Abstract
Rubella remains a social and economic burden due to the high incidence of congenital rubella syndrome (CRS) in some countries. For this reason, an accurate and efficient high-throughput measure of antibody response to vaccination is an important tool. In order to measure rubella-specific neutralizing antibodies in a large cohort of vaccinated individuals, a high-throughput immunocolorimetric system was developed. Statistical interpolation models were applied to the resulting titers to refine quantitative estimates of neutralizing antibody titers relative to the assayed neutralizing antibody dilutions. This assay, including the statistical methods developed, can be used to assess the neutralizing humoral immune response to rubella virus and may be adaptable for assessing the response to other viral vaccines and infectious agents.
INTRODUCTION
Acute infection with rubella virus usually results in a mild fever and rash. However, complications can arise when infection occurs during pregnancy. In this case, rubella virus is able to establish infection in the fetus, where there is up to a 90% risk of developing congenital rubella syndrome (CRS) if infection occurs during the first trimester (1, 2). Although vaccination programs have decreased the number of rubella cases and CRS in most countries, the burden of rubella and CRS is still high in many developing countries. Improved surveillance is needed to evaluate the progress of control programs (3).
Traditional assays, like enzyme immunoassays (EIA), hemagglutination inhibition assays (HAI), and neutralization assays, have been successfully implemented for laboratory diagnostics and screening techniques. However, issues such as false positives, lack of sensitivity, standardization between platforms, and the time and resources needed to efficiently measure responses in a large number of samples still exist (4–10). Thus, a standard and efficient method that resolves some or all of these issues is needed. Our laboratories previously used a rubella virus-specific chemiluminescent immunoassay (Beckman Coulter Access, Brea, CA) and in-house colorimetry- and fluorescence-based immunoassays (Centers for Disease Control and Prevention [CDC]) to measure antibody titers to rubella virus. The chemiluminescent immunoassay produced highly sensitive and reproducible antibody titer data and revealed a large spectrum of antibody responses in individuals vaccinated against rubella virus (11, 12). Although this quantifies rubella virus antibodies, the focus of our current study was to measure functional neutralizing antibodies against rubella virus.
The first aim of this study was to adapt an immunocolorimetric system for the large-scale, high-throughput quantitation of rubella virus-neutralizing antibody in a cohort of 2,091 subjects and to test the level of precision between repeated results. Next, statistical approaches were used to interpolate these data into quantitative estimates of individual subject titer values that can be used as continuous measurements in subsequent analyses. Finally, correlations between titers measured using a standardized, rubella virus-specific IgG EIA and our functional neutralizing antibody assay were investigated.
MATERIALS AND METHODS
Study participants.
Blood samples from eligible subjects were collected from healthy children, older adolescents, and adults (aged 11 to 40 years), consisting of Olmsted County residents (Rochester, MN, cohort) and armed forces personnel (San Diego, CA, cohort). The clinical details and demographic characteristics have been previously reported (13–15). The Rochester cohort consists of 1,092 individuals from three independent, age-stratified random cohorts of healthy schoolchildren and young adults from all socioeconomic strata in Rochester, MN. All participants had written records of having received two doses of measles-mumps-rubella vaccine (MMR II) (Merck). Of 1,092 subjects, 1,082 met our inclusion criteria.
In addition, we enrolled 1,076 healthy older adolescents and adults (18 to 40 years old) in the San Diego cohort. Subject enrollment for this study has been previously described in detail (14, 15). As members of the U.S. military, these subjects represent a cross section of the U.S. population. Of 1,076 subjects, 1,009 met our inclusion criteria for this study. The Institutional Review Boards of the Mayo Clinic and Naval Health Research Center (NHRC) approved the study, and written informed consent was obtained from each subject and/or from the parents of all children who participated, as well as written assent from age-appropriate participants. The measurement of neutralization antibodies was done at CDC anonymously without personal identifiable information of the study subjects.
sICNA.
A modified version of the indirect immunocolorimetric assay (ICA)-based neutralization method described by Chen et al., i.e., a soluble immunocolorimetric neutralization assay (sICNA), was used in this study (16). Each 96-well test plate contained virus-infected control (VIC; 4 wells with no serum), uninfected control (4 wells with no serum or virus), and two control sera: CDC anti-rubella human serum reference preparation (low positive) (IS2153) and a seronegative serum (SNS) collected from an individual prior to rubella vaccination (RP-011 panel member 1; Biomex GmbH, Heidelberg, Germany). The positive control was used undiluted, and the negative control was diluted 1:20 before testing. Both control sera had been extensively characterized and demonstrated reproducible results (our unpublished data).
All sera were heat inactivated for 1 h at 56°C prior to testing. Sera were serially diluted in a 2-fold series beginning from 1:12.5 through 1:100 in 96-well plates using phosphate-buffered saline (PBS) supplemented with 1% fetal bovine serum (FBS) (diluent) to yield a final volume of 30 μl per dilution. Study sera were diluted in triplicate, and control sera were diluted in duplicate. Rubella virus HPV-77, diluted to a final concentration of 1.2 × 103 PFU/ml, was mixed with an equal volume of each diluted serum (or diluent as in the case of VIC), yielding a final serum dilution series of 1:25 through 1:200, after which the plate was incubated for 1.5 h at 37°C, 5% CO2. Fifty microliters of each mixture was used to inoculate confluent Vero cells cultured in 96-well plates and incubated for 1 h at 37°C, 5% CO2, 100% humidity. Note that the 1:25 dilution contained 2 μl of serum in the final inoculant and each infected well received approximately 30 PFU of virus. Uninfected control wells received 50 μl of diluent.
