Abstract
The sets of compounds that can support growth of an organism are defined by the presence of transporters and metabolic pathways that convert nutrient sources into cellular components and energy for growth. A collection of known nutrient sources can therefore serve both as an impetus for investigating new metabolic pathways and transporters and as a reference for computational modeling of known metabolic pathways. To establish such a collection for Escherichia coli K-12, we have integrated data on the growth or nongrowth of E. coli K-12 obtained from published observations using a variety of individual media and from high-throughput phenotype microarrays into the EcoCyc database. The assembled collection revealed a substantial number of discrepancies between the high-throughput data sets, which we investigated where possible using low-throughput growth assays on soft agar and in liquid culture. We also integrated six data sets describing 16,119 observations of the growth of single-gene knockout mutants of E. coli K-12 into EcoCyc, which are relevant to antimicrobial drug design, provide clues regarding the roles of genes of unknown function, and are useful for validating metabolic models. To make this information easily accessible to EcoCyc users, we developed software for capturing, querying, and visualizing cellular growth assays and gene essentiality data.
INTRODUCTION
The diversity of chemical environments that will support organismal growth is a fundamental corpus of scientific knowledge. However, despite decades of research and large amounts of accumulated knowledge, no compendium of the set of nutrients Escherichia coli K-12 is able to utilize exists. Such a compendium is not only of basic interest but also would serve as a reference for computational metabolic modeling, which requires a comprehensive set of reference growth data sets for evaluating the accuracy of growth predictions.
Also of interest for validating computational metabolic models are experimental data describing the essentiality of an organism's genes, because metabolic models can predict the phenotypes of gene knockouts. An essential gene is a gene that is indispensable to support life under a specific set of growth conditions. Further, gene essentiality data are useful for predicting antibiotic targets for pathogenic bacteria, for guiding the design of minimal genomes, and for providing clues regarding the roles of genes of unknown function.
A large number of high-throughput data sets—from gene expression to metabolomics to phenotypic measurements—are becoming available for a variety of organisms. To maximize their value, these data sets should be captured and integrated within a database and published on a website, along with tools for querying and analysis to ensure that the data are maximally available to the scientific community. However, integration requires more than simply collecting multiple data sets together in a common repository. For some data types, integration should include identifying and resolving conflicts between the data sets when possible in order to extract as much knowledge as possible from noisy data.
We integrated the following growth data into a single collection available in the EcoCyc database (1) and in the supplemental files: (i) a collection of observations of the growth or nongrowth of laboratory strains of E. coli K-12 on a variety of media that had been obtained through low-throughput methods and had been previously published in the literature; (ii) low-throughput growth data generated by our group; (iii) previously published high-throughput phenotype microarray (PM) data (2); and (iv) PM data generated by our group. PMs measure cellular respiration as a proxy for growth (henceforth, we refer to growth for simplicity) across several sets of 96-well plates, with each well containing a different combination of nutrients. We also integrated into EcoCyc six data sets that describe the growth of single-gene knockout mutants of E. coli K-12 that have been published in recent years.
We developed software tools for capturing, querying, and visualizing cellular growth assays, the corresponding nutrients and growth conditions, and gene essentiality data. These tools have been integrated within the Pathway Tools software (3), thus enabling their use in conjunction both with the more than 2,000 genomes contained in the BioCyc database collection (BioCyc.org) and with organism databases created by other Pathway Tools users.
MATERIALS AND METHODS
Bacterial strains.
E. coli K-12 MG1655 was obtained from both the Yale Coli Genetic Stock Center (CGSC; strain 7740) and from the American Type Culture Collection (ATCC; strain 700926).
Laboratory evaluation of individual growth media.
To evaluate growth in soft agar, the following protocol was used based on a modification of an earlier Salmonella nutrient evaluation method (4). Bacterial cells were grown overnight in LB medium at 37°C. The overnight culture was then washed three times with phosphate-buffered saline (PBS), resuspended in PBS, and diluted 10:1 into cooling liquefied 0.6% agar solution before it was plated on top of the appropriate base plate. Fully cooled plates were then used for subsequent nutrient testing. Base plates were made with minimal salts medium (1.9 mM potassium sulfate, 25.8 mM dipotassium phosphate, 11.8 mM monopotassium phosphate, 0.13 mM magnesium sulfate heptahydrate) supplemented with 1.5% agar and either 0.5% succinate (for nitrogen source testing) or 2.5 mM NH4Cl (for carbon source testing). Each nutrient was tested on two plates (containing either succinate or NH4Cl) by placing 2 to 20 mg of the nutrient in the center of each plate. The plates were incubated at 37°C, and growth was evaluated visually at 24, 48, and 72 h. A plate was scored as positive for growth if a bacterial lawn of ≥1 cm2 was present during one of these scoring checks. All growth conditions were tested in duplicate.
