Abstract
The objective of this study was to characterize fungal communities in a subsurface environment cocontaminated with uranium and nitrate at the watershed scale and to determine the potential contribution of fungi to contaminant transformation (nitrate attenuation). The abundance, distribution, and diversity of fungi in subsurface groundwater samples were determined using quantitative and semiquantitative molecular techniques, including quantitative PCR of eukaryotic small-subunit rRNA genes and pyrosequencing of fungal internal transcribed spacer (ITS) regions. Potential bacterial and fungal denitrification was assessed in sediment-groundwater slurries amended with antimicrobial compounds and in fungal pure cultures isolated from the subsurface. Our results demonstrate that subsurface fungal communities are dominated by members of the phylum Ascomycota, and a pronounced shift in fungal community composition occurs across the groundwater pH gradient at the field site, with lower diversity observed under acidic (pH <4.5) conditions. Fungal isolates recovered from subsurface sediments, including cultures of the genus Coniochaeta, which were detected in abundance in pyrosequence libraries of site groundwater samples, were shown to reduce nitrate to nitrous oxide. Denitrifying fungal isolates recovered from the site were classified and found to be distributed broadly within the phylum Ascomycota and within a single genus of the Basidiomycota. Potential denitrification rate assays with sediment-groundwater slurries showed the potential for subsurface fungi to reduce nitrate to nitrous oxide under in situ acidic pH conditions.
INTRODUCTION
The subsurface of U.S. Department of Energy (U.S. DOE) nuclear legacy waste sites is often cocontaminated with uranium and nitrate (1). Nitrate is a prevalent groundwater contaminant on a global scale (2) and, in particular, represents a major contaminant at nuclear waste sites because of the use of nitric acid during uranium processing (3). The natural attenuation and remediation of nitrate (NO3−) found in subsurface environments are believed to be largely determined by microbial denitrification coupled with dilution (3, 4). Denitrification in the subsurface is likely regulated by pH and electron donor concentration (5).
Extensive geochemical and microbiological evidence points to denitrification as a major microbial respiration process in the subsurface of the Oak Ridge Integrated Field Research Challenge (OR-IFRC) site, a nuclear legacy waste site managed by the U.S. DOE (e.g., see references 6 to 9). High nitrate concentrations in the OR-IFRC site subsurface, coupled with periodic anoxia, provide selective pressures that favor nitrate-reducing organisms (8, 10, 11). The stable nitrogen isotopic composition of the gaseous products of denitrification, nitrous oxide (N2O) and nitrogen gas (N2), indicates that substantial denitrification is occurring (Juske Horita, personal communication). For example, the dissolved gas composition of groundwaters closest to the contaminant source zone is enriched in N2O relative to the atmospheric composition, and these levels ameliorate down from the plume, along with the pH and nitrate and uranium levels (12).
Fungi mediate key reactions in the biogeochemical cycles of carbon, nutrients, and metals (13). Fungi are known to be metal resistant and occur in abundance in acidic, metal-rich environments observed in terrestrial ecosystems, such as aquifers (e.g., see references 14 and 15); thus, they could play an important role in contaminant transformation in the subsurface of U.S. DOE sites. Traits that favor the survival of fungi under the extreme conditions found in the radionuclide-contaminated acidic subsurface include facultative anaerobic growth, resistance to acidic pH, resistance to toxic metals, spore formation, and in some cases, radiation-enhanced growth (14, 16). Additionally, previous research has also demonstrated a role of fungi in denitrification (17, 18).
Prior research on fungal nitrogen transformation has demonstrated the nitrate and nitrite reduction capabilities of a very limited number of fungal taxa within the major phyla Ascomycota and Basidiomycota, including both filamentous and yeast forms (17, 19–22). Fungi can denitrify nitrate or nitrite via nitric oxide to nitrous oxide or reduce nitrite to ammonium via dissimilatory nitrate reduction to ammonium (DNRA) coupled to the oxidation of organic compounds (17, 18, 23). There is no evidence of the presence of nitrous oxide reductase and complete denitrification to N2 in fungi, but fungi may produce N2 through codenitrification (17, 19, 24).
Although fungi have been shown to mediate denitrification in surficial soils (25–27), little information on fungal denitrification in subsurface environments is available. To our knowledge, the occurrence of fungi has been described for few pristine and contaminated aquifers (28, 29), and the community composition of these microbial eukaryotes in subsurface environments remains poorly characterized. We hypothesized that the structure and composition of the subsurface fungal community are impacted by the subsurface pH gradient across the contaminated watershed and that this group of organisms contributes significantly to in situ denitrification in the contaminated subsurface, in part due to a greater tolerance of acidity by many fungi relative to that by bacteria (14, 15). Thus, the objective of this study was to interrogate subsurface fungal communities through a close coupling of microbiological and biogeochemical approaches. Fungal communities were characterized at the watershed scale using quantitative and semiquantitative molecular analyses, including quantitative PCR (qPCR) and pyrosequencing of PCR-amplified fungal internal transcribed spacer (ITS) regions. Potential denitrification activity was determined in sediment slurry experiments and in pure cultures of fungi isolated from the subsurface.
MATERIALS AND METHODS
Site and sampling description.
Sampling was conducted at the Oak Ridge Integrated Field Research Challenge (OR-IFRC) site, Oak Ridge, TN, where the subsurface has been widely contaminated with a diverse array of mixed contaminants, including uranium and nitrate (http://www.esd.ornl.gov/orifrc/). Groundwater was collected from wells in November 2008 and May 2009 across a contaminant gradient (e.g., see reference 30). Briefly, approximately 2 liters of groundwater was collected separately for geochemical analysis (for which the water was filtered); microbiological studies, including cultivation (for which the water was unfiltered); and molecular analysis (for which the water was collected on filters). Groundwater was collected after purging of the wells until pH, redox (Eh), and temperature were stabilized, pumped through a peristaltic pump, and passed sequentially through two 142-mm Geotech filter holders (Geotech, Denver, CO) using a 3.0-μm-pore-size Versapore prefilter and a 0.2-μm-pore-size Supor sample filter (Pall, Port Washington, NY). A 3.0-μm-pore-size prefilter was used to prevent clogging of the 0.2-μm-pore-size filter membrane due to suspended minerals and sediment particles. Filters bearing biomass were stored frozen on dry ice in the field and then at −80°C until nucleic acid extraction. In addition, groundwater wells were surged and sediment-groundwater slurries were collected from near the contaminant source zone in October 2011. Slurries were collected in the field in sterile preevacuated and sealed serum vials and shipped overnight on ice to a laboratory at Florida State University (FSU; Tallahassee, FL). Samples were stored at 4°C until potential rate incubations were established. Biogeochemical analyses for pH, nitrate, nitrite, and ammonium were conducted on groundwater samples as described previously (31). Other geochemical parameters (uranium [U], total organic carbon [TOC], N2O, oxygen [O2]) were measured during sample collection and are recorded at the OR-IFRC site database (http://www.esd.ornl.gov/orifrc/).
DNA extraction, pyrosequencing, and qPCR.
Genomic DNA (gDNA) was extracted from filtered groundwater samples (0.2-μm-pore-size filters) with a Mo Bio PowerWater DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's recommendations. Genomic DNA recovered from 39 groundwater samples from two sampling seasons and 20 sites across the watershed was subject to pyrotag sequencing of the fungal ITS region to examine fungal community structure and distribution. Fungal amplicons were sequenced on a Roche 454 pyrosequencing instrument by Research and Testing Laboratories (Lubbock, TX); amplifications were performed using the primer pair ITS1-F and ITS4 (32, 33). The abundance of small-subunit (SSU) rRNA genes from fungi and other eukaryotic organisms in groundwater DNA extracts was determined using qPCR. Genomic DNAs extracted from groundwater samples were used as the templates for the TaqMan 18S rRNA endogenous control assay (Life Technologies, Foster City, CA) according to the manufacturer's instructions. Briefly, the PCR was performed in a total volume of 20 μl using TaqMan gene expression master mix (Life Technologies, Foster City, CA), 100 μM each forward and reverse primers, and a 6-carboxyfluorescein (6-FAM) fluorophore-labeled probe. The reaction conditions for amplification were 50οC for 2 min, 95οC for 10 min, and 40 cycles of 95οC for 15 s and 60οC for 1 min. Similarly, gene abundance data for bacterial SSU rRNA genes were generated as described previously (31, 34). Standards for the eukaryotic assay were PCR products generated by amplification of nearly complete fungal SSU rRNA genes using the general fungal primer set NS1/FR1 (33) and the primer set 27F/1492R for bacteria (35). qPCR assays were performed in triplicate using an ABI StepOne real-time PCR system.
Processing of pyrosequencing data.
