Abstract
The Ti plasmid in Agrobacterium tumefaciens strain 15955 carries two alleles of traR that regulate conjugative transfer. The first is a functional allele, called traR, that is transcriptionally induced by the opine octopine. The second, trlR, is a nonfunctional, dominant-negative mutant located in an operon that is inducible by the opine mannopine (MOP). Based on these findings, we predicted that there exist wild-type agrobacterial strains harboring plasmids in which MOP induces a functional traR and, hence, conjugation. We analyzed 11 MOP-utilizing field isolates and found five where MOP induced transfer of the MOP-catabolic element and increased production of the acyl-homoserine lactone (acyl-HSL) quormone. The transmissible elements in these five strains represent a set of highly related plasmids. Sequence analysis of one such plasmid, pAoF64/95, revealed that the 176-kb element is not a Ti plasmid but carries genes for catabolism of MOP, mannopinic acid (MOA), agropinic acid (AGA), and the agrocinopines. The plasmid additionally carries all of the genes required for conjugative transfer, including the regulatory genes traR, traI, and traM. The traR gene, however, is not located in the MOP catabolism region. The gene, instead, is monocistronic and located within the tra-trb-rep gene cluster. A traR mutant failed to transfer the plasmid and produced little to no quormone even when grown with MOP, indicating that TraRpAoF64/95 is the activator of the tra regulon. A traM mutant was constitutive for transfer and acyl-HSL production, indicating that the anti-activator function of TraM is conserved.
INTRODUCTION
Ti plasmids, found in members of the genus Agrobacterium, encode two conjugative-transfer systems, both of which are regulated at the transcriptional level in response to compounds produced by the host plant. The vir system, responsible for transfer of the T strand of the Ti plasmid to plant cells, is induced by, among other factors, phenolic compounds produced by wounded plant cells (reviewed in references 1, 2, and 3). The transferred strand is subsequently integrated into the plant cell nuclear genome (reviewed in reference 4) and results in production of plant growth hormones responsible for the crown gall tumors (reviewed in reference 5). Additionally, the integrated transfer DNA (T-DNA) carries the genes for synthesis by the neoplasias of novel compounds called opines (reviewed in references 5 and 6). Opines are specific conjugates of sugars or, more frequently, amino acids and α-ketoacids or sugars, and they serve as carbon and sometimes as nitrogen or phosphorus sources for the bacteria that induced the tumor (reviewed in reference 6). Remarkably, the Ti plasmid also carries the genes for uptake and catabolism by the bacteria of the specific opines produced by the tumor. The second transfer system, encoded by the tra regulon, is induced by certain opines that are produced by the tumors induced by the bacterium. This system is responsible for conjugative transfer of the plasmid from one bacterium to another (7, 8).
Ti plasmids generally code for uptake and catabolism of two or three of the eight known opine families (7, 9–11). However, in the vast majority of such Agrobacterium megaplasmids studied to date, only one opine type induces transfer and thus is dubbed the conjugative opine (7). For instance, pTiC58 codes for the uptake and catabolism of two opine types, nopaline and agrocinopines A and B, but only the agrocinopines induce plasmid transfer (12). The conjugative opines, however, do not directly induce transfer of pTiC58; rather, they control the transcription of traR, which codes for the quorum-sensing (QS) activator TraR (13, 14). TraR directly activates the Ti plasmid tra and trb operons, responsible for DNA metabolism and mating-pair formation, respectively (15–20).
In the studied systems, the link between the opine signal and the quorum-sensing system results from the inclusion of traR in an operon whose expression is itself regulated by the conjugative opine. For example, traR of pTiC58 is located in the arc operon, which is adjacent and in opposite orientation to the agrocinopine A and B uptake and catabolism operon (acc) (14). The divergently oriented acc and arc operons are both transcriptionally repressed by the opine-responsive repressor AccR (13, 14). When the agrocinopines are available, repression by AccR is lifted, and traR, as part of the arc operon, is expressed (14). Opine-mediated expression of TraR, however, is not sufficient for induction of conjugative transfer. The active form of TraR requires its ligand, N-(3-oxo-octanoyl)-l-homoserine lactone (3-oxo-C8-HSL), the Agrobacterium autoinducer (AAI) (21–23). This acyl-HSL quormone is synthesized by TraI, which is encoded by the first gene of the Ti plasmid trb operon (18, 24). QS-dependent induction of transfer is additionally modulated by TraM, an antiactivator that interacts with TraR, thereby inhibiting activation of the tra regulon (25–27).
Although in most cases only one opine serves as the conjugative signal, this is not always the case. For instance, Agrobacterium radiobacter strain K84 harbors an opine-catabolic megaplasmid, pAtK84b, that is inducible for transfer by two opine types, agrocinopines A and B and nopaline (12, 28). The plasmid carries two copies of traR, and each is independently transcribed. One, traRnoc, is in an operon induced by nopaline, while the other, traRacc, is in an operon similar to arc of pTiC58 and is inducible by agrocinopines A and B (28). The two alleles function independently. For example, in a traRnoc mutant, conjugative transfer is still inducible by agrocinopines. Likewise, if traRacc is mutated, conjugative transfer remains inducible by nopaline, but not by the agrocinopines (28).
The octopine-mannityl opine-type Ti plasmids, pTi15955 and pTiR10, provide another example of opine-inducible conjugative transfer. Tumors induced by strains harboring these Ti plasmids produce two opine types, octopine and the mannityl opines. In turn, the bacteria can catabolize octopine and all four of the mannityl opines: agropine (AGR), mannopine (MOP), mannopinic acid (MOA), and agropinic acid (AGA) (reviewed in reference 7). These plasmids carry two alleles of traR. One, traRoct, is a member of an operon that is activated by octopine, and this opine induces conjugative transfer (29). The second, called trlR, is the terminal gene in the MOP transport operon, motABCDtrlR, located in the mannityl opine-catabolic region of these Ti plasmids (30). Expression of this operon, including trlR, is inducible by MOP, but not by octopine (30, 31). Importantly, the trlR gene is a dominant-negative frame-shifted mutant of traR and encodes a protein with functional N-terminal dimerization and AAI-binding domains, but not the C-terminal DNA-binding domain (30, 31). The protein product can dimerize with the full-size, functional TraR, induced by octopine, thereby inhibiting octopine-inducible conjugative transfer (30–32).
Based on the presence of trlR in an operon inducible by MOP, we hypothesized that there exist in nature Agrobacterium plasmids in which conjugative transfer is induced by this mannityl opine. We also hypothesized that in such plasmids, the functional traR gene would be associated with the mot operon. In this study, while we provide evidence concerning a family of plasmids that supports the first hypothesis, the allele of traR associated with quorum-dependent transfer is not associated with a mot-like operon.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. The mannopine-utilizing field isolates of Agrobacterium spp. were obtained from the laboratory of the late Larry Moore, Oregon State University, Corvallis, OR, and have not been previously described in the literature. The methods for collection of the field tumors, isolation of bacteria, determination of opine catabolism, and pathogenicity testing were all described previously (33).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or referenceb |
|---|---|---|
| Agrobacterium sp. | ||
| AF1/95 | Wild-type mannopine-utilizing strain | L. Moore |
| AR11N/71 | Wild-type mannopine-utilizing strain | L. Moore |
| B24/93 | Wild-type mannopine-utilizing strain | L. Moore |
| B26/94 | Wild-type mannopine-utilizing strain | L. Moore |
| B98/94 | Wild-type mannopine-utilizing strain | L. Moore |
| B155/93 | Wild-type mannopine-utilizing strain | L. Moore |
| F64/95 | Wild-type mannopine-utilizing strain | L. Moore |
| F265/93 | Wild-type mannopine-utilizing strain | L. Moore |
| J62/95 | Wild-type mannopine-utilizing strain | L. Moore |
| J84/95 | Wild-type mannopine-utilizing strain | L. Moore |
| M200/94 | Wild-type mannopine-utilizing strain | L. Moore |
| 15955 | Wild-type octopine strain; MOP+ MOA+ AGR+ AGA+ | OC |
| C58 | Wild-type nopaline strain; pAtC58 pTiC58 | OC |
| C58C1RS | Ti plasmidless derivative of C58; pAtC58 Smr Rifr | OC |
| NTL4 | Ti plasmidless derivative of C58; pAtC58 ΔtetAR | 98 |
| NTL6 | Plasmidless derivative of NTL4 | OC |
| E. coli | ||
| DH5α | λ− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Invitrogen |
| LE392 | hsdR514(rk− mk+) glnV(supE44) tryT (supF58) lacY1 or Δ(lacIZY)6 galK2 galT22 metB1 trpR55 | Promega |
| S17-1 λpir | Tpr, Smr recA thi pro hsdR-M+ RP4::2-Tc::Mu::Km Tn7 λpir | 99 |
| Plasmids | ||
| pAgK84-A1 | Agrocin 84 producer | 100 |
| pArA4 | MOP+ MOA+ AGA+ | J. Tempé |
| pCP13/B | Deletion derivative of pCP13; Tcr | 62 |
| pKD46 | Ampr; λ Red helper plasmid | 63 |
| pSRKGm | Gmr; IPTG-inducible expression vector | 66 |
| pViK107 | Kanr; promoterless lacZY; pir dependent | 65 |
| pWM91 | Ampr sacB | 64 |
| pZLR4 | Gmr; indicator for detection of acyl-HSL | 44 |
TraI+, production of AAI; Ampr, ampicillin resistance; Gmr, gentamicin resistance; Kanr, kanamycin resistance; Rifr, rifampin resistance; Smr, streptomycin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance.
OC, our collection.
Media and growth conditions.
Strains of Escherichia coli were grown in L broth (Fischer Scientific) and in SOB or SOC medium (34). Strains of Agrobacterium spp. were grown in L broth (Fisher Scientific) or MG/L (35) or on Nutrient Agar (Difco). Stonier's medium (36) was used for agrocin 84 sensitivity assays, and AB (35) and AT (37) media were used as minimal media. Liquid AB medium was supplemented with 0.005% yeast extract (Difco) unless otherwise noted. Minimal media were supplemented with mannitol to 0.2% or with mannopine or mannopinic acid at 500 μg/ml as a carbon source. Mannopine and mannopinic acid were the kind gifts of Yves Dessaux (Institut des Sciences du Végétal, Gif-sur-Yvette, France). Antibiotics were used at the following concentrations (μg/ml): rifampin, 50 or 100; streptomycin, 100; kanamycin, 25 or 50; ampicillin, 100; carbenicillin, 50 or 100; and gentamicin, 25. Cultures of E. coli were incubated at 30 or 37°C, and cultures of Agrobacterium spp. were incubated at 28°C.
Biovar determination.
The biovar assignments of the wild-type strains were provided to us by the Moore laboratory. We verified these assignments for strains F64/95, F265/93, J62/95, J84/95, and M200/94 using three tests described by Moore et al. (38): 3-ketolactose production, growth on AB agar with erythritol as the primary carbon source, and growth on Nutrient Agar supplemented with 0.25% glucose and 2% NaCl.
Virulence assays.
The Moore laboratory described the virulence properties of all wild-type strains as tested on tomato plants (Table 2). We repeated these assays with the five MOP-inducible strains using the stem-wound inoculation method on tomato plants, essentially as described previously (39, 40).
TABLE 2.