To evaluate growth in liquid culture, strains were grown overnight in M9 minimal media with 0.2% (vol/vol) glycerol. The overnight culture was washed three times with modified M9 minimal media containing no carbon or nitrogen [48 mM sodium phosphate dibasic, 22 mM monopotassium phosphate, 8.5 mM sodium chloride, 2 mM magnesium sulfate, 0.1 mM calcium chloride, 0.01 mM Fe(II) sulfate] before transfer to a new M9 minimal medium with a starting optical density at 600 nm of 0.05. Carbon sources were tested in M9 minimal medium at a final concentration of 0.2% (wt/vol), while nitrogen sources were tested in modified M9 minimal medium at a final concentration of 0.2% (wt/vol). The carbon source in these latter assays was 0.2% sodium succinate. Cultures were incubated at 37°C for 48 h, and growth rates are reported as means ± the standard deviations for three replicates. The appropriate positive and negative controls were included in all experiments. For nitrogen source assays, the positive control was ammonium chloride; for carbon source assays, the positive control was sodium succinate.
Biolog PMs.
Biolog phenotype microarrays (PMs) are a commercially available (Biolog, Inc., Hayward, CA) high-throughput system for the global analysis of microbial respiration phenotypes (2). PMs use a colorimetric reaction (reduction of a tetrazolium dye by NADH) as a sensitive indicator of microbial respiration in response to the presence of individual nutrients or chemicals. PM plates 1 to 4, which contained 190 sole carbon sources, 95 sole nitrogen sources, 59 sole phosphate sources, and 35 sole sulfur sources, were used in this analysis. E. coli K-12 MG1655 was pregrown on nutrient agar and then used to inoculate the plates according to Biolog instructions. The data were collected and analyzed using the OmniLogH PM system, which records the color change every 15 min for each well in the 96-well assay plates. All incubations were performed at 37°C over 48 h.
To assess the effect of pregrowth conditions on carbon-source utilization, the bacterial strain was pregrown on three types of nutrient agar plates—LB agar, R2A agar, and BUG-S (Biolog universal growth agar with 5% sheep blood)—before the inoculation of PM plates 1 and 2. For the nitrogen, phosphate, and sulfur utilization plates (PM3 and PM4), bacteria were pregrown on LB agar and, in the first instance, inoculated according to the manufacturer's instructions. The user-supplied carbon source in these assays was 20 mM sodium succinate and 2 μM ferric citrate. Under these conditions, a positive response was recorded in the negative-control well for both the nitrogen assays and the sulfur assays. Thereafter, all bacterial pregrowth for these assays was done on the nutrient-limited R2A agar and, after discussion with the manufacturer, the concentration of the cell inoculum was decreased by a factor of 10. The use of R2A agar for bacterial pregrowth as the preferred method for minimizing the response seen in the negative-control well for nitrogen and sulfur assays has been reported previously (2).
Data integration and arbitration.
Respiration data obtained from PMs were used as a proxy for growth. Growth observations can be recorded in Pathway Tools as growth, no growth, or low (borderline) growth. No attempt was made to unify the criteria used to distinguish between these three states in the imported PM data because original growth/respiration curve data were not available from most of the data sets; rather, we used the determinations as specified by the providers of each data set. For our own PM experiments (Mackie12), kinetic data were analyzed using the Omnilog PM software and response calls of growth (G), no growth (NG), and low growth (LG) were made based on parameters of maximum height (the 10th percentile highest value among all values over all time points) and area (the sum of all Omnilog values over all time points). No calls were made for 13 of the nitrogen source wells, 10 of the sulfur source wells, and the three phosphate sources, all of which showed ambiguous responses compared to the negative-control well. Data set S6 in the supplemental material includes raw data for all PM experiments conducted during the course of this work. “Mackie12” refers to experiments conducted during 2012, and these data are currently available on the EcoCyc website. The results of later experiments (dated 2013 in File S6 in the supplemental material) conducted to compare Biolog PM responses of different stock cultures of E. coli K-12 MG1655 will be included in a future EcoCyc release.
