Abstract
The DctSR two-component system of Bacillus subtilis controls the expression of the aerobic C4-dicarboxylate transporter DctA. Deletion of DctA leads to an increased dctA expression. The inactivation of DctB, an extracellular binding protein, is known to inhibit the expression of dctA. Here, interaction between the sensor kinase DctS and the transporter DctA as well as the binding protein DctB was demonstrated in vivo using streptavidin (Strep) or His protein interaction experiments (mSPINE or mHPINE), and the data suggest that DctA and DctB act as cosensors for DctS. The interaction between DctS and DctB was also confirmed by the bacterial two-hybrid system (BACTH). In contrast, no indication was obtained for a direct interaction between the transporter DctA and the binding protein DctB. Activity levels of uptake of [14C]succinate by bacteria that expressed DctA from a plasmid were similar in the absence and the presence of DctB, demonstrating that the binding protein DctB is not required for transport. Thus, DctB is involved not in transport but in cosensing with DctS, highlighting DctB as the first example of a TRAP-type binding protein that acts as a cosensor. The simultaneous presence of DctS/DctB and DctS/DctA sensor pairs and the lack of direct interaction between the cosensors DctA and DctB indicate the formation of a tripartite complex via DctS. It is suggested that the DctS/DctA/DctB complex forms the functional unit for C4-dicarboxylate sensing in B. subtilis.
INTRODUCTION
C4-dicarboxylates are, next to sugars, one of the most important and preferred substrates in bacteria such as Escherichia coli and Bacillus subtilis, and in B. subtilis l-malate even exerts catabolite repression (1). Consequently, many bacteria possess sensors for detecting extracellular and intracellular C4-dicarboxylates and controlling the expression of target genes in response to C4-dicarboxylates (for a review, see reference 2). The molecular details for C4-dicarboxylate sensing have been studied in E. coli. E. coli uses the DcuS/DcuR two-component system for sensing extracellular C4-dicarboxylates (2–5). DcuS is a member of the CitA family of sensor kinases. It constitutes an extracytoplasmic sensing kinase and contains a periplasmic PASP (or PDC) domain that binds C4-dicarboxylates (2, 6–9). In addition, DcuS requires the C4-dicarboxylate transporter DctA or DcuB for function, and possibly also DauA plays an essential role in sensing by DcuS; in dctA- or dcuB-deficient strains, DcuS is deregulated and in the permanent ON state even in the absence of C4-dicarboxylates (10, 11). DcuS and the transporter DctA interact physically, suggesting the formation of a DctA/DcuS sensor complex (12).
In Bacillus subtilis, the DcuS/DcuR homologous system for C4-dicarboxylate sensing is represented by the DctS/DctR two-component system. DctS/DctR is responsible for the detection of fumarate and succinate (13). The sensor kinase DctS resembles in domain structure and arrangement the DcuS sensors and is also a member of the CitA family (2). The dctB, dctS, dctR, and dctA genes are clustered on the B. subtilis chromosome (13). The dctA gene, encoding the C4-dicarboxylate transporter DctA, has the same orientation as dctSR but is transcribed independently (13). Deletion of dctA causes a strong general increase in expression of dctA reporter genes (13), suggesting that DctA has an inhibitory effect on DctS function similar to that of DctA on DcuS in E. coli. The dctB gene, which encodes an extracellular binding protein, is located upstream of the dctSR operon in reverse orientation. DctB shows a high sequence identity (37%) with the DctP binding protein from Rhodobacter capsulatus. In R. capsulatus, DctP together with the membrane components DctQ and DctM constitutes a high-affinity TRAP (tripartite ATP-independent) transport system for C4-dicarboxylates (14, 15). In contrast to R. capsulatus, DctB of B. subtilis is an orphan binding protein without the DctQM membrane components. In B. subtilis, genetic deletion showed that DctB is (in addition to DctS and DctR) required for the expression of dctA and for growth on C4-dicarboxylates (13). It was suggested that DctB functions in sensing by DctS and that it has no direct function in C4-dicarboxylate transport (13).
Overall, the genetic studies by Asai et al. (13) indicate that the sensor kinase DctS of B. subtilis requires DctA as a cosensor for function like the DcuS sensor kinase of E. coli. The TRAP-type DctB binding protein was proposed to act as an additional cosensor, even though TRAP-type binding proteins are known so far only as binding proteins for transport. To provide clear evidence for whether DctA or DctB or both act as cosensors of DctS, we tested physical interaction between DctS and DctA or DctB, which is a prerequisite for the formation of sensor complexes. Therefore, coelution of DctB and DctA after affinity purification of the membrane protein DctS was tested in two independent experimental setups, both either with or without previous in vivo cross-linking. In an alternative approach, interaction of DctS with DctB or DctA was tested in vivo by a bacterial two-hybrid system. Complementary to these approaches, it was shown directly that DctB is not required for the transport process.
MATERIALS AND METHODS
Bacteria and molecular genetic methods.
The E. coli K-12 and B. subtilis 168 strains and plasmids used in this study are listed in Table 1. Molecular genetic methods were performed according to standard procedures (16). Plasmids were isolated with the GeneJET Plasmid Miniprep kit (Fermentas, Germany). For plasmid preparation from B. subtilis, 4 mg/ml lysozyme was added prior to plasmid isolation. PCR products were amplified from B. subtilis 168 genomic DNA using Pfu polymerase (Fermentas, Germany). PCR products were purified with the GeneJET PCR purification kit. Genomic DNA from B. subtilis was isolated using the DNeasy blood and tissue kit (Qiagen, Hilden, Germany). Oligonucleotides were synthesized by Eurofins. E. coli strains were transformed by electroporation (17). B. subtilis strains were transformed with plasmid or linear DNA according to the two-step protocol as described previously (18). Where appropriate, antibiotics were used for cultivation of E. coli strains at the following concentrations: 100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, and 20 μg ml−1 chloramphenicol. When two or more antibiotics were used simultaneously, the concentrations were halved. For cultivation of B. subtilis, the following concentrations of antibiotics were used: 100 μg ml−1 spectinomycin and 2 μg ml−1 erythromycin.
TABLE 1.
Strains and plasmids used in the study
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli K-12 strains | ||
| JM109 | recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ68(lac-proAB) F′ [traD36 proAB+ lacIq lacZΔM15] | 49 |
| BTH101 | F− cya-99 araD139 galE15 galK16 rpsL1 (Strr) hsdR2 mcrA1 mcrB1 | 20 |
| C43(DE3) | F′ hsdS gal1 DE3; spontaneous mutation of BL21(DE3) | 50 |
| B. subtilis 168 strains and plasmids | ||
| IMW610 | dctB::Specr | This study |
| pBQ200 | B. subtilis/E. coli shuttle vector, expression of genes via the degQh promoter, Ampr in E. coli, Eryr in B. subtilis | 51 |
| pDG1726 | Plasmid with Specr for long flanking homology PCR | 52 |
| pMW1927 | dctA-His6 expression plasmid, pBQ200 derivative, Ampr in E. coli, Eryr in B. subtilis | This study |
| Plasmids for mSPINE and mHPINE | ||
| pASK-IBA3+ | Expression plasmid for proteins with a C-terminal Strep tag, AHT-inducible promoter, Ampr | IBA, Göttingen, Germany |
| pET28a | Expression plasmid for proteins with an N- or C-terminal His6 tag, T7 promoter, Kanr | Novagen |
| pBAD33 | Expression plasmid for proteins under the control of the arabinose-inducible pBAD promoter, Camr | 53 |
| pMW1788 | dctS-Strep expression plasmid, pASK-IBA3+ derivative (Ampr) | This study |
| pMW1791 | dctB-His expression plasmid, pET28a derivative (Kanr) | This study |
| pMW1793 | dctA-His expression plasmid, pET28a derivative (Kanr) | This study |
| pMW1795 | dctB-His expression plasmid, pBAD33 derivative (Camr) | This study |
| pMW1797 | dctA-His expression plasmid, pBAD33 derivative (Camr) | This study |
| pMW1800 | His6-dctP expression plasmid, pET28a derivative (Kanr) | This study |
| pMW1974 | dctA-Strep expression plasmid, pASK-IBA3+ derivative (Ampr) | This study |
| pMW1975 | dctA-His and dctB-His expression plasmid, pBAD33 derivative (Camr) | This study |
| Plasmids for BACTH measurements | ||
| pKNT25 | Plasmid for the expression of C-terminal T25-fusion proteins (Kanr) | 20 |
| pKT25 | Plasmid for the expression of N-terminal T25-fusion proteins (Kanr) | 20 |
| pUT18 | Plasmid for the expression of C-terminal T18-fusion proteins (Ampr) | 20 |
| pUT18C | Plasmid for the expression of N-terminal T18-fusion proteins (Ampr) | 20 |
| pKT25-Zip | T25-Zip expression plasmid, pKT25 derivative (Kanr) | 19 |
| pUT18C-Zip | T18-Zip expression plasmid, pUT18C derivative (Ampr) | 19 |
| pMW1695 | DctS-T18 expression plasmid, pUT18 derivative (Ampr) | This study |
| pMW1696 | T18-DctS expression plasmid, pUT18C derivative (Ampr) | This study |
| pMW1697 | T25-DctS expression plasmid, pKT25 derivative (Kanr) | This study |
| pMW1699 | DctR-T18 expression plasmid, pUT18 derivative (Ampr) | This study |
| pMW1704 | T18-DctA expression plasmid, pUT18C derivative (Ampr) | This study |
| pMW1705 | T25-DctB expression plasmid, pKT25 derivative (Kanr) | This study |
| pMW1708 | T18-DctB expression plasmid, pUT18C derivative (Ampr) | This study |
Construction of plasmids and strains.
