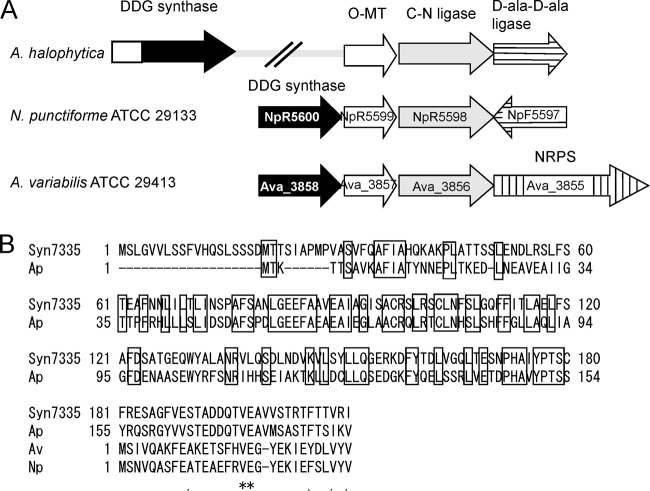
Using the shotgun sequence of A. halophytica, we searched for genes homologous to the clusters of Anabaena (Ava_3855 to Ava_3858) and Nostoc (NpR5600 to NpR5598 and NpF5597). Three identified genes, Ap3857, Ap3856, and Ap3855, were homologous to Ava_3857/NpR5599, Ava_3856/NpR5598, and NpF5597, respectively. As shown in Fig. 3A, these three genes are closely located. A. halophytica does not contain a gene homologous to Ava_3855 encoding nonribosomal peptide synthetase (NRPS) but contains a gene homologous to NpF5597 encoding d-Ala-d-Ala ligase. We designated the three genes of A. halophytica the Ap-OMT, Ap-CNligase, and Ap-AAligase genes, which were the same as those of Nostoc. It should be mentioned that the three genes (Ap3857, Ap3856, and Ap3855) of A. halophytica were transcribed in the same direction, whereas in Nostoc, NpF5597 was transcribed in the direction opposite that of NpR5599 and NpR5598 (Fig. 3A).
FIG 3.
(A) Gene organization for MAAs from A. halophytica, N. punctiforme ATCC 29133, and Anabaena variabilis ATCC 29413. (B) Alignment of N-terminal region of DDG synthase. The same amino acid residues in the upper region in A. halophytica (1 to 154) and Synechococcus sp. PCC 7335 (1 to 180) are boxed. The same amino acid residues among four DDGs after the first Met of Anabaena variabilis ATCC 29413 DDG are shown by asterisks.
The homology of the Ap-CNligase (Ap3856) and Ap-OMT (Ap3857) genes to the corresponding Anabaena and Nostoc genes was very high. The amino acid identities of the product of the Ap-OMT gene to those of Ava_3857 and NpR5599 were 63% and 61%, respectively. The amino acid identities of the product of the Ap-CNligase gene to those of Ava_3856 and NpR5598 were 61% and 60%, respectively. However, the homology of the Ap-AAligase (Ap3855) gene to NpR5597 was quite low; the amino acid identity of the product of the Ap-AAligase gene to that of NpR5597 was only 38%.
We could not find a sequence homologous to Ava_3858/NpR5600 in the upper region of the Ap-OMT gene (Ap3857). In contrast, a sequence similar to Ava_3858/NpR5600 was found at the distant end and is called the Ap-DDG gene (Ap3858). The protein Ap-DDG has an additional functionally unknown N-terminal domain similar to a homologue of Synechococcus sp. PCC 7335 (Fig. 3B) but does not show similarity with any known protein (15). Homology of the Ap-DDG gene to Ava_3858/NpR5600 was low. The amino acid sequence identities of Ap-DDG to the products of Ava_3858 and NpR5600 were 39% and 38%, respectively. In contrast, homology of Ap-DDG to that of Synechococcus sp. PCC 7335 was high. The amino acid identity of these two proteins was 65% if compared with the full-length sequence including the N-terminal extension, whereas it was 76% if compared excluding the N-terminal extension. The homology of N-terminal region between Ap-DDG and the Synechococcus sp. PCC 7335 protein is high, and its identity was 42%. Hitherto, it has not been examined whether DDG containing an extended N-terminal domain is really involved in MAA synthesis.