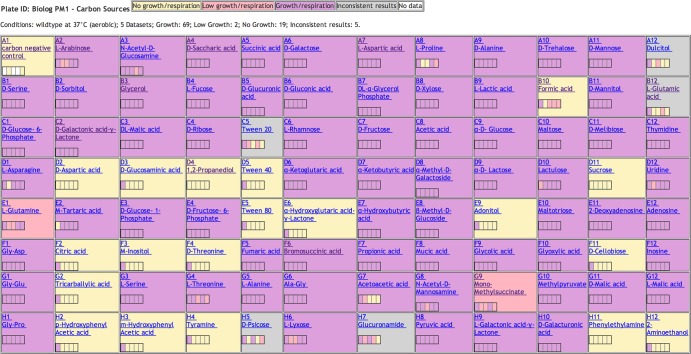
After the PM data sets had been added to EcoCyc, we reviewed the growth observations where at least one difference in observations between the five PM data sets existed, and we resolved the conflict when significant evidence regarding the actual growth outcome was found. The factors considered included the number of available observations of growth versus no growth, whether low-throughput observations could be found in the literature for a given growth condition (in several cases they were), and whether the minority data set(s) were generally judged to be less reliable based on their total pairwise conflicts with other data sets. When we did resolve a conflict, a comment was usually created in EcoCyc to summarize the rationale, with literature citations recorded if appropriate. For example, aerobic cell PM1:F2, which tests for growth on citric acid as a sole carbon source, was resolved in favor of no growth (5), whereas aerobic cell PM1:G7, which tests for growth on acetoacetic acid as a sole carbon source, was resolved in favor of growth (6) (mouse over these cells in the online EcoCyc version of Fig. 1 at http://ecocyc.org/ECOLI/NEW-IMAGE?object=Growth-Media to see the explanation for the resolution).
FIG 1.
Table showing the current display of integrated growth observations in EcoCyc for the PM1 carbon sources under aerobic conditions. Colors indicate consensus and conflicts (see color legend at top). The colored bar within each cell depicts individual observations, e.g., cell H5 (d-psicose) shows there were two observations of no growth, two observations of growth, and one observation of low growth. Mousing over the bar on the BioCyc.org version of this diagram provides a literature reference for each of the growth observations.
RESULTS
Table 1 summarizes the sources of nutrient utilization data present in EcoCyc version 17.5 that was released in October/November 2013. The results of the low-throughput growth experiments reported here will appear in a future EcoCyc release.
TABLE 1.
Numbers of aerobic and anaerobic growth observations described here, grouped by data source
| Type of data | No. of growth observations |
|
|---|---|---|
| Aerobic | Anaerobic | |
| Low-throughput data from the literature | 23 | 0 |
| Low-throughput data generated by our group | 49 | 0 |
| High-throughput PM data from the literature | 1,244 | 191 |
| High-throughput PM data generated by our group | 353 | 0 |
Table 1 includes a total of 1,860 growth observations under 552 growth conditions. The 552 growth conditions include 456 aerobic conditions and 96 anaerobic conditions and include 384 PM media (including 5 negative controls) and 72 other media. For example, if growth was assessed on one set of nutrients, under both aerobic and anaerobic conditions, we count that situation as two growth conditions.
The growth data that we integrated from the literature included the following commonly used media: AB, LB medium enriched, LB Lennox, 3-(N-morpholino)propanesulfonic acid (MOPS), M9, M56, and M63. These and other media are listed in the first table at the EcoCyc.org All Growth Media site (http://biocyc.org/ECOLI/NEW-IMAGE?object=Growth-Media).
Integration of five PM data sets.
We integrated five aerobic phenotype microarray (PM) data sets that had been produced independently by five different laboratories that we refer to as Bochner12 (B. Bochner, unpublished data), AbuOun09 (7), Baumler11 (8), Yoon12 (9), and Mackie12. These data sets consisted of measurements on the four PM plates designated PM1 (95 carbon sources), PM2 (95 carbon sources), PM3 (95 nitrogen sources), and PM4 (59 phosphorus plus 35 sulfur sources), except for Baumler11, which covers PM1 only.
For each data set, we compared the result for each well with the result for the same well from each of the other data sets in turn. Table 2 summarizes the degree of pairwise agreement and disagreement among these data sets. The percent pairwise agreement is the number of matches expressed as a percentage of the total number of comparisons (for example, 86% of the pairwise comparisons between AbuOun09 and all other data sets for the carbon source PM plates were in agreement). Data set S1 in the supplemental material lists all pairwise comparisons among the individual data sets. Table 2 combines all pairwise comparison results into a single number that reflects the percentage agreement of each data set with all four other data sets. We found that pairwise agreement among these data sets for individual growth observations ranged from 72 to 87%. When conflicts exist, we distinguish between the cases in which one data set records growth and the other shows no growth (we call these “Y/N” conflicts) and the cases in which one data set records low or borderline growth and the other shows either growth or no growth. For the 379 growth conditions surveyed by the PM data sets, a conflict existed between at least one pair of data sets for 119 of the wells, of which 57 wells had Y/N conflicts.