All Roche 454 sequencing data were analyzed within the software package QIIME (36). Pyrotag sequences were first binned and filtered to remove low-quality sequences shorter than 300 bases in length and with average quality scores below 25. Sequences without the correct primer sequences were excluded. A work-flow script was used to cluster sequences into operational taxonomic units (OTUs) at a 97% sequence identity level, ultimately building OTU tables. Taxonomy was assigned to each representative sequence by using the BLAST program with a maximum E value of 0.001 against the BLAST database (http://www.emerencia.org/fungalitspipeline.html) within QIIME. For further OTU-based analyses, the original OTU table was rarefied to a depth of 2,500 fungal sequences per sample, to avoid the distorting effects of variable sampling depth (37).
Statistical analyses and diversity estimation.
Geochemical parameters (NO3−, U, TOC, N2O, O2) were log transformed to meet assumptions of normality using the Ryan-Joiner normality test (38). Transformed geochemical data, along with log-transformed quantitative data of microbial SSU rRNA gene abundance, were analyzed using principal component analysis (PCA) within the software package Primer 6 (39, 40). Fungal community composition was also related to geochemical variables using canonical correspondence analysis (CCA). CCA was performed using the XLSTAT-ADA (Addinsoft, New York, NY) add-in for the Microsoft Excel program. Shannon indices (41) were calculated for bacterial and fungal communities on the basis of rarefied OTU tables of SSU rRNA gene and ITS region sequences, respectively. Pairwise comparisons of microbial community structure were computed as Bray-Curtis distances and were visualized using PCA within the software package Primer 6 (40). To test for significant differences in means, t tests were implemented within the software package Sigma Plot (v12; SyStat, San Jose, CA).
Isolation of subsurface fungi.
Highly contaminated groundwater and sediment samples were selected for isolation of fungi (Table 1). For this purpose, 0.1 ml of groundwater or suspended sediment (a 1:10 [wt/vol] mixture of sediment in a minimal salts solution) was aseptically spread onto a variety of fungal cultivation media, including potato dextrose agar (PDA), Czapek Dox agar, cornmeal agar, Sabouraud dextrose agar, and malt extract agar (HiMedia Laboratories, India). Neutral pH was adjusted to 5.0 using HCl to approximate the pH of the OR-IFRC site subsurface. In order to inhibit bacterial growth, media were supplemented with 1 mg/ml streptomycin and 0.05 mg/ml chloramphenicol. Plates were incubated in the dark at room temperature. After visible growth appeared, morphologically distinct colonies were transferred to fresh medium using sterile toothpicks. During each transfer, hyphae from the leading edge of the fungal colonies were transferred in order to reduce the possibility of cross contamination, and pure cultures were obtained from repeated transfers. The purity of the cultures was determined by visual inspection of the colonies for dimorphic growth and reconfirmed by PCR amplification with fungal SSU rRNA gene primers (NS1/FR1 [33]) and direct amplicon sequencing. Pure cultures of the isolated fungal strains were maintained in 20% glycerol at −80°C and on plates for future study.
TABLE 1.
Description of isolated fungi, including isolation source, geochemistry of the site, and fungal taxonomy based on SSU rRNA genes
| Closest BLAST class (phylum), genus match | Accession no. | % identity | Isolate(s) | Sampling wella | pH | % abundance from wells with pH ofb: |
Denitrification potential (nmol N2O-N liter−1 day−1) | |
|---|---|---|---|---|---|---|---|---|
| <4.5 | >4.5 | |||||||
| Eurotiomycetes (Ascomycota) | ||||||||
| Penicillium sp. strain BCC 17468 | GU809208 | 99 | ORNL9 | FW106 | 3.46 | 0.01 | 9,852.64 | |
| ORNL15 | FW025 | 5.14 | ||||||
| ORNL10 | FW116 | 5.51 | ||||||
| ORNL21 | FW410 | 3.30 | ||||||
| ORNL19 | FW115 | 3.20 | ||||||
| Aspergillus fumigatus DAOM 215394 | JN938984 | 99 | ORNL2 | FW116 | 5.51 | 0.02 | 0.002 | 7.50 |
| Sordariomycetes (Ascomycota) | ||||||||
| Coniochaeta velutinac | GQ154626 | 100 | ORNL3,d ORNL4, ORNL8 | FW106 | 3.46 | 18.5 | 119.24 | |
| ORNL5 | FW126 | 3.57 | ||||||
| Fusarium oxysporum ZW-21 | JQ926985 | 99 | ORNL7 | FWB124 | 5.14 | 0.01 | 523.59 | |
| Pochonia suchlasporia | AB214658 | 99 | ORNL11 | PTMW02 | 4.70 | |||
| Apiospora montagnei | AB220230 | 99 | ORNL17 | FWB129 | 3.89 | 0.30 | 83.08 | |
| Paecilomyces lilacinus CBS 284.36 | AY526475 | 99 | ORNL16 | FWB129 | 4.15 | 0.01 | 397.44 | |
| Neolinocarpon globosicarpum | DQ810258 | 100 | ORNL6 | FW025 | 5.14 | 19.36 | ||
| Leotiomycetes (Ascomycota) | ||||||||
| Teberdinia hygrophilac | JQ780647 | 99 | ORNL1,d ORNL12 | FW126 | 3.57 | 0.01 | 256.08 | |
| Sclerotinia sclerotiorum | AY187065 | 98 | ORNL20 | FW410 | 3.30 | |||
| Phialocephala repens MUCL1849 | EU434874 | 99 | ORNL18 | FWB129 | 3.90 | 0.01 | 8.47 | |
| Collophora capensis | GQ154631 | 98 | ORNL13 | FW106 | 3.46 | |||
| Microbotryomycetes (Basidiomycota), Rhodosporidium toruloides | DQ647614 | 99 | ORNL14 | FW106 | 3.46 | 11.5 | ||
FW and PTMW are OR-IFRC groundwater sources, and FWB is an OR-IFRC sediment source.
Percent abundance of the genus in the pyrosequencing library.
Multiple isolates were recovered.
Isolate tested for denitrification potential.
Measurement of potential denitrification rates.
Potential denitrification rates were determined for fungal isolates and for sediment-groundwater slurries. Isolates were grown on half-strength Czapek Dox agar (adjusted to pH 5.0) for 36 to 48 h at room temperature (20°C). Once sufficient growth was achieved, a 1-cm-diameter circular subsample was cored from the edge of the growing culture using a cork borer under aseptic technique. The cored agar was immersed in 100 ml half-strength Czapek Dox broth, and the culture was grown for 7 days with constant shaking at room temperature (20°C). For denitrification assays, cultures were inoculated in triplicate into 60 ml Czapek Dox broth at 10% (vol/vol) with 10 mM nitrate. The vials were purged with 100% N2, sealed using gastight rubber stoppers, and incubated at room temperature (20°C). Autoclaved control cultures were prepared and incubated under similar conditions. The headspace in each slurry bottle was then sampled with a gastight 100-μl syringe, and N2O concentrations were determined with a Shimadzu GC-8a gas chromatograph (GC) equipped with an electron-capture detector (Shimadzu Scientific Instruments, MD). Dissolved oxygen was determined with Shimadzu TCD 8A GC employing a Mol Sieve 5A column and argon as the carrier gas. Dissolved N2O and oxygen concentrations were calculated as described above and by use of the solubility coefficients described previously (42), and total N2O and O2 was calculated as the sum of the headspace and dissolved N2O and O2, respectively. Microcosms were also sampled for dissolved inorganic nitrogen (DIN; nitrate, nitrite, and ammonium) and biomass at the beginning and end of the incubations. Nitrate plus nitrite (NOx) and nitrite were reduced with vanadium (43) and iodide (44), respectively, and the resulting nitric oxide gas concentration was determined with a Thermo model 42i chemiluminescence analyzer (Thermo Fisher Scientific, WA). The nitrate concentration was calculated as the difference between the NOx and nitrite concentrations. Dissolved ammonium was determined colorimetrically (45). For biomass estimates, triplicate cultures were filtered onto preweighed 0.45-μm-pore-size nitrocellulose filters and weighed after drying for 24 h.
Sediment slurry microcosms were established in triplicate 60-ml serum vials. Thirty milliliters of sediment-groundwater mix was dispensed aseptically after vigorous shaking. Each microcosm was supplemented with 1 mM sodium nitrate and glucose, purged with high-purity N2 (Airgas, GA), and sealed using gastight rubber stoppers. Microcosm bottles were shaken well and incubated statically at room temperature (20°C). Sediment slurries were treated with both streptomycin and chloramphenicol (1 mg ml−1) to inhibit bacterial activity or cycloheximide (1.5 mg ml−1) to inhibit fungal activity. Control treatment mixtures without microbial inhibitors (control) were incubated under identical conditions. Potential denitrification rates in sediment slurries were estimated using the acetylene block technique (46). Each microcosm was amended with 10% (vol/vol) acetylene in the headspace. The N2O headspace concentrations for the cultures were determined as described above. The dry weight of the sediment was calculated for each vial after the incubations were completed by drying in an oven at 80οC. All rates were standardized to the dry weight of the sediment (see Table S2 in the supplemental material).
Nucleotide sequence accession numbers.