Heterologous expression of four putative genes of A. halophytica in Escherichia coli and a freshwater cyanobacterium, Synechococcus elongatus PCC 7942, resulted in the biosynthesis of mycosporine-2-glycine.
We further constructed a cluster of four putative MAA synthesis genes consisting of the Ap-DDG, Ap-OMT, Ap-CNligase, and Ap-AAligase genes in E. coli and Synechococcus elongatus PCC 7942. Both host cells were unable to synthesize any MAAs. Western blot analysis of the transformed cells showed the cross-reacting band of Ap-DDG in the E. coli cells (Fig. 4A, band 1). An induced and strongly cross-reacting band was clearly observed using the cells growing under the high-salinity condition (Fig. 4A). The relative intensity of bands increased to about 2.5-fold for Ap-DDG (Fig. 4B). A cross-reacting band for Ap-AAligase (Fig. 4A, band 2), could be observed only in the cells grown at 0.5 M NaCl with a relative faint intensity.
FIG 4.
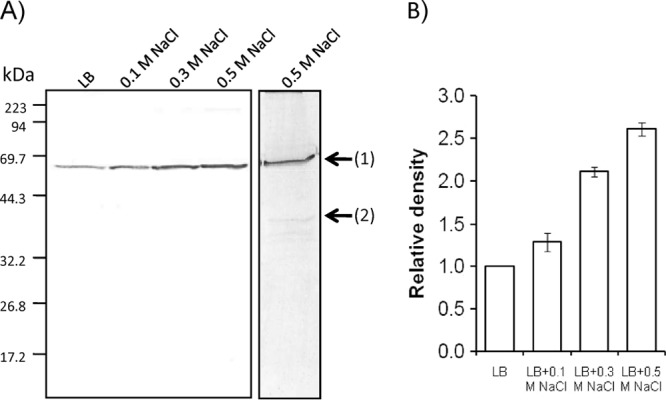
Western blot analysis of the Ap3858 and Ap3855 products. (A) Soluble fractions of E. coli grown at the indicated NaCl concentrations were prepared. A cross-reacting band, band 1, corresponded to Ap3858-His6, and band 2 corresponded to Ap3855-His6. Equal amounts of proteins (20 μg) were applied on SDS-PAGE and visualized by using the antibodies raised against the 6×His tag. Numerals on the left indicate molecular mass. (B) Quantification of immunoreactive bands. The intensity of the specific immunoreactive bands was analyzed by the GelQuant program.
HPLC analysis was carried out on the extract from the transformed cells. We were able to verify an identical retention time for MAAs as we could observe from A. halophytica (data not shown). Thus, we further analyzed the MAA accumulation level in E. coli cells. As shown in Fig. 5A, MAAs remained almost undetectable when E. coli cells were cultured only in LB medium. However, the MAA level increased significantly when cells were grown under the high-salinity condition. The MAA level reached to 85.2 ± 0.7 μmol/g (dry weight) when expressing cells were grown in LB medium supplemented with 0.6 M NaCl (Fig. 5A). Exposure to UV-B also induced the accumulation of MAAs, but the level was extremely low (approximately 0.56 ± 0.02 μmol/g [dry weight]), even after exposure for 3 days (Fig. 5B).
FIG 5.
