Abstract
Mycoplasma genitalium is the smallest self-replicating bacterium and an important human pathogen responsible for a range of urogenital infections and pathologies. Due to its limited genome size, many genes conserved in other bacteria are missing in M. genitalium. Genes encoding catalase and superoxide dismutase are absent, and how this pathogen overcomes oxidative stress remains poorly understood. In this study, we characterized MG_427, a homolog of the conserved osmC, which encodes hydroperoxide peroxidase, shown to protect bacteria against oxidative stress. We found that recombinant MG_427 protein reduced organic and inorganic peroxide substrates. Also, we showed that a deletion mutant of MG_427 was highly sensitive to killing by tert-butyl hydroperoxide and H2O2 compared to the sensitivity of the wild type. Further, the fully complemented mutant strain reversed its oxidative sensitivity. Examination of the expression pattern of MG_427 during osmotic shock, oxidative stress, and other stress conditions revealed its lack of induction, distinguishing MG_427 from other previously characterized osmC genes.
INTRODUCTION
Mycoplasma genitalium is a cell wall-free bacterium with a genome encoding only 482 protein genes (1). Despite its limited genetic content, M. genitalium is a highly successful self-replicating pathogen responsible for human diseases (2). M. genitalium has been recognized as one of the leading urogenital pathogens, being responsible for about 20% to 35% of cases of Chlamydia-negative, nongonococcal urethritis in men (3). Also, M. genitalium has been implicated in a range of reproductive tract diseases in women, including cervicitis, endometritis, and pelvic inflammatory disease (2, 4–7). M. genitalium colonizes the surface of urogenital epithelial cells and has the capability of invading host cells and establishing long-term persistence, indicating its remarkable ability to circumvent host defense mechanisms (8–10).
As with other pathogens colonizing mucosal epithelium, M. genitalium must overcome oxidative stress caused by reactive oxygen species (ROS) released as part of the host's innate immune response. ROS, which include oxidative radicals, such as superoxide, hydroxyl radicals, H2O2, and organic hydroperoxides (OHPs), are highly toxic and cause severe and sometimes irreversible damage to cellular macromolecules, such as DNA, proteins, and lipids (11, 12). In addition to oxidative radicals delivered by host immune cells, mycoplasmas are known to produce ROS as part of their virulence mechanism, leading to damage to host tissues (13, 14). Therefore, the strategies by which M. genitalium protects itself from oxidative stress are crucial in terms of its survival and pathogenicity. Previously, we reported that the oxidative repair enzyme methionine sulfoxide reductase A (MsrA) protected M. genitalium against oxidative stress and thus contributed to virulence (15). Still, it remains poorly understood how M. genitalium copes with the toxic effects of ROS, as it possesses no genes encoding catalase and superoxide dismutase.
Recently, organic hydroperoxide resistance (Ohr) protein and osmotically inducible protein C (OsmC) have been identified as a new family of peroxidases with similar structures and functions (16–18). Biochemical studies have revealed that Ohr/OsmC proteins are cysteine-based, thiol-dependent peroxidases that preferentially detoxify OHPs (17, 18). The crystal structures of Ohr/OsmC indicate a homodimer containing two active sites located at the monomer interface on opposite sides of the molecule (16, 17). Each active site has two highly conserved, redox-reactive cysteine residues directly involved in reducing OHPs. The catalytic mechanism involves the formation of sulfenic acid intermediates (—SOH) after the first cysteine is oxidized by a hydroperoxide, which then reduce hydroperoxide to a corresponding alcohol. Subsequently, the sulfenic acid intermediate forms a disulfide bond with the second cysteine, and the enzyme is reduced back to its active state via dihydrolipoic acid (16, 17, 19). The role of ohr in bacterial defense against OHP-induced stress has been investigated in many bacteria, such as Xanthomonas campestris (20), Pseudomonas aeruginosa (21), Bacillus subtilis (22), and Enterococcus faecalis (23). Compared to their parental strains, ohr deletion mutants exhibit hypersensitivity to killing by OHPs, such as tert-butyl hydroperoxide (t-BHP) and cumene hydroperoxide (CHP). Similar findings were also reported for osmC deletion mutants of Escherichia coli (24) and Mycobacterium smegmatis (25).
Interestingly, M. genitalium contains two ohr and osmC homologs (MG_454 for ohr; MG_427 for osmC). The conservation of these two genes in this genome-streamlined bacterium suggests important biological functions. Previously, a mutant of MG_454 was shown to be hypersensitive to OHPs, and the ectopic expression of MG_454 complemented a P. aeruginosa ohr mutant (11), indicating that MG_454 provides resistance to oxidative stress. MG_427 may also possess peroxidase activity, but its role is unclear. Previous studies suggested that MG_427 is essential for the in vitro growth of M. genitalium (1, 26), which predicts the ineffective use of targeted gene disruption/deletion for functional analysis of this gene. In this paper, we report the successful generation of a deletion mutant of MG_427 and characterization of MG_427 relative to oxidative stress resistance.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. genitalium type strain G37 (ATCC 33530) was grown to exponential phase (72 h) in 50 ml of SP-4 medium (27) at 37°C in 75-cm2 tissue culture flasks. Surface-attached mycoplasma cells were washed three times with phosphate-buffered saline (PBS; pH 7.4) and harvested by scraping and pelleting at 10,000 × g for 20 min at 4°C. The solid medium used for the disk inhibition assays was SP-4 agar containing 0.85% Noble agar. To obtain growth curves, G37 and ΔMG_427 mutant strains were grown in SP-4 medium containing 0.5% (wt/vol) glucose or 0.5% (wt/vol) glycerol as carbon sources (50 ml/flask). At various time points (48, 72, and 96 h), mycoplasma cells were harvested as described above, and the genome numbers were determined by quantitative real-time PCR. E. coli TOP10 cells (Invitrogen) and BL21(DE3) cells (Stratagene) were grown in Luria-Bertani (LB) broth and used as the host for cloning and recombinant protein expression.
Cloning, overexpression, and purification of rMG_427 protein.