After the 1-h incubation, Dulbecco's modified Eagle medium (DMEM) supplemented with 5% FBS and 50 μg/ml gentamicin (Life Technology, Grand Island, NY) was added to each well, and the plate was incubated for 3 days at 37°C, 5% CO2. To measure the amount of rubella virus used in each test run, a back titration of the diluted virus using Vero cells cultured in a 48-well plate was performed (in triplicate), as previously described (16).
At 3 days postinfection, plates were washed once with PBS, fixed with methanol, and blocked with Blotto (PBS supplemented with 5% skim milk [Becton, Dickinson and Company, Franklin Lakes, NJ] and 0.1% Tween 20). The procedure for primary and secondary antibody detection was as described earlier by Chen et al. (16) with minor modifications: an in-house rubella virus E1 mouse monoclonal antibody (5 μg/ml) was used as a primary antibody, followed by three washes using PBS supplemented with 0.05% Tween 20 (PBS-T). Next, a goat anti-mouse horseradish peroxidase (HRP)-conjugated antibody (0.5 μg/ml; Life Technologies) in Blotto was used as a secondary antibody, followed by an identical washing step. A soluble TMB (3,3′,5,5′-tetramethylbenzidine) substrate (NeA-Blue TMB substrate; Clinical Science Products, Mansfield, MA) was added for 10 min, and then the reaction was stopped using 0.5 M sulfuric acid. Optical density (OD) values were determined by spectrophotometry (Eon Microplate; Biotek, Winooski, VT) using a measurement/reference wavelength pair of 450 nm/630 nm. Each plate was considered a unit and was independently assessed. We considered a plate valid if (i) at least three of the four uninfected control wells fell within 2 standard deviations of the mean and the OD of the mean was between 0 and 0.3; (ii) at least three of the four virus-infected control wells fell within 2 standard deviations of the mean and the OD of the mean was between 0.6 and 2; (iii) the in-plate positive control gave a positive neutralization result; and (iv) the in-plate negative control gave a negative neutralization result. The mean OD value of the uninfected control wells was subtracted before signals from replicates were averaged. The neutralization titer (NT50) was considered the highest dilution at which the input virus signal was reduced by at least 50%. Sera that exhibited a reduction of virus greater than 50% at the last dilution (i.e., 1:200) were retested using a higher dilution series. Following titration, the corresponding integer titer value was recorded as an initial estimate of the NT50 for each assayed sample. For convenience in calculation, a value of 0 was assigned to all sera with NT50 less than 1:25. To test for reproducibility, a total of 491 sera were retested on separate days from the original tests. These included the first 300 sera and 191 randomly selected sera from the remaining 1,791 sera.
High-throughput assay optimization.
Conventional rubella neutralization assays using a plaque reduction method require anywhere from 6 to 11 days to complete and are difficult to standardize (17–22). The sICNA allowed neutralizing antibodies to be detected in as few as 3 days and eliminated viewer subjectivity by employing automated data acquisition (enzyme-linked immunosorbent assay [ELISA] plate reader), and the detection portion of the assay (steps following the 3-day incubation) could be completed by anyone with an enzyme immunoassay skill set. Furthermore, the microformat reduced the amount of serum and reagents required compared to other neutralization formats.
To further enhance the assay, the detection portion was optimized to a high-throughput format using an EL406 combination microplate washer dispenser, an Eon microplate spectrophotometer, and two BioStack microplate stackers (Biotek, Winooski, VT). This enhanced throughput by a factor of about 3 and reduced technician hands-on time by a factor of about 6.
Statistical analysis.
The purpose of the statistical analyses was to interpolate the observed titration values in order to estimate the magnitude of the NT50, which could be used as a quantitative outcome in future studies. We considered two different analytical approaches in our efforts to interpolate the assay results, seeking more accurate determinations of the rubella-specific antibody present in each sample. The first approach was to fit a logistic model to the assay results, and the second was to use loess, a statistical smoother.
The logistic model describes an s-shaped curve that exponentially increases as it deviates, with the growth of the curve slowing as it approaches an upper limit (u). The function can be written as: f(x) = u/{1 + exp[(h − x)/s]}, where s is a scale parameter that reflects the rate of growth of the curve and h reflects the value of x at which the value of the function is halfway between the minimum and maximum value. This model has the benefit of directly estimating the NT50 from the observed data.
The loess approach is a robust statistical procedure that fits a line through pairs of data points (23). The method is insensitive to outlying values and creates a smooth curve along the range of x values by combining a series of locally linear line segments into a single curve. In order to use this approach to estimate the NT50 values, we estimated a curve that predicted the observed assay results with the titer values that were assessed. We estimated the interpolated titer value by projecting down to the x axis from the point where the fitted curve was midway between the minimum and maximum control values.