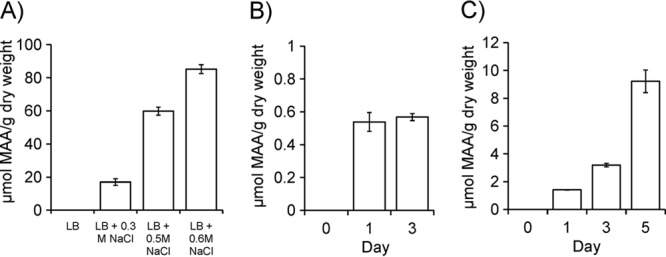
Accumulation levels of MAAs in E. coli and Synechococcus sp. PCC 7942 cells. (A) MAAs in E. coli cells grown with the different concentrations of NaCl. (B) MAAs in E. coli cells grown with exposure to UV-B. (C) MAAs in Synechococcus cells grown in BG11 plus 0.3 M NaCl. Data are the means ± standard deviations of three independent experiments.
Expression of Ap-MAA synthetase genes was analyzed in S. elongatus. Ap-DDG protein could be observed by Western blotting in the transformed cells expressing Ap-MAA synthetase genes but not in the cells expressing an empty vector (data not shown). Using HPLC analysis, we could detect MAA accumulation in S. elongatus cells transformed with Ap-MAA genes. Under the high-salinity condition (BG11 plus 0.3 M NaCl), the MAA level was approximately 9.2 ± 0.4 μmol/g (dry weight) (Fig. 5C).
Salt-dependent accumulation of mRNA for Ap-MAA.
The mRNA accumulation of the Ap-DDG, Ap-OMT, Ap-CNligase, and Ap-AAligase genes was examined in A. halophytica cells prepared under the salt upshock condition. Semiquantitative RT-PCR was performed on total RNA isolated at 0, 2, and 6 h after treatment with gene-specific primers. The obtained results are presented in Fig. 6, for which the relative expression of each gene was set to 1 at time zero. Expression of the four genes was found to be induced by salt upshock. An increase up to 8-fold was found for the Ap-DDG gene, while 4-, 3.5-, and 3-fold increases were observed for the Ap-OMT, Ap-CNligase, and Ap-AAligase genes, respectively, after salt upshock for 6 h.
FIG 6.
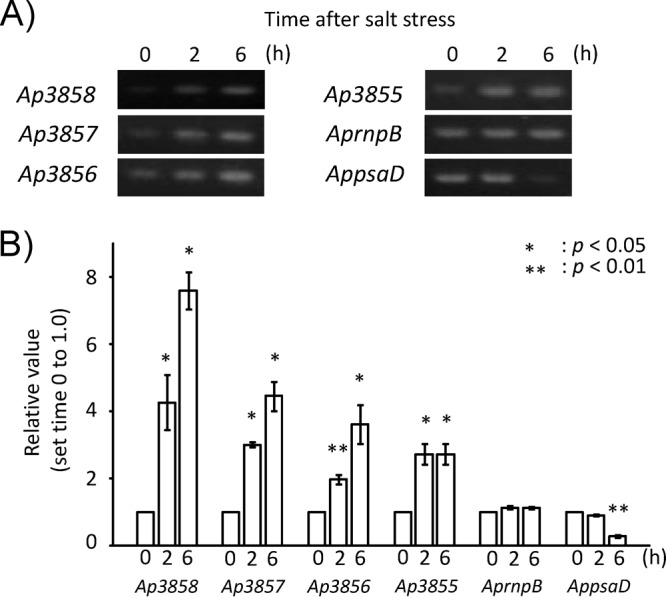
Expression of genes Ap3858 to Ap3855 in A. halophytica cells. Cells were collected at 0, 2, and 6 h of exposure to salinity stress. Semiquantitative RT-PCR analysis was performed as described in Materials and Methods. PCR products were subjected to electrophoresis (A), followed by calculation of relative values of the amount of DNA fragments (B). RNase P (rnpB) and photosystem I subunit II (psaD) genes were used as controls whose mRNA abundance remained unchanged under salinity stress. The values at time zero of each gene were set to 1. Data are the means ± standard deviations of three independent experiments. Asterisks indicate significant difference (P < 0.05 or P < 0.01) from the values at time zero.