For cloning, restriction sites were introduced by amplifying the genes using specific primer pairs (Table 2). Unless otherwise noted, the genes were amplified from B. subtilis 168 chromosomal DNA. For protein interaction studies by the bacterial two-hybrid system (BACTH) (19, 20), the T25 and T18 domains were separately fused to the N or C termini of the target proteins (DctS, DctR, DctB, and DctA). The fusions were produced by cloning the dctS, dctR, dctB, and dctA genes into the pKT25, pKNT25, pUT18, or pUT18C vector. For protein interaction studies using the mSPINE or mHPINE assays, the C termini of DctS and DctA were each fused to a streptavidin (Strep) tag, and a His tag was fused to the C terminus of DctA and DctB, respectively. Therefore, dctS and dctA were each cloned into the vector pASK-IBA3plus, and the dctA and dctB genes were each cloned into the pBAD33 vector. For the construction of pMW1795 and pMW1797, dctB and dctA were amplified from pMW1791 and pMW1793, respectively. pMW1975 was constructed by amplification of dctA with the upstream pBAD promoter sequence from pMW1797 and cloning into pMW1795. pMW1800 was obtained by amplifying dctP from Aromatoleum aromaticum chromosomal DNA and cloning into the plasmid pET28a. To introduce the spectinomycin resistance cassette into the dctB locus on the B. subtilis chromosome, we applied the long flanking homology PCR technique (21). DNA fragments of about 1,000 bp flanking the dctB region at its 5′ (upstream fragment) and 3′ (downstream fragment) ends were amplified from the B. subtilis 168 chromosome, adding sequences at the 3′ end of the upstream fragment and the 5′ end of the downstream fragment complementary to the spectinomycin resistance cassette. The spectinomycin resistance cassette was obtained from plasmid pDG1726. The joining of the two fragments to the spectinomycin resistance cassette was performed using the primer pair dctBBS_up_F and dctBBS_down_R. The PCR product was directly used to transform B. subtilis. The integrity of the regions flanking the integrated resistance cassette was verified by sequencing PCR products of about 1,000 bp amplified from chromosomal DNA of the resulting mutant IMW610.
TABLE 2.
Primers used in this studya
| Primer pair | Sequence (5′→3′) | Resulting plasmid |
|---|---|---|
| dctS-BS-2518-F + | TATGGATCCGAACAAAAAGAAGCTCTCAATCCG (BamHI) | pMW1695 |
| dctS-BS-2518N-R | ATAGGTACCTGCGAGCCATGCTGTGCTTC (KpnI) | |
| dctS-BS-2518-F + | TATGGATCCGAACAAAAAGAAGCTCTCAATCCG (BamHI) | pMW1696 |
| dctS-BS-2518C-R | ATAGGTACCTTACGAGCCATGCTGTGC (KpnI) | |
| dctR-BS-2518-F | TATGGATCCGATGGCTCGTAAAGAATGGAAGG (BamHI) | pMW1699 |
| dctR-BS-2518N-R | ATAGGTACCAGTCCTTTTAACACGTAGCGGTTG (KpnI) | |
| dctB-BS-2518-F | TATGGATCCGATGAAGAGTTTGCTTGCATGTCTCG (BamHI) | pMW1705 |
| dctB-BS-2518C-R | TATGGTACCCTATGAATCTTTTCGCAGCTCC (KpnI) | pMW1708 |
| dctA-BS-2518-F | TATGGATCCGATGAAACTGTTTAAAAATTTAACAGTTCAG (BamHI) | pMW1704 |
| dctA-BS-2518C-R | ATAGGTACCTCAGACTGCTGTTTTCATTTTTTTCATGC (KpnI) | |
| dctS-pASK-For | TATGGTACCATGAACAAAAAGAAGCTCTCAATCCGTTGG (KpnI) | pMW1788 |
| dctS-pASK3-Rev | TATGTCGACCGAGCCATGCTGTGCTTCCTCC (SalI) | |
| dctA-1797-F | GAATTCAGGAGATATACCATGGTAATGAAACTG (EcoRI) | pMW1974 |
| dctA-1797-R | CGGAGCTCGAATTCTATGACTGC (EcoRI) | |
| pBAD_Umk-For | CACTATAGGGGTACCGTGAGCG (KpnI) | pMW1795 |
| pBAD_Umk-Rev | GCTTTGTTAGGTACCGGATCTCAGTGG (KpnI) | pMW1797 |
| C-dctBBs_pET-F | TATCCATGGTAATGAAGAGTTTGCTTGCATGTCTCGCAC (NcoI) | pMW1791 |
| C-dctBBs_pET-R | TATAAGCTTTGAATCTTTTCGCAGCTCCAACAGTTCCC (HindIII) | |
| C-dctABs_pET-F | TATCCATGGTAATGAAACTGTTTAAAAATTTAACAGTTCAGG (NcoI) | pMW1793 |
| C-dctABs_pET-R | TATGAATTCATGACTGCTGTTTTCATTTTTTTCATGCC (EcoRI) | |
| Psy-For | CACTTTGCTATGCGACCGGGTCTTTATCC (PsyI) | pMW1975 |
| Psy-Rev | TCCCGGCGACCCGGTCCTACTCAG (PsyI) | |
| DctP2_for | TCACCGGATCCAGGAGAGTAATG (BamHI) | pMW1800 |
| DctP2_rev | CGTTCAGCGGAATTCATGGATTA (EcoRI) | |
| dctBBS_up_F | GGCCGCTTCCGTTCTGTCC | Upstream fragment |
| dctBBS_up_R | CCTATCACCTCAAATGGTTCGCTGGCCGGTATGATCATGGAATCCG | |
| dctBBS_down_F | CGAGCGCCTACGAGGAATTTGTATCGGCGTCTTGACCCTGTTTACC | Downstream fragment |
| dctBBS_down_R | GGTACGAACCAAACTGGACGG | |
| spec_fwd | CAGCGAACCATTTGAGGTGATAGGGACTGGCTCGCTAATAACGTAACGTGACTGGCAAGAG | Spectinomycin resistance |
| spec_rev | CGATACAAATTCCTCGTAGGCGCTCGGCGTAGCGAGGGCAAGGGTTTATTGTTTTCTAAAATCTG | |
| dctA_F | ATATAGGATCCTTTAAAGGAGGAAACAATCATGAAACTGTTTAAAAATTTAACAGTTCAGG (BamHI) | pMW1927 |
| dctA_R | ATATAGCATGCTCAGTGGTGGTGGTGGTGGTGGACTGCTGTTTTCATTTTTTTCATGC (PaeI) |
The introduced restriction site is underlined and shown in parentheses. The plasmid resulting after cloning of the PCR product is indicated.
Protein interaction studies using BACTH and mSPINE/mHPINE assays.
The β-galactosidase activities for the BACTH measurements were determined as described previously (22). The mSPINE in vivo interaction studies were performed as described with some modifications (23). E. coli C43(DE3) was cotransformed with the plasmids encoding the potential interaction partners. Two flasks of 400-ml cell culture were grown in LB medium supplemented with 100 mM glucose to an optical density at 578 nm (OD578) of 1.3 to 1.5. The protein expression was induced at an OD578 of 0.5 with 200 ng/ml anhydrotetracycline (AHT; for the pASK-IBA3plus derivatives) and 0.002% (wt/vol) arabinose (for the pBAD33 derivatives). For the in vivo cross-link of two potential interaction partners, formaldehyde was added to a final concentration of 0.6% (wt/vol) and the cells were incubated at 37°C for 20 min. The cells were harvested immediately and resuspended in 2 amounts of 20 ml of 20 mM Tris-HCl buffer, pH 8. The bacteria were broken by three passages through the French press, and after removal of debris (9,000 × g), the membrane fraction was pelleted by ultracentrifugation (200,000 × g for 65 min). The membrane fraction was homogenized with 20 mM Tris-HCl buffer, pH 8, and the protein concentration was adjusted to a concentration of 5 mg/ml. To solubilize membrane proteins, the zwitterionic detergent Empigen BB (Sigma-Aldrich; 35% [wt/vol]) was added at a final concentration of 2% (wt/vol) and gently stirred for 60 min in an ice bath. After ultracentrifugation (40 min at 300,000 × g), the supernatant was run by gravity through a Strep-Tactin column (1-ml Superflow Strep-Tactin Sepharose; IBA, Göttingen, Germany) at 4°C. The column was washed two times with 5 ml buffer W (100 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA) supplemented with 0.05% (wt/vol) lauryldimethylamine oxide (LDAO) and eluted with buffer E (100 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, 0.05% LDAO, 2.5 mM desthiobiotin). The first three eluted protein fractions were pooled, concentrated at a final volume of 100 μl, separated by SDS-PAGE, and analyzed by immunoblotting. The proteins were detected with antibodies raised against the respective protein tag (anti-Strep horseradish peroxidase [HRP] conjugate [IBA Life Sciences, Germany] and anti-penta-His HRP conjugate [Qiagen, Germany]). To separate cross-linked proteins, the samples were incubated for 20 min at 95°C.