In order to utilize either of the two approaches, it is necessary to use the results from control values from the plate on which the sample was run. In order to make the estimation of the interpolated values possible, we used the results from the uninfected control to represent the background noise of the assay. We used the results from three different conditions to represent the maximum expected value of the titration curve. These three conditions were as follows: (i) those obtained when assessing the maximum dilution assayed (MAX) for the specific sample; (ii) plate-specific values from VIC; and (iii) plate-specific values from SNS. Under a successful titration, the values from all three conditions should represent the maximum expected value of the titration curve. However, there are often differences among them. In order to obtain interpolated estimates for a given sample, we used the values from each of these three conditions to represent the true maximum value and estimated three different NT50 values.
To obtain estimates from the logistic model, we first subtracted the median value from the uninfected control from all observed values and then estimated the parameters of the logistic model using nonlinear least squares, one of which is explicitly the value of the NT50.
To obtain estimates from the loess smoother, we set the uninfected control as a titration of 0 and fit the loess trend describing the relationship between the titration values and the observed assay results. In order to predict an NT50 value in the first experimental condition, we identified the mean of the titration values, which resulted in loess-predicted assay results that were in a symmetric region around 0.5, after rescaling the data such that the uninfected control had a median of 0 and the median assay result at the maximum titration had a value of 1.0. Because the value at the maximum titration was not always equal to the SNS control or the virus-infected control states (virus-infected control; maximum dilution), we produced two additional NT50 estimates by rescaling this original estimate to correspond to an NT50 value relative to the maximum assay results from these two control conditions. The final estimate of the NT50 was set as equal to the average of the interpolated values obtained when using the VIC and SNS estimates of the maximum absorbance. When possible, the NT50 obtained using the MAX estimate of maximum absorbance was included in this average. For the MAX dilution assay result to accurately reflect the upper limit of the assay activity, the titration curve should be observed to approach an upper threshold. Therefore, in order to use the MAX value in the final estimation of the NT50, we tested for evidence of negative curvature across the observed titers using a quadratic contrast following an analysis of variance, with activity levels as the response variable and titration values as the categorical predictor variable. For experiments with evidence that the observed titration approached a threshold for the absorbance values (P value for negative curvature, <0.05), the MAX value was used in the computation of the NT50. For the remaining experiments, only estimates using the SNS and VIC controls were used.
We assessed the performance of these estimation techniques by summarizing their results within the categorical titration value estimated via laboratory observation and computing the percentage of variability in the interpolation estimates, which is explained by the categorical values obtained by laboratory observation. Additionally, we applied linear mixed-models approaches with per-subject random effects to the NT50 values for those subjects who were assayed twice. This enabled additional assessments of the assay's overall performance. These assessments include estimates of intraclass correlation coefficients to measure assay reproducibility and model residuals that assess the degree by which the estimated NT50 values deviated from their expected values. All statistical analyses were performed using the R statistical software package version 2.15.0 (http://cran.us.r-project.org/).
RESULTS
Characterization of neutralizing antibody titers.
This assay was applied to data from two cohorts of individuals. Table 1 illustrates the demographic information of the subjects from whom the rubella virus-neutralizing antibody data were obtained. A total of 2,091 subjects were assayed, and Fig. 1 illustrates the initial NT50 estimates that were obtained by selecting the specific dilution that best agreed with a reduction of 50% in viral activity levels. Approximately 20% of subjects were deemed to have NT50 estimates of <25. The bulk of subjects had estimates of 25, 50, or 100 (22%, 32%, and 18%, respectively), and roughly 1% had NT50 values greater than 400. Of the 491 subjects who were assayed twice, the distribution was similar between the two repeated assessments (Table 2). There was reasonably strong agreement between the two measurements as measured by weighted kappa, which was equal to 0.76 (95% confidence interval [CI], 0.69 to 0.82). Among those who had repeat measurements, the initial values had a median of 50 and an interquartile range (IQR) ranging from 12.5 to 50. The repeat values had a median of 50 and an IQR ranging from 0 to 50.
TABLE 1.
Demographics of study participantsa
| Characteristic | Rochester (n = 1,082) | San Diego (n = 1,009) | Total (n = 2,091) |
|---|---|---|---|
| Gender | |||
| Male | 596 (55.1) | 738 (73.1) | 1,334 (63.8) |
| Female | 486 (44.9) | 271 (26.9) | 757 (36.2) |
| Age at enrollment (yr) | |||
| Median | 15.0 | 24.0 | 18.0 |
| Range | 11.0–22.0 | 19.0–40.0 | 11.0–40.0 |
| Race | |||
| American Indian, Alaska native | 4 (0.4) | 19 (1.9) | 23 (1.1) |
| Asian, Hawaiian or Pacific Islander | 27 (2.5) | 49 (4.9) | 76 (3.6) |
| Black or African American | 86 (7.9) | 169 (16.7) | 255 (12.2) |
| White | 922 (85.2) | 538 (53.3) | 1,460 (69.8) |
| Multiple | 29 (2.7) | 86 (8.5) | 115 (5.5) |
| Other | 7 (0.6) | 127 (12.6) | 134 (6.4) |
| Unknown | 7 (0.6) | 21 (2.1) | 28 (1.3) |
| Ethnicity | |||
| Not Hispanic | 1,054 (97.4) | 752 (74.5) | 1,806 (86.4) |
| Hispanic | 21 (1.9) | 215 (21.3) | 236 (11.3) |
| Do not know | 7 (0.6) | 42 (4.2) | 49 (2.3) |
Except for age, values are numbers of individuals; values in parentheses are percentages.