DISCUSSION
The extremophilic cyanobacterium A. halophytica exists in highly salty and alkaline environments. We have previously shown that this microorganism possesses several adaptive mechanisms to thrive under hostile conditions (11, 16, 17, 18). For instance, A. halophytica has evolved a unique biosynthetic pathway for glycine betaine via glycine methylation, not the choline oxidation which occurs in most organisms (11). A. halophytica NhaP-type Na+/H+ antiporter also contains a novel ion specificity (17). The transfer of A. halophytica glycine betaine biosynthetic genes resulted in much higher glycine betaine biosynthesis and conferred greater salt tolerance on the freshwater S. elongatus PCC 7942 and nitrogen-fixing Anabaena sp. PCC 7120 as well as on Arabidopsis (11, 14, 19).
In this report, we provide evidence for MAA accumulation in A. halophytica. Using HPLC, amino acid, and TOF-MS analyses, mycosporine-2-glycine was identified in A. halophytica (Fig. 1). It has been reported that shinorine, mycosporine-glycine, porphyra-334, and asterina-330 were the most abundant MAAs found in freshwater and marine cyanobacteria (2, 20). To date, mycosporine-2-glycine has been identified as the major MAA only in the halophilic cyanobacterium Euhalothece (21), marine organisms such as the sea slug Aplysia dactylomela (22), and the green sea urchin Strongylocentrotus droebachiensis (23). The reason for the occurrence of mycosporine-2-glycine in halophilic strains remains unknown.
We also identified the genes involved in biosynthesis of MAAs in A. halophytica (Fig. 3). It was found that A. halophytica contains a unique organization of MAA biosynthetic genes. The Ap-DDG gene was separated from the other three genes for Ap-OMT, Ap-CNligase, and Ap-AAligase. Hitherto, functional expression of MAA genes has been reported for Anabaena variabilis ATCC 29413 (7) and Nostoc punctiforme ATCC 29133 (8). In both cases, four genes are arranged consecutively. Therefore, the present study is the first example to show that a far-separated gene, the Ap-DDG gene, was involved in the biosynthesis of an MAA.
Expression of Ap-MAA genes in E. coli produced mycosporine-2-glycine as a main MAA, whereas the expression of MAA synthesis genes of Anabaena variabilis ATCC 29413 produced shinorine as the main MAA (7, 8). Accumulation of different MAAs, shinorine in Anabaena variabilis ATCC 29413 and Nostoc punctiforme ATCC 29133 and mycosporine-2-glycine in A. halophytica, is interesting and needs to be discussed in relation to the homology of MAA synthesis genes.
The first gene has been reported to encode an enzyme, DDG synthase, which catalyzes the synthesis of 4-DG from SHP (7, 8, 15). Ap-DDG has an additional functionally unknown N-terminal domain (154 amino acid residues) and the homology of the full-length region of DDG (155 to 587 in Ap-DDG) to the Ava_3855 and NpR5600 products is low, with identities of only 38 to 39% (Fig. 3). We did not test the in vitro activity of purified Ap-DDG or the mutant with a deletion of the extended N-terminal region. These might be important to clarify the function of Ap-DDG, especially the role of the novel extended N-terminal region for the regulation of enzyme activity and/or substrate specificity.
The second gene encodes an O-MT catalyzing the methylation of DDG to 4-DG, while the third gene encodes a C-N ligase catalyzing the addition of glycine to 4-DG to form mycosporine-glycine. Mycosporine-glycine is believed to be the precursor of shinorine and mycosporine-2-glycine (8). Therefore, it is plausible that the Ap-OMT and Ap-CNligase genes catalyze the same reactions as Ava_3856/NpR5599 and Ava_3857/NpR5598, respectively. Indeed, the homologies of these enzymes, O-MT and C-N ligase, are very high among A. halophytica, Anabaena, and Nostoc; the identities are about 60 to 63% in these enzymes.
