Abstract
Ralstonia solanacearum, an economically important plant pathogen, must attach, grow, and produce virulence factors to colonize plant xylem vessels and cause disease. Little is known about the bacterial metabolism that drives these processes. Nitrate is present in both tomato xylem fluid and agricultural soils, and the bacterium's gene expression profile suggests that it assimilates nitrate during pathogenesis. A nasA mutant, which lacks the gene encoding the catalytic subunit of R. solanacearum's sole assimilatory nitrate reductase, did not grow on nitrate as a sole nitrogen source. This nasA mutant exhibited reduced virulence and delayed stem colonization after soil soak inoculation of tomato plants. The nasA virulence defect was more severe following a period of soil survival between hosts. Unexpectedly, once bacteria reached xylem tissue, nitrate assimilation was dispensable for growth, virulence, and competitive fitness. However, nasA-dependent nitrate assimilation was required for normal production of extracellular polysaccharide (EPS), a major virulence factor. Quantitative analyses revealed that EPS production was significantly influenced by nitrate assimilation when nitrate was not required for growth. The plant colonization delay of the nasA mutant was externally complemented by coinoculation with wild-type bacteria but not by coinoculation with an EPS-deficient epsB mutant. The nasA mutant and epsB mutant did not attach to tomato roots as well as wild-type strain UW551. However, adding either wild-type cells or cell-free EPS improved the root attachment of these mutants. These data collectively suggest that nitrate assimilation promotes R. solanacearum virulence by enhancing root attachment, the initial stage of infection, possibly by modulating EPS production.
INTRODUCTION
Although often overlooked, pathogen physiology is a key component of virulence. Pathogens rarely cause disease without the physiological capacity to grow and multiply (1). Nitrogen metabolism, in particular assimilation (the constructive metabolism by which nutrients are used for biosynthesis), is underexplored as it relates to virulence in pathogenic bacteria (1). Genomic and transcriptomic studies have suggested the possible importance of nitrate assimilation in virulence or fitness, but few have functionally investigated these hypotheses (2, 3). Based on genomic analyses, it has been proposed that Xanthomonas campestris pv. campestris uses nitrate as a nitrogen source while infecting crucifer plants (2). The presence and expression of genes encoding nitrate assimilation has been noted in another plant pathogen, Ralstonia solanacearum (4). R. solanacearum, the causal agent of bacterial wilt disease, is a betaproteobacterial plant pathogen that costs the global potato industry over $1 billion per year and can cause up to 90% yield losses of tomato (5, 6). There is no generally effective strategy for controlling bacterial wilt of tomato or potato, so understanding the biology of this host-pathogen interaction is important for both industrial and subsistence agriculture.
The R. solanacearum infection process begins with attachment to host roots, followed by formation of microcolonies on the root surface. The bacterium then enters host plants through wounds or natural openings in the roots. Once beyond the root surface, the bacterium moves through the cortex and colonizes the water-transporting xylem elements in the vascular system (7, 8). R. solanacearum then moves up into the stem with the plant's transpirational flow and multiplies rapidly in the xylem fluid. Bacterial population sizes in infected plants can surpass 109 CFU/g of stem tissue (8). Wilting symptoms are believed to result from physical blockage of the xylem by R. solanacearum cells and their cellular products (9). As infected plants wilt and die, the bacteria spread out of the xylem and return to the soil through the roots of the decayed plant. They persist in the environment until another host becomes available. The nutrients that support the growth, virulence, and survival of R. solanacearum over its life cycle are not known.
R. solanacearum produces extracellular polysaccharide (EPS), a complex polymer of N-acetylated sugars (10). EPS is a major bacterial wilt virulence factor; mutants lacking EPS colonize plants poorly and rarely cause symptoms (11, 12). It has been suggested that EPS contributes to virulence by blocking xylem vessels (6, 13). In addition, EPS was recently found to specifically trigger defense responses in bacterial wilt-resistant tomato plants (14). However, the role of EPS in pathogenesis prior to xylem colonization has not been explored. EPS production is regulated by a complex environmentally responsive network that is not fully characterized (15, 16).