FIG 1.
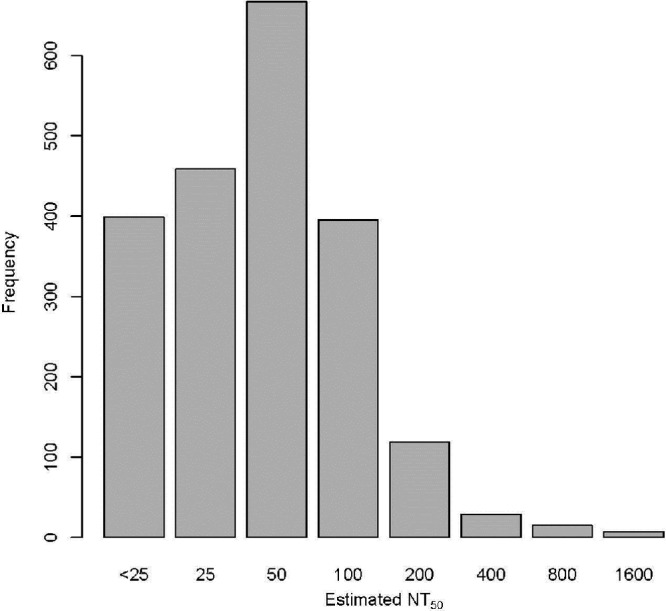
Distribution of initial NT50 values, estimated as the first dilution that resulted in a 50% reduction in observed activity from the positive control. Rubella virus-specific neutralizing antibodies were measured in 2,091 vaccinated subjects using a high-throughput ICA. The broad spectrum of observed NT50 (0 to 1,600) demonstrates that there is a large range in the levels of neutralizing antibodies in vaccinated cohorts.
TABLE 2.
Agreement between initial and repeated neutralizing antibody titers, evaluated without interpolationa
| Initial value | No. (%) of subjects with indicated NT50 value |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Repeat value |
Total | ||||||||
| 0 | 25 | 50 | 100 | 200 | 400 | 800 | 1,600 | ||
| 0 | 123 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 123 (25.1) |
| 25 | 1 (1.0) | 60 (57.7) | 42 (40.4) | 1 (1.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 104 (21.2) |
| 50 | 0 (0.0) | 42 (24.6) | 94 (55.0) | 35 (20.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 171 (34.8) |
| 100 | 0 (0.0) | 1 (1.7) | 25 (43.1) | 27 (46.6) | 5 (8.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 58 (11.8) |
| 200 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 7 (29.2) | 14 (58.3) | 3 (12.5) | 0 (0.0) | 0 (0.0) | 24 (4.9) |
| 400 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (40.0) | 1 (20.0) | 2 (40.0) | 0 (0.0) | 5 (1.0) |
| 800 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 4 (100.0) | 0 (0.0) | 4 (0.8) |
| 1,600 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (50.0) | 1 (50.0) | 2 (0.4) |
| Total | 124 | 103 | 161 | 70 | 221 | 4 | 7 | 1 | 491 |
Values are shown as n (% of the row), with initial NT50 estimates shown for rows and repeat NT50 estimates shown for columns. Those subjects with an initial NT50 of <1:25 were assigned a value of 0. A weighted kappa of 0.76 with a 95% confidence interval between 0.69 and 0.82 suggests that this assay produces reproducible data.
Interpolation of neutralizing antibody titers.
Loess interpolation was performed on all of the observed data as described above to obtain three NT50 values that corresponded to the three different reference conditions, the MAX, the VIC, and the SNS. We chose to use averages of either 2 or 3 of these three values to represent our best estimate of rubella virus-specific neutralizing antibody titers. We estimated the intraclass correlation (ICC) from the log-transformed estimates obtained from the 491 subjects with repeated measurements for the NT50 values in order to measure the reproducibility of our interpolation approach when applied to results from repeated assays. The ICC was equal to 0.89, suggesting a high degree of reproducibility in the assay, as measured by our interpolated NT50 values. Figure 2 illustrates the distribution of these interpolated estimates. We compared these interpolated values to the initial NT50 estimates, and the relationship between these values suggests that the interpolated values are broadly in agreement with the uninterpolated values. Figure 3 illustrates the agreement of initial observed NT50 levels and those calculated using the loess approach of interpolation. The most visible discrepancy between the interpolated and uninterpolated values is for those originally read as having a titer of 25. There are outlying points where the interpolation procedure estimated a value that was somewhat higher than typical. Upon inspection, these differences tended to arise when a much lower absorbance was observed in one or both of the SNS- or VIC-positive controls than the absorbance observed at the maximum dilution of 1:200.
FIG 2.