Recently, a fourth gene has been shown to catalyze the condensation of serine onto mycosporine-glycine, yielding shinorine for both Ava_3856 encoding NRPS and NpF5597 encoding d-Ala-d-Ala ligase (7, 8). The A. halophytica Ap-AAligase gene is homologous to that of Nostoc, NpF5597, but the product was different, shinorine for NpF5597 and mycosporine-2-glycine for the Ap-AAligase gene. This finding suggests that the substrate specificities of NpF5597 and the Ap-AAligase gene are different, namely, the condensation of serine onto mycosporine-glycine for NpF5597 and condensation of glycine onto mycosporine-glycine for the Ap-AAligase gene. Low homology between the Ap-AAligase gene and NpF5597, 38% identity, is compatible with different substrate specificities.
The concentration of MAAs generally shows a positive relationship with UV exposure (6, 24). However, our in vivo analysis showed that the accumulation of mycosporine-2-glycine was stimulated more under high salinity rather than under UV-B exposure (Fig. 2B). It has been reported MAA synthesis and accumulation can be induced by photosynthetically active radiation (PAR), UV-A, UV-B, osmotic stress, and nutrient composition (nitrogen and sulfur) (6, 25–27). Therefore, the present results support the notion that MAAs can play a role not only as an UV-absorbing compound but also as an osmoprotectant in response to salt stress (5). A. halophytica is a halotolerant cyanobacterium and accumulates a large amount of glycine betaine under high-salinity conditions such as 2.5 M NaCl. Glycine betaine is synthesized from glycine by a three-step methylation (11). Therefore, significant production of glycine is required in A. halophytica cells under high-salinity conditions. Molecular mechanisms of regulation of biosynthesis of glycine betaine and mycosporine-2-glycine are interesting topics for further investigation and clarification. In this study, we have successfully generated E. coli and Synechococcus strains to produce mycosporine-2-glycine via the introduction of A. halophytica MAA biosynthetic genes. A set of recombinant plasmids containing five different promoters, PT7, Plac, Ptrc, PcspA, and Pnative, was generated to facilitate expression of foreign genes. Among them, a native Aphanothece promoter seems to be most suitable for high accumulation of mycosporine-2-glycine. Heterologous expression in E. coli showed the significant increase of mycosporine-2-glycine under salt stress (Fig. 5). In Synechococcus, we could detect the expression of Ap-DDG by Western blotting with a similar trend as in E. coli (data not shown), but the product mycosporine-2-glycine was less produced (Fig. 5). Although the reason for this is unclear, availability of precursor and substrate might be one factor.
To summarize, we report the accumulation of MAAs in A. halophytica and identification of its biosynthetic genes. Heterologous expression of Aphanothece MAA biosynthetic genes in E. coli and Synechococcus cells produced a large amount of MAAs (85.2 ± 0.7 μmol/g [dry weight]) (Fig. 5). The results not only highlighted the unique genes for MAAs in A. halophytica but also will offer a new opportunity to provide alternative routes for commercial production.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Science and Culture of Japan and the International Center for Green Biotechnology of Meijo University to T.T.
We thank K. Ishihara (National Research Institute of Fisheries Science) for the generous gift of shinorine, porphyra-334, and palythine.
We have no conflict of interest to declare.
Footnotes
Published ahead of print 27 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03729-13.