For the mHPINE in vivo interaction studies, the cells were grown and the membranes were isolated as described for the mSPINE assay. The solubilized membrane fraction was run by gravity through a Ni2+-nitrilotriacetic acid (NTA)-agarose column (2 ml; Qiagen), equilibrated in buffer WP (50 mM Tris-HCl, pH 8, 10% [wt/vol] glycerol, 0.5 M NaCl, 20 mM imidazole, 0.05% LDAO). The column was rinsed with 20 ml buffer WP, and bound protein was eluted with 3 ml of buffer EB (buffer WP with 500 mM imidazole). The first three eluted protein fractions were pooled, concentrated at a final volume of 0.5 to 1 ml, separated by SDS-PAGE, and analyzed by immunoblotting. The proteins were detected as in the mSPINE assay.
Transport of [14C]succinate by Bacillus subtilis strains.
For transport measurements, B. subtilis strains were grown overnight at 37°C in 5 ml White medium (24), supplemented with 0.05% (wt/vol) yeast extract, 0.01% (wt/vol) l-tryptophan, 0.1% (wt/vol) acid-hydrolyzed casein (AHC), and 20 mM succinate as carbon source. Two hundred milliliters of the same medium in a 2-liter flask was then inoculated with an overnight culture to an OD600 of 0.1 and incubated at 37°C under vigorous shaking to an OD600 of about 1. The cells were harvested, washed with 20 ml ice-cold buffer (38 mM NaH2PO4, 62 mM K2HPO4, 1 mM MgSO4, pH 7), resuspended at an OD600 of 8 in the same buffer, and kept on ice until use. Transport assays were performed at 37°C in 1.5-ml reaction tubes by mixing 360 μl of cell suspension (preheated to 37°C for 10 min) with 360 μl preheated 40 μM [14C]succinate (Moravec Biochemicals, Brea, CA; 44 μCi/mmol). One-hundred-microliter samples were taken at different times, mixed with 900 μl ice-cold stop solution (0.1 M LiCl), and transferred onto glass fiber filters (APFF; 25-mm diameter, 0.7-μm pore size; Millipore, Billerica, MA), followed by vacuum filtration (FH 225V Ten-Place filter manifold; Hoefer Pharmacia Biotech, San Francisco, CA). Immediately after sample transfer, the filters were washed twice with 1 ml stop solution and, after the last sampling, transferred into measuring vials (Minivial C, low-density polyethylene [LDPE]; Roth, Karlsruhe, Germany) containing 4 ml scintillation fluid (Rotiszint Eco Plus LSC universal cocktail; Roth, Karlsruhe, Germany). Radioactivity Γ (dpm) was determined in a 1414 liquid scintillation counter (Wallac WinSpectral). Unspecific binding of [14C]succinate was accounted for by mixing 50 μl of cell suspension with 50 μl of [14C]succinate and immediate filtration and measurement as described above (Γ0). Activities of [14C]succinate solutions were determined prior to each assay by measuring 10 μl solution in 4 ml scintillation fluid; the resulting activities α lay at 4,000 to 5,000 dpm/nmol. Transport assays were performed in at least 3 independent experiments with two repeats each. Succinate uptake u was calculated as follows: u = (Γ − Γ0)/(α × OD600 × 281 g/liter × 100 μl).
RESULTS
The binding protein DctB interacts with the sensor kinase DctS but not with the transporter DctA.
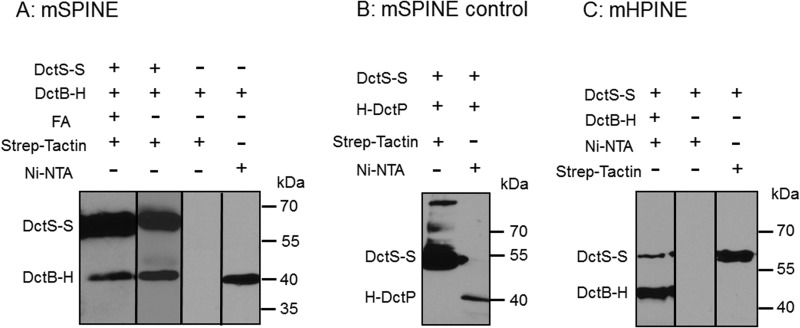
DctB of B. subtilis shows 37% sequence identity to the C4-dicarboxylate binding protein DctP of the high-affinity transport system DctPQM of Rhodobacter capsulatus. Gene deletion in B. subtilis, on the other hand, suggested that DctB is required for efficient induction of dctA expression and to increase the specificity of the two-component system DctSR (13). The direct interaction of DctB with the sensor kinase DctS was studied using mSPINE (membrane Strep protein interaction experiment) (22) assays. The mSPINE assay is based on the coelution of two potentially interacting proteins in E. coli cells, with only one of the proteins carrying a Strep tag for affinity purification. Binding of the nontagged or His-tagged protein to a Strep-Tactin affinity column is then tested in the presence and the absence of the Strep-tagged protein. The latter mediates binding of the nontagged protein to the column (or retardation) when the proteins interact. The membranes from an E. coli strain coexpressing DctS-Strep and DctB-His were purified after treatment of the cells with formaldehyde for cross-linking interacting proteins. The membrane proteins were solubilized with the detergent Empigen BB, and the membrane extract was applied to the Strep-Tactin column. After extensive washing, the bound proteins were eluted specifically with a buffer containing desthiobiotin and the detergent LDAO. The eluate contained DctS-Strep and in addition significant amounts of DctB as tested by Western blotting (Fig. 1A). Similar amounts of DctB were retained on the column when the proteins were extracted without previous cross-linking by formaldehyde. On the other hand, when DctB-His was prepared from bacteria that were deficient in DctS-Strep, no DctB-His was retained on the Strep-Tactin column by the same procedure, demonstrating that Strep-tagged DctS is required for the copurification of DctB-His (Fig. 1A). DctB-His was used in addition for testing the extracts on a Ni-NTA column, proving that the extracts all contained DctB-His (Fig. 1A). The interaction between DctS and DctB was specific, and a similar binding protein, DctPAa from Aromatoleum aromaticum, was unable to bind to a Strep-Tactin column via DctS-Strep (Fig. 1B). For verification, a similar approach was used in a second set of experiments. Here, nontagged or Strep-tagged proteins were tested for binding to a Ni-NTA column, where binding was mediated by interaction with a His-tagged (Ni-NTA-bound) protein (mHPINE [membrane His protein interaction experiment]). In the mHPINE assay, it was tested if DctS-Strep was retained on a Ni-NTA column in the presence of DctB-His. The copurification was tested without previous cross-linking of the samples. Significant amounts of DctS-Strep bound to the Ni-NTA column when DctB-His was coexpressed (Fig. 1C). On the other hand, when DctS-Strep was expressed in the bacteria without DctB-His, DctS-Strep did not bind to the Ni-NTA column. Overall, the experiments in Fig. 1A and C show that DctS and DctB interact with each other in complexes of sufficient stability to bind each other mutually to affinity columns.
FIG 1.
In vivo binding of DctB-His6 (DctB-H) to DctS-Strep (DctS-S) and copurification using a Strep-Tactin column (mSPINE) (A) and a Ni-NTA column (mHPINE) (C). (A) The two proteins were coexpressed in E. coli C43(DE3). Formaldehyde (FA) was added at an OD578 of 1.3 to 1.6 where indicated. The membrane fraction was isolated and solubilized with 2% (wt/vol) Empigen. DctS-S was isolated from the membrane fraction using a Strep-Tactin column, and 20 μl of the eluate was subjected to SDS gel electrophoresis and Western blotting. As a control, DctB-H was expressed without DctS-S. (B) For testing the specificity of the mSPINE assay, the two noninteracting proteins DctS-S and His-DctP from Aromatoleum aromaticum were coexpressed in E. coli C43(DE3). At an OD578 of 1.3 to 1.6, the cells were incubated with formaldehyde. The membrane fraction was solubilized as described above, and DctS-S was isolated using a Strep-Tactin column. (C) For the mHPINE assay, DctB-H was isolated from the solubilized membrane fraction using a Ni-NTA column. As a control, DctS-S was expressed without DctB-H.