SSU rRNA gene sequences were submitted to GenBank under accession numbers KF971835 to KF971855. Sequence data were submitted to the Sequence Read Archive (SRA) under the study accession number SRP034562.
RESULTS
Composition of fungal community across the watershed.
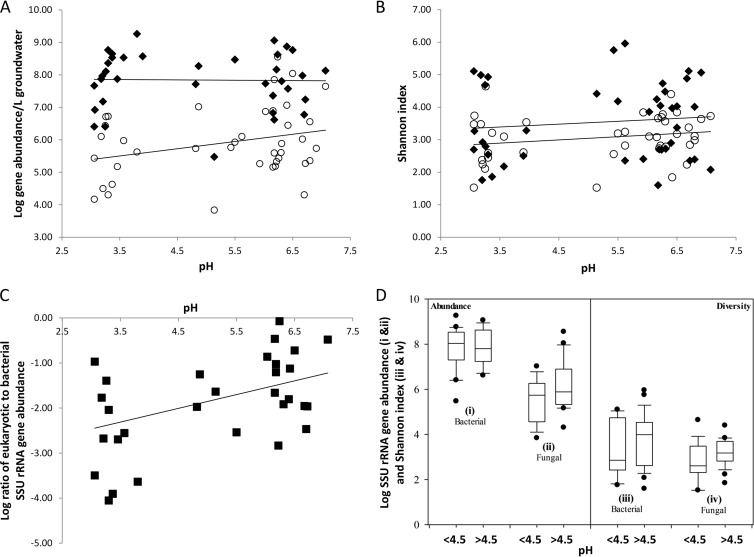
The distribution and abundance of fungi were examined by quantitative analysis of SSU rRNA gene abundance and ITS amplicon sequencing performed on gDNA extracts of groundwater sampled from 20 wells across the contaminant gradient and during two sampling seasons. Eukaryotic and bacterial SSU rRNA gene abundance was quantified using qPCR. The sampling sites ranged from highly acidic and nitrate rich close to the contaminant source zone (area 3) to circumneutral with lower contaminant levels downgradient from the source zone (area 2). Total eukaryote SSU rRNA gene abundance ranged from 1.48 × 104 to 1.04 × 107 copies per liter of groundwater for acidic sites (pH <4.5) and 6.87 × 103 to 3.55 × 108 copies per liter of groundwater for circumneutral sites (pH >4.5) (Fig. 1A). Total bacterial SSU rRNA gene abundance ranged from 2.5 × 106 to 1.8 × 109 copies per liter of groundwater for acidic sites (pH <4.5) and 2.98 × 105 to 1.14 × 109 copies per liter of groundwater for circumneutral sites (pH >4.5) (Fig. 1A) (31). An increase in total eukaryotic SSU rRNA gene abundance was observed with increasing pH (Fig. 1A). This occurred within the context of relatively stable bacterial SSU rRNA gene abundance across the pH gradient, resulting in a trend of increasing relative abundance of eukaryotic to bacterial SSU rRNA genes across the pH gradient (Fig. 1C). Bacterial SSU rRNA gene abundance exceeded eukaryotic SSU rRNA gene abundance by approximately 1 to 4 orders of magnitude at low pH (pH <4.5), while in most wells with a pH of >4.5, this ratio was generally between 1 and 2 orders of magnitude.
FIG 1.
Groundwater microbial diversity and abundance across the watershed. (A) Comparison of SSU rRNA gene abundance of total eukaryotes (open circles) and total bacteria (filled diamonds) in groundwater samplings from November 2008 and May 2009 plotted against pH. The slope of the regression line of total eukaryote SSU rRNA gene abundance is 0.22 (R2 = 0.08), and the slope of the regression line for bacterial SSU rRNA gene abundance is 0.003 (R2 = 0.01). (B) Fungal and bacterial diversity across the watershed compared on the basis of the number of OTUs determined from amplicon sequencing of fungal ITS regions (open circles) and SSU rRNA from bacteria (black diamonds). The linear regression for the analysis is shown (for fungi, slope = 0.03 and R2 = 0.05; for bacteria, slope = 0.1 and R2 = 0.01). (C) Ratio of eukaryotic to bacterial SSU rRNA gene abundance in groundwater samples across the watershed plotted against pH. The slope of the regression line is 0.3 (R2 = 0.2). (D) Box plots of the gene abundances of bacterial and total eukaryotic SSU rRNA (i and ii) and bacterial and fungal diversity (Shannon index) (iii and iv) by pH. Two plots were generated for each analysis, one for groundwater samples with pH values below 4.5 and one for samples with pH values above 4.5. The data are plotted as the 10th, 25th 75th, and 95th percentiles. No significant differences for gene abundance values (for bacterial SSU rRNA, P = 0.959; for eukaryotic SSU rRNA, P = 0.058) and diversity indices (for fungi, P = 0.087; for bacteria, P = 0.256) of low- and neutral-pH sites were observed.
Diversity (Shannon) indices were calculated for fungal OTUs from each groundwater sample classified at 97% sequence similarity. Fungal ITS sequence libraries from acidic (pH <4.5) groundwater samples had an average Shannon index value of 2.89 (n = 13; range, 1.52 to 4.64), while groundwater samples with a pH of >4.5 had an average Shannon index value of 3.15 (n = 26; range, 1.53 to 3.84). A slight trend of increasing diversity was observed with increasing pH (Fig. 1B and D) for fungal communities (slope = 0.03, R2 = 0.05). A significant difference (P = 0.049, Student's t test) in the ratio between total eukaryotic and bacterial abundances was observed by pH and total eukaryotic abundance (P = 0.05, Student's t test). No significant difference, however, in total SSU rRNA gene abundances (for bacterial gene abundance, P = 0. 959; for eukaryotic gene abundance, P = 0.058) or diversity indices (for fungi, P = 0.087; for bacteria, P = 0.256) for sites with low and neutral pHs was observed over the range of pHs sampled and the fungal (P = 0. 087) and bacterial (P = 0.256) diversity detected (Fig. 1D).
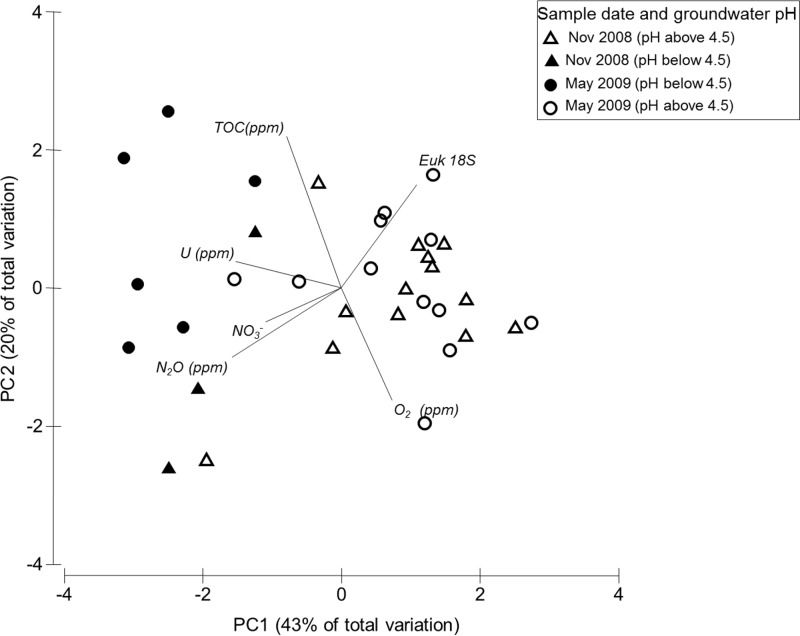
PCA was performed to examine the relationship between total eukaryotic SSU rRNA gene abundance with measured variables in the groundwater (U, TOC, O2, NO3−, and N2O) (Fig. 2; see Table S1 in the supplemental material). Strong separation of samples by pH was observed, despite substantial subsurface heterogeneity. The first principal component (PC) axis accounted for 43% of the measured variation; the U concentration load was primarily on PC axis 1 (PC1). TOC and O2 loads were primarily on PC axis 2 (PC2; 20% of the measured variation), while eukaryotic SSU rRNA gene abundance and NO3− and N2O loads were on PC axes 1 and 2.
FIG 2.
Principal component analysis of select groundwater geochemical variables with total eukaryotic SSU rRNA gene abundance. Vectors represent variables used to generate Euclidean distance. Samples are coded with groundwater pH ranges and season, as indicated in the key. PC1 and PC2 explain 43% and 20% of the variation, respectively. Euk 18S, eukaryotic 18S rRNA.