REFERENCES
- 1.Shick JM, Dunlap WC. 2002. Mycosporine-like amino acids and related gadusols: biosynthesis, acumulation, and UV-protective functions in aquatic organisms. Annu. Rev. Physiol. 64:223–262. 10.1146/annurev.physiol.64.081501.155802 [DOI] [PubMed] [Google Scholar]
- 2.Carreto JI, Carignan MO. 2011. Mycosporine-like amino acids: relevant secondary metabolites. Chemical and ecological aspects. Mar. Drugs 9:387–446. 10.3390/md9030387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llewellyn CA, White DA, Martinez-Vincente V, Tarran G, Smyth TJ. 2012. Distribution of mycosporine-like amino acids along a surface water meridional transect of the Atlantic. Microb. Ecol. 64:320–333. 10.1007/s00248-012-0038-6 [DOI] [PubMed] [Google Scholar]
- 4.Gao Q, Garcia-Pichel F. 2011. Microbial ultraviolet sunscreens. Nat. Rev. Microbiol. 9:791–802. 10.1038/nrmicro2649 [DOI] [PubMed] [Google Scholar]
- 5.Oren A, Gunde-Cimerman N. 2007. Mycosporines and mycosporine-like amino acids: UV protectants or multipurpose secondary metabolites? FEMS Microbiol. Lett. 269:1–10. 10.1111/j.1574-6968.2007.00650.x [DOI] [PubMed] [Google Scholar]
- 6.Singh SP, Klisch M, Sinha RP, Häder DP. 2008. Effects of abiotic stressors on synthesis of the mycosporine-like amino acid shinorine in the cyanobacterium Anabaena variabilis PCC 7937. Photochem. Photobiol. 84:1500–1505. 10.1111/j.1751-1097.2008.00376.x [DOI] [PubMed] [Google Scholar]
- 7.Balskus EP, Walsh CT. 2010. The genetic and molecular basis for sunscreen biosynthesis in cyanobacteria. Science 329:1653–1656. 10.1126/science.1193637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao Q, Garcia-Pichel F. 2011. An ATP-grasp ligase involved in the last biosynthetic step of the iminomycosporine shinorine in Nostoc punctiforme ATCC 29133. J. Bacteriol. 193:5923–5928. 10.1128/JB.05730-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oren A. 1997. Mycosporine-like amino acids as osmotic solutes in a community of halophilic cyanobacteria. Geomicrobiol. J. 14:231–240. 10.1080/01490459709378046 [DOI] [Google Scholar]
- 10.Laloknam S, Tanaka K, Buaboocha T, Waditee R, Incharoensakdi A, Hibino T, Tanaka Y, Takabe T. 2006. Halotolerant cyanobacterium Aphanothece halophytica contains a betaine transporter active at alkaline pH and high salinity. Appl. Environ. Microbiol. 72:6018–6026. 10.1128/AEM.00733-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waditee R, Tanaka Y, Aoki K, Hibino T, Jikuya H, Takano J, Takabe T, Takabe T. 2003. Isolation and functional characterization of N-methyltransferases that catalyze betaine synthesis from glycine in a halotolerant photosynthetic organism Aphanothece halophytica. J. Biol. Chem. 278:4932–4942. 10.1074/jbc.M210970200 [DOI] [PubMed] [Google Scholar]
- 12.Stochaj WR, Dunlap WC, Shick JM. 1994. Two new UV-absorbing mycosporine-like amino acids from the sea anemone Anthopleura elegantissima and the effects of zooxanthellae and spectral irradiance on chemical composition and content. Mar. Biol. 118:149–156. 10.1007/BF00699229 [DOI] [Google Scholar]
- 13.Nomura M, Ishitani M, Takabe T, Rai AK, Takabe T. 1995. Synechococcus sp. PCC7942 transformed with Escherichia coli bet genes produces glycine betaine from choline and acquires resistance to salt stress. Plant Physiol. 107:703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waditee R, Bhuiyan MN, Rai V, Aoki K, Tanaka Y, Hibino T, Suzuki S, Takano J, Jagendorf AT, Takabe T, Takabe T. 2005. Genes for direct methylation of glycine provide high levels of glycinebetaine and abiotic-stress tolerance in Synechococcus and Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 102:1318–1323. 10.1073/pnas.0409017102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SP, Häder DP, Sinha RP. 2012. Bioinformatics evidence for the transfer of mycosporine-like amino acid core (4-deoxygadusol) synthesizing gene from cyanobacteria to dinoflagellates and an attempt to mutate the same gene (YP_324358) in Anabaena variabilis PCC 7937. Gene 500:155–163. 10.1016/j.gene.2012.03.063 [DOI] [PubMed] [Google Scholar]