Characteristics of MOP-utilizing wild-type isolates
| Straina | Opine(s) utilizedb | Conjugation frequencyc |
Isolation |
Tumorigenic | Agrocin sensitivity | Biovar | ||
|---|---|---|---|---|---|---|---|---|
| Uninduced | Induced with MOP | Host | Location | |||||
| AF1/95 | MOP, OCT | <10−8 | <10−8 | Lilac | Visalia, CA | Yes | No | 2 |
| AR11N/71 | MOP, NOP, OCT | <10−8 | <10−8 | Apple | Canby, OR | Yes | NDd | ND |
| B24/93 | MOP | 3.3 × 10−6 | 9.0 × 10−7 | Quince | Portland, OR | No | No | 2 |
| B26/94 | MOP, NOP | <10−8 | <10−8 | Walnut | Portland, OR | Yes | No | 2 |
| B98/94 | MOP | 1.5 × 10−5 | 2.6 × 10−6 | Walnut | Gridley, CA | Yes | No | 2 |
| B155/93 | MOP, NOP | <10−8 | <10−8 | Quince | Portland, OR | No | Yes | ND |
| F64/95 | MOP | <10−8 | 6.6 × 10−5 | Apple | Modesto, CA | No | Yes | 2 |
| F265/93 | MOP | <10−8 | 4.4 × 10−5 | Apple | Yamhill, OR | No | Yes | 2 |
| J62/95 | MOP, NOP | <10−8 | 4.6 × 10−5 | Apple | Modesto, CA | Yes | Yes | 2 |
| J84/95 | MOP, NOP | <10−8 | 3.1 × 10−5 | Apple | Modesto, CA | Yes | Yes | 2 |
| M200/94 | MOP | <10−8 | 3.6 × 10−5 | Apple | Woodburn, OR | No | ND | 2 |
The wild-type isolates listed all utilize mannopine and were obtained from Larry Moore, Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR.
NOP, nopaline; OCT, octopine.
Conjugation frequency was measured as the number of transconjugates recovered per input donor cell.
ND, not determined.
Monitoring growth.
Strains F64/95 and NTL6(pAoF64/95) were precultured by growth in LB. In the morning, the cells were collected by centrifugation, washed three times with 0.9% NaCl, and suspended in 1 ml of sterile 0.9% NaCl. The washed cells were inoculated into liquid AB mannitol (ABM), AB plus MOP, or AB with no additional carbon source to a population density of about 107 cells/ml. Strains were monitored for growth by turbidity as measured with a Klett colorimeter using a red filter. Periodically, volumes were removed, part of which was frozen at −20°C and used later for AAI detection and quantification. The remainder was immediately used for conjugative-transfer assays and viable-cell determinations.
Conjugative-transfer efficiency.
Conjugative transfer of the Agrobacterium plasmids to strain C58C1RS (Table 1) was determined using the drop plate method (8, 41, 42). Transconjugants were selected for using opine catabolism or resistance to an appropriate antibiotic, and donors were counterselected using rifampin and streptomycin.
Detection and quantification of AAI.
AAI was extracted from the culture supernatant essentially as described previously (8, 43, 44). For chromatographic analysis, the concentrated samples were spotted on a C18 reversed-phase thin-layer chromatography (TLC) plate and separated using methanol-H2O (60:40 [vol/vol]) (8, 44). For quantification, 5-μl volumes of a 2-fold dilution series of each sample were spotted in a grid pattern onto a TLC plate. The samples were allowed to dry, and the plates were overlaid with 1% agar containing 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and the acyl-HSL indicator strain NTL4(pZLR4) (8, 44) and incubated overnight at 28°C. The plates were dried and scanned, and ImageJ (version 1.44o; National Institutes of Health [http://imageJ.nih.gov/ij]) (45) was used to measure the area of the blue spots. These values were compared to those of a dilution series of an AAI standard (Sigma) spotted on the same plate (8).
Agrocin 84 sensitivity assays.
A modified version of the procedure of Hayman and Farrand (46) was used to assess sensitivity to agrocin 84. Briefly, two colonies of the agrocin 84 producer strain, NT1(pAgK84-A1) (Table 1), were suspended in 0.25 ml of 0.9% NaCl, and 15 μl of the suspension was spotted onto the centers of Stonier's agar (1.5% agar) plates (36) and incubated at 28°C for 3 days. The producer strain was killed by exposure of the plate to chloroform vapor for 15 min. The cultures to be tested for sensitivity were inoculated into MG/L for overnight growth. Two or three drops of these overnight cultures were mixed with 3 ml of melted 0.7% agar and overlaid onto the plates. The cultures were incubated at 28°C, and zones of growth inhibition were assessed daily.
Isolation of mannityl opines from tumors.
Crude preparations of the mannityl opines were isolated from plant tumors as described previously (10, 47). Briefly, tomato tumors induced by Agrobacterium tumefaciens strain 15955 were minced and placed in 50-ml sterile conical tubes. Enough Milli Q water was added to cover the tops of the minced tissue, and the tubes were placed in boiling water for 10 min. The mixture was ground with a mortar and pestle, and the solids were removed by decanting the mixture over cheesecloth. The remaining particulate matter was removed by centrifugation, and the tumor extract was sterilized by passage through a 0.22-μm filter.
Separation and detection of mannityl opines.
Samples containing mannityl opines were spotted on a pencil line drawn across the middle of a sheet of Whatman 3MM paper and allowed to dry. The opines were separated by high-voltage paper electrophoresis (HVPE), using an acetic acid-formic acid buffer with a pH between 1.7 and 1.9 according to methods described previously (10, 39, 48). The opines were visualized using an alkaline silver nitrate reagent, as described previously (35, 39, 49).
Mannityl opine utilization studies.
Growth on solid medium was assessed by visual inspection of strains inoculated on AB agar medium with MOP or MOA as the sole source of carbon. The plates were observed daily over a 7-day period. Growth was compared to appropriate positive and negative controls and recorded as follows: −, no growth; +/−, very poor growth; +, good growth; and ++, very good growth. Growth in liquid medium was assessed turbidimetrically as described above. We additionally assessed mannityl opine utilization by the disappearance of opines from liquid culture medium (50). Sterilized tumor extract prepared as described above was added to 2× AT medium in a 1:1 volume ratio. Volumes of 0.5 ml were inoculated with a single colony of the strain to be tested or left uninoculated. The cultures were incubated at 28°C with shaking for 5 days, at which time the cells were removed by centrifugation. A 6-μl volume of each of the culture supernatants was spotted onto Whatman paper and analyzed for the disappearance of mannityl opines by high-voltage paper electrophoresis, as described above.
MOP cyclase activity.
Five-microliter volumes of ABM were inoculated with a single colony of the strain to be tested and allowed to grow overnight with aeration at 28°C. Subsequently, MOP was added to the cultures at a concentration of 20 μg/ml. After incubation for an additional 3 h, the cells were harvested by centrifugation, and MOP cyclase activity was determined by the conversion of MOP to agropine as assessed by high-voltage paper electrophoresis, as previously described (51).
Preparation and purification of plasmid DNA.
Small-scale plasmid isolation from E. coli was done by the alkaline lysis methods of Sambrook and Russell (52). Large Agrobacterium plasmids to be used for restriction enzyme analysis were isolated and purified using ethidium bromide (EtBr) and phenol, as described by Zhang and Kerr (53). In all other cases, large- and small-scale isolations of plasmids from Agrobacterium strains were performed as described by Hayman and Farrand (54). Plasmid preparations for DNA sequence analysis were further purified by two rounds of centrifugation in cesium chloride-EtBr. The EtBr was removed by extraction with equal volumes of isopropanol saturated with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and the samples were dialyzed against LTE (10 mM Tris, 1 mM EDTA, pH 8.0).
Sequencing of plasmid DNA.
The complete nucleotide sequence of pAoF64/95 was determined at the Keck Center for Comparative and Functional Genomics at the University of Illinois at Urbana-Champaign (UIUC). Briefly, the purified plasmid DNA was sheared with a nebulizer, and the ends were repaired and dephosphorylated. The fragmented DNA was separated on an agarose gel, and fragments with sizes between 1.5 and 5.0 kb were selected for and cloned into pCR-Blunt II-TOPO (Invitrogen). Sequencing was done from both ends of the insert in vectors, using the Sanger dideoxy method. Sequences were assigned a quality score with Phred (55, 56), and clean sequences were assembled using PHRAP (57, 58). After final assembly, open reading frames (ORFs) were called using Glimmer (version 3.02 [59, 60; http://www.ncbi.nlm.nih.gov/genomes/MICROBES/glimmer_3.cgi]). We used BLAST to search the NCBI nonredundant protein sequence database. All other sequencing was done by the UIUC Core sequencing facility or by ACGT, Inc. Primers were ordered from Integrated DNA Technologies, Inc., or from the UIUC Core sequencing facility.
DNA manipulations.
Agrobacterium strains were transformed using the freeze-thaw method (61), and E. coli was made competent using the calcium chloride method (52). In some cases, plasmids were mobilized from E. coli into Agrobacterium using a filter-mating technique (8). An overlapping cosmid bank of pAoF64/95 was constructed by cloning a Sau3AI partial digest of purified plasmid DNA into the cosmid vector pCP13b (Table 1) digested with BamHI and calf intestinal alkaline phosphatase (CIP). The clones were then packaged into λ using the Packagene λ DNA-packaging system (Promega) and transfected into E. coli strain LE392. Cosmid clones were mapped by restriction digestion, and the ends of appropriate inserts were sequenced using primers complementary to vector sequences.
Construction of mutant strains.
All PCRs described here used Pfu polymerase (Stratagene). An in-frame deletion of traR was constructed on cosmid clone pMWS110 by the method of Datsenko and Wanner (63). The kanamycin cassette of pKD4 was amplified using the following primers, which contained 5′ overhang sequences of traR: forward primer, 5′ GTGGACGGTGACCTTCGCTCACTCATCGACATGACAGAAGTGTAGGCTGGAGCTGCTTCG 3′, and reverse primer, 5′ CTACAGCAGGCCGTGGTCCTTGGCGATCGCGACGAGGTGCATATGAATATCCTCCTTAGT 3′. The PCR product was transformed into E. coli(pKD46, pMWS110) following the published protocol (63), and alleles of traR in which the cassette was inserted into pMWS110 were selected for by resistance to kanamycin. Mutations were confirmed by restriction analysis and PCR amplification from the regions upstream and downstream of traR using the following primers: traRcheckup, 5′ AGCTCGCGAGGACTTGAATACCCGG 3′, and traRcheckdwn, 5′ GATCGCTGCGATCAGAGAGCACCG 3′. Cosmid clone pMWS110 contains repABC of pAoF64/95, which poses a plasmid incompatibility problem with pAoF64/95. Therefore, a 6.9-kb SpeI fragment containing the kanamycin-marked indel mutation of traR was subcloned into pWM91 (64). The resulting plasmid was transformed into E. coli S17-1/λpir, and the plasmid was transferred into NTL4(pAoF64/95) by filter mating (8). Transconjugants were selected on solid ABM medium containing kanamycin. Strains with double crossovers were screened for by their ability to grow on sucrose and by sensitivity to carbenicillin while retaining resistance to kanamycin. Mutants were confirmed by PCR amplification using the traRcheckup and traRcheckdwn primers. The mutant megaplasmid pAoF64/95ΔtraR was genetically purified by isolation and subsequent transformation into strain NTL4.