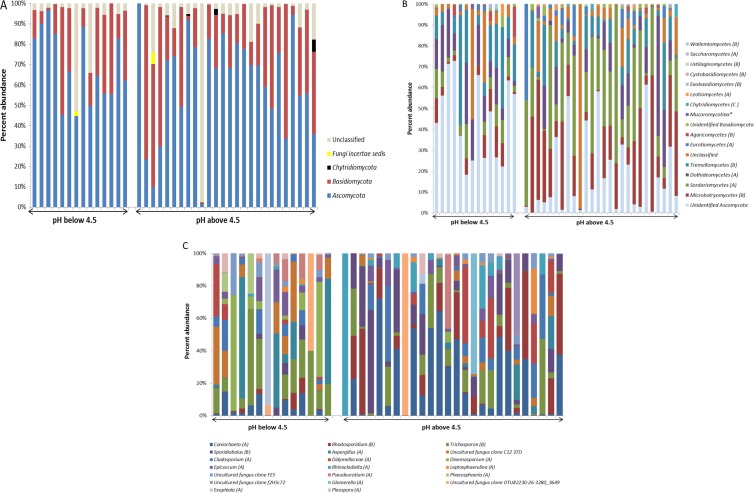
Sequencing of fungal ITS amplicons using the Roche 454 pyrosequencing platform was performed on 39 groundwater samples from two sampling seasons and 20 wells across the watershed. In total, 204,886 sequences were analyzed (average number of sequences per sample = 3,260; range = 2,500 to 7,653). The sequences were clustered at a 97% similarity threshold into 3,379 OTUs. The distributions of the most abundant taxa identified are shown in Fig. 3. Ninety-three percent of the recovered ITS sequences could be classified at the phylum level, while the remainder could not be classified within the QIIME pipeline. Sequences from fungi of the phyla Ascomycota and Basidiomycota were the most frequently recovered (64% and 29% of all classified sequences, respectively). Sequences derived from members of the Chytridiomycota were also detected, but these represented a very small fraction of the total sequence data set (Fig. 3A). Sequences belonging to the Ascomycota were derived from fungi of the classes Sordariomycetes, Dothideomycetes, Eurotiomycetes, Leotiomycetes, and Saccharomycetes Abundant Basidiomycota classes included the Microbotryomycetes, Tremellomycetes, Agaricomycetes, Exobasidiomycetes, Cystobasidiomycetes, Ustilaginomycetes, and Wallemiomycetes (Fig. 3B). A substantial portion of the sequences (28% of Ascomycota sequences and 7% of Basidiomycota sequences) could be classified only to the phylum level (Fig. 3B). The fungal community structure varied substantially from well to well, and striking differences were observed between communities from circumneutral (pH >4.5) and acidic (pH <4.5) wells (Fig. 3C). For example OTU_434, derived from fungi of the genus Coniochaeta, was the most abundant OTU in neutral pH groundwater, whereas OTU_1654, derived from fungi of the genus Trichosporon, was abundant in many low-pH groundwater samples (Fig. 3C).
FIG 3.
Fungal community structure at various levels of taxonomic resolution. Taxa (i.e., OTUs) were classified and grouped into phyla, classes, and genera. The relative abundance of the taxa is shown for each of the 39 groundwater samples (well identifiers are described in Table S1 in the supplemental material). Groundwater samples are plotted with increasing pH and split into two broad categories on the basis of the distribution of groundwater pH values (i.e., pHs below 4.5 and pHs above 4.5). (A and B) Phylum-level (A) and class-level (B) classification of the detected taxa. In panel B, the phylum classification is indicated in parentheses (A, Ascomycota; B, Basidiomycota; C, Chytridiomycota). (C) The most abundant OTUs in the low-pH and neutral-pH groundwater samples. Percent abundance is the percentage of the classified taxa. The phylum is indicated in parentheses, as defined for panel B.
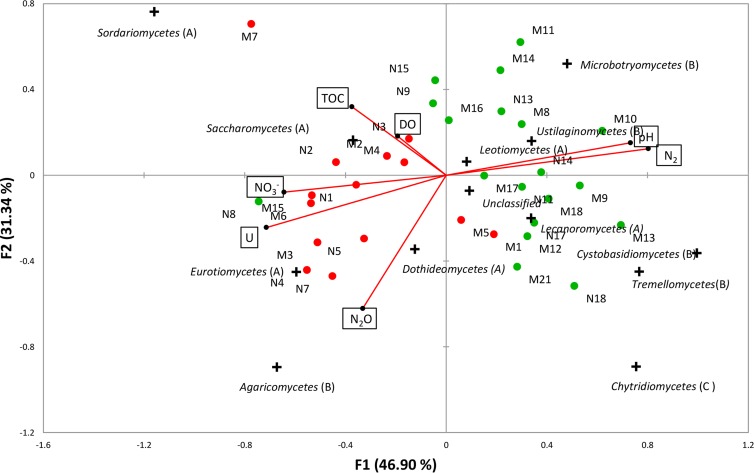
CCA ordination was used to delineate the effects of geochemical variables on fungal community composition plotted at the class level (Fig. 4; see Table S1 in the supplemental material). Fungal communities in low-pH groundwater were nearly completely separated from those in the high-pH sites along CCA axis 1 (F1). The first canonical axis explained 47% of the detected fungal diversity and variance in pH and NO3−, N2, and U loads primarily on this axis. The second axis (F2) represented 31% of the variance in the TOC, dissolved oxygen, and N2O loads partially on this axis. No effect of sampling season was observed. Fungi from the classes Sordariomycetes, Saccharomycetes, Eurotiomycetes, and Agaricomycetes were abundant in acidic groundwater samples from the highly contaminated zone near the source, while those from the Microbotryomycetes, Ustilaginomycetes, Leotiomycetes, Lecanoromycetes, Tremellomycetes, and Chytridiomycetes were abundant in neutral-pH groundwater. Taxa abundant under circumneutral conditions were positively correlated with N2, and N2 was observed as the terminal product of denitrification in wells with circumneutral pH.
FIG 4.
Ordination of canonical correspondence analysis (CCA) of the fungal community composition based on pyrosequencing of the fungal ITS region (plus symbols) with groundwater geochemistry (vectors). The percentages of variation explained by each axis are shown. Groundwater samples are abbreviated and referenced in Table S1 in the supplemental material. Solid red circles, groundwater samples with pHs below 4.5; solid green circles, groundwater samples with pHs above 4.5. Red vectors are the variables, and plus symbols represent the ordination of taxa at the taxonomic level of class.
Isolation of subsurface fungi.
More than 50 colonies were recovered, and 21 isolates were chosen for further analysis on the basis of unique colony morphology, growth pattern, or pigmentation. Analysis of the SSU rRNA gene sequences of these organisms revealed that the isolates belonged to 13 genera from six orders and four classes within the phyla Ascomycota and Basidiomycota. Isolates belonged to the genera Penicillium and Aspergillus within the Eurotiomycetes, the genera Fusarium, Neolinocarpon, Apiospora, Pochonia, and Coniochaeta within the Sordariomycetes, the genus Rhodosporidium within the Microbotryomycetes, and the genera Lachnum, Teberdinia, Sclerotinia, Phialocephala, Collophora, and Heterodermia within the Leotiomycetes (Table 1). Although the majority of the isolates were also detected in the pyrosequencing libraries, these organisms were not present in great abundance in these cultivation-independent assays. The ITS sequences of fungi from the genus Coniochaeta, however, represented approximately 18% of all fungal ITS sequences recovered from pyrosequence analyses of site groundwater and were recovered multiple times in cultivation assays (Fig. 3C; Table 1).
Potential denitrification rates in isolates and sediment-groundwater slurries.