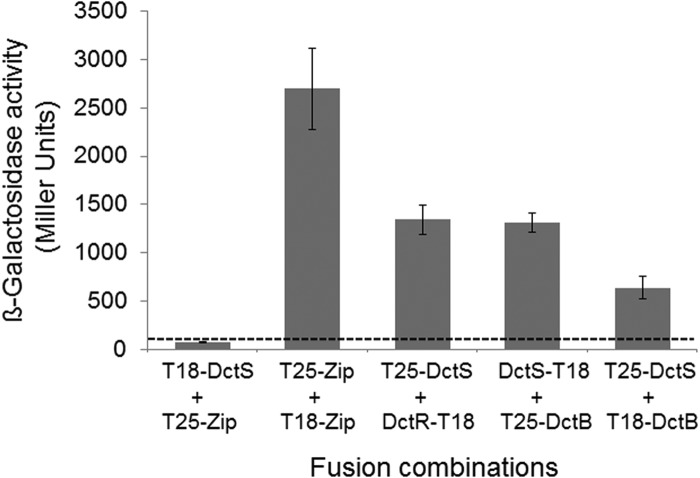
Next, the interaction between DctB and DctS was analyzed in an independent approach by a bacterial two-hybrid assay (BACTH) in vivo. The BACTH assay is based on the reconstitution of adenylate cyclase activity from separate T18 and T25 domains of the enzyme (19). In this assay, the T18 and T25 domains are fused to two separate proteins. If these two proteins interact with each other, the T18 and T25 domains are brought together and adenylate cyclase activity can be restored. Restored adenylate cyclase forms cyclic AMP (cAMP), which is detected in an adenylate cyclase-negative E. coli reporter strain by the expression of the cAMP/cAMP receptor protein (CRP)-dependent lacZ gene (19). Fusion of DctS and DctB C terminally or N terminally to the T18 and T25 domains, respectively, and expression in the reporter strain showed a high response of β-galactosidase expression in various combinations (Fig. 2). The expression was well (at least 5-fold) above the interaction of nonrelated proteins (pair DctS/Zip), which represents the background level of interaction. The β-galactosidase activity of the DctS-T18/T25-DctB amounted to nearly 50% of the positive control (Leu zipper pair) and was the same as the activity of the sensor/regulator pair DctS/DctR. (Fig. 2). Thus, the BACTH studies confirm the interaction between DctS and DctB.
FIG 2.
Interaction of DctS with DctB in the bacterial two-hybrid system (BACTH). E. coli BTH101 was cotransformed with plasmid pairs encoding the T25 fusions and T18 fusions of the proteins as indicated: T25-DctS (pMW1697), T25-DctB (pMW1705), T18-DctS (pMW1696), DctS-T18 (pMW1695), DctR-T18 (pMW1699), and T18-DctB (pMW1708). The β-galactosidase activity was determined after growth to an OD578 of 0.5 to 0.7. The strain expressing the pair of leucine zipper proteins T25-Zip and T18-Zip was used as a positive control for two interacting cytosolic proteins. The activity of the noninteracting proteins T18-DctS plus T25-Zip (80 Miller units, dashed line) represents the background. The data are from four or more independent biological replicates. Error bars represent the standard deviations.
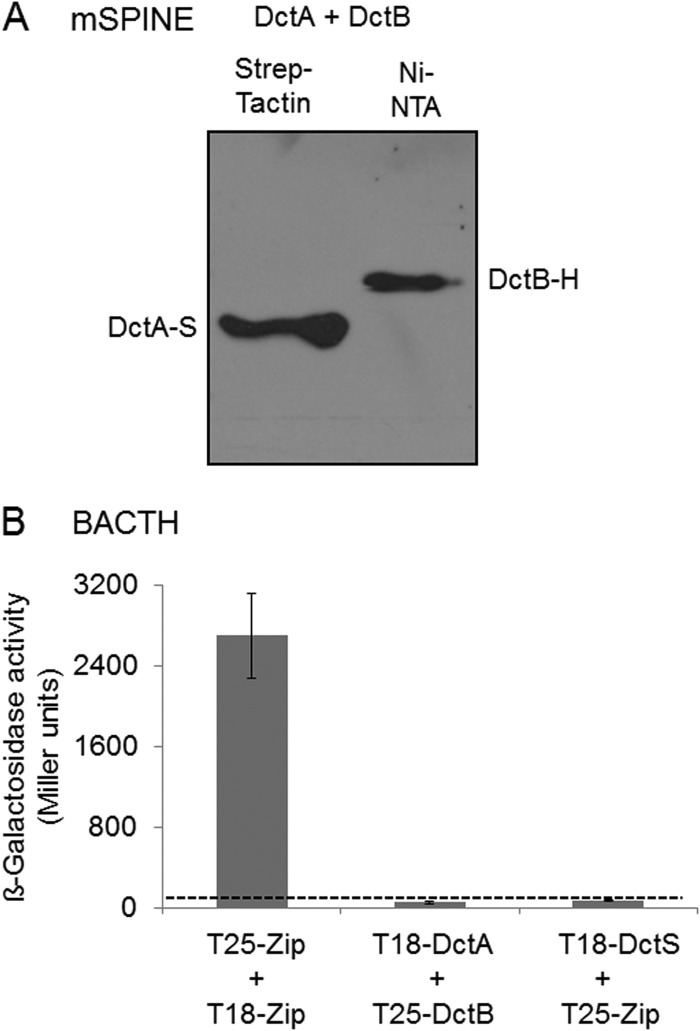
Potential physical interaction of the binding protein DctB with the transporter DctA was tested using mSPINE and BACTH assays. In the mSPINE assay, DctB-His was not copurified on a Strep-Tactin column (Fig. 3A), even when the cell extract contained DctA-Strep, suggesting that the two proteins do not interact with each other to an extent sufficient for mutual binding to a column. Cross-linking by formaldehyde cannot be used for DctA due to the formation of aggregates of DctA when boiling the samples for reversal of cross-linking before SDS-PAGE. In the BACTH assay, all C- and N-terminal fusions of DctA and DctB with T18 and T25 were expressed and tested in each combination. None of the DctA/DctB pairs showed a β-galactosidase activity above background activity (Fig. 3B), which supports the assumption that DctA and DctB do not interact directly.
FIG 3.
Lack of interaction of DctA with the binding protein DctB: studies using the mSPINE assay followed by Western blotting (A) and the BACTH assay (B). For the mSPINE assay, DctA-Strep (DctA-S) and DctB-His (DctB-H) were coexpressed in E. coli C43(DE3) and the membranes were solubilized using 2% (wt/vol) Empigen BB detergent. DctA-S was isolated from the solubilized membrane fraction using a Strep-Tactin column. For the BACTH assays, E. coli BTH101 was cotransformed with plasmid pairs. In each pair, one plasmid encoded a T25 fusion and the other encoded a T18 fusion: T25-DctS (pMW1697) and T25-DctB (pMW1705) were the T25 fusions, and DctR-T18 (pMW1699), T18-DctA (pMW1704), and T18-DctS (pMW1696) were the T18 fusions. The β-galactosidase activity was determined after growth to an OD578 of 0.5 to 0.7. The strain expressing the pair of leucine zipper proteins was used as a positive control for two interacting cytosolic proteins. The background activity obtained for noninteracting proteins like T18-DctS plus T25-Zip was below 80 Miller units (dashed line). All results are the averages of the measurements of at least four biological replicates. Error bars represent the standard deviations.
Overall, the data consistently show that DctB interacts with DctS but not with DctA.
The transporter DctA interacts with the sensor kinase DctS in vivo.