TABLE 2.
Concordance for aerobic datasetsa
| Data set | % Agreement |
% Y/N conflicts |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Carbon | Nitrogen | Phosphorus | Sulfur | All | Carbon | Nitrogen | Phosphorus | Sulfur | All | |
| AbuOun09 | 86 | 75 | 89 | 84 | 84 | 5 | 11 | 5 | 1 | 4 |
| Baumler11 | 72 | NA | NA | NA | 72 | 21 | NA | NA | NA | 21 |
| Bochner12 | 80 | 75 | 91 | 81 | 81 | 5 | 13 | 3 | 1 | 6 |
| Mackie12 | 87 | 80 | 93 | 91 | 87 | 4 | 8 | 4 | 0 | 5 |
| Yoon12 | 86 | 75 | 92 | 75 | 84 | 4 | 3 | 2 | 1 | 3 |
| All combined | 83 | 76 | 91 | 82 | 83 | 6 | 9 | 3 | 1 | 6 |
The left half of this table shows the percent pairwise agreement of the different datasets for sources of carbon, nitrogen, phosphorus, and sulfur, respectively. The right half of the table shows the percent pairwise Y/N (growth/no-growth) conflicts for the same data. NA, not applicable.
The Baumler11 data set is the outlier among the five PM data sets in that it shows significantly more pairwise conflicts with the other data sets than does any other data set. Further, in 15 of the 16 cases in which four data sets recorded the same growth observation (meaning Y [growth] or N [no growth], ignoring low growth) and one data set recorded the opposite observation, the outlier was Baumler11. If we remove the Baumler11 data set when computing the “all combined” agreement score in Table 2, the percent agreement for carbon sources increases from 82 to 87%; similarly, the Y/N conflict percentage for carbon sources in Table 2 drops from 6 to 2%.
Note that the PM3 (nitrogen source) plate shows significantly more disagreement than do the other plates. The different data sets show similar degrees of disagreement. After a systematic investigation of the differences among these data sets and a consideration of low-throughput E. coli growth data from the literature, we could resolve 73 of these differences (24 Y/N). As a result, consensus growth calls now exist for 333 of the 379 conditions surveyed. Unresolved differences prevent growth calls for the remaining 45 (of which 34 represent Y/N differences). Table 3 summarizes the number of conflicts that we resolved for each of the four nutrient types under aerobic conditions, and the number of conflicts remaining in each case. Table 4 lists the compounds that are involved in the remaining conflicts.
TABLE 3.
Summary of PM data after integration and conflict resolutiona
| Parameter or status | No. of wells |
|||
|---|---|---|---|---|
| Carbon | Nitrogen | Phosphorus | Sulfur | |
| No. of wells | 190 | 95 | 59 | 35 |
| No growth | 99 | 38 | 5 | 4 |
| Low growth | 2 | 0 | 0 | 0 |
| Growth | 80 | 32 | 50 | 23 |
| Remaining conflicts | 8 (9)b | 25 | 4 | 8 |
| Resolved conflicts | 52 | 10 | 6 | 5 |
Row 1 indicates the total numbers of wells for each nutrient type. Rows 2 to 4 indicate the numbers of wells for each nutrient type showing different growth levels for which growth can be assessed. Row 5 indicates the number of wells for which the growth state cannot be assessed because of conflicting observations that cannot be arbitrated. Row 6 gives the number of wells that originally contained conflicts that were resolved by our arbitration procedure. The sum of rows 2 to 5 equals row 1.
The conflicting PM result recorded for dulcitol (galactitol) is due to genetic variation between stock cultures of E. coli K-12 MG1655.
TABLE 4.