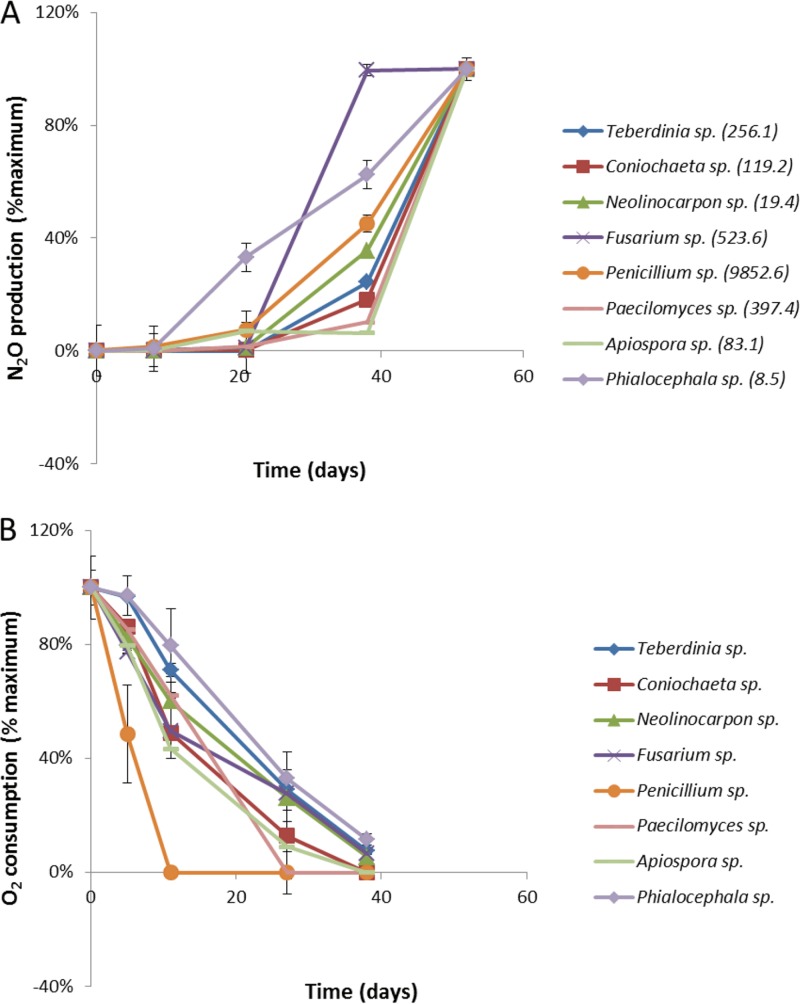
The denitrification potential of fungal isolates recovered from the OR-IFRC site and the groundwater and sediment microbial communities was assessed in microcosm studies. Incubations were established and inoculated with mycelia of representative strains of fungi from the OR-IFRC site, including those belonging to the genera Teberdinia, Coniochaeta, Neolinocarpon, Fusarium, Penicillium, Apiospora, Phialocephala, and Paecilomyces. Incubations were initially microaerophilic, as preliminary results (data not shown) suggested that the presence of some O2 was necessary for site fungi to denitrify. Molecular oxygen and nitrous oxide concentrations were monitored concurrently in the microcosms. No N2O was detected during the initial 10 days of incubations until the initial O2 concentrations had declined to an average of 20% (vol/vol) of the initial concentration in all microcosms (Fig. 5). O2 was depleted from all microcosms between days 11 and 27 of the incubation. All isolates produced N2O from NO3−, with the rates ranging from approximately 0.09 to 10 μmol N2O-N liter−1 day−1 (Fig. 5). Concentrations of N2O remained below the limit if detection in autoclaved control cultures, indicating that fungi were the source of N2O in the microcosms. In addition, treatment with the antibacterial compounds streptomycin and chloramphenicol had little to no effect on the denitrification of fungal pure cultures (data not shown). The Penicillium strain (ORNL9) produced the most N2O and depleted oxygen the most rapidly. Fungi of this genus were not detected in great abundance in site groundwater sequence libraries (Table 1). Mass balance analyses of the nitrogen species showed conversion (i.e., to N2O and biomass) of 10 to 15% of the 5 mM NO3− added in all microcosms, with the exception of the vigorously growing Penicillium spp., where 75% of the nitrate was converted. No NO2− or NH4+ was detected in any of the microcosms.
FIG 5.
N2O production (A) and O2 production (B) by fungal isolates grown in pure culture. Denitrification was assayed in microcosms with fungal isolates recovered from the OR-IFRC site subsurface. N2O production (as a percentage of the maximum) and O2 consumption (as a percentage of the maximum) were measured concurrently. Maximum rates of N2O production (nmol liter−1 day−1) are indicated in parentheses adjacent to each species name in the key to the figure.
Fungal and bacterial contributions toward N2O production from nitrate in the groundwater and sediment slurry were evaluated in slurry incubation experiments with microcosms amended with antibiotics, according to the substrate-induced respiration method (47). Potential denitrification rates were the highest in groundwater samples collected from moderately acidic wells FW122 and FW123. Fungal denitrification (i.e., denitrification in the presence of antibacterial compounds) was measured in 6 of 11 groundwater wells analyzed (see Table S2 in the supplemental material). No correlation between the rates of potential fungal or bacterial denitrification and source groundwater pH was observed. The maximum measured rate of bacterial denitrification was 54 nmol N2O-N g (dry weight)−1 day−1, and the maximum measured potential fungal denitrification rate in microcosms amended with streptomycin and chloramphenicol was 32 nmol N2O-N g (dry weight)−1 day−1.
DISCUSSION
The OR-IFRC site is representative of nuclear legacy waste sites in the U.S. Department of Energy complex, where the subsurface is often contaminated with uranium and nitrate due to the use of nitric acid during uranium processing. At the OR-IFRC site, long-term exposure to contamination has resulted in a highly distorted microbial community (e.g., see reference 31).
Despite the fact that fungi are known to be metal resistant and pH tolerant (48), their distribution and activity in the subsurface of nuclear legacy waste sites have not been studied. Furthermore, molecular surveys targeting functional genes have been unable to detect the presence of denitrifying organisms at the OR-IFRC site (e.g., see references 7 and 8). We previously demonstrated that commonly used PCR primers targeting nitrite reductase genes do not (and cannot) amplify genes from the most dominant bacterial denitrifying lineage at the site and, likewise, cannot amplify fungal genes (8). Thus, knowledge of fungi and their contribution to subsurface nitrogen cycling at the site is lacking.
This study provides a systematic characterization of the fungal diversity and community structure in a subsurface environment and directly addresses the impact of contamination and pH on fungal community abundance, diversity, and composition. Results demonstrate the presence across the watershed of a diverse and dynamic fungal community that is substantially impacted by pH and that has the potential for nitrous oxide production through denitrification.
Fungal community structure in groundwater.
The abundance, distribution, and diversity of fungi in subsurface groundwater samples were determined using quantitative and semiquantitative molecular techniques, including qPCR of eukaryotic SSU rRNA genes and pyrosequencing of fungal ITS regions. Our study targeted the ITS region for sequencing because of a higher sensitivity to fungal diversity than the SSU rRNA gene (49, 50). Statistical analysis of groundwater geochemical variables indicates that pH has a moderate influence on the diversity of fungal communities, while a strong effect of pH on fungal community composition and abundance was observed. Although many fungi are acid tolerant and can survive under a diverse range of pH conditions and elsewhere such acid tolerance has been shown to lead to increased fungal dominance (e.g., in acidic soils [51]), bacteria dominated the groundwater microbial communities at all pHs in this study. Furthermore, the relative dominance of bacteria was especially pronounced in the acidic and highly contaminated groundwaters near the source zone of contamination, where bacterial SSU rRNA gene abundance exceeded eukaryotic SSU rRNA gene abundance by up to 4 orders of magnitude. This may represent an underestimate of the true relative abundance of fungi to bacteria in the subsurface groundwater samples, as qPCR primers target all eukaryotes, not just fungi, and fungi have many more copies of rRNA operons per genome than bacteria (52). We note that the relative abundance of eukaryotic to bacterial SSU rRNA gene abundance, as measured through qPCR analysis, is partially confounded by the variability in gene copy number in organisms from these different domains. Bacteria, for example, can have from 1 to 15 copies (53), while fungi typically have many more and may have in the 10s to 100s, though there is much more uncertainty regarding the exact number (54). A further confounding factor is the use of a domain-level eukaryotic primer set for quantitative analysis of SSU rRNA genes in the subsurface samples. The value of the eukaryotic SSU rRNA gene abundance should be taken as an upper limit for fungal SSU rRNA abundance, and the true abundance of fungal SSU rRNA genes could be lower if nonfungal eukaryotes are abundant in the sample. This observation is consistent with observations from prior studies that have reported negative effects of contaminants on microbial diversity and abundance (29, 55, 56). Nonetheless, the data indicate that low-pH conditions in the OR-IFRC site groundwater are highly limiting for fungal abundance and activity, while bacterial abundance is less impacted by the low-pH and highly contaminated conditions. This appears to be in large part due to the abundance of bacteria from the genus Rhodanobacter in the acidic source zone; here, few other bacteria are active and diversity is very low (31, 57). Further research will examine more carefully the relationship between total eukaryotic DNA and fungal DNA in the OR-IFRC site subsurface.
At a coarse taxonomic level, fungi from the phyla Ascomycota and Basidiomycota were the predominant taxa detected, with a low relative abundance of taxa from the phylum Chytridiomycota being observed. Comprehensive fungal community analyses of this type are limited, and accordingly, it is difficult to place these data into context. In previous studies, fungi from the phyla Ascomycota and Chytridiomycota were found to dominate in some freshwater aquatic ecosystems (58, 59), while Basidiomycota were found to dominate in others (29, 60). At the taxonomic level of order, fungi from the Coniochaetales were the most abundant taxa detected across the field site and were particularly abundant in the circumneutral wells. Members of the genus Rhodosporidium, abundant in site circumneutral groundwaters (but with a representative isolated from groundwater with a pH of <4.5), have been detected in other acidic environments (61) but were not previously shown to denitrify. Prior studies of cultivated members of the order Coniochaetales have demonstrated that these organisms can be acid and metal tolerant (62); in addition, we demonstrate that members of this taxon are capable of denitrification. Fungi from the genera Trichosporon and Aspergillus were detected with cultivation-independent analyses in an acid mine drainage lake but were not isolated in pure culture (63). Likewise, a number of phylotypes affiliated with the classes Dothideomycetes and Eurotiomycetes (notably, Aspergillus spp.) were observed in abundance in acid mine drainage samples (64). Overall, the majority of the taxa present were generally associated with soil, and many of the taxa have been observed in other contaminated environments. In addition, many of the OTUs identified in this study could not be taxonomically resolved due to the limited number of representative sequences available for comparison in the fungal ITS database (65).