To grow and produce virulence factors that contain nitrogen, like EPS, R. solanacearum requires nitrogen input. The bacterium encounters high concentrations of nitrate in both agricultural soils and in tomato plant xylem sap (17, 18). Nitrate is consistently the most abundant potential nitrogen source in both environments. High-pressure liquid chromatography (HPLC) analyses confirmed that the xylem sap of our tomato plants contains high levels (40 mM) of nitrate (Jean-Claude Davidian, unpublished results). All 11 sequenced R. solanacearum genomes in the MaGE RalstoniaScope database encode putative nitrate assimilation capacity (http://www.genoscope.cns.fr/agc/microscope/home/index.php) (2). Transcriptional analyses revealed that the nitrate assimilatory network is expressed by the bacterium during tomato pathogenesis (4). Based on these preliminary observations, we sought to determine the role of assimilatory nitrate reduction in bacterial wilt pathogenesis.
We found that the deletion of nasA, which encodes the catalytic subunit of the pathogen's only apparent assimilatory nitrate reductase, caused defects in virulence and plant colonization, but only when bacteria infected unwounded plant roots. The virulence defect was exacerbated after a period of soil survival. Once bacteria reached xylem tissue, nitrate assimilation was dispensable for growth, virulence, and competitive fitness, indicating that nasA contributes to virulence prior to xylem entry. Unexpectedly, nasA was also required for the normal production of EPS and for attachment to tomato roots. Our data suggest a previously unrecognized role for R. solanacearum EPS in attachment to host roots and that this EPS-mediated root attachment is enhanced by nitrate metabolism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All bacterial strains and plasmids used in the present study are listed in Table 1. Unless otherwise noted, E. coli strains were grown in Luria-Bertani (LB) medium at 37°C and R. solanacearum strains were grown in CPG medium (19) at 28°C. When appropriate, media were supplemented with antibiotics (15 μg of gentamicin/ml, 25 μg of kanamycin/ml, and 50 μg of rifampin/ml). Boucher's minimal medium (BMM) buffered with 20 mM MES (morpholineethanesulfonic acid) was used when minimal medium was needed (20), with either nitrate or ammonium as sole nitrogen sources at concentrations noted. Chemicals were from Difco Laboratories (Detroit, MI), Sigma-Aldrich (St. Louis, MO), or Fisher Scientific (Hanover Park, IL).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TOP10 | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| R. solanacearum | ||
| UW551 (WT) | Wild-type race 3, biovar 2 | 8 |
| UW551Rif (WTRif) | UW551; Rifr | 25 |
| epsB mutant | UW551 ΔepsB::Km; Kmr | 14 |
| nasA mutant | UW551 ΔnasA::Gm; Gmr | This study |
| nasA mutant + pNascomp | UW551 ΔnasA::Gm + pNascomp; Gmr Kmr | This study |
| nasA mutant + pUFJ10 | UW551 ΔnasA::Gm + pUFJ10; Gmr Kmr | This study |
| Plasmids | ||
| pSTBlue-1 | Cloning vector; Ampr Kmr | EMD Bioscience |
| pCR-Blunt | Cloning vector; Kmr | Invitrogen |
| pUFJ10 | Gmr Kmr; stable in R. solanacearum | 23 |
| pSTBlueΔnasA | pSTBlue-1 + nasA::Gm deletion construct in EcoRV MCS | This study |
| pNascomp | pUFJ10 + narK3 nirB-nirD-nasB-nasA DNA subcloned from pCR-Blunt | This study |
Gmr, gentamicin resistance; Rifr, rifampin resistance; Ampr, ampicillin resistance; Strr, streptomycin resistance; Kmr, kanamycin resistance.
Measuring gene expression.
Bacterial cells from overnight CPG cultures were centrifuged (6,000 rpm; 10 min) and resuspended to a cell density of 2 × 108 CFU/ml in 5 ml of Boucher's minimal medium (described above) containing either no nitrogen, 5 mM ammonium, 5 mM nitrate, 5 mM ammonium plus 5 mM nitrate, or a 1:10 final concentration of xylem sap (collected and pooled from 30 3-week-old Bonny Best tomato plants as described previously [4]). After 3 h of incubation at 228 rpm and 28°C, 1.25 ml of transcriptional stop solution of 5% phenol in ethanol was added to the cells. Cells were immediately centrifuged (12,000 rpm for 5 min at 4°), and the supernatant was discarded. Pellets were frozen in liquid nitrogen and stored at −80°C. RNA was extracted using a phenol-chloroform extraction method as described previously (4), followed by four DNase I treatments to ensure complete DNA removal. cDNA was synthesized using a SuperScript VILO cDNA Synthesis kit according to manufacturer's instructions (Invitrogen). Quantitative real-time PCR (qRT-PCR) was used to determine differences in expression of nasA under the various treatments using the constantly expressed rplM gene for normalization (Fanhong Meng, unpublished data). Relative quantification qRT-PCR was carried out as described previously (4) and analyzed using the ΔΔCT method (14). Nine biological replicate experiments were performed under each condition.