The traM mutant was constructed using vectors described by Kalorgeraki and Winans (65). Briefly, an internal fragment of traM was amplified by PCR using primers that contained EcoRI and SalI sites (underlined): traMleft, 5′ GATCCAGAATTCGTCAGAACTGGAGGCTCTGG 3′, and traMright, 5′ GATCCAGTCGACTACAGAACTCGACACCGCAG 3′. The PCR fragment was directionally cloned into pVik107, resulting in an internal portion of traM fused in frame to lacZ on pVik107. This construct was transformed into E. coli strain S17-1/λpir, and the resulting strain was mated with NTL4(pAoF64/95) to yield NTL4(pAoF64/95::traMpVik107). The single-crossover mutation was confirmed by sequence analysis, and the mutated plasmid was genetically isolated by transformation into NTL4.
To complement the mutant strains, traR was amplified by PCR with traRpSRKGmsense (5′ GCCGGAATTCATATGGACGGTGACCTTCGCTC 3′) and traRpSRKGmantisense (5′ CGCGAATTCGGATCCTACAGCAGGCCGTGGTCCT 3′. The traM gene was separately amplified by PCR using the following primers: traMpSRKGmsense (5′ GCCCGAGCTCATATGAGCGACGTGAACTCGTCTG 3′) and traMpSRKGmantisense (5′ GCGGAGCTCGGATCCTCAATCACCGACTTCGGGGGC 3′). The traR and traM genes were directionally cloned into pSRKGm (66) using NdeI and BamHI (the sites are underlined). In each case, the cloning results in fusion of the native start codon of the gene to the start codon of lacZα in the vector. This arrangement puts expression of the cloned gene solely under the control of LacI. Cloning was confirmed by sequence analysis, and the constructs were transformed into the appropriate mutant strain. In complementation experiments, expression of the cloned gene was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM to the cultures.
Nucleotide sequence accession number.
The full sequence of pAoF64/95 is available in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank/) under accession number JX683454.
RESULTS
Identification and characterization of Agrobacterium isolates in which mannopine induces conjugation.
We assessed a collection of mannopine-utilizing field isolates of Agrobacterium spp. for strains in which MOP induced conjugative transfer of the opine utilization trait. Of the isolates we tested, five transferred the trait to A. tumefaciens C58C1RS in an opine-inducible manner (Table 2). These strains were part of a collection of 11 isolates obtained mostly from apple and nut orchards in California and Oregon (the Corvallis strains). Two of the remaining six strains exhibited constitutive low-frequency transfer of MOP utilization, while four strains failed to transfer the trait under the culture conditions tested (Table 2).
All but 2 of the 11 Corvallis isolates were classified as biovar 2 agrobacteria. Six of the isolates induced tumors on at least one of three plant hosts tested (Table 2). Of the nine isolates tested, five were susceptible to agrocin 84, an antiagrobacterial antibiotic susceptibility to which is conferred by particular plasmid-encoded opine-catabolic systems (Table 2) (67, 68). Detailed growth studies of the five MOP-inducible isolates indicated that all of them also utilize MOA and AGA, but none utilize AGR (see Fig. S1A in the supplemental material). Consistent with this pattern, none of the five isolates expressed detectable levels of mannopine cyclase activity (see Fig. S1B in the supplemental material).
Mannopine catabolism and its inducible transfer are associated with a family of closely related plasmids.
Gel electrophoretic analysis indicated that all five strains in which MOP induces conjugative transfer harbor at least one plasmid between 150 and 200 kb in size (Fig. 1A). Given the possibility that one or more of the isolates harbors more than one plasmid migrating in a single band, we genetically isolated the plasmids responsible for MOP catabolism by transforming strain NTL6, a plasmidless derivative of A. tumefaciens strain NTL4 (Table 1), with total plasmid DNA isolated from each strain, selecting directly for MOP utilization. MOP-utilizing transformants were isolated in each case, and individual colonies from each transformation tested also utilized MOA and AGA, but not AGR (see Fig. S1A in the supplemental material).
FIG 1.
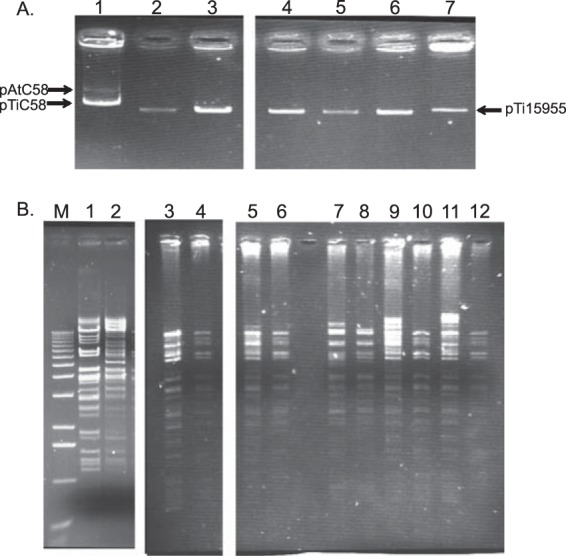
Isolates that transfer MOP catabolism harbor related plasmids. (A) Plasmid profiles of the MOP-inducible isolates. Plasmid DNA was isolated from the wild-type isolates. The lanes contain DNA from NTL4(pTiC58) (lane 1), F64/95 (lane 2), F265/93 (lane 3), J62/95 (lane 4), J84/95 (lane 5), M200/94 (lane 6), and 15955 (lane 7). The standards are pAtC58 (543 kb) and pTiC58 (214 kb) of strain C58 (lane 1) and pTi15955 (194 kb) of strain 15955 (lane 7). (B) BamHI fragment patterns of total plasmid DNA isolated from the wild-type isolates and the genetically isolated MOP catabolism plasmid from each wild-type strain. Total plasmid DNA was extracted from each wild-type isolate and an NTL6 transformant of the MOP catabolism plasmid from each strain and digested with BamHI, and the fragments were separated on a 0.8% agarose gel. Lanes: M, 1-kb ladder (Invitrogen); 1, NTL4(pAoF64/95); 2, C58C1RS(pArA4); 3, F64/95; 4, NTL6(pAoF64/95); 5, J84/95; 6, NTL6(pAoJ84/95); 7, F265/93; 8, NTL6(pAoF265/93); 9, J62/95; 10, NTL6(pAoJ62/95); 11, M200/94; 12, NTL6(pAoM200/94).
To gauge the plasmid complement of the field isolates and the nature of the plasmids encoding opine utilization and MOP-inducible conjugation, we subjected plasmid samples from each of the five field isolates and one corresponding NTL6 transformant of each to restriction enzyme analysis. In all cases, the plasmids in the transformants yielded a fragment pattern that formed a subset of the pattern seen in the plasmid preparations from the corresponding field isolates (Fig. 1B, compare odd- and even-numbered lanes). Moreover, the fragment patterns of plasmids from the five different transformants were very similar to each other (Fig. 1B, odd-numbered lanes). These results strongly suggest that most, if not all, of the five field isolates tested harbor at least two plasmids of about the same size and that one of these plasmids, which is strongly conserved among the five isolates, is conjugative and encodes utilization of MOP, MOA, and AGA.
Given the apparent relatedness of the mannopine-catabolic plasmids, we focused on one wild-type field isolate, F64/95, and its MOP-catabolic plasmid, which we named pAoF64/95.
Growth with mannopine induces production of an acyl-homoserine lactone, as well as conjugative transfer of pAoF64/95.
In all conjugative Ti and opine-catabolic plasmids studied to date, transfer is controlled by opines through regulation of an acyl-homoserine lactone-dependent quorum-sensing system (21, 23, 28, 29, 47). Preliminary studies indicated that growth of strain F64/95 with mannopine resulted in the production of an acyl-HSL that has the chromatographic properties of 3-oxo-C8-HSL (see Fig. S2 in the supplemental material). We assessed the conjugative-transfer characteristics of pAoF64/95 and whether opine-induced transfer was associated with induction of production of this acyl-HSL quormone. The field isolate and one NTL6 transformant harboring pAoF64/95 were cultured in minimal medium containing either mannitol or MOP as the primary source of carbon. At intervals, samples were removed and assayed for growth by viable-cell counts, and the cells were tested for conjugative competence. In addition, the culture supernatants were assayed quantitatively for acyl-HSLs, as described in Materials and Methods.
Strain NTL6 lacking pAoF64/95 grew well with mannitol but failed to grow with MOP (data not shown). In contrast, the field isolate and NTL6(pAoF64/95) grew almost as well with MOP as with mannitol (Fig. 2). Either plasmid-containing strain produced only barely detectable levels of 3-oxo-C8-HSL when grown with mannitol but accumulated steadily increasing amounts of the quormone as growth proceeded in medium containing MOP (Fig. 2). Concomitant with the increasing levels of the acyl-HSL signal, donors grown with MOP transferred pAoF64/95 at increasing frequencies as growth continued (Fig. 2). Donors grown with mannitol failed to transfer the plasmid at a detectable level at any stage of growth tested.
FIG 2.
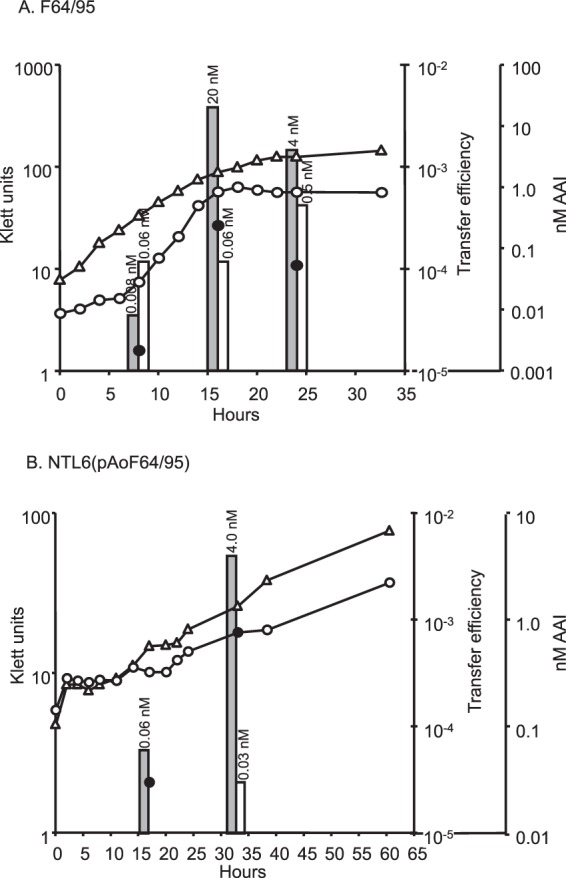
Growth with mannopine induces production of AAI and conjugative transfer of pAoF64/95. Strains F64/95 (A) and NTL6(pAoF64/95) (B) were grown in AB medium with 0.005% yeast extract and mannitol (open triangles and open bars) or MOP (open circles and gray bars). Growth was followed turbidometrically using a Klett colorimeter. At approximately 8-h intervals, a portion of each culture was removed and assayed for conjugative-transfer efficiency (black circles) and accumulation of AAI (bars). The amount of extractable AAI in the culture supernatant is indicated over the corresponding bar. The transfer frequency is expressed as the number of transconjugates recovered per input donor cell.
Plasmid pAoF64/95 is an opine-catabolic element.