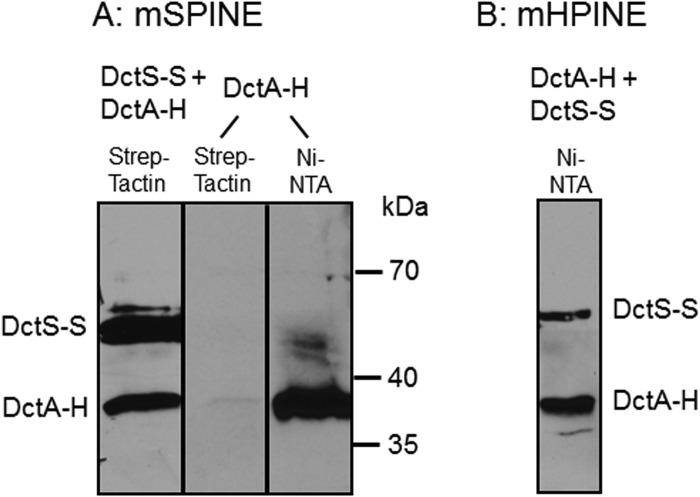
The transport of C4-dicarboxylates in B. subtilis is accomplished by DctA, which mediates the proton-coupled symport of fumarate, malate, succinate, and oxaloacetate into the cell (25). To test the role of DctA as a binding partner of the DctS sensor kinase, it was investigated whether DctA and DctS interact in vivo by mSPINE and mHPINE interaction studies. The experiments were performed with strains of E. coli expressing DctS and DctA in different combinations of tagged variants as described for DctS and DctB. In the mSPINE assay, DctA-His was retained on a Strep-Tactin column when DctS-Strep was coexpressed in the bacterial cells (Fig. 4A). On the other hand, DctA-His did not bind to the column when no DctS-Strep was present in the cell homogenate. The data indicate a strong interaction between the two proteins even without cross-linking, which cannot be used in samples with DctA due to the formation of aggregates in the subsequent boiling step as described above. In the mHPINE assay, DctS-Strep was copurified on a Ni-NTA column when the membrane extract was gained from bacteria coexpressing DctS-Strep and DctA-His (Fig. 4B). Again, in the absence of DctA-His no binding of DctS-Strep to the Ni-NTA column was observed (Fig. 1C), confirming the interaction between DctS and the transporter DctA.
FIG 4.
Interaction of DctA with DctS using the mSPINE (A) and mHPINE (B) methods followed by Western blotting. DctS-Strep (DctS-S) and DctA-His (DctA-H) were coexpressed in E. coli C43(DE3), and the membranes were solubilized using 2% Empigen BB detergent. For the mSPINE assay, DctS-S was isolated from the solubilized membrane fraction using a Strep-Tactin column. As a control, DctA-H was expressed without DctS-S and the membrane fraction was applied to a Strep-Tactin column. For the mHPINE assay, DctA-H was isolated from the solubilized membrane fraction using a Ni-NTA column.
Overall, by the mSPINE and mHPINE assays, a strong and specific interaction between DctS and the transporter DctA was demonstrated, suggesting that DctA and DctS interact.
Presence of a DctS/DctA/DctB sensory unit.
The previous experiments demonstrated that DctS interacts directly with both DctB and DctA. To study whether the two proteins interact with DctS in one complex, the simultaneous binding of DctA and DctB to DctS was tested. In an mSPINE assay from a cell homogenate of B. subtilis coexpressing DctS-Strep, DctB-His, and DctA-His, the membrane proteins were solubilized with Empigen BB and applied to a Strep-Tactin column. After washing, the proteins bound to the column were eluted with a buffer containing the detergent LDAO and analyzed by Western blotting (Fig. 5). The eluate contained large amounts of DctS-Strep and in addition DctB-His and DctA-His. Thus, binding of DctA-His and DctB-His to the Strep-Tactin column is DctS-Strep dependent, suggesting a tripartite sensory unit, DctS/DctA/DctB, or a mixture of two bipartite complexes, DctA/DctS and DctB/DctS, which might be present in parallel.
FIG 5.
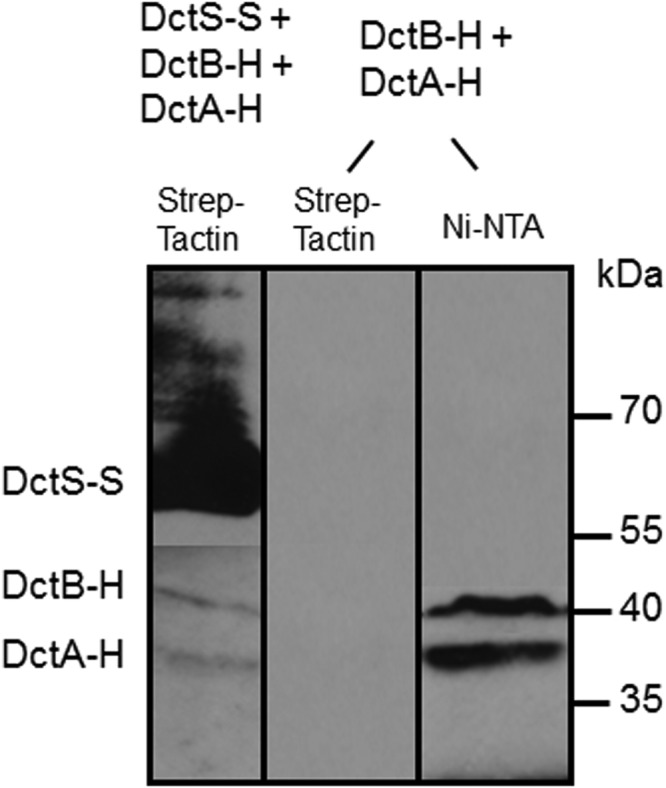
Binding of DctB-His (DctB-H) and DctA-His (DctA-H) on DctS-Strep (DctS-S) and copurification by mSPINE. The three proteins were coexpressed in E. coli C43(DE3). DctS-S was isolated from the solubilized membrane fraction using a Strep-Tactin column. The proteins were detected in Western blots by antibodies directed against the His6 or Strep tag, respectively. After blotting of the proteins, the nitrocellulose membrane was divided and the antibodies were used separately for detection of proteins. As a control, DctA-H and DctB-H were expressed without DctS-S and the membrane fraction was applied to a Strep-Tactin column.
The binding protein DctB is not involved in C4-dicarboxylate transport.
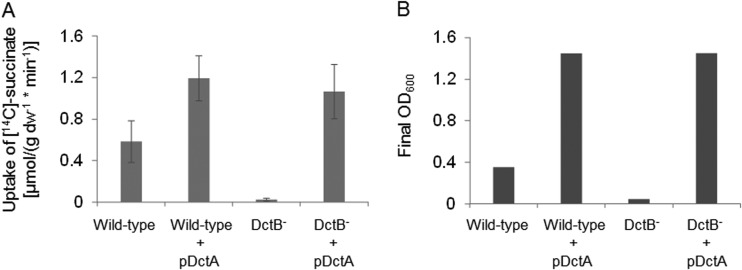
In R. capsulatus, the DctB homologue DctP functions as a binding protein in the uptake of C4-dicarboxylates by the DctPQM TRAP transporter (15, 26, 27). The genetic arrangement of dctB, which is located next to dctSR and dctA in B. subtilis, could be a further indication for a role of DctB not only in cosensing with DctS but also in transport of C4-dicarboxylates by B. subtilis. For testing the role of DctB in succinate uptake, e.g., as a binding protein for the transporter DctA, [14C]succinate uptake into cells of B. subtilis that were either deficient or proficient for DctB was measured. Succinate uptake in B. subtilis is known to depend on the DctA transporter (25). Strain B. subtilis 168, which is wild type for dctA and dctB, showed efficient uptake of [14C]succinate, which was completely lost in the isogenic dctB mutant (Fig. 6A). This effect can be explained in two ways: first, DctB is required for the transport process, or second, dctA expression is reduced because DctB is an essential component for inducing dctA expression (13), probably by the DctB/DctS sensor complex. To differentiate between the two possibilities, the dctB mutant was transformed with a plasmid carrying dctA (pMW1927) under the control of a DctSR-independent promoter. This strain that is able to produce DctA, but not DctB, regained full capacity for the uptake of [14C]succinate, and the activities exceeded those of the wild-type strain without additional plasmid-encoded DctA (Fig. 6A). Furthermore, the succinate transport-positive strains were able to grow on succinate, even when dctB was lacking (Fig. 6B). On the other hand, growth on succinate of the transport-deficient dctB mutant was strongly impaired (Fig. 6B). These findings demonstrate that DctA has full activity for succinate uptake in the absence of DctB. The high transport activities were observed at low [14C]succinate concentrations (20 μM), which demonstrates that the transport is DctB independent even at low succinate concentrations.
FIG 6.
Uptake of [14C]succinate by B. subtilis strains with various genetic backgrounds for dctB and dctA after aerobic growth of the strains on succinate. For transport measurements and growth experiments, the cells were grown in White minimal medium (24), which was enriched with 0.05% or 0.01% (wt/vol) yeast extract, respectively, and supplemented with 20 mM succinate. The transport rates and growth capabilities on succinate were determined for the B. subtilis 168 wild-type strain as a positive control, the wild-type strain plus plasmid-encoded DctA (pDctA, pMW1927) as a control for the addition of pDctA to B. subtilis cells, the DctB-deficient strain (DctB−), and the DctB− strain supplemented with pMW1927. (A) Transport assays were performed at 37°C with 20 μM [14C]succinate. The transport rates are given in μmol × g [dry weight]−1 × min−1 and are the averages of at least three biological replicates. Error bars represent the standard deviations. (B) The bacterial growth is given as the final OD600 after incubation in enriched White minimal medium at 37°C for 16 h.