The full-length MG_427 gene was amplified with primers (5′MG_427 and 3′MG_427; Table 1) by overlapping fusion PCR using M. genitalium genomic DNA as the template. Overlapping primers (overlap F and overlap R) were designed to perform codon correction from TGA to TGG to facilitate the expression of MG_427 in E. coli. The final PCR product was digested with NdeI and XhoI and cloned into the pET19b expression vector to yield pET19b-MG_427. After the UGA codon change was verified by DNA sequencing, pET19-MG_427 was transformed into E. coli BL21(DE3), and positive colonies were screened for resistance to ampicillin and expression of recombinant MG_427 protein (rMG_427). To overexpress rMG_427, strain BL21(DE3) transformed with pET19b-MG_427 was cultured at 37°C overnight in 25 ml of LB medium containing ampicillin (100 μg/ml). Then, the culture was transferred to 1 liter of fresh LB plus ampicillin (100 μg/ml) and grown until the optical density at 600 nm (OD600) reached 0.6. After isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added to a final concentration of 0.5 mM, growth was continued at 30°C for 4 h and cells were harvested by centrifugation. To purify rMG_427, the bacterial pellet was resuspended in lysis buffer (20 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 7.4) and sonicated on ice for 5 cycles (1 min/cycle). The suspension was centrifuged at 12,000 × g for 30 min, and the supernatant was passed through 0.45- and 0.22-μm-pore-size filters. The cleared supernatant was loaded onto a nickel-nitrilotriacetic acid (Ni-NTA) affinity column (Qiagen), and protein purification was performed by fast protein liquid chromatography (AKTAexplorer; Amersham). Bound proteins were washed with 20 column volumes of wash buffer (20 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 7.4) and eluted by a linear gradient reaching 500 mM imidazole. Purified proteins were stored in HEPES buffer (50 mM HEPES, pH 7.5, 100 mM NaCl, 10% glycerol) at −70°C.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′ to 3′)a |
|---|---|
| 5′MG_427 (NdeI) | GGAATTCCATATGGATAAAAAATACGATATCACAG |
| 3′MG_427 (XhoI) | CCGCTCGAGTTAGTGGACTAAAGTAACGCTAA |
| Overlap F | ACATTCAC(TGG)GAAATTCACTC |
| Overlap R | GAGTGAATTTC(CCA)GTGAATGT |
| 5′UMG_427 (BamHI) | CGCGGATCCTGGGATCTTTATTGGATCAC |
| 3′UMG_427 (NotI) | TAAGAATGCGGCCGCATATGCTTATGGATTAAGTTTAACTTAAAT |
| 5′DMG_427 (XhoI) | CCGCTCGAGTAGCGTTACTTTAGTCCACTAAAAC |
| 3′DMG_427 (XbaI) | GCTCTAGAGATAGGATGAATAAAAACGTTAGTTATC |
| 5′MG_427C (NotI) | ATAAGAATGCGGCCGCACTAATGGTAAAGTAACACGGATTC |
| 3′MG_427C (BamHI) | CGCGGATCCTTAGTGGACTAAAGTAACGCTAAT |
| MG_427SP1 | CCACGTTTACTTACAAAATCAATG |
| MG_427SP2 | GAGTGAATTTCTCAGTGAATGT |
Underlined sequences are the restriction enzyme recognition sequences. The codon change from TGA to TGG is included in parentheses.
Assessment of rMG_427 peroxidase activity.
A ferrous oxidation xylenol (FOX) assay was used to measure the peroxidase activity of rMG_427 (12, 16). Briefly, purified rMG_427 protein was preincubated on ice with 5 mM dithiothreitol (DTT) for 1 h. The subsequent enzymatic reaction was carried out at room temperature in 1 ml of buffer (100 mM potassium phosphate, pH 7.0, 1 mM DTT, various amounts of substrate peroxides) plus rMG_427 protein. At various time points, 50-μl aliquots were transferred to new Eppendorf tubes containing 20 μl of 0.25 M H2SO4 to stop the reaction. The peroxide concentrations were determined colorimetrically by adding 930 μl of freshly prepared FOX reagent, and the optical density at 560 nm was read after 1 h, when the color reaction reached equilibrium. The removal of peroxides was calculated by comparing the OD560 to the values on standard curves generated with respective peroxides.
Construction of ΔMG_427 mutant.
The in-frame deletion of the MG_427 gene was achieved by homologous recombination using the tetracycline resistance gene (tetM438) as the selectable marker. The tetM438 gene was excised from pMTnTetM438 (28) by digestion with XhoI and NotI, and the gel-purified fragment was cloned into the pCRII-TOPO vector (Invitrogen) to create plasmid pCR_tetM438. To construct the MG_427-knockout vector, DNA fragments that included 1 kb of sequence upstream or downstream of MG_427 were PCR amplified from the genomic DNA of M. genitalium using primer pairs 5′UMG_427/3′UMG_427 and 5′DMG_427/3′DMG_427, respectively. These PCR products were double digested with the corresponding restriction enzymes and successively cloned into pCR_tetM438, yielding plasmid pCR-MG_427KO. This plasmid was used to transform M. genitalium by electroporation, as previously described (29). Mutants were selected on SP-4 agar containing 2 μg/ml of tetracycline, and single colonies were picked and propagated.
Southern blot analysis.
Genomic DNA was isolated from M. genitalium wild-type (WT) and ΔMG_427 strains using an Easy-DNA kit (Invitrogen), digested with HindIII, and separated on a 1% agarose gel by electrophoresis. The resolved DNA fragments were transferred to Zeta-Probe membranes according to the manufacturer's instructions (Bio-Rad). After transfer, membranes were baked at 80°C for 2 h in a hybridization oven to fix DNA fragments onto membranes. For hybridization, membranes were prehybridized at 42°C for 30 min in a prehybridization solution containing 50% formamide, 0.12 M Na2HPO4, 0.25 M NaCl, 7% (wt/vol) sodium dodecyl sulfate (SDS), and 1 mM EDTA. The probe for Southern hybridization was generated by labeling the linearized vector pCR-MG_427KO with [α-32P]dCTP by random priming. After prehybridization, membranes were soaked in fresh prehybridization buffer, and labeled probes were added. Hybridization was carried out at 42°C for 12 h. Then, membranes were washed successively by vigorous agitation at room temperature for 15 min with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS, with 0.5× SSC–0.1% SDS, and finally, with 0.1× SSC–0.1% SDS, before exposure to X-ray film for autoradiography.