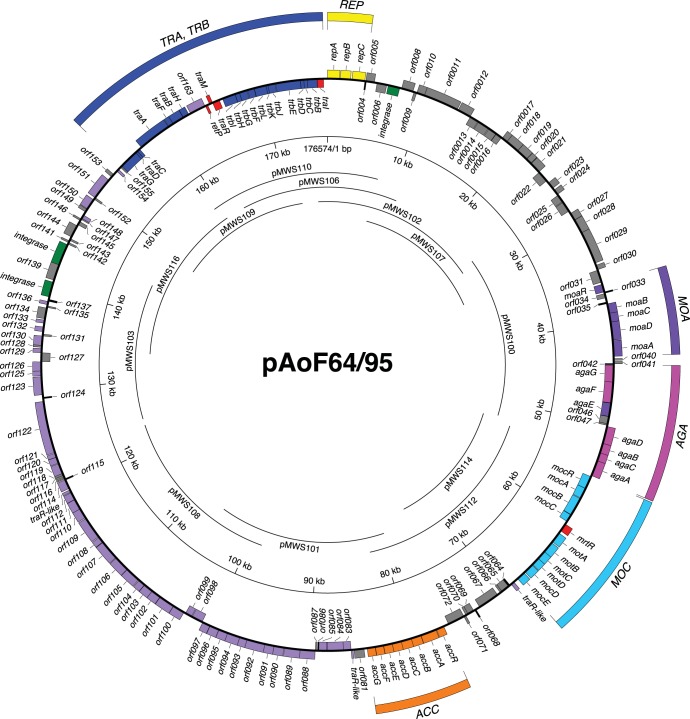
We determined the nucleotide sequence of pAoF64/95. The plasmid, with a size of 176,574 bp, codes for 178 annotatable open reading frames (Fig. 3). We also constructed an overlapping cosmid clone bank of the plasmid (Fig. 3). Like most, if not all, large plasmids in the family Rhizobiaceae, pAoF64/95 encodes a repABC-type replication system. Consistent with our observation that F64/95 fails to induce tumors, pAoF64/95 lacks a T region and all known components of the vir regulon.
FIG 3.
Physicogenetic map of pAoF64/95. The complete 176,574-bp sequence of pAoF64/95 was determined, and the sequence was annotated as described in Materials and Methods. Significant open reading frames are shown as boxes, with those on the outside oriented in a clockwise direction and those on the inside oriented in a counterclockwise direction. The genes are color coded with respect to known or putative functions as follows: yellow, replication; violet, uptake and catabolism of MOA; magenta, uptake and catabolism of AGA; cyan, uptake and catabolism of MOP; orange, uptake and catabolism of agrocinopines; blue, conjugative transfer; red, regulation of MOP catabolism or conjugative transfer; green, three putative integrase genes; lavender, a segment that is largely syntenic in gene composition and order with similarly sized regions of two Ri plasmids, pRi1724 and pRi2659; gray, other genes that have a returned hit in the NCBI nonredundant sequence database; black, ORFs with no significant similarities to genes in the databases. The extents of the inserts in each cosmid in the ordered library are represented by the arcs within the rings. The annotated sequence is available in GenBank (see the text).
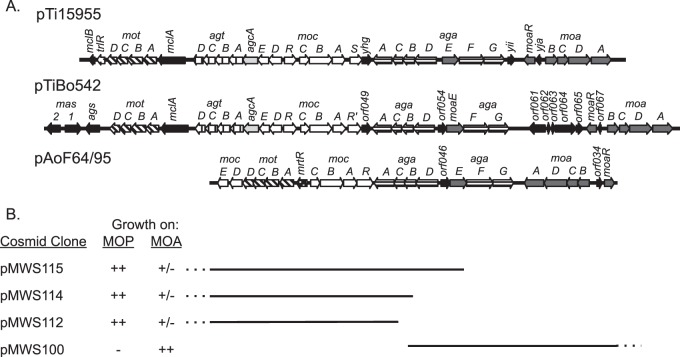
Consistent with the observation that pAoF64/95 confers catabolism of three of the four mannityl opines, the plasmid contains a contiguous, approximately 30-kb region comprising 22 genes organized in six groups that is closely related to regions of Ti plasmids known to confer catabolism of MOP, MOA, and AGA (Fig. 3 and 4). The plasmid does not contain agcA, encoding catabolic mannopine cyclase (69–71), or the four-gene agt operon required for agropine uptake (51, 69), consistent with our observation that strains harboring pAoF64/95 do not catabolize the lactone opine. The organization of the two aga operons is virtually identical to those of pTi15955 and pTiBo542 (Fig. 4). However, the region comprising the moa transport genes and moaR is inverted in comparison to that of the two Ti plasmids. The 10 genes of pTi15955 comprising the MOP-catabolic regulon all are present in pAoF64/95. However, while they are grouped in similar units, their orientations and fine structures differ (Fig. 4). In pAoF64/95, the mocDE gene pair, which is organized as a two-gene operon in pTi15955, comprises the distal portion of the four-gene mot operon that encodes the MOP transporter. Notably, the mocDE gene pair replaces trlR at the end of the mot operon, and there is no other gene encoding a full-size TraR homolog in this region of the plasmid. Moreover, while the moc region of pTi15955 carries two closely related putative regulatory genes, mocR and mocS (72, 73), the corresponding region of pAoF64/95 carries only one such gene, which we call mocR (Fig. 4).
FIG 4.
The mannityl opine catabolism region of pAoF64/95 is related to those of other Agrobacterium plasmids. (A) Alignment of the regions coding for the catabolism of the mannityl opines from two Ti plasmids, pTi15955 and pTiBo542, and from pAoF64/95. Orthologous genes and gene systems are depicted as arrows with identical fill patterns. Genes with common shading are involved in the same opine-catabolic pathways. mot, mannopine transport; agt, agropine transport; acgA, mannopine cyclase; moc, mannopine catabolism; aga, transport and catabolism of agropinic acid and mannopinic acid; moa, transport of mannopinic acid. All genes not directly involved in the catabolism and transport of the mannityl opines are in black. mrtR, a novel gene in pAoF64/95, codes for a member of the GntR family of regulators. trlR is a frame-shifted allele of traR in pTi15955 (30, 31). (B) A set of cosmids with overlapping inserts define the catabolic region of pAoF64/95. Derivatives of strain NTL4 harboring each cosmid were tested for the ability to grow on AB minimal agar containing either MOP or MOA as the sole carbon source. ++, growth as good as that of a known mannityl opine utilizer; +/−, very poor growth; −, no significant growth. The dotted lines indicate where the cosmid extends beyond the scope of the map.
We confirmed that this region of pAoF64/95 is responsible for catabolism of the mannityl opines by analysis of the cosmid clone bank using two strategies. In the first, we electroporated an unordered pool of the cosmid bank into A. tumefaciens NTL4 and selected for progeny that could utilize MOP. Such transformants contained three cosmids, pMWS112, pMWS114, and pMWS115 (Fig. 3 and 4B). Restriction enzyme and sequence analyses showed that these three cosmids overlap the putative moc and mot genes and parts of the AGA transport and catabolism operons (Fig. 4B). In the second strategy, we transformed a cosmid, pMWS100, in which the insert overlapped the putative MOA catabolic region and parts of the AGA transport and catabolism operons, but not the MOP catabolism region (as depicted in Fig. 4B), into strain NTL4, selecting for resistance to tetracycline. We screened these transformants for the ability to utilize MOP and MOA. NTL4(pMWS100) utilized MOA but not MOP (Fig. 4B), which is consistent with our bioinformatics predictions for this cosmid clone.
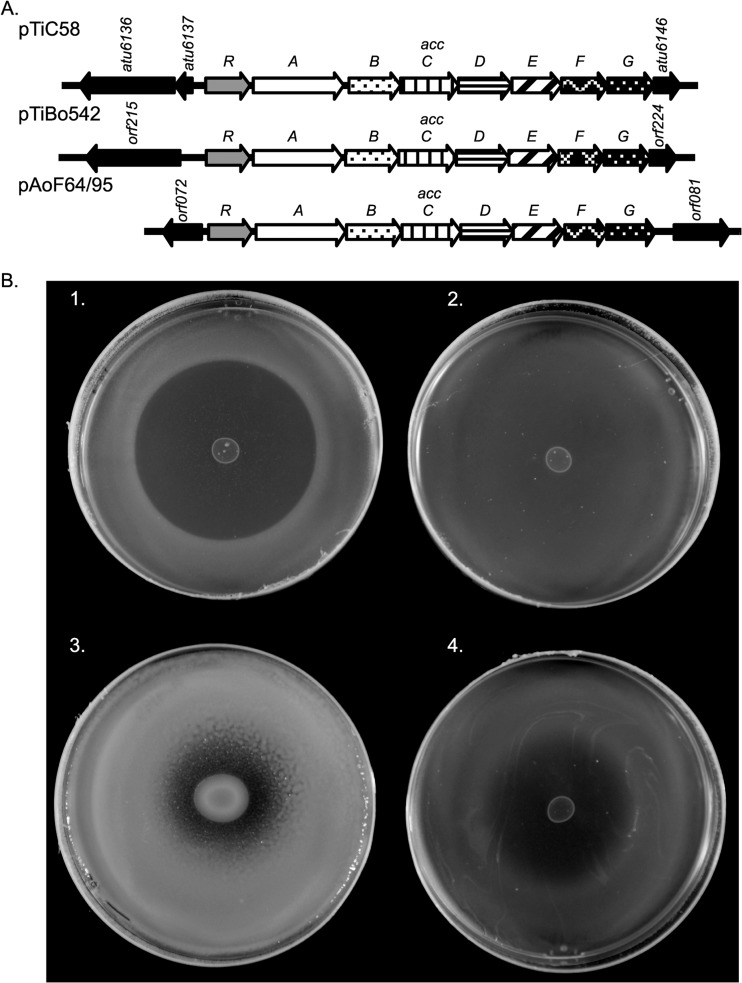
Plasmid pAoF64/95 carries a second putative opine-catabolic region, acc, mapping between coordinates 77 and 84 kb (Fig. 3 and 5). On certain Ti and Ri plasmids, orthologous loci code for uptake and catabolism of the agrocinopines, a family of sugar phosphodiester opines (67, 74), as well as susceptibility to agrocin 84, the unique antiagrobacterial antibiotic (67, 68). Of the five strains with pAoF64/95-like plasmids, four were described to us by the Moore laboratory as being susceptible to agrocin 84, while one was not tested. In our assays, the field isolate F64/95 showed weak susceptibility to the antibiotic, only sometimes producing cloudy zones of growth inhibition (Fig. 5B). Strain NTL4(pAoF64/95) produced similar, although often more defined, zones of growth inhibition (Fig. 5B).
FIG 5.
pAoF64/95 carries a locus related to the agrocinopine A and B catabolism operon. (A) Alignment of the acc operons from pTiC58, pTiBo542, and pAoF64/95. accR genes of pTiC58 and pTiBo542 encode the repressor that coregulates opine catabolism and conjugative transfer. Genes encoding orthologous functions are shown with the same fill pattern, while genes not directly involved in catabolism and transport are in black. (B) Agrocin 84 sensitivity assays. Strains were assessed for sensitivity to agrocin 84 as described in Materials and Methods. 1, C58; 2, NTL4; 3, F64/95; 4, NTL4(pAoF64/95).