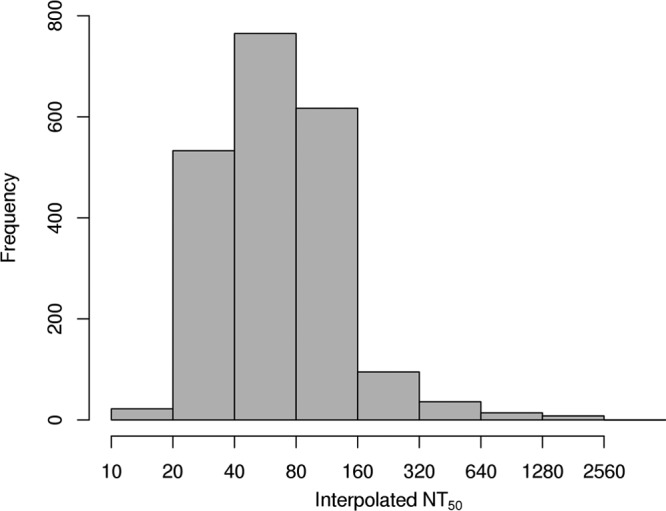
Distribution of interpolated NT50 values, estimated via the loess-based interpolation approach. Rubella virus-specific neutralizing antibodies were interpolated for 2,090 vaccinated subjects, using a high-throughput ICA. This distribution agrees with that generated by the uninterpolated values, with the exception that fewer interpolated values were estimated to be less than a titer of 1:25.
FIG 3.
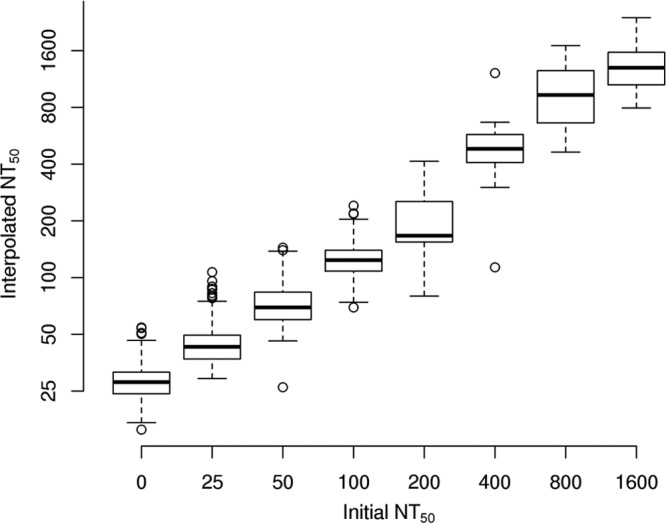
Box plots of NT50 values, estimated via the loess-based interpolation approach, within categories of initial NT50 values, estimated without interpolation. A high degree of agreement is reflected between the two methods, as the box plots of the interpolated values demonstrate tight distributions within uninterpolated NT50 values and they increase in a regular pattern as a function of the uninterpolated NT50 values.
That our assessment of the reproducibility of rubella virus-specific neutralizing antibodies resulted in an ICC of 0.89 suggests that the loess-interpolated values are highly replicable and that the approach is more than adequate for research purposes, although it doesn't quite meet the standard of 0.9 required for clinical use (24). This compares favorably to the weighted kappa of 0.76 for the categorical lab-based measurements, which suggests a substantial, but not outstanding, degree of reproducibility (25). Importantly, when we compared the categorical titration values estimated via laboratory observation to the interpolated NT50 values using analysis of variance, nearly 90% of the variability in the interpolated values was explained by the categorical lab-based estimates of rubella virus-specific antibody levels.
In certain cases, we had to apply omission thresholds and custom titer definitions in response to observed performance of the interpolation approach. For example, if the data were consistent with the neutralizing antibody titer being lower than the lowest dilution (1:25), then the interpolation approach would typically estimate the neutralizing titer to be just outside the range of observed dilutions (e.g., 1:24 or 1:23). When we use these interpolated values in analyses, we use the interpolated titer value. However, for descriptive purposes we define these subjects as having an NT50 below minimum detection (1:25) and would consider them “nonresponders.” This is in keeping with the manually estimated values, where the observed value was recorded as 0 if the observed NT50 was below a titer of 1:25. Other quality control thresholds allow us to flag the assay results from subjects whose data do not follow a traditional titration curve. Upon review of the data flagged by these thresholds, we are able to assess whether the interpolated value is likely to be an appropriate estimate of an individual's rubella virus-specific neutralizing antibody titers and therefore to decide whether or not it is appropriate to include the data in analyses.
We have previously reported rubella virus-specific IgG levels using Beckman Coulter's Access Rubella IgG assay (12, 26–30). This assay calculates anti-rubella virus (strain HPV-77) titers based on a standard curve calibrated against the WHO 2nd International Standard Preparation for Anti-Rubella Serum. In order to demonstrate the accuracy of our interpolated values, we examined the correlation between rubella virus-specific IgG levels and NT50 values in 732 subjects with both assessments (Fig. 4). The degree of agreement between the results of the two assays decreases for increasing estimates of antibody levels, and the rubella virus-specific IgG levels have some ability to distinguish antibody levels below the lower limit of detection in a neutralization assay. However, the Spearman correlation coefficient between the two measurements was 0.76, suggesting a strong, but not perfect, association between the results of the two assays. Although the major focus of this study was to test the precision of the sICNA high-throughput assay and to develop a statistical interpolation method, the strong association observed between NT50 and rubella IgG EIA could be considered a step toward standardizing our assay.