Nutrients that generate conflicting responses in phenotype microarrays
| Source | Nutrients |
|---|---|
| Carbon | l-Glutamic acid, Tween 20, d-psicose, glucuronamide, α-methyl-d-glucoside, pectin, α-keto-valeric acid, l-alaninamide |
| Nitrogen | l-Cysteine, l-glutamic acid, l-lysine, l-methionine, l-phenylalanine, l-threonine, d-asparagine, d-lysine, d-valine, l-homoserine, putrescine, agmatine, d-mannosamine, N-acetyl-d-mannosamine, adenine, guanosine, uracil, uridine, xanthine, xanthosine, uric acid, alloxan, dl-α-amino-N-butyric acid γ-amino-N-butyric acid, dl-α-amino-caprylic acid |
| Phosphorus | Cytidine 3′,5′-cyclic mono-P, phosphono acetic acid, 2-aminoethyl phosphonic acid, methylene diphosphonic acid |
| Sulfur | l-Cysteic acid, cysteamine, S-methyl-l-cysteine, dl-ethionine, thiourea, 1-thio-6-d-glucose, dl-lipoamide, taurocholic acid |
Nutrients in boldface have growth/no-growth (as opposed to growth/low-growth or low-growth/no-growth) conflicts.
Baumler11 and Bochner12 also include measurements under anaerobic observations, for PM1 only. Their data sets agreed for only 45% of the wells, with a Y/N conflict rate of 49%. We did not attempt to resolve these conflicts.
Low-throughput growth experiments.
After integration of the PM results into EcoCyc, the ability of 33 compounds to support growth of E. coli K-12 as a sole carbon or nitrogen source remained unresolved (Table 4). We tested 19 of these compounds in both minimal salts soft agar and M9 minimal medium to determine whether they could support aerobic growth as a sole nitrogen source (Table 5), and we tested six compounds to determine whether they could support aerobic growth as a sole carbon source (Table 6). This information will be included as conflict resolutions in a future release of EcoCyc.
TABLE 5.
Growth of E. coli K-12 MG1655 in minimal salts agar or M9 minimal medium with sole nitrogen sourcesa
| Nitrogen source | Growth in: |
|
|---|---|---|
| Minimal salts agar | M9 minimal medium | |
| l-Cysteine | Y | Y |
| l-Glutamic acid | Y | Y |
| l-Lysine | Y | Y |
| Putrescine | Y | Y |
| N-Acetyl-d-mannosamine | Y | Y |
| Adenine | Y | Y |
| Guanosine | Y | NT |
| Uric acid | Y | N |
| dl-α-Amino-N-butyric acid | Y | N |
| l-Threonine | Y | Y |
| l-Methionine | N | Y |
| l-Phenylalanine | N | Y |
| d-Valine | N | N |
| Agmatine | N | Y |
| Uracil | N | Y |
| Uridine | N | Y |
| Xanthine | N | N |
| γ-Amino-N-butyric acid | N | Y |
| Alloxan | N | N |
Growth curves in M9 minimal medium are provided in Fig. S4 and S5 in the supplemental material. Y, yes; N, no; NT, not tested.
TABLE 6.
Growth of E. coli K-12 MG1655 in minimal salts agar or M9 minimal medium with sole carbon sourcesa
| Carbon source | Growth in: |
|
|---|---|---|
| Minimal salts agar | M9 minimal medium | |
| l-Glutamate | N | N |
| α-Keto-valeric acid | N | N |
| Tween 20 | Y/N | N |
| Pectin | N | N |
| α-Methyl-d-glucoside | N | N |
| l-Alaninamide | N | N |
Growth curves in M9 minimal medium are provided in Fig. S4 and S5 in the supplemental material. N, no; Y/N, ambiguous.
Integration of gene essentiality data.
Table 7 lists the gene essentiality data sets that are present in version 17.5 of EcoCyc. These data sets include a total of 16,119 growth observations for knockout strains of E. coli. Note that the numbers listed below reflect the number of observations incorporated into EcoCyc and not necessarily the numbers reported in the original studies. Discrepancies between these numbers can be caused by subsequent corrections to the data (which have been reflected in the numbers below), gene merges, deletions, etc.
TABLE 7.
Gene essentiality datasets present in EcoCyca
| Data set | Medium | No. of knockouts |
||
|---|---|---|---|---|
| No growth | Growth | Data unavailable | ||
| Gerdes03 | LB medium enriched, aerobic | 3,082 | 1,419 | |
| Baba06 | LB Lennox, medium, aerobic | 324 | 3,844 | 333 |
| Baba06 | MOPS + 0.4% glucose, aerobic | 15 | 3,828 | 658 |
| Feist07 | MOPS + 0.4% glucose, aerobic | 107 | 996 | 3,398 |
| Patrick07 | M9 + 0.4% glucose, aerobic | 106 | 4,395 | |
| Joyce06 | M9 + 1% glycerol, aerobic | 113 | 3,704 | 684 |
For each data set, we list the one or more growth conditions under which gene essentiality was determined, and the number of knockouts exhibiting no growth, growth, or an indeterminate level of growth is indicated. MOPS, morpholinepropanesulfonic acid.