Complementation of the ΔMG_427 strain.
A fragment that harbored the full-length MG_427 gene and 100 bp of its upstream sequence was amplified by PCR (primer pair 5′MG_427C/3′MG_427C) using M. genitalium genomic DNA as the template. This fragment was digested with NotI and BamHI and cloned into pMTn4001-puro, which carries the selectable marker of the puromycin resistance gene (puromycin N-acetyltransferase) (30). The plasmid was used to transform the ΔMG_427 mutant, and single colonies were obtained after growth in the presence of 5 μg/ml puromycin for 2 weeks. Immunoblotting analysis was used to confirm the expression of MG_427 in the complemented ΔMG_427 mutant strain.
Immunoblotting analysis.
Equal amounts (5 μg/well) of M. genitalium whole-cell lysate (used for confirming the ΔMG_427 and complemented strains) or membrane and cytoplasmic fractions (used for determining the subcellular location of MG_427) were separated on 4 to 12% NuPAGE gradient gels (Invitrogen). After electrophoresis, proteins were transferred to nitrocellulose membranes. Immunoblotting was carried out using individual rabbit antisera reactive against rMG_427, elongation factor G (EF-G; a kind gift from Richard Herrmann), and P140 adhesin (MG_191), followed by alkaline phosphatase-conjugated goat anti-rabbit IgG antibody.
Disk inhibition assay.
A disk inhibition assay was used to determine the sensitivity of M. genitalium to peroxide stress. M. genitalium strains (WT, ΔMG_427, and ΔMG_427 complemented strains) were grown to exponential phase, and surface-attached cells were washed twice with PBS. Then, cells were scraped into 10 ml PBS, and cell suspensions were passed successively through 22.5-gauge and 25.5-gauge syringes. The OD600 was measured, the cells were diluted in PBS, and the OD600 was adjusted to 0.45. Subsequently, cell suspensions were transferred to SP-4 agar plates (100 μl per plate) and evenly spread, followed by brief drying under a laminar flow hood. Then, sterile filter paper disks (Sigma) were placed onto each plate in triplicate, and 10 μl of peroxide (1 M H2O2, 1 M CHP, or 7 M t-BHP) was pipetted to the center of individual disks. Plates were inverted and incubated at 37°C in a 5% CO2, humidified incubator. After 7 days, zones of inhibition were examined with a dissecting microscope, and diameters were recorded.
Quantitative real-time PCR.
Genomic DNA of mycoplasma cells collected from cultures used for growth curve assays was extracted with an Easy-DNA kit (Invitrogen) according to the manufacturer's instruction. Quantification of M. genitalium genome numbers was performed with a StepOnePlus real-time PCR system (Life Technologies) and SYBR green chemistry (Applied Biosystems). Primers used for amplification were specific for MG_149, as previously described (29). Serial 100-fold dilutions of M. genitalium genomic DNA (1010 to 106 copies/reaction mixture) were included in the amplification to establish a standard curve.
M. genitalium membrane purification.
Membrane fractions of M. genitalium were obtained by glycerol-osmotic lysis as we previously described (31). Briefly, exponential-phase cultures (400 ml) of M. genitalium were washed three times with 0.02 M Tris-HCl, and the harvested cell pellets were resuspended in 5 ml glycerol (2 M) and passed through a 25.5-gauge syringe. An aliquot of the cell suspension was saved for determining total cell proteins. The remaining cell suspension was rapidly injected into 250 ml of prewarmed, deionized water and incubated for 15 min at 37°C and 90 rpm, which resulted in the complete lysis of the mycoplasmas. Membrane fractions were collected by centrifugation at 60,000 × g for 45 min and washed once with 0.25 M NaCl and once with deionized water. Membranes were further purified by centrifugation through linear 30 to 60% (wt/vol) sucrose gradients at 160,000 × g for 4 h at 4°C and collected by needle puncture. Cytoplasmic fractions were concentrated using Amicon concentrators (Millipore). Protein determinations were performed on total, cytoplasmic, and membrane samples.
RNA isolation and Northern blot analysis.
RNA isolation was performed as previously described (32). Briefly, surface-attached mycoplasma cells were washed twice with ice-cold PBS, and cells were lysed by the addition of TRI Reagent (Sigma). The extraction of total RNA was performed according to the instructions provided by the manufacturer. For Northern blot analysis, 3 μg of RNA from each sample was resolved using formaldehyde denaturing gels and transferred by capillary action to a Zeta-Probe GT nylon membrane (Bio-Rad). The probe was generated by random priming of the full-length MG_427 PCR product with the DNA polymerase I large (Klenow) fragment (New England BioLabs) in the presence of [α-32P]dATP (PerkinElmer). Membranes were hybridized at 42°C overnight, washed twice for 10 min each time at 59°C as described previously (33), and scanned using a Typhoon 9400 PhosphorImager (Molecular Dynamics). To quantify changes in the levels of RNA, the intensity of bands on digital images of Northern blots was analyzed by using Image Studio Lite (version 3.1) software (Li-Cor). Values were expressed in arbitrary units, and the fold change in the signal intensity was determined after the background value was subtracted.
5′ RACE.
5′ rapid amplification of cDNA ends (5′ RACE) was performed with a 5′/3′ RACE kit (Roche) designed for amplification of the 5′/3′ ends of mRNAs by reverse transcription-PCR. Briefly, 2 μg of total mycoplasma RNA was used to synthesize the first-strand cDNA by Transcriptor reverse transcriptase using MG_427-specific primer SP1 (Table 1). The cDNA was enriched by QIAquick PCR purification column chromatography (Qiagen) and further treated with terminal transferase to add a homopolymeric A tail at the 3′ end in the presence of dATP. The obtained cDNA was used as the template for the amplification of targeted cDNA with an oligo(dT) anchor primer and MG_427-specific primer SP2. Finally, the PCR product was TA cloned into plasmid pCRII (Invitrogen), and plasmids from five individual clones were sequenced to map the 5′ end.
RESULTS
Recombinant MG_427 protein responds to treatment with DTT and oxidants.