FIG 4.
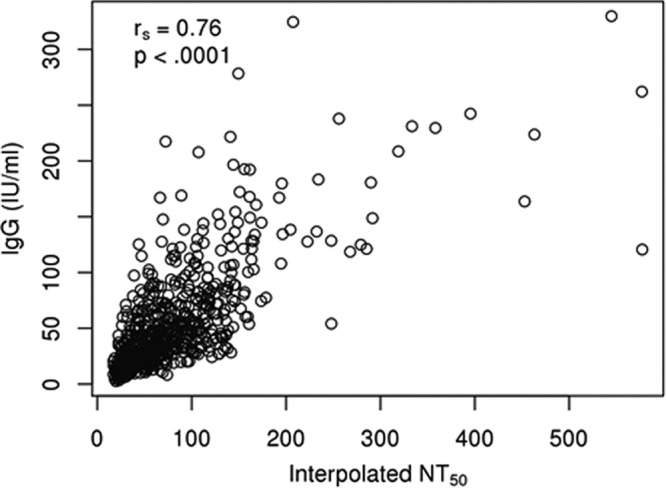
Scatter plot demonstrating the association between IgG levels calculated using a rubella virus-specific EIA and the interpolated values obtained from our high-throughput ICA for 732 participants with both measurements. Open circles represent the assay results from both assays, measured for each individual, and rs is the IgG/NT50 Spearman rank correlation coefficient. Although agreement between the two methods decreases with increasing antibody levels and the interpolated NT50 values have a somewhat higher lower limit of detection, the correlation coefficient of 0.76 suggests a strong correspondence between results from the two assay methods.
DISCUSSION
The high-throughput neutralizing antibody assay described here was adapted from an indirect immunocolorimetric assay (ICA) used for the detection and quantification of rubella virus (16). The advantages of the first generation of the ICA are its low cost, sensitivity of detection (as few as 10 PFU), adaptability to other virus models, and the ability to measure neutralizing antibodies. Because the detection of virus-infected cells relies on the naked eye to visualize results by ICA, it is not feasible to measure neutralizing titers in a large number of sera. Our goal was to build upon the ICA to develop an automated, high-throughput assay that would eliminate human error in the quantification of neutralizing antibodies in a large cohort of vaccinated subjects. This assay measures the total amount of virus in infected cells rather than the number of individual foci of infection. Since foci of rubella infection can vary in size, this assay should be more reproducible than counting individual foci visually. We successfully deployed this approach to assess neutralizing antibody titers in a total of 2,091 subjects. The high-throughput format allowed testing for the current study to be accomplished with ∼15 person months' worth of investment. After optimization of the statistical methods, the NT50 interpolation is now automated and can be run over a very short time.
The initial serum titers were reported as the value nearest to the dilution that decreased rubella virus levels by 50% (NT50). These results were efficient enough to categorize the extremes of our cohort, such as the highest and lowest responders (Fig. 1). However, our laboratory focuses on the influence of individual genetic variants on immunity to rubella virus vaccine. We have previously used statistical methods to estimate neutralizing antibodies to vaccinia virus in more than 3,000 samples from military personnel vaccinated with ACAM2000 (Acambis) (31). Our first approach was to fit the initial antibody titers to a logistic model. Although this model was successful in interpolating values between titers and has the advantage of directly modeling the NT50 value as one of its parameters, it was successful for only 95% of samples. We found that the logistic model failed either when there was no evidence of a rubella virus antibody response or when there were large differences between the measured absorbance at the maximum titration and the values observed for the positive virus controls. As the data from the assay did not show issues with monotonicity, we opted to use a different statistical estimation model in order to obtain estimates of NT50 from such samples rather than to repeat the assay in this high-throughput setting. The loess approach that we ultimately used successfully returned an interpolated NT50 estimate for all subjects.
Traditionally, immunity to rubella is considered to occur when antibody levels are at or above 10 IU/ml (32). It is unclear at present how neutralizing antibody titers measured using this method compare to established methodologies. The gold standard to compare novel methods of rubella virus-specific antibody quantification is the HAI test (33). By developing a standardized and highly reproducible neutralization testing protocol, our work has likely made statistically significant studies correlating international units (IU/ml) with neutralization titers possible, as has been done for measles virus (34).
In conclusion, we have described the adaption of an established ICA to quantify the levels of neutralizing antibodies against rubella virus in vaccinated individuals and used statistical methods to interpolate titers. This assay is highly sensitive and exhibits acceptable levels of variability between repeats. There is also a positive correlation between this assay, which measures functional, neutralizing antibody levels, and an accepted, standardized assay for the measurement of all rubella virus-specific antibody levels. The automation of a portion of the assay has decreased the potential for human error and allows for rapid assessment of large numbers of samples. It is substantially lower in cost than other high-throughput systems and may be adaptable to other viral diseases. Another great strength of the assay is the ability to quantify the functional (neutralizing antibody) humoral response to rubella vaccine.
The interpolation of rubella virus-specific neutralizing antibodies produces results that are nearly equivalent to the measurements obtained without interpolation. This finding, taken together with the high reproducibility of the assay, suggests that the use of this interpolation approach produces estimates of rubella-specific neutralizing antibody levels that appropriately estimate the levels actually present in the serum samples. Although this assay still needs to be standardized, its usefulness as a functional measurement may be applied to investigating interindividual differences in vaccine response or, perhaps, as a tool in clinical trials of novel rubella vaccine candidates and in determining postvaccination immunity in a population for rubella control and elimination activities.