The Gerdes03 (10) study used a genetic footprinting technique with a Tn5-based transposon system and reported assessment of ca. 87% of E. coli open reading frames for essentiality. Although that study identified 620 genes as essential for aerobic growth in rich media, the actual experimental result is that no transposon insertions were identified for those genes. We did not consider that sufficient evidence for essentiality and have thus incorporated into EcoCyc only observations for the 3,082 genes where transposon insertions have been identified. Because a transposon insertion may not result in a null mutation, genes identified as nonessential in this data set may in fact be required for growth.
The Baba06 (11) knockouts were made using the lambda Red recombinase system, replacing target genes with in-frame kanamycin resistance genes. It was later found that a subset of these mutants contained partial duplications and that others were cross-contaminated (12). We combined the data from both publications in EcoCyc. A total of 324 genes were unable to be disrupted and were predicted to be essential for growth in rich media at 37°C. The 3,844 strains containing knockouts of genes that were found to be nonessential on rich media (henceforth referred to as the Keio collection) were further tested for growth on glucose minimal medium (11).
Joyce et al. (13) profiled the Keio collection for growth on minimal medium with glycerol as the sole source of carbon and energy. A total of 113 genes that were nonessential on rich medium were identified as essential for growth on glycerol. Joyce et al. also combined these observations with those made by Baba et al. regarding the conditional essentiality of the mutants when grown on glucose-supplemented minimal media and were able to identify a conserved conditionally essential core of 94 genes that are required for E. coli K-12 to grow under minimal nutritional supplementation but are not essential for growth under rich conditions.
Feist et al. (14) used the experimental data regarding conditional gene essentiality from earlier studies (11, 13) and compared them to the computationally predicted essential genes in their genome-scale metabolic reconstruction of E. coli K-12. This data set (which includes only the genes used in their model) is included in EcoCyc to facilitate benchmarking of computational predictions of essentiality from the EcoCyc model with predictions from the model of Feist et al.(14). Based on the ability of the Keio collection single-gene knockout mutants to grow on rich versus defined media, Baba et al. and Joyce et al. (11, 13) identified a set of conditionally essential genes. Patrick et al. (15) tested this subset of Keio mutants for their ability to form colonies on M9-glucose agar.
Growth and essentiality data within the Pathway Tools software.
We have extended the Pathway Tools software (3) to handle several aspects of growth observation data and gene essentiality data. A pathway/genome database (PGDB) such as EcoCyc can now capture growth observations of wild-type or mutant strains and can capture descriptions of the media in which growth was assayed. The new schema class “Growth-Media” lists the set of chemical compounds that comprise a single growth medium. The new schema class “Growth-Observations” defines one or more measurements of growth or nongrowth on one or more specified growth media. This class also specifies other experimental conditions such as the temperature, aerobicity, and any gene knockouts within the strain that was studied.
Growth media and growth observations can both be entered using interactive editing windows within Pathway Tools. In addition, growth observations can be imported from data files, such as importing PM data. Once such data sets have been imported, the user can inspect and compare growth data in several ways through a Pathway Tools-based web site such as EcoCyc.org. The web site command “Search → Growth Media” enables searching for growth media according to their chemical composition or growth outcome. In addition, the user can find a growth medium by navigating to a page listing all growth observations recorded for that organism (http://biocyc.org/ECOLI/NEW-IMAGE?object=Growth-Media), which contains a series of diagrams, including that shown in Fig. 1. Clicking on a PM well image in such a page will take the user to a page describing the composition of the corresponding growth medium (see Fig. S1 in the supplemental material). The PGDB pages for chemical compounds include links to all growth media that contain that compound.
The growth-medium page (see Fig. S1 in the supplemental material) lists the composition of a growth medium in two forms: as a recipe of substances that can be used to create the medium (e.g., ions are listed as salts) and as a listing of all of the constituents of the medium (e.g., each ion is listed once, its concentration summed from multiple constituents if necessary). When growth data are available, the growth medium page lists whether wild-type E. coli grows on that medium, and the growth status of the single-gene knockouts are listed when available.
EcoCyc gene pages include a table listing the growth status of the knockouts of the gene on all growth media for which data are available (see Fig. S2 in the supplemental material). From the page listing all growth media, it is possible to generate a heatmap comparing gene essentiality with growth on different nutrient sources. Currently, in EcoCyc, gene essentiality data are available for growth on glucose, glycerol, and a small number of other substrates. As more PM data for gene knockouts become available, the power of this tool should become more apparent. Figure S3 in the supplemental material shows an example heatmap with the data currently available in EcoCyc.