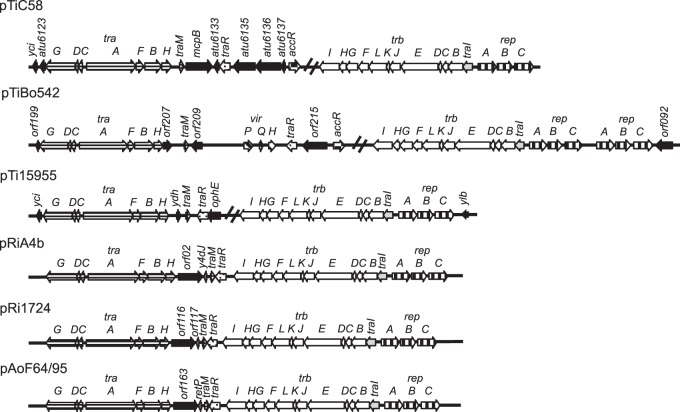
pAoF64/95 encodes a Ti plasmid-like conjugative-transfer system and all of the components of a TraR-TraI quorum-sensing regulatory system.
By bioinformatics analysis, pAoF64/95 encodes a single identifiable conjugative-transfer system, and this system is virtually identical in gene content and operon organization to those of other Ti, Ri, and opine-catabolic plasmids (Fig. 6). The 12-gene trb operon, encoding an IncP-like type IV secretion system, is adjacent to and oriented divergently from the repA gene of the replication operon (Fig. 3 and 6). The two DNA-processing operons, traAFBH and traCDG, are adjacent to and divergently oriented with respect to each other, although the latter carries an additional putative small open reading frame, orf155, not present in the traCDG operons of other Ti plasmid-like systems (Fig. 3 and 6). The traA and traC genes are separated by a 254-bp intergenic region that contains a sequence similar to that of the oriT regions of pTiC58 and pTiR10 (75, 76) (Fig. 7).
FIG 6.
The tra and trb operons of Ti, Ri, and opine-catabolic plasmids are conserved. The regions coding for DNA metabolism (tra) and mating-pair formation (trb) from three Ti plasmids (pTiC58, pTiBo542, and pTi15955), two Ri plasmids (pRiA4b and pRi1724), and pAoF64/95 were aligned. Orthologous genes and gene systems are depicted as arrows with identical fill patterns. The genes known to be involved in regulation of Ti plasmid transfer are traM, traR, and traI. The plasmid replication genes are denoted rep. In pTiC58 and pTiBo542, the product of accR represses the acc and arc operons (13). The two Ri plasmids and pAoF64/95 carry a small helix-turn-helix xre family gene labeled y4dJ, orf117, and retP. Novel genes not yet characterized are filled in black. The double slash lines on the Ti plasmids indicate where the tra and trb regions are separated on those plasmids.
FIG 7.

The oriT region of pAoF64/95 is strongly conserved with those of two Ti plasmids, pTiC58 (75) and pTiR10 (76). Nucleotides identical in all three regions are shaded black. Nucleotides conserved in two of the three regions are shaded gray. The nic site of pTiR10 (76) is indicated by an arrowhead.
The conjugative-transfer region also carries the three genes traR, traI, and traM, which in studied Ti, opine-catabolic, and some Rhizobium plasmids are responsible for regulating conjugative transfer in a quorum-dependent manner (Fig. 6). Moreover, the general locations and orientations of the three genes in pAoF64/95 closely resemble those in other known conjugative elements. traI, which encodes the acyl-HSL synthase (24), is the first gene of the trb operon, and traR and traM, encoding the activator and antiactivator, are closely linked, convergently oriented, and located just distal to the traAFBH operon. However, unlike the gene organization in systems in which conjugative transfer is opine inducible, traR appears to be monocistronic and is certainly not a member of a plasmid-specific opine-regulated operon (Fig. 6).
We assessed the activity of traI by examining a set of overlapping cosmid clones that comprise the tra, trb, traI, and repABC regions of pAoF64/95 for production of the acyl-HSL quormone. Derivatives of strain NTL4 harboring cosmids pMWS106 and pMWS110, both of which carry traI, produced low but detectable levels of the signal, while the strain harboring pMWS109, which maps to the same region but does not overlap traI, failed to produce the acyl-HSL (see Fig. S3 in the supplemental material).
The traR gene product is required for the MOP-dependent induction of AAI production and conjugative transfer.
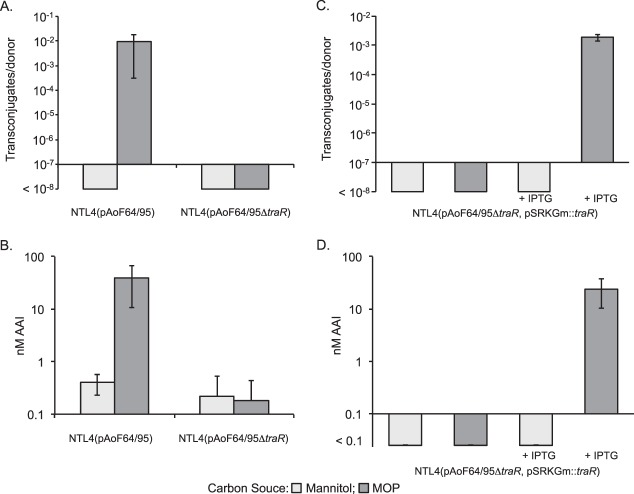
In the known Ti plasmid conjugative-transfer systems, TraR directly activates transcription of the traItrb operon and the two divergently oriented tra operons (15–20). We constructed a traR mutant by creating an in-frame, kanamycin-marked deletion derivative of the gene on the megaplasmid. Donors harboring this construct failed to transfer the plasmid at detectable frequencies, even when grown with MOP (Fig. 8A). Moreover, these donors failed to produce elevated levels of the acyl-HSL signal under any growth conditions tested (Fig. 8B). These results suggest that a functional TraR is required to express traI and the tra and trb genes.
FIG 8.
TraR is essential for induction of acyl-HSL production and conjugative transfer. (A and B) NTL4(pAoF64/95) and the traR mutant, NTL4(pAoF64/95ΔtraR), were grown in AB medium containing mannitol or MOP as the primary carbon source. After overnight growth, samples were removed and tested for conjugative transfer using A. tumefaciens strain C58C1RS as the recipient (A) and levels of AAI accumulation (B). (C and D) The traR mutant was complemented with a wild-type copy of traR cloned into pSRKGm. The resulting strain, NTL4(pAoF64/95ΔtraR, pSRKGm::traR), was grown in AB minimal medium containing mannitol or MOP, each with and without IPTG, and assessed for conjugative transfer (C) and AAI accumulation (D). The experiment was done two or three times, and the mean and standard deviation for each culture condition are shown.
The traR mutation was fully complementable with the wild-type allele cloned into pSRKGm. When this merodiploid construct was pregrown with MOP as the primary carbon source and expression of the recombinant traR was induced with IPTG, conjugative transfer and acyl-HSL production were restored to approximately wild-type frequencies (Fig. 8C and D). Surprisingly, when the strain was grown on mannitol and traR was induced with IPTG, conjugative transfer and induction of AAI production remained undetectable (Fig. 8C and D).
TraM negatively regulates conjugative transfer of pAoF64/95.
In the known conjugative-transfer systems of Agrobacterium, TraM inhibits the activity of TraR by directly binding to the activator (27, 77). We created a disruption mutant of traM on pAoF64/95 [strain NTL4(pAoF64/95::traMpVik107)] as described in Materials and Methods and assayed the mutant for AAI production and conjugative transfer following growth with MOP or with mannitol as the primary carbon source. Consistent with its role as an antiactivator, the traM mutant produced high levels of AAI and transferred the plasmid at high frequency, even when grown with mannitol, a noninducing substrate (Fig. 9A and B).
FIG 9.
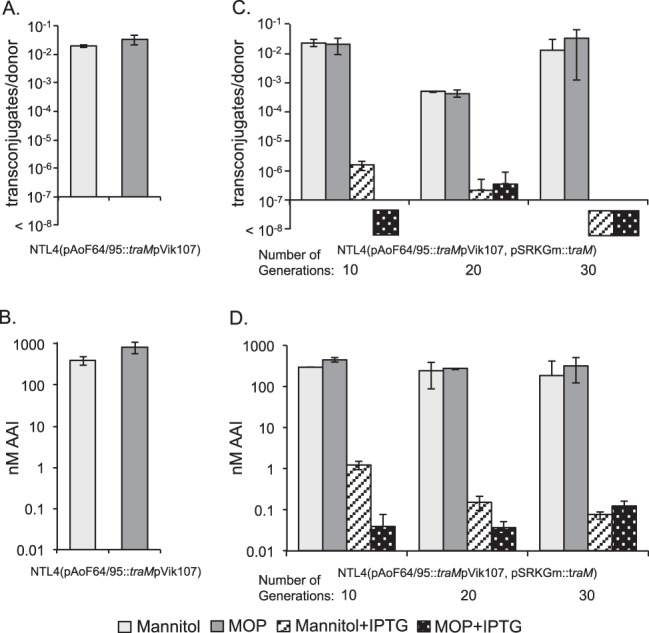
TraM inhibits TraR-dependent induction of AAI production and conjugative transfer. (A and B) The traM mutant, NTL4(pAoF64/95::traMpVik107), was assessed for conjugative transfer (A) and AAI accumulation (B) when grown on AB medium supplemented with either mannitol or MOP. (C and D) The mutant was complemented in trans with a wild-type copy of traM cloned into pSRKGm. This strain, NTL4(pAoF64/95::traMpVik107, pSRKGm::traM), was continuously cultured in AB minimal medium supplemented with either mannitol or MOP and with or without IPTG to induce expression of traM, as described in Materials and Methods. Samples were removed after the indicated numbers of cumulative generations, and the cells were tested for conjugative competence (C) and AAI accumulation (D), as described in Materials and Methods. The numbers of generations are approximate. Each experiment was done two or three times, and the means and standard deviations are shown.
In initial studies, an IPTG-inducible copy of traM cloned into pSRKGm did not fully complement the traM mutant (data not shown). We reasoned that since the mutant is constitutive for transfer, to abolish transfer, traM would have to be expressed over many generations to dilute out the donors that had already produced fully active conjugative-transfer systems (78). To test this, we passaged the complemented mutant in AB minimal medium with either mannitol or mannopine through sequential subcultures, either continually with or without IPTG. We tested samples for conjugative competence before each dilution step. When no IPTG was added, transfer and AAI levels remained high regardless of whether the cells were grown with mannitol or MOP (Fig. 9C and D). Donor cells grown with IPTG continued to transfer the plasmid, though at lower frequency, over the first 10 to 20 generations. However, after passage of the strain through approximately 20 to 30 generations with inducing levels of IPTG, conjugative transfer was reduced to undetectable levels, even when the cells were grown with MOP (Fig. 9C and D). We reasoned that overexpressing TraM should keep conjugative transfer repressed, even in cells grown with the inducing opine (Fig. 9C).
DISCUSSION
Five independently isolated opine-catabolic plasmids confer MOP utilization and are highly similar to each other.