- 16.Hibino T, Kaku N, Yoshikawa H, Takabe T, Takabe T. 1999. Molecular characterization of DnaK from the halotolerant cyanobacterium Aphanothece halophytica for ATPase, protein folding, and copper binding under various salinity conditions. Plant Mol. Biol. 40:409–418. 10.1023/A:1006273124726 [DOI] [PubMed] [Google Scholar]
- 17.Waditee R, Hibino T, Tanaka Y, Nakamura T, Incharoensakdi A, Takabe T. 2001. Halotolerant cyanobacterium Aphanothece halophytica contains an Na+/H+ antiporter, homologous to eukaryotic ones, with novel ion specificity affected by C-terminal tail. J. Biol. Chem. 276:36931–36938. 10.1074/jbc.M103650200 [DOI] [PubMed] [Google Scholar]
- 18.Fukaya F, Promden W, Hibino T, Tanaka Y, Nakamura T, Takabe T. 2009. An Mrp-like cluster in the halotolerant cyanobacterium Aphanothece halophytica functions as a Na+/H+ antiporter. Appl. Environ. Microbiol. 75:6626–6629. 10.1128/AEM.01387-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waditee-Sirisattha R, Singh M, Kageyama H, Sittipol D, Rai AK, Takabe T. 2012. Anabaena sp. PCC7120 transformed with glycine methylation genes from Aphanothece halophytica synthesized glycine betaine showing increased tolerance to salt. Arch. Microbiol. 194:909–914 [DOI] [PubMed] [Google Scholar]
- 20.Volkmann M, Gorbushina AA. 2006. A broadly applicable method for extraction and characterization of mycosporines and mycosporine-like amino acids of terrestrial, marine and freshwater origin. FEMS Microbiol. Lett. 255:286–295. 10.1111/j.1574-6968.2006.00088.x [DOI] [PubMed] [Google Scholar]
- 21.Kedar L, Kashman Y, Oren A. 2002. Mycosporine-2-glycine is the major mycosporine-like amino acid in a unicellular cyanobacterium (Euhalothece sp.) isolated from a gypsum crust in a hypersaline saltern pond. FEMS Microbiol. Lett. 208:233–237. 10.1111/j.1574-6968.2002.tb11087.x [DOI] [PubMed] [Google Scholar]
- 22.Corredor JE, Bruckner AW, Muszynski FZ, Armstrong RA, García R, Morell JM. 2000. UV-absorbing compounds in three species of Caribbean zooxanthellate corals: depth distribution and spectral response. Bull. Mar. Sci. 67:821–830 [Google Scholar]
- 23.Adams NL, Shick JM. 2001. Mycosporine-like amino acids prevent UVB-induced abnormalities during early development of the green sea urchin Strongylocentrotus droebachiensis. Mar. Biol. 138:267–280. 10.1007/s002270000463 [DOI] [Google Scholar]
- 24.Carefoot TH, Karentz D, Pennings SC, Young CL. 2000. Distribution of mycosporine-like amino acids in the sea hare Aplysia dactylomela: effect of diet on amounts and types sequestered over time in tissues and spawn. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 126:91–104 [DOI] [PubMed] [Google Scholar]
- 25.Litchman E, Neale PJ, Banaszak AT. 2002. Increased sensitivity to ultraviolet radiation in nitrogen-limited dinoflagellates: photoprotection and repair. Limnol. Oceanogr. 47:86–94. 10.4319/lo.2002.47.1.0086 [DOI] [Google Scholar]
- 26.Portwich A, Garcia-Pichel F. 2003. Biosynthetic pathway of mycosporines (mycosporine-like amino acids) in the cyanobacterium Chlorogloeopsis sp. strain PCC 6912. Phycologia 42:384–392. 10.2216/i0031-8884-42-4-384.1 [DOI] [Google Scholar]
- 27.Singh SP, Klisch M, Sinha RP, Häder DP. 2010. Sulfur deficiency changes mycosporine-like amino acid (MAA) composition of Anabaena variabilis PCC 7937: a possible role of sulfur in MAA bioconversion. Photochem. Photobiol. 86:862–870. 10.1111/j.1751-1097.2010.00736.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.