Taken together, our data provide clear evidence that DctB is not required for the uptake of succinate by DctA, which is consistent with the finding that DctA and DctB show no direct interaction. Therefore, it is concluded that DctB has no direct role in DctA-dependent succinate transport.
DISCUSSION
DctA and DctB are required for the function of the sensor kinase DctS: a tripartite DctS/DctA/DctB sensor unit.
The functionality of the DctSR system depends on the presence of the binding protein DctB (13). Here, we show that DctB directly interacts with DctS, which was demonstrated by two independent assays in vivo (BACTH and mSPINE/mHPINE) (Fig. 7), suggesting a DctB/DctS complex for sensing. The PASP domain of DctS shows a high similarity to PASP of E. coli DcuS (2), and the residues known for effector binding in DcuS are well conserved (7, 9). From 11 essential residues at the C4-dicarboxylate binding site in PASP of DcuS, seven residues are conserved in PASP of DctS (R107 DcuS/R98 DctS; H110/H101; G140/G131; L142/I133; A143/G134; R147/R138; F149/F140). This suggests that DctS is able to bind C4-dicarboxylates, although Asai et al. (13) concluded from their experiments that DctS seems to be incapable of functioning as a standalone C4-dicarboxylate receptor. It is suggested that DctB represents the primary binding site in the DctS/DctB complex that increases the specificity or affinity of DctS for C4-dicarboxylates.
FIG 7.
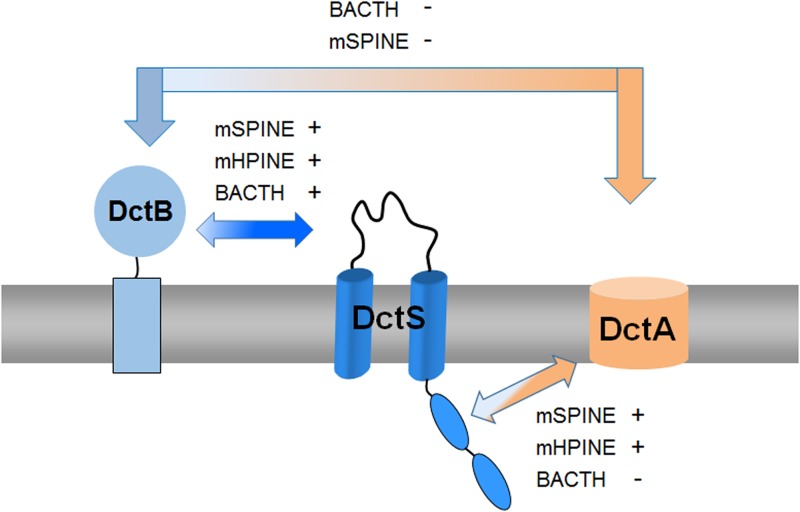
Interaction of the DctB, DctA, and DctS proteins. The arrows between the different proteins are labeled to show whether the interaction experiments using the BACTH, mSPINE, and mHPINE experiments were positive (+) or negative (−). There was a strong interaction between DctB and DctS, and the mSPINE and mHPINE assays showed a strong interaction of DctA with the sensor kinase DctS. No interaction was detected between DctB and DctA, suggesting that the transport of C4-dicarboxylates by DctA is independent of the presence of the binding protein DctB. In addition to the BACTH assays shown in Fig. 2 and Fig. 3B, the assay was used to analyze the interaction between DctS and DctA, but none of the T18 and T25 fusion pairs showed β-galactosidase activity above background levels (data not shown). In the BACTH assay, sterical hindrances of potential interaction partners can lead to a false-negative result (11), which is a common problem with membrane proteins which can move and rotate only in two dimensions.
Binding proteins typically bind their ligands with KD (equilibrium dissociation constant) values in the submicromolar range (27–32). DcuS of E. coli, which lacks a substrate binding protein, has a low affinity for C4-dicarboxylates which is in the submillimolar range (7). It is conceivable that DctB forms a sensory unit with DctS to increase in this way the affinity of the sensor DctS (Fig. 8). By this, the function of DctB would be clearly different from that of DctA, which represents a second signal input site (see references 12 and 33).
FIG 8.
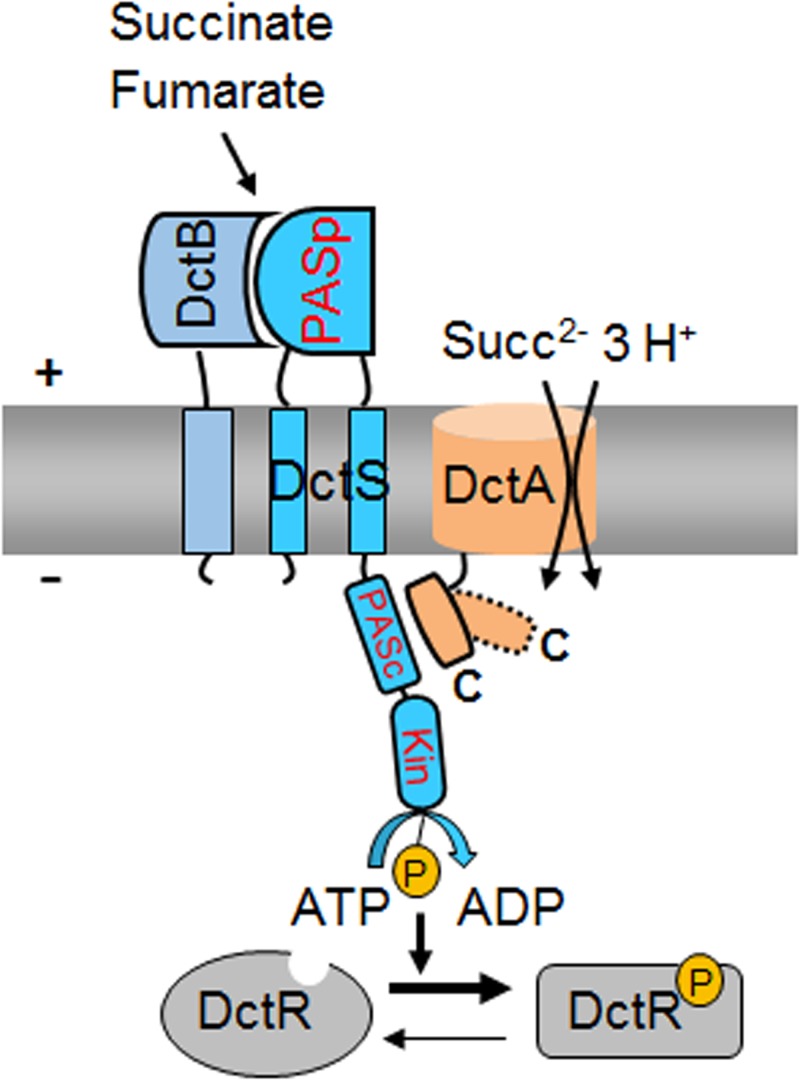
Model for the functional tripartite DctS/DctA/DctB complex for sensing fumarate and succinate in B. subtilis. According to the model, DctB is the primary site for perception of fumarate and succinate, which are then transmitted to PASP of DctS. The putative sites for DctA/DctS interaction and the proposed routes for signal transfer are shown. Transport of fumarate and succinate modifies the interaction between DctA and DctS (dashed and kinked C-terminal helix in DctA), which affects activity or signal transmission to the kinase domain. Abbreviations: PASP, periplasmic PAS (Per-ARNT-Sim) domain; PASC, cytoplasmic PAS (Per-ARNT-Sim) domain; Kin, kinase domain; P, phosphoryl group; Succ, succinate.
Using mSPINE and mHPINE studies, a strong interaction was also detected between DctS and the transporter DctA (Fig. 7), confirming the functional interdependence of DctA and DctS in B. subtilis (13). The physical and functional interaction suggests that DctA functions as a cosensor of DctS similarly to the DctA/DcuS complex in E. coli (12, 33). DctA of E. coli interacts with DcuS by the C-terminal α-helix 8b (12). This helix is DctA specific and not present in structurally similar transporters like the glutamate transporter GltPh of Pyrococcus horikoshii (12, 33). Helix 8b contains an LX4LX3L signature for the interaction with DcuS. DctA of B. subtilis contains a predicted helix 8b, and the LX4LX3L signature is conserved in parts.