Isolation and identification of subsurface fungi.
A wide diversity of fungi was isolated from the subsurface in this study, including members of the genera Coniochaeta, Aspergillus, Penicillium, Fusarium, Sclerotinia, and Paecilomyces. These isolates are representatives of genera that were also detected in abundance (>37.5% of all pyrosequences belonged to these genera) in pyrosequencing libraries from OR-IFRC site groundwaters. All 21 strains isolated were capable of growing under acidic, aerobic, and microaerophilic conditions in the presence of a high contaminant load, consistent with the OR-IFRC site subsurface environment. Fungi from the genus Coniochaeta were particularly abundant in pyrosequencing libraries of circumneutral groundwaters (18.5%) as well as in a few acidic groundwater samples. Multiple isolates from the genus capable of denitrification were isolated, and the SSU rRNA gene sequences from these isolates were most similar by BLAST analysis to the SSU rRNA gene sequence of Coniochaeta velutina (100% sequence similarity), a Saccharomyces cerevisiae yeast-like fungus (62). Fungi affiliated with C. velutina are lignocellulose degraders and are commonly found in terrestrial habitats, such as soil, trees, and leaves (62), but this group has also been detected in other acidic, heavy metal-contaminated aquatic systems. For example, members of this family were detected in waters of the contaminated Tinto River, which is characterized by extremely low pH (2.0 to 2.5) and high concentrations of heavy metals (e.g., Fe, Cu, Zn) (15).
Other isolated denitrifiers belonged to the common soil fungal genera Penicillium and Aspergillus and were also detected at the OR-IFRC site by ITS pyrosequencing analysis. These organisms could be of ecological importance, as some species of Penicillium and Aspergillus produce organic acids and can detoxify heavy metals through the formation of insoluble organic acid-metal complexes (66, 67). Penicillium species are capable of growing under extreme conditions and have been isolated from hydrocarbon-contaminated aquatic environments as well as from metal-contaminated systems, such as acid mine drainage waters, uranium mines, and aquifers (68–70). In this study, the Penicillium sp. isolate demonstrated the highest rates of oxygen consumption, removing oxygen within 11 days, a rate more than twice that of the other isolates.
An improved understanding of denitrifying microbial communities is required to predict and control denitrification mechanisms for the remediation of contaminated groundwater. Detection and isolation of fungi from low-pH and high-nitrate groundwater samples suggest that the denitrifying phenotype is beneficial in the subsurface at the OR-IFRC site or that the true abundance of environmental denitrifying fungi is greatly underestimated. In this study, subsurface isolates from a large number of fungal genera (Teberdinia, Aspergillus, Coniochaeta, Neolinocarpon, Fusarium, Penicillium, Apiospora, Phialocephala, and Paecilomyces) were all demonstrated to reduce NO3− to N2O. Prior to this study, the only fungal pure cultures shown to denitrify belonged to the genera Fusarium, Cylindrocarpon, and Trichosporon (71). This study, therefore, greatly expands the fungal taxa known to conduct denitrification. We note that many of the denitrifying strains of fungi isolated in this study, in addition to isolates from the genera Fusarium and Cylindrocarpon previously described, belong to the Sordariomycetes (also known as Pyrenomycetes), a large class of ascomycete fungi. The current database of known denitrifying fungi is currently too limited to determine if this is a common physiological characteristic within the class.
Our analysis shows that N2O production started before O2 was depleted in the microcosms, in agreement with previous work that observed fungi to carry out denitrification and O2 respiration simultaneously under aerobic conditions (22). The potential rates of N2O production observed here for subsurface fungi (approximately 0.09 to 10 μmol N2O-N liter−1 day−1) were comparable to the rates measured in previous studies of fungal isolates from soil, such as the study of Bleakley and Tiedje (72), who reported rates in the range of 0.4 to 1.2 μmol N2O-N liter−1 day−1 for a variety of fungal isolates. N2O production by soil isolate Fusarium oxysporum was estimated to be 1.56 μmol N2O-N liter−1 day−1 (23). Low rates of growth and metabolism as well as growth under a range of oxygen concentrations could well reflect the adaptation of fungi to the nutrient-poor and oscillating redox conditions observed in the subsurface habitat.
Evidence for fungus-mediated denitrification.
While a few prior studies have demonstrated the potential for fungus-mediated denitrification in surficial soils and wetlands (26, 27), fungal denitrification has not been explored in subsurface environments. Here we demonstrate the potential for fungal denitrification in subsurface groundwater and sediment microcosms, with a maximum measured rate of 32 nmol N2O-N g−1 day−1 in the presence of antibacterial compounds. The potential rates of fungus-mediated denitrification from the OR-IFRC subsurface lie in the lower range of fungal denitrification rates reported previously for agricultural soils and grasslands. In mildly acidic grassland soils (pH 6.3) in Ireland, the fungal contribution to denitrification was estimated to be 24 to 240 nmol N2O-N g−1 day−1 (26), while Long et al. reported a rate of 24.14 nmol N2O-N g−1 day−1 in agricultural surface soil in the United States in 2013 (73). These data must be considered within the context that each study was performed using different cultivation conditions and growth periods and, further, that some of the potential denitrification activity might represent activity from spore-forming fungi, which might be favored by cultivation conditions. This finding is consistent with the low abundance of fungi in the acidic source zone, and the low measured activity of fungi is generally below that measured for bacteria. Denitrifying bacteria, particularly from the genus Rhodanobacter, are abundant and active in the zone near the source and are likely the dominant denitrifiers in these systems, with the possibility of a modest contribution by fungal denitrifiers (31). Further studies are needed to delineate the in situ contribution of bacterial and fungal denitrifiers in this heterogeneous contaminated, low-organic-matter, and low-pH subsurface environment.
Currently, all known denitrifying fungi lack nitrous oxide reductase genes, and therefore, fungal denitrification cannot produce dinitrogen gas as a terminal product of nitrate/nitrite respiration (72). Thus, fungal denitrification, regardless of pH, could contribute to significantly higher N2O release if this excess product is not consumed by denitrifying bacteria. For acidic sites such as the OR-IFRC site, the sensitivity of N2O reductase enzymes to low pH could contribute to N2O production by bacteria as well as fungi (74). The presence of isotopically light N2, in addition to isotopically light N2O, in groundwater in the zone near the source, however, indicates the presence of some complete (i.e., bacterial) denitrification. Some denitrification activity could be contributed by Archaea (75, 76), and inhibition experiments conducted in this study would not have inactivated archaeal respiration. A possible archaeal nirK gene sequence was identified in a metagenome derived from an OR-IFRC acidic site, but no significant contribution of Archaea was observed in the community shotgun metagenome (8, 57), suggesting that bacteria and, to a lesser extent, fungi are the primary denitrifiers at the site.
In summary, this study demonstrates that the contaminated subsurface hosts a diverse fungal community and groundwater geochemical variables, particularly pH, play a role in structuring that community. Isolation of a broad range of denitrifying fungi greatly increases the known fungal lineages capable of denitrification, but whether the abundance of denitrifying lineages in this study is site specific or, rather, represents an unexplored general capability of fungi is not known. The lack of a nitrous oxide reduction capability in fungi and the detection of denitrification potential in slurry microcosms point to their role in site biogeochemistry, with implications for greenhouse gas emissions and remediation strategies. Future research should be directed toward the development of molecular techniques for elucidating the metabolically active fungi that mediate denitrification in a range of ecosystems.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Office of Science (Biological and Environmental Research [BER]), U.S. Department of Energy, grants DEFG02-07ER64373, -97ER62469, and -97ER64398 and by the Oak Ridge Integrated Field Research Challenge, operated by the Environmental Sciences Division, Oak Ridge National Laboratory (ORNL). ORNL is managed by UT-Battelle LLC for the U.S. Department of Energy under contract no. DE-AC05-00OR22725.
We kindly acknowledge assistance from Tonia Mehlhorn and Kenneth Lowe for sampling and uranium measurements and thank Ashvini Chauhan for critical comments on the manuscript during preparation.
Footnotes
Published ahead of print 3 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03423-13.