ACKNOWLEDGMENTS
We thank Lijuan Hao and Adebola Adebayo of the Centers for Disease Control and Prevention (CDC) for their assistance with neutralization tests. We thank Caroline L. Vitse for her editorial assistance.
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R37 AI048793-11 (which recently received a MERIT award). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing interests: G. A. Poland is the chair of a Safety Evaluation Committee for novel investigational vaccine trials being conducted by Merck Research Laboratories. G. A. Poland offers consultative advice on vaccine development to Merck & Co. Inc., CSL Biotherapies, Avianax, Sanofi Pasteur, Dynavax, Novartis Vaccines and Therapeutics, PAXVAX Inc., and Emergent Biosolutions. G. A. Poland holds two patents related to vaccinia peptide research. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. This research has been reviewed by the Mayo Clinic Conflict of Interest Review Board and was conducted in compliance with Mayo Clinic Conflict of Interest policies.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the U.S. Department of Health and Human Services.
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Katow S. 1998. Rubella virus genome diagnosis during pregnancy and mechanism of congenital rubella. Intervirology 41:163–169. 10.1159/000024931 [DOI] [PubMed] [Google Scholar]
- 2.Katow S. 2004. Molecular epidemiology of rubella virus in Asia: utility for reduction in the burden of diseases due to congenital rubella syndrome. Pediatr. Int. 46:207–213. 10.1046/j.1442-200x.2004.01866.x [DOI] [PubMed] [Google Scholar]
- 3.Strebel PM, Gacic-Dobo M, Reef S, Cochi SL. 2011. Global use of rubella vaccines, 1980–2009. J. Infect. Dis. 204(Suppl 2):S579–S584. 10.1093/infdis/jir447 [DOI] [PubMed] [Google Scholar]
- 4.Dimech W, Arachchi N, Cai J, Sahin T, Wilson K. 2013. Investigation into low-level anti-rubella virus IgG results reported by commercial immunoassays. Clin. Vaccine Immunol. 20:255–261. 10.1128/CVI.00603-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castellano GA, Madden DL, Hazzard GT, Cleghorn CS, Vails DV, Ley AC, Tzan NR, Sever JL. 1981. Evaluation of commercially available diagnostic test kits for rubella. J. Infect. Dis. 143:578–584. 10.1093/infdis/143.4.578 [DOI] [PubMed] [Google Scholar]
- 6.Dimech W, Panagiotopoulos L, Francis B, Laven N, Marler J, Dickeson D, Panayotou T, Wilson K, Wootten R, Dax EM. 2008. Evaluation of eight anti-rubella virus immunoglobulin g immunoassays that report results in international units per milliliter. J. Clin. Microbiol. 46:1955–1960. 10.1128/JCM.00231-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meegan JM, Evans BK, Horstmann DM. 1982. Comparison of the latex agglutination test with the hemagglutination inhibition test, enzyme-linked immunosorbent assay, and neutralization test for detection of antibodies to rubella virus. J. Clin. Microbiol. 16:644–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimech W, Bettoli A, Eckert D, Francis B, Hamblin J, Kerr T, Ryan C, Skurrie I. 1992. Multicenter evaluation of five commercial rubella virus immunoglobulin G kits which report in international units per milliliter. J. Clin. Microbiol. 30:633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Truant AL, Barksdale BL, Huber TW, Elliott LB. 1983. Comparison of an enzyme-linked immunosorbent assay with indirect hemagglutination and hemagglutination inhibition for determination of rubella virus antibody: evaluation of immune status with commercial reagents in a clinical laboratory. J. Clin. Microbiol. 17:106–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson JC, Hadler SC, Dykewicz CA, Reef S, Phillips L. 1998. Measles, mumps, and rubella—vaccine use and strategies for elimination of measles, rubella, and congenital rubella syndrome and control of mumps: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend. Rep. 47:1–57 [PubMed] [Google Scholar]
- 11.Tosh PK, Kennedy RB, Vierkant RA, Jacobson RM, Poland GA. 2009. Correlation between rubella antibody levels and cytokine measures of cell-mediated immunity. Viral Immunol. 22:451–456. 10.1089/vim.2009.0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenwood NP, Ovsyannikova IG, Vierkant RA, O'Byrne MM, Poland GA. 2010. A qualitative and quantitative comparison of two rubella virus-specific IgG antibody immunoassays. Viral Immunol. 23:353–357. 10.1089/vim.2010.0026 [DOI] [PubMed] [Google Scholar]