DISCUSSION
Differences among PM data sets.
The addition of five high-throughput Biolog PM data sets to EcoCyc highlighted a number of apparent conflicts in the data regarding E. coli K-12 MG1655's ability to use various compounds as the sole source of carbon, nitrogen, phosphate, or sulfur. This integration exercise was successful both in identifying conflicting information and in resolving some of these conflicts based on low-throughput growth data from the literature and from our own experimental results. It is interesting to briefly discuss possible reasons for the disparity in growth observation and PM data since this highlights considerations that arise when attempting to unify data from various sources.
One clear difficulty in integrating such data sets is the confounding effect that may result from slight variations in experimental method used by each laboratory. Potential reasons for disagreement among the PM data sets added to EcoCyc include (i) genetic differences in the E. coli K-12 MG1655 stock cultures, (ii) variation in the pregrowth conditions of the bacteria (LB, R2A, or BUG-S agar), (iii) variation in the length of incubation times used for the assay (26 to 48 h), and (iv) variability in the choice of cutoff values for assigning positive/negative results for respiration in the PM assays.
Genetic variation between stock cultures of E. coli K-12 MG1655 is documented in the literature (16, 17) and is likely to have implications for physiological experiments. It is predicted, for example, that a frameshift in gatC that is present in some stock cultures of E. coli K-12 MG1655 will result in a nonfunctional product and an inability to transport d-galactitol (dulcitol), and it is likely that differences in PM data for this compound are due to genetic variation in stock cultures. We were able to identify three different stock cultures of E. coli K-12 MG1655 that were used to generate the data sets added to EcoCyc: two came from central repositories (ATCC and CGSC), and one was a laboratory-held strain. We tested two of these stock cultures (ATCC 700926 and CGSC7740) on PM plates 1 to 4 and compared their responses (see Fig. S6 in the supplemental material). Overall, there was little variation in response to the majority of compounds (note that in this case both stock cultures contain the gatC frameshift), although it was noted that ATCC 700926 did show a slightly greater response than CGSC7740 when l-glutamate was present as either a sole carbon source or a sole nitrogen source. Since l-glutamate was also one of the compounds that had generated conflicting results among the data sets integrated into EcoCyc, we conducted further tests comparing growth of the two stock cultures in M9 minimal medium to test whether the small variation in PM response reflected a difference in the ability of the two strains to use l-glutamate as either sole N or sole C (see Fig. S6 in the supplemental material). No difference between the two strains was observed; in both cases and for both stock cultures, l-glutamate was able to serve as a sole nitrogen source but not as a sole carbon source. This is in agreement with the experimental literature, which records that E. coli K-12 MG1655 is unable to use l-glutamate as a sole carbon source due to insufficient transport of this compound (18). Although our results suggest there is no variation in PM responses between these two particular stock cultures, they do not preclude the possibility that genetic variation in other stock cultures may contribute to variation in PM data.
We also tested the three different pregrowth agars used by the experimenters (LB, R2A, and BUG-S) and compared the response on PM plates 1 and 2 (see Fig. S5 in the supplemental material). Despite a difference in colony appearance on the three agar plates, no difference could be seen in any of the kinetic curves, and thus we conclude that the variation in PM response to carbon sources between the data sets collated in the present study is not due to differences in pregrowth conditions. Another potential contributor to disagreement among PM data sets is that different laboratories use different criteria to distinguish between growth, no growth, and low growth. We were unable to investigate this factor because original growth curve data are not available for most of the data sets. The existence of an international repository for PM data that included growth curves would facilitate such data reanalysis.
Growth tests in minimal salts agar and in M9 minimal medium identified 15 compounds that could support growth when supplied as a sole nitrogen source despite generating conflicting results between the PM data sets (Table 5). Varying the incubation time is likely a confounding factor for nitrogen source PM assays. Long lag times are particularly noticeable in response to nitrogen compounds (see Fig. S4 in the supplemental material, PM3) and may contribute to the higher conflict rate observed for the nitrogen source plate. For example, in our nitrogen source PM assays, 6 compounds had a lag time of >20 h, and 10 additional compounds had a lag time of >24 h (see Data set S6 in the supplemental material). This suggests that it is important to incubate Biolog nitrogen source plates for the full 48 h as recommended by the manufacturer. Within the nitrogen compounds we tested, there was also some variation between detecting growth on minimal salts agar compared to M9 minimal medium. For example, growth of E. coli K-12 using α-amino-N-butyrate and uric acid as the sole nitrogen source could be detected in soft agar but not in liquid culture, whereas growth on l-methionine, l-phenylalanine, agmatine, uracil, uridine, and amino-N-butyrate as sole nitrogen sources could be detected in liquid minimal media but not in soft agar. The agar used for plate media may contain impurities, and contact with a solid surface and neighboring cells may result in differential regulation of enzymes.