MG_427 was annotated as a gene encoding an OsmC-like protein in M. genitalium. A notable feature in the primary sequence of MG_427 is two highly conserved cysteine residues (Cys52 and Cys119) which are typical of Ohr/OsmC proteins and shown to be involved in detoxifying peroxides (34). To study whether MG_427 possesses peroxidase activity, rMG_427 from M. genitalium was overexpressed in E. coli and purified to homogeneity using an Ni-NTA affinity column. As shown in Fig. 1, two predominant bands (bands a and b) corresponding to MG_427 were observed, and both of these migrated close to 16 kDa, as expected for a monomer. Moreover, two weak bands corresponding to a dimer of MG_427 were observed at ∼35 kDa. In the presence of DTT, the bands corresponding to dimers disappeared, while the bands corresponding to monomers were reduced to one band (see the increased intensity of band b). Therefore, the lower band (band a) of the monomer represents oxidized MG_427, whereas the upper band (band b) represents reduced MG_427 (confirmed with mass spectrometry analysis; data not shown). Treatment of MG_427 with t-BHP and a mild concentration of H2O2 resulted in oxidized MG_427, shown by the augmented intensity of the lower band (band a). The increased intensity of the upper band (band b) after treatment with a high dose of H2O2 may represent a mixture of reduced and oxidized states of MG_427, as previously observed for Ohr (18). Additionally, a single band corresponding to dimers of MG_427 appeared after treatment of MG_427 with a high dose of H2O2 (band c), suggesting the formation of intermolecular disulfide bonds. Together, these results indicate that the MG_427 protein responds to redox treatment, consistent with previously reported properties of Ohr/OsmC proteins.
FIG 1.
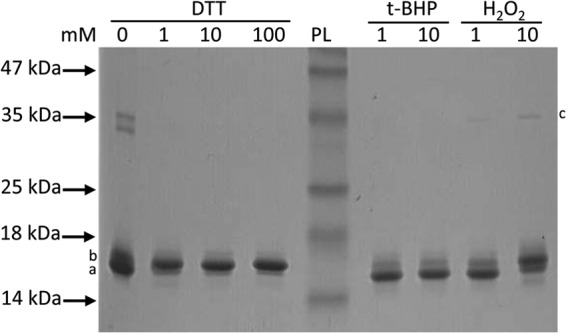
SDS-PAGE analysis of recombinant MG_427 protein. Purified recombinant MG_427 protein was treated with DTT or peroxides for 1 h at room temperature. Proteins were resolved by nonreducing 12% SDS-PAGE and visualized by Coomassie blue staining. PL, protein ladder. a and b, the lower and upper band, respectively; c, the band corresponding to dimers of MG_427.
Recombinant MG_427 protein reduces peroxides.
Ohr/OsmC proteins were shown to reduce peroxides in the presence of DTT in vitro (16–18). To characterize whether MG_427 possesses peroxidase activity, we measured its ability to reduce various peroxide substrates by a FOX assay. Figure 2 shows that rMG_427 reduced t-BHP, CHP, and H2O2 in a concentration-dependent manner. The reduction of organic hydroperoxides t-BHP and CHP by rMG_427 is consistent with the known activities of Ohr/OsmC proteins, which preferentially detoxify OHPs. In addition, the observation that MG_427 was able to reduce H2O2 is consistent with the reported property of OsmC from several bacteria (25, 35, 36).
FIG 2.
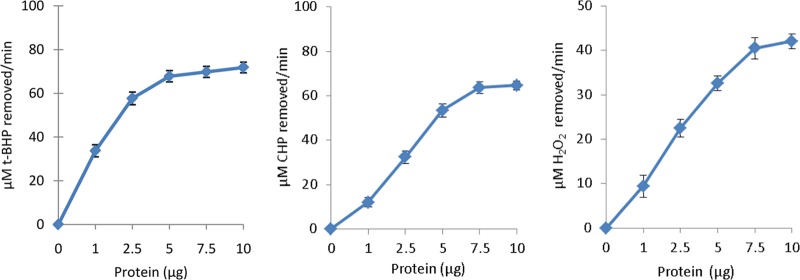
rMG_427 reduces t-BHP, CHP, and H2O2 in a protein concentration-dependent manner. The reduction of peroxides was measured by FOX assay, as described in Materials and Methods. Each point represents a mean value from three separate experiments, and bars represent standard errors.
Generation of ΔMG_427 mutant and complementation strain.
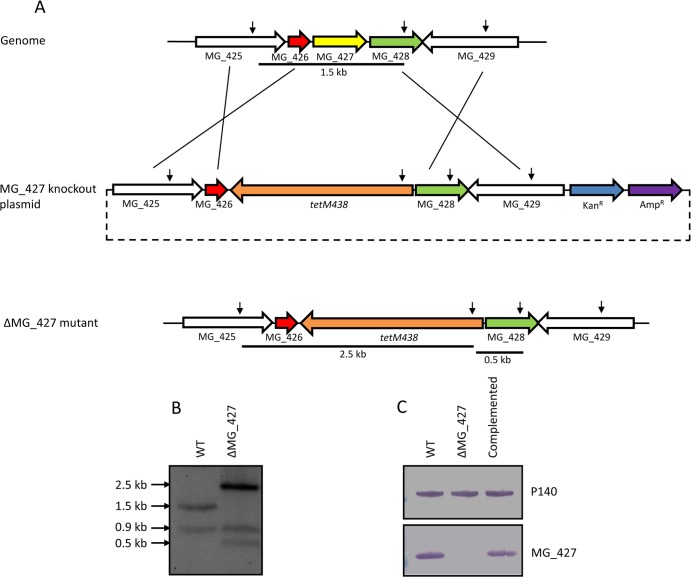
Since rMG_427 exhibited peroxidase activity, we investigated whether MG_427 truly provides protection against oxidative stress in M. genitalium. To this end, we sought to delete MG_427 from the chromosome of M. genitalium. Although MG_427 was considered an essential gene for in vitro growth of M. genitalium, as suggested by studies using Tn4001 transposon mutagenesis (1, 26), we successfully generated a deletion mutant of MG_427 by homologous recombination (Fig. 3). To delete the MG_427 gene, we generated a knockout construct in which the open reading frame of MG_427 was replaced by tetM438 derived from pMTnTet438 (28) (Fig. 3A). This knockout plasmid was electroporated into wild-type M. genitalium, and mycoplasma cells were plated onto SP-4 agar and selected by tetracycline resistance. Single colonies were picked, propagated, and screened. The deletion of MG_427 was confirmed by the Southern blotting profile (Fig. 3B), as well as immunoblotting (Fig. 3C), which clearly demonstrated that the mutant strain failed to produce the MG_427 protein. To complement the ΔMG_427 mutant, we transformed the mutant with a plasmid that carries a fragment harboring the MG_427 gene and its 100-bp upstream sequence. Single colonies were obtained, and the expression of MG_427 was confirmed by immunoblotting (Fig. 3C).