The existence of trlR, a mutant allele of traR associated with the mot operon of octopine-type Ti plasmids, led us to the hypothesis that Agrobacterium plasmids in which quorum-dependent conjugative transfer is induced by MOP would exist. The prediction proved accurate; of 11 wild isolates of mannopine-utilizing Agrobacterium spp. obtained from Oregon State University, five conjugatively transferred the catabolic trait only when grown with MOP (Table 2). In all five cases, the transmissible trait is associated with a large conjugative plasmid, and as judged by restriction enzyme analysis (Fig. 1), these five MOP-inducible plasmids are closely related. All five of the isolates are biovar 2 strains, but only two are demonstrably pathogenic (Table 2). Of the remaining six isolates examined, four failed to transfer MOP utilization at a detectable frequency under the culture conditions tested, and two transferred the trait at a low opine-independent constitutive level (Table 2).
pAoF64/95 is an opine-catabolic plasmid that carries a conserved set of genes for transport and catabolism of the mannityl opines.
Consistent with the phenotype, sequence analysis of one such opine-inducible plasmid, pAoF64/95, indicated that these elements are not Ti plasmids; pAoF64/95 lacks a T region and the genes of the vir regulon (Fig. 3). Instead, it is an opine-catabolic plasmid and carries genes for the uptake and catabolism of at least two families of these tumor-specific substrates, the agrocinopines and three of the four mannityl opines. Both loci confer the expected phenotypes; the acc operon confers sensitivity to agrocin 84, a marker for catabolism of the agrocinopine opines (67) (Fig. 5), while the moc locus confers utilization of MOP, MOA, and AGA (Fig. 4; see Fig. S1 in the supplemental material). Strain F64/95 does not utilize agropine, the fourth member of the mannityl opine family (see Fig. S1 in the supplemental material). Consistent with this observation, pAoF64/95 lacks agcA, which codes for the enzyme that converts agropine to MOP (69–71), and also the agt operon that encodes the agropine transporter (51, 69). While the overall organization of the moc locus is similar to that of the octopine-type Ti plasmids, there are some rearrangements (Fig. 4). Most significantly, the mocDE genes in pAoF64/95 are coupled with the 3′ end of the mot operon, and instead of two mocR-like genes, there is only one (Fig. 4). pAoF64/95 carries an additional novel gene in the mannopine catabolism region, mrtR, which codes for a product that aligns with the GntR family of transcriptional regulators. Perhaps MrtR contributes to the regulation of mannopine catabolism, MOP-inducible transfer, or both.
Given that growth with MOP induces transfer, we expected that, like trlR, traR of pAoF64/95 would be linked to the mot operon. However, on pAoF64/95, traR is monocistronic, is located between the contiguous tra and trb regions, and is approximately 66 kb removed from the closest gene in the moc operon (Fig. 3 and 6). The absence of a functional traR linked to a MOP-associated operon on pAoF64/95 does not preclude the existence of a plasmid with a gene organization similar to that of the motABCDtrlR operon of octopine-type Ti plasmids. However, there currently is no evidence of such a mot-associated functional form of the traR gene on an Agrobacterium megaplasmid.
pAoF64/95 encodes a conserved conjugative-transfer system that is most similar in gene organization to Ri and Rhizobium plasmids.
The pAoF64/95 plasmid additionally shares a set of core genes that are strongly conserved in a large group of plasmids found in Agrobacterium and Rhizobium isolates. These genes code for replication and conjugative transfer, as well as components associated with the quorum-dependent regulation of these functions. In a typical Ti plasmid, the two tra operons, oriT, traR, and traM, are located near a conjugative opine catabolism region of the plasmid. In these plasmids, traR is, without exception, located in an opine-inducible operon (28; reviewed in reference 7). The trb operon, which codes for the type IV mating bridge, as well as TraI, the acyl-HSL synthase, is invariably directly linked and oriented divergently from repA of the repABC operon (reviewed in reference 7). Moreover, in Ti plasmids, the tra complex and the trb-rep locus are separated by 60 to 85 kb of sequence.
The organization of these loci in pAoF64/95 more closely resembles that of megaplasmids from Agrobacterium rhizogenes isolates, including pRi1724, pRiA4 (Fig. 7), and pRi2659, as well as some Rhizobium plasmids, including pRL1JI (79) and p42a (80). In these plasmids, the trb-repABC complex and the two divergently oriented tra operons all are located in close association, with traM and a monocistronic traR located between the two conjugative-transfer regions (Fig. 6).
TraR is an activator but may be regulated differently from TraR in the Ti plasmid systems.
Like the identified conjugative-transfer systems of the Ti plasmids, transfer of pAoF64/95 is regulated by a TraR-dependent mechanism. Mutational analysis clearly indicates that, as in Ti plasmid systems, traR is required for opine-mediated induction of conjugative transfer (Fig. 8). However, complementation analysis showed some differences from opine-mediated regulation of traR in Ti plasmids. When their cognate traR genes are overexpressed in strains harboring Ti plasmids, the opine signals are no longer required to induce transfer (23, 29, 47). Intriguingly, donors containing pAoF64/95ΔtraR were complemented in trans with a cloned, fully induced copy of wild-type traR, but only when grown with MOP (Fig. 8). This continued requirement for the conjugative opine suggests that the opine-dependent regulatory system that controls TraR-mediated activation of conjugative transfer is novel and may require an additional MOP-dependent element.
Activation of transfer of pAoF64/95 is further modulated by the antiactivator, TraM.
In strains harboring the archetypical Ti plasmids pTiC58 and pTiR10, the product of the traM gene modulates TraR activity. TraM functions as an antiactivator by direct interaction with TraR (27, 77); mutants lacking traM transfer constitutively, but at frequencies lower than those observed in wild-type strains induced by growth with their conjugative opine (25, 26). Apparently, TraR expressed at its basal level is sufficient to induce transfer, albeit at a low frequency. One role of TraM, then, is to sequester this low level of TraR, thereby preventing plasmid transfer in the absence of the conjugative opines (25, 26).
pAoF64/95 also carries a traM gene, and a traM mutant of the plasmid is constitutive for transfer (Fig. 9). When this mutant is complemented by overexpression of traM, transfer is abolished, even when the cells are grown with mannitol (Fig. 9). These results support a role for TraM as an antiactivator of TraR. In contrast to the Ti plasmid system, the traM mutant of pAoF64/95 exhibits constitutive transfer at higher than anticipated frequencies (Fig. 9). This observation, coupled with the data concerning complementation of the traR mutant (Fig. 8) discussed above, suggests that TraR is expressed at a basal level higher than that of the Ti plasmid systems and that perhaps the ratio of TraM to TraR is higher in pAoF64/95 than it is in strains harboring the Ti plasmids.
The pAoF64/95 group of plasmids and pArA4 of A. rhizogenes A4 are related.
Based on our restriction enzyme analysis, pAoF64/95 defines a family of closely related opine-catabolic plasmids that are widely distributed among the agrobacteria. In addition to their presence in the subset of the Corvallis isolates in which MOP induces transfer, our results suggest that the members of this group are distributed among the classical mannityl-opine-utilizing strains of A. rhizogenes. Wild-type strain A4, isolated from the hairy roots of naturally infected rose plants (81) in California (82), has three well-studied plasmids: pRiA4, pArA4, and a cointegrate of the two (83). Based on restriction enzyme cleavage patterns, pArA4 is similar, but not identical, to pAoF64/95 and the other four MOP-inducible plasmids in this family (Fig. 2). Consistent with this physical relatedness, like pAoF64/95, the plasmid codes for catabolism of MOP, MOA, and AGA, but not agropine (48), as well as utilization of agrocinopines (54).
These related plasmids are all found in independent isolates of Agrobacterium spp. While the plasmids are not virulence elements, based on the pathogenicity properties of the host isolates (Table 2), they are present in strains that harbor Ri plasmids, Ti plasmids, and possibly other opine-catabolic plasmids. In strain A4, whose two plasmids have been studied in detail, the T-right region of the Ri plasmid carries the genes for synthesis by the plant neoplasia of all four mannityl opines and the agrocinopines (84, 85). pRiA4, however, confers utilization only of AGR, while pArA4 codes for the uptake and catabolism of AGA, MOA, MOP, and the agrocininopines (48). Thus, the catabolic properties of pArA4 expand the range of the mannityl opines utilizable by strains harboring pRiA4. Additionally, opine-catabolic plasmids can provide an advantage to avirulent Agrobacterium strains, such as F64/95, by enabling the bacteria to utilize opines produced by neoplasias induced by a virulent strain of Agrobacterium. Such opine-utilizing, nonpathogenic isolates of Agrobacterium have been repeatedly cultured from pathogen-induced neoplasias (38, 86–89). The fact that these opine-catabolic plasmids responsible for the cheater phenotype of these strains so closely resemble Ti and Ri plasmids of the pathogen illustrates the genetic plasticity of the core replicon structure of these rhizobial elements.
A large region of pAoF64/95 is syntenic with a region from two Ri plasmids, indicating that pAoF64/95 is chimeric and that this region may encode advantageous functions.
Including the core replication and transfer regions, a large portion of pAoF64/95 is highly syntenic with regions of two Ri plasmids, pRi1724 and pRi2659 (Fig. 3, lavender-colored genes). This region, about 94 kb in size, includes ORFs that are annotated as putative unknown opine transport and metabolic genes (90, 91). Likewise, the region contains sequences that align with sugar transporters and glycerol metabolism genes (90, 91), as well as a large number of hypothetical genes or genes of unknown function.
An 8.8-kb region located in the middle of this syntenic region is unique to pAoF64/95. This segment consists mainly of ORFs coding for hypothetical proteins and two putative phage integrase proteins (Fig. 3, genes in green around 140 kb). The presence of such genes in pAoF64/95 suggests that this small region was acquired by an insertion event.
All told, fully one half of pAoF64/95 is synonymous with syntenic regions of two Ri plasmids. The conservation of this large region is particularly interesting considering the varied nature of the plasmids and the geographical locations from which the parental strains were isolated. MAFF 301724, a biovar 1 strain from which pRi1724 was identified, was isolated in Japan from a melon plant with hairy root disease (92, 93). The parental strain of pRi2659, also a biovar 1 strain, was isolated in the United Kingdom from a similarly diseased cucumber plant (91). Based on sequence analysis, pRi1724 and pRi2659 are closely related plasmids (90). Strain F64/95, on the other hand, is a nonpathogenic biovar 2 isolate that was cultured from an apple crown gall in California (Table 2). The varied locations and strain backgrounds of the three isolates and the fact that this region is located on Ri and opine-catabolic plasmids suggest that the genes located on this segment confer some unknown but advantageous functions in the habitats occupied by these bacteria. Additionally, the fact that this region is conserved in three different plasmids with at least two different gene contexts supports the notion that the large megaplasmids in both Rhizobium and Agrobacterium are chimeric and evolved by recombination with other rhizobial plasmids (90, 94–97).
The mechanism of opine-inducible transfer of pAoF64/95 may be novel.
In Ti plasmid systems, traR is transcriptionally controlled by opines through the simple fact that it is a member of an operon that is regulated by the opine-responsive regulatory element. However, in pAoF64/95, traR is monocistronic and is not located near the mannopine catabolism region. In addition, our mutational and complementation analyses of traR and traM suggest that traR expression may not be controlled directly at the transcriptional level by a MOP-responsive regulatory element. It is possible, for example, that MOP controls the expression of some other, still unidentified component of the regulatory circuitry.
This accumulating evidence supports a novel mode of regulation of transfer of pAoF64/95, and perhaps other plasmids of Agrobacterium spp. with transfer and regulatory genes organized in a similar fashion. A more detailed understanding of the regulation of the quorum-sensing system of pAoF64/95 may be useful, for example, in understanding the regulation of conjugative transfer among the Ri group of plasmids, which have all of the components of other conjugative plasmids.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grant no. R01 GM52465 from the NIH to S.K.F.; by grant no. SC0006642 from the U.S. Department of Energy Office of Biological and Environmental Research to J. Sweedler, P. Bohn, and S.K.F.; and by funds from the Oregon Association of Nurserymen and the Oregon Department of Agriculture Center for Applied Agricultural Research to Larry Moore and M.M.