Therefore, variation in the growth response highlights the need to consider all growth and PM data in the context of all experimental conditions. We therefore strive to ensure that the EcoCyc website records the specific conditions under which all growth observations are made, including, for example, whether the assay was performed in liquid culture or on solid surface. Regulation of enzyme expression, such as due to temperature or catabolite repression, as well as regulation of enzyme activity, all play a prominent role in an organism's ability to utilize nutrient sources. The EcoCyc database curates information on both the metabolic and the regulatory networks of E. coli K-12, and we hope that this effort will assist E. coli biologists and other users to evaluate the growth observations that we have integrated in a meaningful way.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by award U24GM077678 from the National Institute of General Medical Sciences, grant IIS-0513857 from the National Science Foundation, and by SRI International.
The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences, the National Institutes of Health, or the National Science Foundation.
Footnotes
Published ahead of print 20 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01209-13.
REFERENCES
- 1.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muniz-Rascado L, Ong Q, Paley S, Schroder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res. 41(Database Issue):D605–D612. 10.1093/nar/gks1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bochner BR, Gadzinski P, Panomitros E. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246–1255. 10.1101/gr.186501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karp P, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee T, Kaipa P, Gilham F, Spaulding A, Popescu L, Altman T, Paulsen I, Keseler IM, Caspi R. 2010. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief Bioinform. 11:40–79. 10.1093/bib/bbp043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutnick D, Calvo JM, Klopotowski T, Ames BN. 1969. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J. Bacteriol. 100:215–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall B. 1982. Chromosomal mutation for citrate utilization by Escherichia coli K-12. J. Bacteriol. 151:269–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauli G, Overath P. 1972. ato Operon: a highly inducible system for acetoacetate and butyrate degradation in Escherichia coli. Eur. J. Biochem. 29:553–562. 10.1111/j.1432-1033.1972.tb02021.x [DOI] [PubMed] [Google Scholar]
- 7.Oun MA, Suthers PF, Jones GI, Carter BR, Saunders MP, Maranas CD, Woodward MJ, Anjum MF. 2009. Genome scale reconstruction of a salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain. J. Biol. Chem. 284:29480–29488. 10.1074/jbc.M109.005868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumler DJ, Peplinski RG, Reed JL, Glasner JD, Perna NT. 2011. The evolution of metabolic networks of Escherichia coli. BMC Syst. Biol. 5:182. 10.1186/1752-0509-5-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon SH, Han MJ, Jeong H, Lee CH, Xia XX, Lee DH, Shim JH, Lee SY, Oh TK, Kim JF. 2012. Comparative multi-omics systems analysis of Escherichia coli strains B and K-12. Genome Biol. 13:R37. 10.1186/gb-2012-13-5-r37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabasi AL, Oltvai ZN, Osterman AL. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673–5684. 10.1128/JB.185.19.5673-5684.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, Datsenko KA, Nakayashiki T, Tomita M, Wanner BL, Mori H. 2009. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 5:335. 10.1038/msb.2009.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BØ, Agarwalla S. 2006. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J. Bacteriol. 188:8259–8271. 10.1128/JB.00740-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feist A, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BØ. 2007. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1,260 ORFs and thermodynamic information. Mol. Syst. Biol. 3:121–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patrick WM, Quandt EM, Swartzlander DB, Matsumura I. 2007. Multicopy suppression underpins metabolic evolvability. Mol. Biol. Evol. 24:2716–2722. 10.1093/molbev/msm204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freddolino PL, Amini S, Tavazoie S. 2012. Newly identified genetic variations in common Escherichia coli MG1655 stock cultures. J. Bacteriol. 194:303–306. 10.1128/JB.06087-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soupene E, van Heeswijk WC, Plumbridge J, Stewart V, Bertenthal D, Lee H, Prasad G, Paliy O, Charernnoppakul P, Kustu S. 2003. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185:5611–5626. 10.1128/JB.185.18.5611-5626.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halpern YS, Lupo M. 1965. Glutamate transport in wild-type and mutant strains of Escherichia coli. J. Bacteriol. 90:1288–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.