REFERENCES
- 1.Riley RG, Zachara JM. 1992. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface research. Report DOE/ER-0547T U.S. Department of Energy, Oak Ridge, TN [Google Scholar]
- 2.Rivett MO, Buss SR, Morgan P, Smith JWN, Bemment CD. 2008. Nitrate attenuation in groundwater: a review of biogeochemical controlling processes. Water Res. 42:4215–4232. 10.1016/j.watres.2008.07.020 [DOI] [PubMed] [Google Scholar]
- 3.Kostka JE, Green SJ. 2011. Microorganisms and processes linked to uranium reduction and immobilization, p 117–138 In Stolz JF, Oremland RS. (ed), Microbial metal and metalloid metabolism. Advances and applications. ASM Press, Washington, DC [Google Scholar]
- 4.Gasperikova E, Hubbard SS, Watson DB, Baker GS, Peterson JE, Kowalsky MB, Smith M, Brooks SC. 2012. Long-term electrical resistivity monitoring of recharge-induced contaminant plume behavior. J. Contam. Hydrol. 142–143:33–49. 10.1016/j.jconhyd.2012.09.007 [DOI] [PubMed] [Google Scholar]
- 5.Wallenstein MD, Myrold DD, Firestone M, Voytek M. 2006. Environmental controls on denitrifying communities and denitrification rates: insights from molecular methods. Ecol. Appl. 16:2143–2152. 10.1890/1051-0761(2006)016[2143:ECODCA]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 6.Akob DM, Mills HJ, Kostka JE. 2007. Metabolically active microbial communities in uranium-contaminated subsurface sediments. FEMS Microbiol. Ecol. 59:95–107. 10.1111/j.1574-6941.2006.00203.x [DOI] [PubMed] [Google Scholar]
- 7.Yan T, Fields MW, Wu L, Zu Y, Tiedje JM, Zhou J. 2003. Molecular diversity and characterization of nitrite reductase gene fragments nirK and nirS from nitrate- and uranium-contaminated groundwater. Environ. Microbiol. 5:13–24. 10.1046/j.1462-2920.2003.00393.x [DOI] [PubMed] [Google Scholar]
- 8.Green SJ, Prakash O, Gihring TM, Akob DM, Jasrotia P, Jardine PM, Watson DB, Brown SD, Palumbo AV, Kostka JE. 2010. Denitrifying bacteria from the terrestrial subsurface exposed to mixed waste contamination. Appl. Environ. Microbiol. 76:3244–3254. 10.1128/AEM.03069-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spain AM, Krumholz L. 2012. Cooperation of three denitrifying bacteria in nitrate removal of acidic nitrate- and uranium-contaminated groundwater. Geomicrobiol. J. 29:830–842. 10.1080/01490451.2011.635757 [DOI] [Google Scholar]
- 10.Fields MW, Yan T, Rhee SK, Carroll SL, Jardine PM, Watson DB, Criddle CS, Zhou J. 2005. Impacts on microbial communities and cultivable isolates from groundwater contaminated with high levels of nitric acid-uranium waste. FEMS Microbiol. Ecol. 53:417–428. 10.1016/j.femsec.2005.01.010 [DOI] [PubMed] [Google Scholar]
- 11.Palumbo AV, Schryver JC, Fields MW, Bagwell CE, Zhou JZ, Yan T, Liu X, Brandt CC. 2004. Coupling of functional gene diversity and geochemical data from environmental samples. Appl. Environ. Microbiol. 70:6525–6534. 10.1128/AEM.70.11.6525-6534.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spalding BP, Watson DB. 2008. Passive sampling and analyses of common dissolved fixed gases in groundwater. Environ. Sci. Technol. 42:3766–3772. 10.1021/es7024005 [DOI] [PubMed] [Google Scholar]
- 13.Gadd MG. 2010. Metals, minerals and microbes, geomicrobiology and bioremediation. Microbiology 156:609–643. 10.1099/mic.0.037143-0 [DOI] [PubMed] [Google Scholar]
- 14.Brake SS, Hasiotis ST. 2010. Eukaryote-dominated biofilms and their significance in acidic environments. Geomicrobiol. J. 27:534–558. 10.1080/01490451003702966 [DOI] [Google Scholar]
- 15.López-Archilla I, González AE, Terrón MC, Amils R. 2004. Ecological study of the fungal populations of the acidic Tinto River in southwestern Spain. Can. J. Microbiol. 50:923–934. 10.1139/w04-089 [DOI] [PubMed] [Google Scholar]
- 16.Hayatsu M, Tago K, Saito M. 2008. Various players in the nitrogen cycle: diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 54:33–45. 10.1111/j.1747-0765.2007.00195.x [DOI] [Google Scholar]
- 17.Shoun H, Kim DH, Uchiyama H, Sugiyama J. 1992. Denitrification by fungi. FEMS Microbiol. Lett. 94:277–281. 10.1111/j.1574-6968.1992.tb05331.x [DOI] [PubMed] [Google Scholar]
- 18.Bollag JM, Tung G. 1972. Nitrous oxide release by soil fungi. Soil Biol. Biochem. 4:271–276. 10.1016/0038-0717(72)90021-1 [DOI] [Google Scholar]
- 19.Tsuruta S, Takaya N, Zhang L, Shoun H, Kimura K, Hamamoto M, Nakase T. 1998. Denitrification by yeasts and occurrence of cytochrome P450nor in Trichosporon cutaneum. FEMS Microbiol. Lett. 168:105–110. 10.1111/j.1574-6968.1998.tb13262.x [DOI] [PubMed] [Google Scholar]
- 20.Usuda K, Toritsuka N, Matsuo Y, Kim DH, Shoun H. 1995. Denitrification by the fungus Cylindrocarpon tonkinense, anaerobic cell growth and two isozyme forms of cytochrome P-450nor. Appl. Environ. Microbiol. 61:883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi M, Matsuo Y, Takimoto A, Suzuki S, Maruo F, Shoun H. 1996. Denitrification, a novel type of respiratory metabolism in fungal mitochondrion. J. Biol. Chem. 271:16263–16267. 10.1074/jbc.271.27.16263 [DOI] [PubMed] [Google Scholar]
- 22.Zhou Z, Takaya N, Sakairi MAC, Shoun H. 2001. Oxygen requirement for denitrification by the fungus Fusarium oxysporum. Arch. Microbiol. 175:19–25. 10.1007/s002030000231 [DOI] [PubMed] [Google Scholar]
- 23.Shoun H, Tanimoto T. 1991. Denitrification by the fungus Fusarium oxysporum and involvement of cytochrome P-450 in the respiratory nitrite reduction. J. Biol. Chem. 266:11078–11082 [PubMed] [Google Scholar]
- 24.Spott O, Russow R, Stange CF. 2011. Formation of hybrid N2O and hybrid N2 due to codenitrification, first review of a barely considered process of microbially mediated N-nitrosation. Soil Biol. Biochem. 43:1995–2011. 10.1016/j.soilbio.2011.06.014 [DOI] [Google Scholar]
- 25.Prendergast-Miller MT, Baggs EM, Johnson D. 2011. Nitrous oxide production by the ectomycorrhizal fungi Paxillus involutus and Tylospora fibrillosa. FEMS Microbiol. Lett. 316:31–35. 10.1111/j.1574-6968.2010.02187.x [DOI] [PubMed] [Google Scholar]
- 26.Laughlin J, Stevens RJ. 2002. Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 66:1540–1548. 10.2136/sssaj2002.1540 [DOI] [Google Scholar]
- 27.Seo DC, DeLaune RD. 2010. Fungal and bacterial mediated denitrification in wetlands: influence of sediment redox condition. Water Res. 44:2441–2450. 10.1016/j.watres.2010.01.006 [DOI] [PubMed] [Google Scholar]
- 28.Bärlocher F, Nikolcheva LG, Wilson KP, Williams DD. 2006. Fungi in the hyporheic zone of a springbrook. Microb. Ecol. 52:708–715. 10.1007/s00248-006-9102-4 [DOI] [PubMed] [Google Scholar]
- 29.Brad T, Braster M, van Breukelen BM, van Straalen NM, Roling WFM. 2008. Eukaryotic diversity in an anaerobic aquifer polluted with landfill leachate. Appl. Environ. Microbiol. 74:3959–3968. 10.1128/AEM.02820-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watson DB, Kostka JE, Fields MW, Jardine PM. 2004. The Oak Ridge Field Research Center conceptual model. NABIR Field Research Center report NABIR Field Research Center, Oak Ridge, TN: http://public.ornl.gov/orifc/FRC-conceptual-model.pdf [Google Scholar]
- 31.Green SJ, Prakash O, Jasrotia P, Overholt WA, Cardenas E, Hubbard D, Tiedje JM, Watson DB, Schadt CW, Brooks SC, Kostka JE. 2012. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microbiol. 78:1039–1047. 10.1128/AEM.06435-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- 33.White TJ, Bruns T, Lee S, Taylor JW. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322 In Innis M, Gelfand DH, Sninsky JJ, White TJ, (ed), PCR protocols—a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 34.Nadkarni MA, Martin FE, Jacques NA, Hunter N. 2002. Determination of bacterial load by real-time PCR using a broad-range universal probe and primers set. Microbiology 148:257–266 [DOI] [PubMed] [Google Scholar]
- 35.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, NY [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gihring TM, Green SJ, Schadt CW. 2012. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ. Microbiol. 14:285–290. 10.1111/j.1462-2920.2011.02550.x [DOI] [PubMed] [Google Scholar]