FIG 3.
Generation of ΔMG_427 mutant and complementation strain. (A) Schematic representation of the strategy for generating the MG_427 deletion mutant by homologous recombination. Downward-pointing arrows represent HindIII cleavage sites. Lines underneath indicate fragments that distinguish the wild type from the MG_427 deletion mutant. (B) Southern hybridization profiles. WT, wild type; ΔMG_427, deletion mutant. (C) Western blot. Whole-cell lysates from the WT, ΔMG_427, and complemented mutant strains were resolved by SDS-PAGE, and proteins were transferred to a nitrocellulose membrane. The membrane was probed with polyclonal antibodies against the adhesin P140 (MG_191) or MG_427.
Sensitivity of the ΔMG_427 strain of M. genitalium to oxidative stress.
To understand the role of MG_427 in relation to oxidative stress, we examined the sensitivity of the ΔMG_427 strain to various peroxides (H2O2, t-BHP, and CHP) by disk inhibition assay. As shown in Table 2, wild-type M. genitalium was relatively resistant to t-BHP and CHP compared to its resistance to H2O2, on the basis of the zones of inhibition. The ΔMG_427 strain exhibited increased sensitivity to H2O2 and hypersensitivity to t-BHP. Interestingly, the ΔMG_427 strain showed no significant change in sensitivity to CHP, despite our observation that the rMG_427 strain reduced CHP. The role of MG_427 in detoxifying t-BHP and H2O2 was validated with the introduction of an intact MG_427 gene into the deletion mutant. The complementation of the ΔMG_427 mutant restored sensitivity to a level similar to that of the wild type (Table 2).
TABLE 2.
Sensitivity of M. genitalium strains to peroxides
| Peroxide species | Zone of inhibition (mm)a |
||
|---|---|---|---|
| WT | ΔMG_427 mutant | ΔMG_427 complemented strain | |
| H2O2 | 25.3 ± 0.6 | 30 ± 2.0 | 26 ± 1.7 |
| t-BHP | 16.7 ± 1.2 | 25 ± 1.7 | 17.3 ± 1.5 |
| CHP | 20 ± 1.0 | 20 ± 2.0 | 20.3 ± 2.3 |
Data are expressed as means ± standard deviations for three independent determinations.
To further test the role of MG_427 in protecting M. genitalium against oxidative stress, we monitored the growth curves of the M. genitalium wild type and ΔMG_427 mutant in SP-4 medium containing either glucose or glycerol as the carbon source. Previously, glycerol oxidation by mycoplasmas was shown to generate H2O2, a major product for cytotoxicity (37, 38). Figure 4 shows that although both strains grew well with glucose, they exhibited slow growth with glycerol, as was previously reported for M. pneumoniae (37), with the ΔMG_427 mutant exhibiting significantly slower growth. These data suggest a protective role of MG_427 against H2O2.
FIG 4.
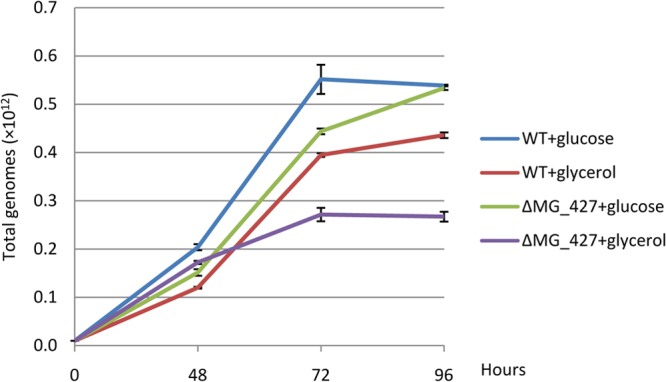
Growth of M. genitalium WT and ΔMG_427 mutant strains in SP-4 medium containing glucose or glycerol as the carbon source. SP-4 medium containing 0.5% (wt/vol) glucose or 0.5% (wt/vol) glycerol was prepared, and equal numbers of mycoplasma cells (2 × 108/ml) were inoculated in 75-cm2 flasks (50 ml/flask) and grown at 37°C. At 48, 72, and 96 h, attached mycoplasma cells were washed and harvested. The total numbers of M. genitalium genomes were determined by quantitative real-time PCR analysis using triplicate samples.
Subcellular localization of MG_427.
We examined the subcellular localization of MG_427 by cell fractionation and immunoblotting. Figure 5 shows that MG_427 was predominantly localized in the cytoplasmic fraction. On the basis of the membrane yield (10%) and band intensity, an estimated 1.4% of total MG_427 protein localized to the membrane fraction.
FIG 5.
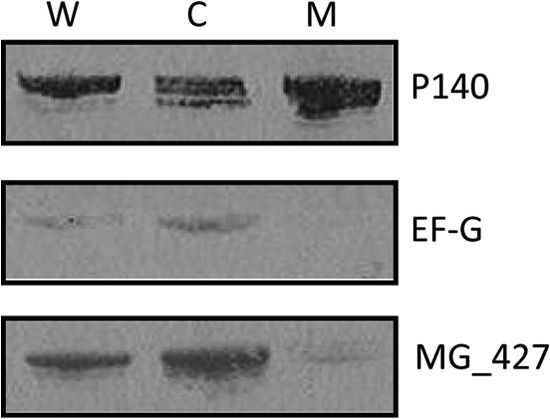
Subcellular location of MG_427 in M. genitalium. Equal amounts (5 μg/well) of whole-cell lysate (W), cytoplasmic (C), and membrane (M) fractions of M. genitalium were resolved by 4 to 12% NuPAGE and transferred to a nitrocellulose membrane. The membrane was cut into three pieces on the basis of the expected protein sizes before exposure to specific antibodies (anti-P140, anti-EF-G, and anti-rMG_427). Adhesin P140 (MG_191) served as the positive control for membrane protein, while EF-G, a cytoplasmic protein, served as the internal cytosolic control.