We thank Sharik Khan, Yinping Qin, Edgaurdo Sepulveda, Michael Barnhart, and Vandana Chakravartty for helpful discussions.
Footnotes
Published ahead of print 20 December 2013
This article is dedicated to the late Larry Moore of Oregon State University.
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01365-13.
REFERENCES
- 1.Charles TC, Jin S, Nester EW. 1992. Two-component sensory transduction systems in phytobacteria. Annu. Rev. Phytopathol. 30:463–484 [DOI] [PubMed] [Google Scholar]
- 2.Hooykaas PJJ, Beijersbergen AGM. 1994. The virulence system of Agrobacterium tumefaciens. Annu. Rev. Phytopathol. 32:157–181 [Google Scholar]
- 3.Winans SC. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelvin SB. 2000. Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:223–256. 10.1146/annurev.arplant.51.1.223 [DOI] [PubMed] [Google Scholar]
- 5.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJJ, Farrand SK, Winans SC. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 182:3885–3895. 10.1128/JB.182.14.3885-3895.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tempé J, Petit A, Farrand S. 1984. Induction of cell proliferation by Agrobacterium tumefaciens and A. rhizogenes: a parasite's point of view, p 271–286 In Verma DPS, Hohn T. (ed), Genes involved in microbe-plant interactions. Springer, Vienna, Austria [Google Scholar]
- 7.Farrand SK. 1998. Conjugal plasmids and their transfer, p 199–233 In Spaink H, Kondorosi A, Hooykaas PJ. (ed), The Rhizobiaceae. Springer, Dordrecht, Netherlands [Google Scholar]
- 8.Farrand SK, Qin Y, Oger P. 2002. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 358:452–484. 10.1016/S0076-6879(02)58074-5 [DOI] [PubMed] [Google Scholar]
- 9.Dessaux Y, Petit A, Farrand SK, Murphy PJ. 1998. Opines and opine-like molecules involved in plant-Rhizobiaceae interactions, p 173–197 In Spaink H, Kondorosi A, Hooykaas PJ. (ed), The Rhizobiaceae. Springer, Dordrecht, Netherlands [Google Scholar]
- 10.Dessaux Y, Petit A, Tempé J. 1992. Opines in Agrobacterium biology, p 109–136 In Verma DPS. (ed), Molecular signals in plant-microbe communications. CRC Press, Boca Raton, FL [Google Scholar]
- 11.Chilton WS, Petit A, Chilton M-D, Dessaux Y. 2001. Structure and characterization of the crown gall opines heliopine, vitopine and ridéopine. Phytochemistry 58:137–142. 10.1016/S0031-9422(01)00166-2 [DOI] [PubMed] [Google Scholar]
- 12.Ellis JG, Kerr A, Petit A, Tempé J. 1982. Conjugal transfer of nopaline and agropine Ti-plasmids: the role of agrocinopines. Mol. Gen. Genet. 186:269–274. 10.1007/BF00331861 [DOI] [Google Scholar]
- 13.Beck von Bodman S, Hayman GT, Farrand SK. 1992. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. U. S. A. 89:643–647. 10.1073/pnas.89.2.643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piper KR, Beck von Bodman S, Hwang I, Farrand SK. 1999. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol. Microbiol. 32:1077–1089. 10.1046/j.1365-2958.1999.01422.x [DOI] [PubMed] [Google Scholar]
- 15.Fuqua C, Winans SC. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cook DM, Li PL, Ruchaud F, Padden S, Farrand SK. 1997. Ti plasmid conjugation is independent of vir: reconstitution of the tra functions from pTiC58 as a binary system. J. Bacteriol. 179:1291–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrand S, Hwang I, Cook D. 1996. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 178:4233–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li P-L, Everhart DM, Farrand SK. 1998. Genetic and sequence analysis of the pTiC58 trb locus, encoding a mating-pair formation system related to members of the type IV secretion family. J. Bacteriol. 180:6164–6172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P-L, Hwang I, Miyagi H, True H, Farrand SK. 1999. Essential components of the Ti plasmid trb system, a type IV macromolecular transporter. J. Bacteriol. 181:5033–5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P-L, Farrand SK. 2000. The replicator of the nopaline-type Ti plasmid pTiC58 is a member of the repABC family and is influenced by the TraR-dependent quorum-sensing regulatory system. J. Bacteriol. 182:179–188. 10.1128/JB.182.1.179-188.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LH, Kerr A. 1991. A diffusible compound can enhance conjugal transfer of the Ti plasmid in Agrobacterium tumefaciens. J. Bacteriol. 173:1867–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Murphy PJ, Kerr A, Tate ME. 1993. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 362:446–448. 10.1038/362446a0 [DOI] [PubMed] [Google Scholar]
- 23.Piper KR, von Bodman SB, Farrand SK. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448–450. 10.1038/362448a0 [DOI] [PubMed] [Google Scholar]
- 24.Hwang I, Li PL, Zhang L, Piper KR, Cook DM, Tate ME, Farrand SK. 1994. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc. Natl. Acad. Sci. U. S. A. 91:4639–4643. 10.1073/pnas.91.11.4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang I, Cook D, Farrand S. 1995. A new regulatory element modulates homoserine lactone-mediated autoinduction of Ti plasmid conjugal transfer. J. Bacteriol. 177:449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuqua C, Burbea M, Winans S. 1995. Activity of the Agrobacterium Ti plasmid conjugal transfer regulator TraR is inhibited by the product of the traM gene. J. Bacteriol. 177:1367–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang I, Smyth AJ, Luo Z-Q, Farrand SK. 1999. Modulating quorum sensing by antiactivation: TraM interacts with TraR to inhibit activation of Ti plasmid conjugal transfer genes. Mol. Microbiol. 34:282–294. 10.1046/j.1365-2958.1999.01595.x [DOI] [PubMed] [Google Scholar]
- 28.Oger P, Farrand SK. 2002. Two opines control conjugal transfer of an Agrobacterium plasmid by regulating expression of separate copies of the quorum-sensing activator gene traR. J. Bacteriol. 184:1121–1131. 10.1128/jb.184.4.1121-1131.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuqua WC, Winans SC. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J. Bacteriol. 176:2796–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oger P, Kim K-S, Sackett RL, Piper KR, Farrand SK. 1998. Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol. Microbiol. 27:277–288. 10.1046/j.1365-2958.1998.00671.x [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Winans SC. 1998. Activity of the quorum-sensing regulator TraR of Agrobacterium tumefaciens is inhibited by a truncated, dominant defective TraR-like protein. Mol. Microbiol. 27:289–297. 10.1046/j.1365-2958.1998.00672.x [DOI] [PubMed] [Google Scholar]
- 32.Chai Y, Zhu J, Winans SC. 2001. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40:414–421. 10.1046/j.1365-2958.2001.02385.x [DOI] [PubMed] [Google Scholar]
- 33.Moore LW, Chilton WS, Canfield ML. 1997. Diversity of opines and opine-catabolizing bacteria isolated from naturally occurring crown gall tumors. Appl. Environ. Microbiol. 63:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580. 10.1016/S0022-2836(83)80284-8 [DOI] [PubMed] [Google Scholar]
- 35.Cangelosi GA, Best EA, Martinetti G, Nester EW. 1991. Genetic analysis of Agrobacterium. Methods Enzymol. 204;384–397 [DOI] [PubMed] [Google Scholar]
- 36.Stonier T. 1956. Labeling crown gall bacteria with P32 for radioautography. J. Bacteriol. 72:259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tempé J, Petit A, Holsters M, Montagu M, von Schell J. 1977. Thermosensitive step associated with transfer of the Ti plasmid during conjugation: possible relation to transformation in crown gall. Proc. Natl. Acad. Sci. U. S. A. 74:2848–2849. 10.1073/pnas.74.7.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore LW, Kado CI, Bouzar H. 1988. Agrobacterium, p 16–36 In Schaad NW. (ed), Laboratory guide for identification of plant pathogenic bacteria, 2nd ed, APS Press, St. Paul, MN [Google Scholar]
- 39.Ellis JG, Ryder MH, Tate ME. 1984. Agrobacterium tumefaciens TR-DNA encodes a pathway for agropine biosynthesis. Mol. Gen. Genet. 195:466–473. 10.1007/BF00341448 [DOI] [Google Scholar]
- 40.Palanichelvam K, Oger P, Clough SJ, Cha C, Bent AF, Farrand SK. 2000. A second T-region of the soybean-supervirulent chrysopine-type Ti plasmid pTiChry5, and construction of a fully disarmed vir helper plasmid. Mol. Plant Microbe Interact. 13:1081–1091. 10.1094/MPMI.2000.13.10.1081 [DOI] [PubMed] [Google Scholar]
- 41.Piper KR, Farrand SK. 2000. Quorum sensing but not autoinduction of Ti plasmid conjugal transfer requires control by the opine regulon and the antiactivator TraM. J. Bacteriol. 182:1080–1088. 10.1128/JB.182.4.1080-1088.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beck von Bodman S, McCutchan JE, Farrand SK. 1989. Characterization of conjugal transfer functions of Agrobacterium tumefaciens Ti plasmid pTiC58. J. Bacteriol. 171:5281–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo Z-Q, Su S, Farrand SK. 2003. In situ activation of the quorum-sensing transcription factor TraR by cognate and noncognate acyl-homoserine lactone ligands: kinetics and consequences. J. Bacteriol. 185:5665–5672. 10.1128/JB.185.19.5665-5672.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Rinehart KL, Farrand SK. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U. S. A. 94:6036–6041. 10.1073/pnas.94.12.6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasband WS. 2012. ImageJ. National Institutes of Health, Bethesda, MD [Google Scholar]
- 46.Hayman GT, Farrand SK. 1988. Characterization and mapping of the agrocinopine-agrocin 84 locus on the nopaline Ti plasmid pTiC58. J. Bacteriol. 170:1759–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oger P, Farrand SK. 2001. Co-evolution of the agrocinopine opines and the agrocinopine-mediated control of TraR, the quorum-sensing activator of the Ti plasmid conjugation system. Mol. Microbiol. 41:1173–1185. 10.1046/j.1365-2958.2001.02584.x [DOI] [PubMed] [Google Scholar]
- 48.Petit A, David C, Dahl GA, Ellis JG, Guyon P, Casse-Delbart F, Tempé J. 1983. Further extension of the opine concept: plasmids in Agrobacterium rhizogenes cooperate for opine degradation. Mol. Gen. Genet. 190:204–214. 10.1007/BF00330641 [DOI] [Google Scholar]
- 49.Trevelyan WE, Procter DP, Harrison JS. 1950. Detection of sugars on paper chromatograms. Nature 166:444–445 [DOI] [PubMed] [Google Scholar]