- 38.Ryan TA, Joiner BL. 1976. Normal probability plots and tests for normality. The Pennsylvania State University, University Park, PA [Google Scholar]
- 39.Clarke KR, Warwick RM. 2001. Change in marine communities, an approach to statistical analysis and interpretation. Primer-E, Lutton, United Kingdom [Google Scholar]
- 40.Warwick RM, Clarke KR. 2006. Primer 6. Primer-E Ltd, Plymouth, United Kingdom [Google Scholar]
- 41.Shannon CE. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:379–423. 10.1002/j.1538-7305.1948.tb01338.x [DOI] [Google Scholar]
- 42.Weiss RF, Price BA. 1980. Nitrous oxide solubility in water and seawater. Mar. Chem. 8:347–359. 10.1016/0304-4203(80)90024-9 [DOI] [Google Scholar]
- 43.Braman RS, Hendrix SA. 1989. Nanogram nitrite and nitrate determination in environmental and biological materials by vanadium (III) reduction with chemiluminescence detection. Anal. Chem. 61:2715–2718. 10.1021/ac00199a007 [DOI] [PubMed] [Google Scholar]
- 44.Garside C. 1982. A chemiluminescent technique for the determination of nanomolar concentrations of nitrate, nitrite, or nitrite alone in seawater. Mar. Chem. 11:159–167. 10.1016/0304-4203(82)90039-1 [DOI] [Google Scholar]
- 45.Bower CE, Holm-Hansen T. 1980. A salicylate-hypochlorite method for determining ammonia in seawater. Can. J. Fish Aquat. Sci. 37:794–798. 10.1139/f80-106 [DOI] [Google Scholar]
- 46.Tiedje JM. 1982. Denitrification, p 1011–1026 In Page AL, Miller RH, Keeney DR. (ed), Methods of soil analysis. Part 2. Chemical and microbiological properties, 2nd ed. American Society of Agronomy, Soil Science Society of America, Madison, WI [Google Scholar]
- 47.Anderson JPE, Domsch KH. 1975. Measurement of bacterial and fungal contributions to respiration of selected agricultural and forest soils. Can. J. Microbiol. 21:314–322. 10.1139/m75-045 [DOI] [PubMed] [Google Scholar]
- 48.Gadd GM, Fomina M. 2011. Uranium and fungi. Geomicrobiol. J. 28:471–482. 10.1080/01490451.2010.508019 [DOI] [Google Scholar]
- 49.Begerow D, Nilsson H, Unterseher M, Maier W. 2010. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 87:99–108. 10.1007/s00253-010-2585-4 [DOI] [PubMed] [Google Scholar]
- 50.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. U. S. A. 109:6241–6246. 10.1073/pnas.1117018109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bååth E, Anderson TH. 2003. Comparison of soil fungal/bacterial ratios in a pH gradient in a pH gradient using physiological and PLFA-based techniques. Soil Biol. Biochem. 35:955–963. 10.1016/S0038-0717(03)00154-8 [DOI] [Google Scholar]
- 52.Chemidlin Prévost-Bouré N, Christen R, Dequiedt S, Mougel C, Lelièvre M, Claudy J, Shahbazkia HR, Guillou L, Arrouays D, Ranjard L. 2011. Validation and application of a PCR primer set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS One 6:e24166. 10.1371/journal.pone.0024166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181–184. 10.1093/nar/29.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Black J, Jean T, Byfield G, Foarde K, Menetrez M. 2013. Determining fungi rRNA copy number by PCR. J. Biomol. Tech. 24:32–38. 10.7171/jbt.13-2401-004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stephenson S, Chariton AA, Holley MP, O'Sullivan M, Gillings MR, Hose GC. 2013. Changes in prokaryote and eukaryote assemblages along a gradient of hydrocarbon contamination in groundwater. Geomicrobiol. J. 30:623–634. 10.1080/01490451.2012.746408 [DOI] [Google Scholar]
- 56.Ludvigsen L, Albrechtsen HJ, Ringelberg DB, Ekelund F, Christensen TH. 1999. Distribution and composition of microbial populations in a landfill leachate contaminated aquifer (Grinsted, Denmark). Microb. Ecol. 37:197–207. 10.1007/s002489900143 [DOI] [PubMed] [Google Scholar]
- 57.Hemme CL, Deng Y, Gentry TJ, Fields MW, Wu LW, Barua S, Barry K, Tringe SG, Watson DB, He Z, Hazen TC, Tiedje JM, Rubin EM, Zhou J. 2010. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 4:660–672. 10.1038/ismej.2009.154 [DOI] [PubMed] [Google Scholar]
- 58.Wurzbacher CM, Bärlocher F, Grossart HP. 2010. Fungi in lake ecosystems. Aquat. Microbiol. Ecol. 59:125–149. 10.3354/ame01385 [DOI] [Google Scholar]
- 59.Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanova L, Padgett D, Porter D, Raja HA, Schmit JP, Thorton HA, Voglymay H. 2007. Fungal biodiversity in aquatic habitats. Biodivers. Conserv. 16:49–67. 10.1007/s10531-006-9120-z [DOI] [Google Scholar]
- 60.Gadanho M, Sampaio J. 2004. Application of temperature gradient gel electrophoresis to the study of yeast diversity in the estuary of the Tagus River, Portugal. FEMS Yeast Res. 5:253–261. 10.1016/j.femsyr.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 61.Álvarez-Pérez S, Blanco JL, Alba P, García ME. 2011. Fungal growth in culture media simulating an extreme environment. Rev. Iberoam. Micol. 28:159–165. 10.1016/j.riam.2011.03.005 [DOI] [PubMed] [Google Scholar]
- 62.Damm U, Fourie PH, Crous PW. 2010. Coniochaeta Lecythophora, Collophora gen. nov. and Phaeomoniella species associated with wood necroses of Prunus trees. Persoonia 24:60–80. 10.3767/003158510X500705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Hao C, Wang L, Li S, Feng C. 2012. Characteristics of the eukaryotic community structure in acid mine drainage lake in Anhui Province, China. Wei Sheng Wu Xue Bao 52:875–884 (In Chinese) [PubMed] [Google Scholar]
- 64.Zettler LA, Messerli MA, Laatsch AD, Smith PJS, Sogin ML. 2003. From genes to genomes, beyond biodiversity in Spain's Rio Tinto. Biol. Bull. 204:205–209. 10.2307/1543560 [DOI] [PubMed] [Google Scholar]
- 65.Nilsson RH, Bok G, Ryberg M, Kristiansson E, Hallenberg N. 2009. A software pipeline for processing and identification of fungal ITS sequences. Source Code Biol. Med. 4:1. 10.1186/1751-0473-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawai F, Zhang D, Sugimoto M. 2000. Isolation and characterization of acid- and Al-tolerant microorganisms. FEMS Microbiol. Lett. 189:143–147. 10.1111/j.1574-6968.2000.tb09220.x [DOI] [PubMed] [Google Scholar]
- 67.Kochian LV. 1995. Cellular mechanisms of aluminium toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46:2237–2260 [Google Scholar]
- 68.Lategan MJ, Torpy FR, Newby S, Stephenson S, Hose GC. 2012. Fungal diversity of shallow aquifers in southeastern Australia. Geomicrobiol. J. 29:352–361. 10.1080/01490451.2011.559306 [DOI] [Google Scholar]
- 69.White C, Gadd GM. 1990. Biosorption of radionuclides by yeast and fungal biomass. J. Chem. Technol. Biotechnol. 49:331–343 [DOI] [PubMed] [Google Scholar]
- 70.Gottlich E, van der Lubbe W, Lange B, Fiedler S, Melchert I, Reifenrath M, Flemming HC, de Hoog S. 2002. Fungal flora in groundwater-derived public drinking water. Int. J. Hyg. Environ. Health 205:269–279. 10.1078/1438-4639-00158 [DOI] [PubMed] [Google Scholar]
- 71.Shoun H, Fushinobu S, Jiang L, Kim SW, Wakagi T. 2012. Fungal denitrification and nitric oxide reductase cytochrome P450nor. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:1186–1194. 10.1098/rstb.2011.0335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bleakley BH, Tiedje JM. 1982. Nitrous oxide production by organisms other than nitrifiers or denitrifiers. Appl. Environ. Microbiol. 44:1342–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Long A, Heitmanb J, Tobias C, Philips R, Song B. 2013. Co-occurring anammox, denitrification, and codenitrification in agricultural soils. Appl. Environ. Microbiol. 79:168–176. 10.1128/AEM.02520-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tiedje J. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p 179–244 In Zehnder AJB. (ed), Biology of anaerobic microorganisms. John Wiley & Sons, Inc, New York, NY [Google Scholar]
- 75.Cabello P, Roldan MD, Moreno-Vivian C. 2004. Nitrate reduction and the nitrogen cycle in Archaea. Microbiology 150:3527–3546. 10.1099/mic.0.27303-0 [DOI] [PubMed] [Google Scholar]
- 76.Laughlin RJ, Rutting T, Muller C, Watson CJ, Stevens RJ. 2009. Effect of acetate on soil respiration, N2O emissions and gross N transformations related to fungi and bacteria in a grassland soil. Appl. Soil Ecol. 42:25–30. 10.1016/j.apsoil.2009.01.004 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.