Expression pattern of MG_427 under various stress conditions.
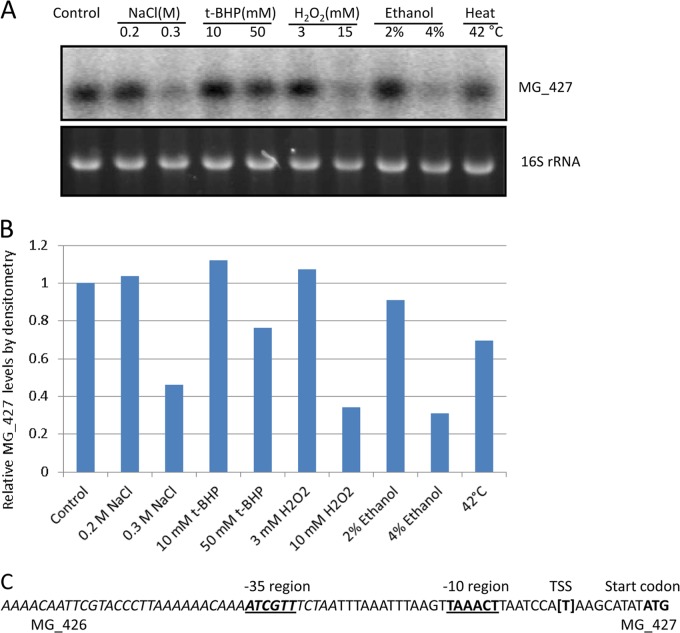
Generally, the expression pattern of osmC differs from that of ohr homologs (39). While osmC is upregulated by osmotic shock and ethanol stress conditions, ohr is induced only by OHPs. Since MG_427 provides resistance to oxidants, we performed Northern blot analysis to determine the expression pattern of MG_427 in response to osmotic shock, oxidative stress, and other stress conditions. Our result shows that MG_427 is significantly downregulated by osmotic shock (0.3 M NaCl) and ethanol (4%) and less so by heat shock (Fig. 6A and B). Under oxidative stress conditions, the transcription of MG_427 appeared to be largely unchanged at mild concentrations of t-BHP and H2O2 and significantly decreased at higher concentrations of t-BHP and H2O2 (Fig. 6A and B).
FIG 6.
Transcription of MG_427 under various stress conditions and identification of the putative promoter. (A) Cultures of M. genitalium were grown in SP-4 broth to logarithmic phase and exposed to various stresses (indicated on the top) for 1 h before total RNA was isolated. Northern blot analysis was performed as described in Materials and Methods. (B) Relative levels of MG_427 determined by densitometry. (C) Putative promoter region of MG_427. The putative −10 region and −35 region for σ70 are bold-faced and underlined. The transcriptional start site (TSS) is shown in brackets. The sequence from MG_426 is italicized.
Identification of the putative promoter region of MG_427.
Since MG_427 was expressed as a monocistronic mRNA, we examined the putative promoter region by determining the transcriptional start site (TSS) using 5′ RACE. The first base was mapped to a T residue 6 bases upstream of the translation start of MG_427 (Fig. 6C). On the basis of this information, a putative −10 region (TAAACT) for σ70 was identified. In addition, a putative −35 region (ATCGTT) which localizes within the upstream open reading frame of MG_426 was also identified (Fig. 6C).
DISCUSSION
In this study, we demonstrated that the M. genitalium OsmC-like protein encoded by MG_427 possesses hydroperoxide reductase activity. We showed that purified rMG_427 protein responded to DTT and oxidant exposures and reduced OHPs and H2O2 in a concentration-dependent manner. These results are consistent with the reported hydroperoxide peroxidase properties of OsmC proteins in E. coli (17, 34), Thermus thermophilus (36), Thermococcus kodakarensis KOD1 (35), as well as M. tuberculosis and M. smegmatis (25). Further, we demonstrated that the ΔMG_427 strain of M. genitalium exhibited hypersensitivity to t-BHP and increased sensitivity to H2O2 and this sensitivity could be reversed in the mutant strain complemented with an intact MG_427 gene, similar to osmC mutants of E. coli (24) and M. smegmatis (25). We also observed a slower growth of the ΔMG_427 mutant than the wild type in SP-4 medium containing glycerol as the carbon source, a growth condition known to generate H2O2 by mycoplasmas (37, 38). Altogether, these data suggest that MG_427 encodes a hydroperoxide reductase which likely plays an important role in protecting M. genitalium against oxidative stress.
Our finding that MG_427 can detoxify H2O2 provides an additional example that OsmC may target substrates different from Ohr. Although OsmC and Ohr are structurally and functionally similar, they may preferentially metabolize different peroxide substrates (16, 17, 34, 36). Compared to OHPs such as t-BHP and CHP, H2O2 is a very poor substrate for Ohr (16, 18). In contrast, H2O2 appears to be a good substrate for OsmC, shown by the similar activity of OsmC toward H2O2 and OHPs, as reported in T. thermophilus (36), T. kodakarensis KOD1 (35), and Mycobacterium species (25). This difference in reactivity to H2O2 could be due to subtle three-dimensional alterations in the active-site clefts between OsmC and Ohr (36). Regarding substrate specificity between OsmC and Ohr, we also observed that the ΔMG_427 strain showed no response to CHP by disk inhibition assay (Table 2), although MG_427 reduced CHP in vitro. Currently, it is not clear what causes the discrepancy. One possibility could be the inhibition of enzyme regeneration, as suggested by the observations for Ohr encoded by MGA1142 in Mycoplasma gallisepticum (12). MGA1142 was observed to reduce CHP at a high rate, but this activity was rapidly lost. Since MGA1142 produced distinct patterns of migration on SDS-polyacrylamide gels upon CHP exposure, a change in the structure or redox status of MGA1142 may inhibit enzyme turnover.
It is interesting that some bacteria express both OsmC and Ohr proteins, given that they have overlapping activity (16, 17). This raises the question of how bacteria employ these two hydroperoxide reductases. It was speculated that each protein may reside in a distinct subcellular location to deal with exogenous peroxides derived from host immune cells or endogenous peroxides produced by bacterial metabolism, or both (16, 17). Our results indicate that the MG_427 protein is predominantly localized in the cytoplasmic fraction, with only a tiny fraction (1.4%) of total MG_427 proteins being associated with the membrane (Fig. 5). Further experiments are needed to validate this result as well as to examine the subcellular localization of MG_454.