Together with previous genetic studies by Asai et al. (13), our results propose a model for C4-dicarboxylate sensing by B. subtilis. The DctS/DctB and DctS/DctA sensor units are present in parallel, which indicates that a tripartite DctS/DctA/DctB complex rather than two bipartite complexes is present and forms the functional unit (Fig. 8). In this complex, DctS might interact with DctB via its PASP domain. DctB is supposed to increase affinity and specificity of C4-dicarboxylate binding by the sensor. DctS might interact with the transporter DctA via its PASC domain, and the interaction is supposed to have a similar role as in E. coli. Here, the transporters have been suggested to link the functional state of the sensor to the presence or lack of the transporters which represent the major targets of DcuS regulation, or to the metabolic flux through the transporter (11, 12, 33). After activation, DctS of B. subtilis transduces the signal by phosphoryl transfer to the response regulator DctR, which induces the expression of dctA (13) and probably dctB. Thus, the tripartite sensor complex of B. subtilis differs substantially from the well-established bipartite DctA/DcuS sensor unit for C4-dicarboxylates of E. coli (12, 33). Interestingly, evidence exists that the alternative acidic DauA succinate transporter of E. coli might also regulate DcuS activity (34), indicating that besides DcuS/DctA and DcuS/DcuB, a DcuS/DauA sensor complex exists (11, 12, 34). Thus, alternative bipartite sensor complexes of DcuS are presumably formed under conditions where the respective (alternative) transporters are expressed, that is, under aerobic, anaerobic, or acidic growth conditions. The sensor complex in B. subtilis, however, appears to be tripartite since the DctS/DctB and DctS/DctA interaction is found at the same time. Moreover, DctB and DctA are supposed to interact on different sites with DctS and on opposite sides of the membrane, which allows binding of the two cosensors at the same time without spatial interference. The tripartite DctS/DctA/DctB sensor unit suggested here represents an extension of the “three-component systems” that are composed of the two-component sensor kinase/response regulators and an additional protein which is required for function (35, 36).
The binding protein DctB has no role in succinate transport by DctA.
DctB exhibits a high sequence similarity to the TRAP (tripartite ATP-independent periplasmic) binding proteins of the DctP type. The DctP proteins are usually part of high-affinity transport systems together with the membrane components DctQ and DctM (31, 32, 37, 38). Unlike genes coding for TRAP binding proteins, the dctB gene does not cluster with genes for dctQM-like membrane components, but with the dctSR and dctA genes. However, as shown here, succinate transport by DctA is independent of DctB, and DctB does not directly interact with DctA. Additionally, in earlier transport experiments (25) there were also no indications that DctA requires additional components for transport. Therefore, the requirement of DctB for the expression of dctA (13) and the direct interaction with DctS indicate that DctB functions as a cosensor for the DctSR system only.
Despite low sequence identity among substrate binding proteins (SBPs) of about 20% in most cases, they are structurally well conserved (32). Based on their tertiary structure, SBPs are classified into six clusters, whereby the TRAP binding proteins form one cluster (39). DctB shows a high sequence identity of 37% to DctP from R. capsulatus, and the predicted structure is very similar to that of the SiaP protein of Haemophilus influenzae. SiaP is part of the SiaPQM TRAP transport system and binds sialic acid (28, 31, 40–42). A common feature of ligand binding in the known DctP-type TRAP SBPs is the electrostatic interaction between a carboxylate group of the substrate and a conserved arginine residue in domain II of the SBP (28, 31, 43). The corresponding residue (Arg188) is also conserved in DctB.
Binding proteins of various types are known to function in cosensing alongside sensor histidine kinases and methyl-accepting chemotaxis proteins (MCPs). Citrate sensing in Bordetella pertussis is accomplished by the interaction of the citrate-liganded SBP BctC with the sensor kinase BctE (44). Together with the membrane components BctBA, BctC is also part of a TTT (tripartite tricarboxylate transporter) system. In Vibrio harveyi, the periplasmic binding protein LuxP senses the signal molecule autoinducer 2 (AI-2) for quorum sensing and interacts directly with the periplasmic domain of the LuxQ sensor kinase for transducing the signal into the cell (45, 46). Furthermore, the periplasmic binding protein ChvE, which is a homologue of the ribose binding protein RbsB and galactose/glucose binding protein MglB of E. coli, mediates a sugar-induced increase in virulence gene expression through the VirA/VirG two-component system in Agrobacterium tumefaciens (47). In addition to its role in transport by the maltose ABC transporter MalFGK2, the maltose binding protein MalE of E. coli mediates the chemotactic response to maltose by interacting with the chemotactic signal transducer Tar (taxis to aspartate and some repellents) (48). Thus, periplasmic binding proteins like LuxP, ChvE, and MalE are known to serve as cosensors (39), but DctB is the first example of a TRAP-type binding protein that is involved in signal perception.
ACKNOWLEDGMENTS
Support by a grant from Deutsche Forschungsgemeinschaft to G.U. is gratefully acknowledged.
We are grateful to Jörg Stülke and Felix Mehne (Göttingen, Germany) for supporting us in the construction of gene inactivation in B. subtilis and providing plasmids pDG1726 and pBQ200. We thank Andrea Ebert-Jung (Mainz, Germany) for supply of plasmid pMW1800 and Patrick Scheu (Mainz) for helpful comments. We are grateful to G. Karimova and D. Ladant (Paris, France) for supply of the strains and plasmids for the BACTH system.
Footnotes
Published ahead of print 27 December 2013
REFERENCES
- 1.Meyer FM, Jules M, Mehne FM, Le Coq D, Landmann JJ, Görke B, Aymerich S, Stülke J. 2011. Malate-mediated carbon catabolite repression in Bacillus subtilis involves the HPrK/CcpA pathway. J. Bacteriol. 193:6939–6949. 10.1128/JB.06197-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheu PD, Kim OB, Griesinger C, Unden G. 2010. Sensing by the membrane-bound sensor kinase DcuS: exogenous versus endogenous sensing of C4-dicarboxylates in bacteria. Future Microbiol. 5:1383–1402. 10.2217/fmb.10.103 [DOI] [PubMed] [Google Scholar]
- 3.Zientz E, Bongaerts J, Unden G. 1998. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J. Bacteriol. 180:5421–5425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golby P, Davies S, Kelly DJ, Guest JR, Andrews SC. 1999. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J. Bacteriol. 181:1238–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheu PD, Liao YF, Bauer J, Kneuper H, Basché T, Unden G, Erker W. 2010. Oligomeric sensor kinase DcuS in the membrane of Escherichia coli and in proteoliposomes: chemical cross-linking and FRET spectroscopy. J. Bacteriol. 192:3474–3483. 10.1128/JB.00082-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pappalardo L, Janausch IG, Vijayan V, Zientz E, Junker J, Peti W, Zweckstetter M, Unden G, Griesinger C. 2003. The NMR structure of the sensory domain of the membranous two-component fumarate sensor (histidine protein kinase) DcuS of Escherichia coli. J. Biol. Chem. 278:39185–39188. 10.1074/jbc.C300344200 [DOI] [PubMed] [Google Scholar]
- 7.Kneuper H, Janausch IG, Vijayan V, Zweckstetter M, Bock V, Griesinger C, Unden G. 2005. The nature of the stimulus and of the fumarate binding site of the fumarate sensor DcuS of Escherichia coli. J. Biol. Chem. 280:20596–20603. 10.1074/jbc.M502015200 [DOI] [PubMed] [Google Scholar]
- 8.Krämer J, Fischer JD, Zientz E, Vijayan V, Griesinger C, Lupas A, Unden G. 2007. Citrate sensing by the C4-dicarboxylate/citrate sensor kinase DcuS of Escherichia coli: binding site and conversion of DcuS to a C4-dicarboxylate- or citrate-specific sensor. J. Bacteriol. 189:4290–4298. 10.1128/JB.00168-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung J, Hendrickson WA. 2008. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 283:30256–30265. 10.1074/jbc.M805253200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies SJ, Golby P, Omrani D, Broad SA, Harrington VL, Guest JR, Kelly DJ, Andrews SC. 1999. Inactivation and regulation of the aerobic C4-dicarboxylate transport dctA gene of Escherichia coli. J. Bacteriol. 181:5624–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleefeld A, Ackermann B, Bauer J, Krämer J, Unden G. 2009. The fumarate/succinate antiporter DcuB of Escherichia coli is a bifunctional protein with sites for regulation of DcuS-dependent gene expression. J. Biol. Chem. 284:265–275. 10.1074/jbc.M807856200 [DOI] [PubMed] [Google Scholar]