- 13.Ovsyannikova IG, Dhiman N, Haralambieva IH, Vierkant RA, O'Byrne MM, Jacobson RM, Poland GA. 2010. Rubella vaccine-induced cellular immunity: evidence of associations with polymorphisms in the Toll-like, vitamin A and D receptors, and innate immune response genes. Hum. Genet. 127:207–221. 10.1007/s00439-009-0763-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy RB, Ovsyannikova IG, Pankratz VS, Vierkant RA, Jacobson RM, Ryan MA, Poland GA. 2009. Gender effects on humoral immune responses to smallpox vaccine. Vaccine 27:3319–3323. 10.1016/j.vaccine.2009.01.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. 2011. Human leukocyte antigen genotypes in the genetic control of adaptive immune responses to smallpox vaccine. J. Infect. Dis. 203:1546–1555. 10.1093/infdis/jir167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen MH, Zhu Z, Zhang Y, Favors S, Xu WB, Featherstone DA, Icenogle JP. 2007. An indirect immunocolorimetric assay to detect rubella virus infected cells. J. Virol. Methods 146:414–418. 10.1016/j.jviromet.2007.08.021 [DOI] [PubMed] [Google Scholar]
- 17.Chen MH, Icenogle JP. 2007. Molecular virology of rubella virus, p 1–18 In Banatvala J, Pekcham C. (ed), Rubella viruses. Elsevier, Amsterdam, Netherlands [Google Scholar]
- 18.Bellini WJ, Icenogle JP, Sever JL. 2009. Measles, mumps, and rubella, p 562–577 In Spector S, Hodinka RL, Young SA, Wiedbrauk DL. (ed), Clinical virology manual, 4th ed. ASM Press, Washington, DC [Google Scholar]
- 19.Steece RS, Talley MS, Skeels MR, Lanier GA. 1985. Comparison of enzyme-linked immunosorbent assay, hemagglutination inhibition, and passive latex agglutination for determination of rubella immune status. J. Clin. Microbiol. 21:140–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuno T. 1984. A rapid simple indirect neutralization test for rubella antibody. Acta Virol. 28:523. [PubMed] [Google Scholar]
- 21.Kobayashi N, Shibuta H, Matumoto M. 1973. Agar-disc neutralization test for rubella virus. Jpn. J. Microbiol. 17:313–316. 10.1111/j.1348-0421.1973.tb00778.x [DOI] [PubMed] [Google Scholar]
- 22.Rawls WE, Desmyter J, Melnick JL. 1967. Rubella virus neutralization by plaque reduction. Proc. Soc. Exp. Biol. Med. 124:167–172. 10.3181/00379727-124-31692 [DOI] [PubMed] [Google Scholar]
- 23.Cleveland WS, Gross E, Shyu WM. 1992. Local regression models, p 309–376 In Chambers JM, Hastie TJ. (ed), Statistical models. Wadsworth & Brooks/Cole, Springer, Berlin, Germany [Google Scholar]
- 24.Shoukri MM, Asyali MH, Donner A. 2004. Sample size requirements for the design of reliability study: review and new results. Stat. Methods Med. Res. 13:251–271. 10.1191/0962280204sm365ra [DOI] [Google Scholar]
- 25.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174. 10.2307/2529310 [DOI] [PubMed] [Google Scholar]
- 26.Ovsyannikova IG, Jacobson RM, Vierkant RA, O'Byrne MM, Poland GA. 2009. Replication of rubella vaccine population genetic studies: validation of HLA genotype and humoral response associations. Vaccine 27:6926–6931. 10.1016/j.vaccine.2009.08.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovsyannikova IG, Haralambieva IH, Dhiman N, O'Byrne MM, Pankratz VS, Jacobson RM, Poland GA. 2010. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. J. Infect. Dis. 201:207–213. 10.1086/649588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. 2010. Extended LTA, TNF, LST1 and HLA gene haplotypes and their association with rubella vaccine-induced immunity. PLoS One 5:e11806. 10.1371/journal.pone.0011806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haralambieva IH, Dhiman N, Ovsyannikova IG, Vierkant RA, Pankratz VS, Jacobson RM, Poland GA. 2010. 2′-5′-Oligoadenylate synthetase single-nucleotide polymorphisms and haplotypes are associated with variations in immune responses to rubella vaccine. Hum. Immunol. 71:383–391. 10.1016/j.humimm.2010.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhiman N, Haralambieva IH, Kennedy RB, Vierkant RA, O'Byrne MM, Ovsyannikova IG, Jacobson RM, Poland GA. 2010. SNP/haplotype associations in cytokine and cytokine receptor genes and immunity to rubella vaccine. Immunogenetics 62:197–210. 10.1007/s00251-010-0423-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kennedy R, Pankratz VS, Swanson E, Watson D, Golding H, Poland GA. 2009. Statistical approach to estimate vaccinia-specific neutralizing antibody titers using a high-throughput assay. Clin. Vaccine Immunol. 16:1105–1112. 10.1128/CVI.00109-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skendzel LP. 1996. Rubella immunity. Defining the level of protective antibody. Am. J. Clin. Pathol. 106:170–174 [DOI] [PubMed] [Google Scholar]
- 33.Weber B. 1997. Current developments in the laboratory diagnosis of rubella. Bull. Soc. Sci. Med. Grand Duche Luxemb. 134:31–41 (In German) [PubMed] [Google Scholar]
- 34.Cohen BJ, Audet S, Andrews N, Beeler J. 2007. Plaque reduction neutralization test for measles antibodies: description of a standardised laboratory method for use in immunogenicity studies of aerosol vaccination. Vaccine 26:59–66. 10.1016/j.vaccine.2007.10.046 [DOI] [PubMed] [Google Scholar]