The expression pattern of MG_427 is significantly different from that of other osmC and ohr homologs (Fig. 6A and B), suggesting that novel mechanisms are used for its regulation. The downregulation of MG_427 in response to physical stresses (osmotic shock and ethanol) contrasts with the strong induction of osmC under these conditions in E. coli and other bacteria with large genomes (39, 40). On the other hand, MG_427 showed no induction upon exposure to t-BHP, a finding which differs considerably from the robust ohr induction by OHPs (20, 39). Interestingly, the expression pattern of MG_427 also differed from that of ohr homologs in mycoplasmas. Unlike MG_427, MG_454 of M. genitalium is strongly induced by osmotic shock (0.4 M NaCl) and heat shock (11). The MGA1142 of M. gallisepticum is induced about 2-fold by ethanol, despite being downregulated by osmotic shock (12). It appears that osmC is regulated differently from ohr in mycoplasmas, even though they have overlapping functions, but the mechanism remains elusive. In E. coli, the induction of osmC is under complex control involving the two-component system rcsB-rcsC, NhaR, and an alternative sigma factor, RpoS (24, 41). However, like other mycoplasmas, M. genitalium lacks genes encoding a two-component system and contains only a housekeeping sigma factor (σ70), which is consistent with the identification of the putative MG_427 promoter for σ70 (Fig. 6C). Nevertheless, a distant homolog of rpoE (σE, MG_022) was identified in M. genitalium, which encodes a putative alternative sigma factor known to regulate the stress response (42). Its relationship in regulating the expression of MG_427 is unknown.
In summary, we have demonstrated that MG_427 encodes a hydroperoxide reductase and have provided evidence that it plays a role in the resistance of M. genitalium to oxidative stress. Further investigation of its regulation and contribution to pathogenesis will shed light on how mycoplasmas colonize and persist in the host to lead to acute and chronic disease.
ACKNOWLEDGMENT
This work was supported by Frost Bank Trusts.
Footnotes
Published ahead of print 20 December 2013
REFERENCES
- 1.Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103:425–430. 10.1073/pnas.0510013103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baseman JB, Tully JG. 1997. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 3:21–32. 10.3201/eid0301.970103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGowin CL, Anderson-Smits C. 2011. Mycoplasma genitalium: an emerging cause of sexually transmitted disease in women. PLoS Pathog. 7:e1001324. 10.1371/journal.ppat.1001324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor-Robinson D, Horner PJ. 2001. The role of Mycoplasma genitalium in non-gonococcal urethritis. Sex. Transm. Infect. 77:229–231. 10.1136/sti.77.4.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor-Robinson D. 2002. Mycoplasma genitalium—an up-date. Int. J. STD AIDS 13:145–151. 10.1258/0956462021924776 [DOI] [PubMed] [Google Scholar]
- 6.Baseman JB, Cagle M, Korte JE, Herrera C, Rasmussen WG, Baseman JG, Shain R, Piper JM. 2004. Diagnostic assessment of Mycoplasma genitalium in culture-positive women. J. Clin. Microbiol. 42:203–211. 10.1128/JCM.42.1.203-211.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korte JE, Baseman JB, Cagle MP, Herrera C, Piper JM, Holden AE, Perdue ST, Champion JD, Shain RN. 2006. Cervicitis and genitourinary symptoms in women culture positive for Mycoplasma genitalium. Am. J. Reprod. Immunol. 55:265–275. 10.1111/j.1600-0897.2005.00359.x [DOI] [PubMed] [Google Scholar]
- 8.Baseman JB, Lange M, Criscimagna NL, Giron JA, Thomas CA. 1995. Interplay between mycoplasmas and host target cells. Microb. Pathog. 19:105–116. 10.1006/mpat.1995.0050 [DOI] [PubMed] [Google Scholar]
- 9.Dallo SF, Baseman JB. 2000. Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb. Pathog. 29:301–309. 10.1006/mpat.2000.0395 [DOI] [PubMed] [Google Scholar]
- 10.Ueno PM, Timenetsky J, Centonze VE, Wewer JJ, Cagle M, Stein MA, Krishnan M, Baseman JB. 2008. Interaction of Mycoplasma genitalium with host cells: evidence for nuclear localization. Microbiology 154:3033–3041. 10.1099/mic.0.2008/020735-0 [DOI] [PubMed] [Google Scholar]
- 11.Saikolappan S, Sasindran SJ, Yu HD, Baseman JB, Dhandayuthapani S. 2009. The Mycoplasma genitalium MG_454 gene product resists killing by organic hydroperoxides. J. Bacteriol. 191:6675–6682. 10.1128/JB.01066-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenkins C, Samudrala R, Geary SJ, Djordjevic SP. 2008. Structural and functional characterization of an organic hydroperoxide resistance protein from Mycoplasma gallisepticum. J. Bacteriol. 190:2206–2216. 10.1128/JB.01685-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch RE, Cole BC. 1980. Mycoplasma pneumoniae: a prokaryote which consumes oxygen and generates superoxide but which lacks superoxide dismutase. Biochem. Biophys. Res. Commun. 96:98–105. 10.1016/0006-291X(80)91186-9 [DOI] [PubMed] [Google Scholar]
- 14.Vilei EM, Frey J. 2001. Genetic and biochemical characterization of glycerol uptake in Mycoplasma mycoides subsp. mycoides SC: its impact on H(2)O(2) production and virulence. Clin. Diagn. Lab. Immunol. 8:85–92. 10.1128/CDLI.8.1.85-92.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhandayuthapani S, Blaylock MW, Bebear CM, Rasmussen WG, Baseman JB. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645–5650. 10.1128/JB.183.19.5645-5650.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesniak J, Barton WA, Nikolov DB. 2002. Structural and functional characterization of the Pseudomonas hydroperoxide resistance protein Ohr. EMBO J. 21:6649–6659. 10.1093/emboj/cdf670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesniak J, Barton WA, Nikolov DB. 2003. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 12:2838–2843. 10.1110/ps.03375603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cussiol JR, Alves SV, de Oliveira MA, Netto LE. 2003. Organic hydroperoxide resistance gene encodes a thiol-dependent peroxidase. J. Biol. Chem. 278:11570–11578. 10.1074/jbc.M300252200 [DOI] [PubMed] [Google Scholar]