- 12.Witan J, Bauer J, Wittig I, Steinmetz PA, Erker W, Unden G. 2012. Interaction of the Escherichia coli transporter DctA with the sensor kinase DcuS: presence of functional DctA/DcuS sensor units. Mol. Microbiol. 85:846–861. 10.1111/j.1365-2958.2012.08143.x [DOI] [PubMed] [Google Scholar]
- 13.Asai K, Baik SH, Kasahara Y, Moriya S, Ogasawara N. 2000. Regulation of the transport system for C4-dicarboxylic acids in Bacillus subtilis. Microbiology 146:263–271 [DOI] [PubMed] [Google Scholar]
- 14.Forward JA, Behrendt MC, Kelly DJ. 1993. Evidence that the high affinity C4-dicarboxylate transport system of Rhodobacter capsulatus is a novel type of periplasmic permease. Biochem. Soc. Trans. 21:343S. [DOI] [PubMed] [Google Scholar]
- 15.Forward JA, Behrendt MC, Wyborn NR, Cross R, Kelly DJ. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179:5482–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17.Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127–6145. 10.1093/nar/16.13.6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunst F, Rapoport G. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karimova G, Pidoux J, Ullmann A, Ladant D. 1998. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U. S. A. 95:5752–5756. 10.1073/pnas.95.10.5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karimova G, Dautin N, Ladant D. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233–2243. 10.1128/JB.187.7.2233-2243.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265 [DOI] [PubMed] [Google Scholar]
- 22.Scheu PD, Witan J, Rauschmeier M, Graf S, Liao YF, Ebert-Jung A, Basché T, Erker W, Unden G. 2012. CitA/CitB two-component system regulating citrate fermentation in Escherichia coli and its relation to the DcuS/DcuR system in vivo. J. Bacteriol. 194:636–645. 10.1128/JB.06345-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller VS, Jungblut PR, Meyer TF, Hunke S. 2011. Membrane-SPINE: an improved method to identify protein-protein interaction partners of membrane proteins in vivo. Proteomics 11:2124–2128. 10.1002/pmic.201000558 [DOI] [PubMed] [Google Scholar]
- 24.White PJ. 1972. The nutrition of Bacillus megaterium and Bacillus cereus. J. Gen. Microbiol. 71:505–514. 10.1099/00221287-71-3-505 [DOI] [PubMed] [Google Scholar]
- 25.Groeneveld M, Weme RG, Duurkens RH, Slotboom DJ. 2010. Biochemical characterization of the C4-dicarboxylate transporter DctA from Bacillus subtilis. J. Bacteriol. 192:2900–2907. 10.1128/JB.00136-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaw JG, Hamblin MJ, Kelly DJ. 1991. Purification, characterization and nucleotide sequence of the periplasmic C4-dicarboxylate-binding protein (DctP) from Rhodobacter capsulatus. Mol. Microbiol. 5:3055–3062. 10.1111/j.1365-2958.1991.tb01865.x [DOI] [PubMed] [Google Scholar]
- 27.Walmsley AR, Shaw JG, Kelly DJ. 1992. The mechanism of ligand binding to the periplasmic C4-dicarboxylate binding protein (DctP) from Rhodobacter capsulatus. J. Biol. Chem. 267:8064–8072 [PubMed] [Google Scholar]
- 28.Müller A, Severi E, Mulligan C, Watts AG, Kelly DJ, Wilson KS, Wilkinson AJ, Thomas GH. 2006. Conservation of structure and mechanism in primary and secondary transporters exemplified by SiaP, a sialic acid binding virulence factor from Haemophilus influenzae. J. Biol. Chem. 281:22212–22222. 10.1074/jbc.M603463200 [DOI] [PubMed] [Google Scholar]
- 29.Thomas GH, Southworth T, León-Kempis MR, Leech A, Kelly DJ. 2006. Novel ligands for the extracellular solute receptors of two bacterial TRAP transporters. Microbiology 152:187–198. 10.1099/mic.0.28334-0 [DOI] [PubMed] [Google Scholar]
- 30.Rucktooa P, Antoine R, Herrou J, Huvent I, Locht C, Jacob-Dubuisson F, Villeret V, Bompard C. 2007. Crystal structures of two Bordetella pertussis periplasmic receptors contribute to defining a novel pyroglutamic acid binding DctP subfamily. J. Mol. Biol. 370:93–106. 10.1016/j.jmb.2007.04.047 [DOI] [PubMed] [Google Scholar]
- 31.Fischer M, Zhang QY, Hubbard RE, Thomas GH. 2010. Caught in a TRAP: substrate-binding proteins in secondary transport. Trends Microbiol. 18:471–478. 10.1016/j.tim.2010.06.009 [DOI] [PubMed] [Google Scholar]
- 32.Mulligan C, Fischer M, Thomas GH. 2011. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 35:68–86. 10.1111/j.1574-6976.2010.00236.x [DOI] [PubMed] [Google Scholar]
- 33.Witan J, Monzel C, Scheu PD, Unden G. 2012. The sensor kinase DcuS of Escherichia coli: two stimulus input sites and a merged signal pathway in the DctA/DcuS sensor unit. Biol. Chem. 393:1291–1297. 10.1515/hsz-2012-0229 [DOI] [PubMed] [Google Scholar]
- 34.Karinou E, Compton EL, Morel M, Javelle A. 2013. The Escherichia coli SLC26 homologue YchM (DauA) is a C4-dicarboxylic acid transporter. Mol. Microbiol. 87:623–640. 10.1111/mmi.12120 [DOI] [PubMed] [Google Scholar]
- 35.Tetsch L, Jung K. 2009. The regulatory interplay between membrane-integrated sensors and transport proteins in bacteria. Mol. Microbiol. 73:982–991. 10.1111/j.1365-2958.2009.06847.x [DOI] [PubMed] [Google Scholar]
- 36.Buelow DR, Raivio TL. 2010. Three (and more) component regulatory systems—auxiliary regulators of bacterial histidine kinases. Mol. Microbiol. 75:547–566. 10.1111/j.1365-2958.2009.06982.x [DOI] [PubMed] [Google Scholar]
- 37.Rabus R, Jack DL, Kelly DJ, Saier MH. 1999. TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology 145:3431–3445 [DOI] [PubMed] [Google Scholar]
- 38.Kelly DJ, Thomas GH. 2001. The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol. Rev. 25:405–424. 10.1111/j.1574-6976.2001.tb00584.x [DOI] [PubMed] [Google Scholar]
- 39.Berntsson RP, Smits A, Sander HJ, Schmitt L, Slotboom DJ, Poolman B. 2010. A structural classification of substrate-binding proteins. FEBS Lett. 584:2606–2617. 10.1016/j.febslet.2010.04.043 [DOI] [PubMed] [Google Scholar]
- 40.Severi E, Randle G, Kivlin P, Whitfield K, Young R, Moxon R, Kelly D, Hood D, Thomas GH. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 58:1173–1185. 10.1111/j.1365-2958.2005.04901.x [DOI] [PubMed] [Google Scholar]
- 41.Allen S, Zaleski A, Johnston JW, Gibson BW, Apicella MA. 2005. Novel sialic acid transporter of Haemophilus influenzae. Infect. Immun. 73:5291–5300. 10.1128/IAI.73.9.5291-5300.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mulligan C, Geertsma ER, Severi E, Kelly DJ, Poolman B, Thomas GH. 2009. The substrate-binding protein imposes directionality on an electrochemical sodium gradient-driven TRAP transporter. Proc. Natl. Acad. Sci. U. S. A. 106:1778–1783. 10.1073/pnas.0809979106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnston JW, Coussens NP, Allen S, Houtman JC, Turner KH, Zaleski A, Ramaswamy S, Gibson BW, Apicella MA. 2008. Characterization of the N-acetyl-5-neuraminic acid-binding site of the extracytoplasmic solute receptor (SiaP) of nontypeable Haemophilus influenzae strain 2019. J. Biol. Chem. 283:855–865. 10.1074/jbc.M706603200 [DOI] [PubMed] [Google Scholar]
- 44.Antoine R, Huvent I, Chemlal K, Deray I, Raze D, Locht C, Jacob-Dubuisson F. 2005. The periplasmic binding protein of a tripartite tricarboxylate transporter is involved in signal transduction. J. Mol. Biol. 351:799–809. 10.1016/j.jmb.2005.05.071 [DOI] [PubMed] [Google Scholar]
- 45.Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 18:507–518. 10.1016/j.molcel.2005.04.020 [DOI] [PubMed] [Google Scholar]
- 46.Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. 2006. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 126:1095–1108. 10.1016/j.cell.2006.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He F, Nair GR, Soto CS, Chang Y, Hsu L, Ronzone E, DeGrado WF, Binns AN. 2009. Molecular basis of ChvE function in sugar binding, sugar utilization, and virulence in Agrobacterium tumefaciens. J. Bacteriol. 191:5802–5813. 10.1128/JB.00451-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardina PJ, Bormans AF, Manson MD. 1998. A mechanism for simultaneous sensing of aspartate and maltose by the Tar chemoreceptor of Escherichia coli. Mol. Microbiol. 29:1147–1154. 10.1046/j.1365-2958.1998.00964.x [DOI] [PubMed] [Google Scholar]
- 49.Yanisch-Perron C, Vieira J, Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119. 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- 50.Miroux B, Walker JE. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289–298. 10.1006/jmbi.1996.0399 [DOI] [PubMed] [Google Scholar]
- 51.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. 1994. Interactions of wild-type and truncated LevR of Bacillus subtilis with the upstream activating sequence of the levanase operon. J. Mol. Biol. 241:178–192. 10.1006/jmbi.1994.1487 [DOI] [PubMed] [Google Scholar]
- 52.Guérout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336. 10.1016/0378-1119(95)00652-4 [DOI] [PubMed] [Google Scholar]
- 53.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]