- 50.Chilton WS, Chilton MD. 1984. Mannityl opine analogs allow isolation of catabolic pathway regulatory mutants. J. Bacteriol. 158:650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong SB, Dessaux Y, Chilton WS, Farrand SK. 1993. Organization and regulation of the mannopine cyclase-associated opine catabolism genes in Agrobacterium tumefaciens 15955. J. Bacteriol. 175:401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 53.Zhang L, Kerr A. 1993. Rapid purification of Ti plasmids from Agrobacterium by ethidium bromide treatment and phenol extraction. Lett. Appl. Microbiol. 16:265–268. 10.1111/j.1472-765X.1993.tb01415.x [DOI] [PubMed] [Google Scholar]
- 54.Hayman GT, Farrand SK. 1990. Agrobacterium plasmids encode structurally and functionally different loci for catabolism of agrocinopine-type opines. Mol. Gen. Genet. 223:465–473 [DOI] [PubMed] [Google Scholar]
- 55.Ewing B, Green P. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 56.Ewing B, Hillier L, Wendl MC, Green P. 1998. Base-calling of automated sequencer traces using Phred. I. accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 57.Green P. 1996. Documentation for PHRAP. Genome Center, University of Washington, Seattle, WA: http://bozeman.mbt.washington.edu/ [Google Scholar]
- 58.de la Bastide M, McCombie WR. 2007. Assembling genomic DNA sequences with PHRAP. Curr. Protoc. Bioinformatics Chapter 11:Unit 11.4. 10.1002/0471250953.bi1104s17 [DOI] [PubMed] [Google Scholar]
- 59.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641. 10.1093/nar/27.23.4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salzberg SL, Delcher AL, Kasif S, White O. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544–548. 10.1093/nar/26.2.544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Holsters M, Waele D, Depicker A, Messens E, Montagu M, Schell J. 1978. Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. 163:181–187. 10.1007/BF00267408 [DOI] [PubMed] [Google Scholar]
- 62.Darzins A, Chakrabarty AM. 1984. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J. Bacteriol. 159:9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Metcalf WW, Jiang W, Daniels LL, Kim S-K, Haldimann A, Wanner BL. 1996. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35:1–13. 10.1006/plas.1996.0001 [DOI] [PubMed] [Google Scholar]
- 65.Kalogeraki VS, Winans SC. 1997. Suicide plasmids containing promoterless reporter genes can simultaneously disrupt and create fusions to target genes of diverse bacteria. Gene 188:69–75. 10.1016/S0378-1119(96)00778-0 [DOI] [PubMed] [Google Scholar]
- 66.Khan SR, Gaines J, Roop RM, Farrand SK. 2008. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Appl. Environ. Microbiol. 74:5053–5062. 10.1128/AEM.01098-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim H, Farrand S. 1997. Characterization of the acc operon from the nopaline-type Ti plasmid pTiC58, which encodes utilization of agrocinopines A and B and susceptibility to agrocin 84. J. Bacteriol. 179:7559–7572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tate ME, Murphy PJ, Roberts WP, Kerr A. 1979. Adenine N6-substituent of agrocin 84 determines its bacteriocin-like specificity. Nature 280:697–699. 10.1038/280697a0 [DOI] [PubMed] [Google Scholar]
- 69.Hong SB, Farrand SK. 1994. Functional role of the Ti plasmid-encoded catabolic mannopine cyclase in mannityl opine catabolism by Agrobacterium spp. J. Bacteriol. 176:3576–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hong S, Farrand S. 1996. Purification and characterization of catabolic mannopine cyclase encoded by the Agrobacterium tumefaciens Ti plasmid pTi15955. J. Bacteriol. 178:2427–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dessaux Y, Guyon P, Farrand SK, Petit A, Tempé J. 1986. Agrobacterium Ti and Ri plasmids specify enzymic lactonization of mannopine to agropine. J. Gen. Microbiol. 132:2549–2559 [DOI] [PubMed] [Google Scholar]
- 72.Kim K, Chilton W, Farrand S. 1996. A Ti plasmid-encoded enzyme required for degradation of mannopine is functionally homologous to the T-region-encoded enzyme required for synthesis of this opine in crown gall tumors. J. Bacteriol. 178:3285–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jung W-H, Baek C-H, Lee J-K, Kim K-S. 1999. Analysis of trans-acting elements for regulation of moc operons of pTi15955 in Agrobacterium tumefaciens. J. Microbiol. Biotechnol. 9:637–645 [Google Scholar]
- 74.Ellis JG, Murphy PJ. 1981. Four new opines from crown gall tumours: their detection and properties. Mol. Gen. Genet. 181:36–43. 10.1007/BF00339002 [DOI] [Google Scholar]
- 75.Cook DM, Farrand SK. 1992. The oriT region of the Agrobacterium tumefaciens Ti plasmid pTiC58 shares DNA sequence identity with the transfer origins of RSF1010 and RK2/RP4 and with T-region borders. J. Bacteriol. 174:6238–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cho H, Winans SC. 2007. TraA, TraC and TraD autorepress two divergent quorum-regulated promoters near the transfer origin of the Ti plasmid of Agrobacterium tumefaciens. Mol. Microbiol. 63:1769–1782. 10.1111/j.1365-2958.2007.05624.x [DOI] [PubMed] [Google Scholar]
- 77.Luo Z-Q, Qin Y, Farrand SK. 2000. The antiactivator TraM interferes with the autoinducer-dependent binding of TraR to DNA by interacting with the C-terminal region of the quorum-sensing activator. J. Biol. Chem. 275:7713–7722. 10.1074/jbc.275.11.7713 [DOI] [PubMed] [Google Scholar]
- 78.Su S, Khan SR, Farrand SK. 2008. Induction and loss of Ti plasmid conjugative competence in response to the acyl-homoserine lactone quorum-sensing signal. J. Bacteriol. 190:4398–4407. 10.1128/JB.01684-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Danino VE, Wilkinson A, Edwards A, Downie JA. 2003. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol. Microbiol. 50:511–525. 10.1046/j.1365-2958.2003.03699.x [DOI] [PubMed] [Google Scholar]
- 80.Tun-Garrido C, Bustos P, Gonzalez V, Brom S. 2003. Conjugative transfer of p42a from Rhizobium etli CFN42, which is required for mobilization of the symbiotic plasmid, is regulated by quorum sensing. J. Bacteriol. 185:1681–1692. 10.1128/JB.185.5.1681-1692.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore L, Warren G, Strobel G. 1979. Involvement of a plasmid in the hairy root disease of plants caused by Agrobacterium rhizogenes. Plasmid 2:617–626. 10.1016/0147-619X(79)90059-3 [DOI] [PubMed] [Google Scholar]
- 82.Bouzar H, Moore LW. 1987. Complementary methodologies to identify specific Agrobacterium strains. Appl. Environ. Microbiol. 53:2660–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.White FF, Nester EW. 1980. Relationship of plasmids responsible for hairy root and crown gall tumorigenicity. J. Bacteriol. 144:710–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Slightom JL, Durand-Tardif M, Jouanin L, Tepfer D. 1986. Nucleotide sequence analysis of TL-DNA of Agrobacterium rhizogenes agropine type plasmid. Identification of open reading frames. J. Biol. Chem. 261:108–121 [PubMed] [Google Scholar]
- 85.Paulus F, Otten L. 1993. Functional and mutated agrocinopine synthase genes on octopine T-DNAs. Mol. Plant Microbe Interact. 6:393–402. 10.1094/MPMI-6-393 [DOI] [PubMed] [Google Scholar]
- 86.Anderson AR, Moore LW. 1979. Host specificity in the genus Agrobacterium. Phytopathology 69:320–323. 10.1094/Phyto-69-320 [DOI] [Google Scholar]
- 87.Nesme X, Michel M-F, Digat B. 1987. Population heterogeneity of Agrobacterium tumefaciens in galls of Populus L. from a single nursery. Appl. Environ. Microbiol. 53:655–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kerr A. 1969. Crown gall of stone fruit. I. isolation of Agrobacterium tumefaciens and related species. Aust. J. Biol. Sci. 22:111–116 [Google Scholar]
- 89.Keane PJ, Kerr A, New PB. 1970. Crown gall of stone fruit. II. identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23:585–596 [Google Scholar]
- 90.Moriguchi K, Maeda Y, Satou M, Hardayani NSN, Kataoka M, Tanaka N, Yoshida K. 2001. The complete nucleotide sequence of a plant root-inducing (Ri) plasmid indicates its chimeric structure and evolutionary relationship between tumor-inducing (Ti) and symbiotic (Sym) plasmids in Rhizobiaceae. J. Mol. Biol. 307:771–784. 10.1006/jmbi.2001.4488 [DOI] [PubMed] [Google Scholar]
- 91.Mankin SL, Hill DS, Olhoft PM, Toren E, Wenck AR, Nea L, Xing L, Brown JA, Fu H, Ireland L, Jia H, Hillebrand H, Jones T, Song H-S, Ozias-Akins P. 2007. Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. In Vitro Cell. Dev. Biol. Plant 43:521–535. 10.1007/s11627-007-9071-4 [DOI] [Google Scholar]
- 92.Kiyokawa S, Kobayashi K, Kikuchi Y, Kamada H, Harada H. 1994. Root-inducing region of mikimopine type Ri plasmid pRi1724. Plant Physiol. 104:801–802. 10.1104/pp.104.2.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shiomi T, Shirakata T, Takeuchi A, Oizumi T, Uematsu S. 1987. Hairy root of melon caused by Agrobacterium rhizogenes biovar 1. Ann. Phytopathol, Soc. Jpn. 53:454–549. 10.3186/jjphytopath.53.454 [DOI] [Google Scholar]
- 94.Cervantes L, Bustos P, Girard L, Santamaría R, Dávila G, Vinuesa P, Romero D, Brom S. 2011. The conjugative plasmid of a bean-nodulating Sinorhizobium fredii strain is assembled from sequences of two Rhizobium plasmids and the chromosome of a Sinorhizobium strain. BMC Microbiol. 11:149. 10.1186/1471-2180-11-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.González V, Bustos P, Ramírez-Romero MA, Medrano-Soto A, Salgado H, Hernández-González I, Hernández-Celis JC, Quintero V, Moreno-Hagelsieb G, Girard L, Rodríguez O, Flores M, Cevallos MA, Collado-Vides J, Romero D, Dávila G. 2003. The mosaic structure of the symbiotic plasmid of Rhizobium etli CFN42 and its relation to other symbiotic genome compartments. Genome Biol. 4:R36. 10.1186/gb-2003-4-6-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Streit WR, Schmitz RA, Perret X, Staehelin C, Deakin WJ, Raasch C, Liesegang H, Broughton WJ. 2004. An evolutionary hot spot: the pNGR234b replicon of Rhizobium sp.strain NGR234. J. Bacteriol. 186:535–542. 10.1128/JB.186.2.535-542.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Freiberg C, Fellay R, Bairoch A, Broughton WJ, Rosenthal A, Perret X. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394–401. 10.1038/387394a0 [DOI] [PubMed] [Google Scholar]
- 98.Luo Z-Q, Clemente TE, Farrand SK. 2001. Construction of a derivative of Agrobacterium tumefaciens C58 that does not mutate to tetracycline resistance. Mol. Plant Microbe Interact. 14:98–103. 10.1094/MPMI.2001.14.1.98 [DOI] [PubMed] [Google Scholar]
- 99.Simon R, Priefer U, Pühler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1:784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 100.Farrand SK, Slota JE, Shim JS, Kerr A. 1985. Tn5 insertions in the agrocin 84 plasmid: the conjugal nature of pAgK84 and the locations of determinants for transfer and agrocin 84 production. Plasmid 13:106–117. 10.1016/0147-619X(85)90063-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.