- 19.Cussiol JR, Alegria TG, Szweda LI, Netto LE. 2010. Ohr (organic hydroperoxide resistance protein) possesses a previously undescribed activity, lipoyl-dependent peroxidase. J. Biol. Chem. 285:21943–21950. 10.1074/jbc.M110.117283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. 1998. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 180:2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochsner UA, Hassett DJ, Vasil ML. 2001. Genetic and physiological characterization of ohr, encoding a protein involved in organic hydroperoxide resistance in Pseudomonas aeruginosa. J. Bacteriol. 183:773–778. 10.1128/JB.183.2.773-778.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. 2001. OhrR is a repressor of ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J. Bacteriol. 183:4134–4141. 10.1128/JB.183.14.4134-4141.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rincé A, Giard JC, Pichereau V, Flahaut S, Auffray Y. 2001. Identification and characterization of gsp65, an organic hydroperoxide resistance (ohr) gene encoding a general stress protein in Enterococcus faecalis. J. Bacteriol. 183:1482–1488. 10.1128/JB.183.4.1482-1488.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conter A, Gangneux C, Suzanne M, Gutierrez C. 2001. Survival of Escherichia coli during long-term starvation: effects of aeration, NaCl, and the rpoS and osmC gene products. Res. Microbiol. 152:17–26. 10.1016/S0923-2508(00)01164-5 [DOI] [PubMed] [Google Scholar]
- 25.Saikolappan S, Das K, Sasindran SJ, Jagannath C, Dhandayuthapani S. 2011. OsmC proteins of Mycobacterium tuberculosis and Mycobacterium smegmatis protect against organic hydroperoxide stress. Tuberculosis (Edinb.) 91(Suppl 1):S119–S127. 10.1016/j.tube.2011.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165–2169. 10.1126/science.286.5447.2165 [DOI] [PubMed] [Google Scholar]
- 27.Tully J, Whitcomb R, Clark H, Williamson D. 1977. Pathogenic mycoplasmas: cultivation and vertebrate pathogenicity of a new spiroplasma. Science 195:892–894. 10.1126/science.841314 [DOI] [PubMed] [Google Scholar]
- 28.Pich OQ, Burgos R, Planell R, Querol E, Piñol J. 2006. Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement. Microbiology 152:519–527. 10.1099/mic.0.28287-0 [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Baseman JB. 2011. Transcriptional regulation of MG_149, an osmoinducible lipoprotein gene from Mycoplasma genitalium. Mol. Microbiol. 81:327–339. 10.1111/j.1365-2958.2011.07717.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Algire MA, Lartigue C, Thomas DW, Assad-Garcia N, Glass JI, Merryman C. 2009. New selectable marker for manipulating the simple genomes of Mycoplasma species. Antimicrob. Agents Chemother. 53:4429–4432. 10.1128/AAC.00388-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dallo SF, Kannan TR, Blaylock MW, Baseman JB. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46:1041–1051. 10.1046/j.1365-2958.2002.03207.x [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Baseman JB. 2011. Transcriptional response of Mycoplasma genitalium to osmotic stress. Microbiology 157:548–556. 10.1099/mic.0.043984-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Church GM, Gilbert W. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991–1995. 10.1073/pnas.81.7.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shin DH, Choi IG, Busso D, Jancarik J, Yokota H, Kim R, Kim SH. 2004. Structure of OsmC from Escherichia coli: a salt-shock-induced protein. Acta Crystallogr. D Biol. Crystallogr. 60:903–911. 10.1107/S0907444904005013 [DOI] [PubMed] [Google Scholar]
- 35.Park SC, Pham BP, Van Duyet L, Jia B, Lee S, Yu R, Han SW, Yang JK, Hahm KS, Cheong GW. 2008. Structural and functional characterization of osmotically inducible protein C (OsmC) from Thermococcus kodakaraensis KOD1. Biochim. Biophys. Acta 1784:783–788. 10.1016/j.bbapap.2008.02.002 [DOI] [PubMed] [Google Scholar]
- 36.Rehse PH, Ohshima N, Nodake Y, Tahirov TH. 2004. Crystallographic structure and biochemical analysis of the Thermus thermophilus osmotically inducible protein C. J. Mol. Biol. 338:959–968. 10.1016/j.jmb.2004.03.050 [DOI] [PubMed] [Google Scholar]
- 37.Hames C, Halbedel S, Hoppert M, Frey J, Stülke J. 2009. Glycerol metabolism is important for cytotoxicity of Mycoplasma pneumoniae. J. Bacteriol. 191:747–753. 10.1128/JB.01103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pilo P, Vilei EM, Peterhans E, Bonvin-Klotz L, Stoffel MH, Dobbelaere D, Frey J. 2005. A metabolic enzyme as a primary virulence factor of Mycoplasma mycoides subsp. mycoides small colony. J. Bacteriol. 187:6824–6831. 10.1128/JB.187.19.6824-6831.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Atichartpongkul S, Loprasert S, Vattanaviboon P, Whangsuk W, Helmann JD, Mongkolsuk S. 2001. Bacterial Ohr and OsmC paralogues define two protein families with distinct functions and patterns of expression. Microbiology 147:1775–1782 [DOI] [PubMed] [Google Scholar]
- 40.Gutierrez C, Devedjian JC. 1991. Osmotic induction of gene osmC expression in Escherichia coli K12. J. Mol. Biol. 220:959–973. 10.1016/0022-2836(91)90366-E [DOI] [PubMed] [Google Scholar]
- 41.Sturny R, Cam K, Gutierrez C, Conter A. 2003. NhaR and RcsB independently regulate the osmCp1 promoter of Escherichia coli at overlapping regulatory sites. J. Bacteriol. 185:4298–4304. 10.1128/JB.185.15.4298-4304.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muto A, Ushida C. 2002. Transcription and translation, p 323–345 In Razin S, Herrmann RE. (ed), Molecular biology and pathogenicity of mycoplasmas. Kluwer/Plenum Publishers, New York, NY [Google Scholar]