Construction of R. solanacearum nasA mutant and nasA-complemented strains.
Overlap extension PCR (21) was used to create the marked nasA deletion construct ΔnasA::Gm. Specifically, a 700-bp region upstream of the nasA gene was amplified from R. solanacearum strain UW551 genomic DNA using the primers nasAupForward and nasAupReverse. A 700-bp region downstream of the nasA gene was amplified using the primers nasAdownForward and nasAdownReverse. A Tn7 Gm plasmid was used as the template for amplification of the gentamicin resistance gene using the primers GmF and GmR. Both nasAupReverse and nasAdownForward primers have gentamicin resistance gene sequence overlap, which allowed construction of a deletion construct containing the gentamicin (Gm) resistance cassette with the three resulting DNA fragments. All primer sequences are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′) | Source |
|---|---|---|
| nasAupForward | GTGGAGGTGCGCTACGACCG | This study |
| nasAupReverse | ATTCGAGCTCGGTACCCGCCTGCCGCTGGATCAGCTC | This study |
| nasAdownForward | AATTGTCACAACGCCGCGGCGTGGGCCGCAACACCATCT | This study |
| nasAdownReverse | GCCGTTCGGGTTGGCGATCA | This study |
| GmF | GGGTACCGAGCTCGAATTG | Jacobs and Allen, unpublished |
| GmR | CCGCGGCGTTGTGACAATTT | Jacobs and Allen, unpublished |
| nasoperonF | TCGCACCATAACCGCACACCG | This study |
| nasoperonR | ACGAGGAGCCGGATTTTAAGTGC | This study |
The resulting 2.5-kb blunt-ended deletion construct was phosphorylated with T4N kinase using standard procedures and cloned into the EcoRV site in the multiple cloning region of pSTBlue-1 (Kmr; Novagen) to create pSTBlueΔnasA. Correct deletion construct insertion was confirmed by sequencing. The deletion construct was introduced into wild-type (WT) R. solanacearum chromosome by double homologous recombination following electroporation, as previously described (22). Correct allelic replacement was confirmed with PCR.
An initial complementation construct contained only the nasA open reading frame (ORF). This plasmid was introduced into the nasA mutant strain and conferred the ability to grow on nitrate, but at a lower rate than the wild-type strain. We therefore constructed a new complementation plasmid, this time including the upstream region to facilitate proper transcriptional control.
This nasA complementation plasmid (pNascomp) was generated using the primers nasoperonF and nasoperonR on R. solanacearum genomic DNA template to amplify a fragment encoding the promoter, narK3, nirB, nirD, nasB, and nasA. This 8.8-kb DNA fragment was cloned into pCR-Blunt (Invitrogen). The full nas operon was excised with XbaI and BamHI and transferred into XbaI/BamHI-digested pUFJ10 (23) to create pNascomp. This plasmid, which is stably maintained in R. solanacearum, was introduced into the nasA mutant R. solanacearum strain via electroporation. The presence of the plasmid was confirmed by plasmid purification and antibiotic resistance. Plasmid pUFJ10 is one of the few complementation vectors that is stable in R. solanacearum (23). However, pUFJ10 confers a significant fitness defect on R. solanacearum (Tuan Tran, unpublished data), so the complemented nasA mutant was not used for in planta studies.
Growth measurements.
Nitrate assimilation capacity was measured using growth curves and imaging of R. solanacearum wild-type and nasA mutant strains growing in BMM (pH 7.0) with equimolar concentrations (5 mM) of either ammonium or nitrate as a nitrogen source and 55 mM glucose as a carbon source. Filter-sterilized tomato xylem sap collected as previously described (4) was also used as a growth medium. Then, 96-well plates were inoculated with 104 CFU of single R. solanacearum strains and incubated in a spectrophotometric plate reader (Bio-Tek Instruments) for 72 h at 28°C with slow shaking. Each treatment was replicated nine times. Growth curves indicated that growth resulting from nitrate assimilation when nitrate was the sole N source can be simplified to a +/− scale. We depicted readings with endpoint images illustrating the qualitative nature of the metabolism across strains. Viability and cell density at the 72 h endpoint were assessed via serial dilution plating.
Plant assays.
A bacterial wilt-susceptible tomato cultivar (Lycopersicon esculentum Mill cv. Bonny Best) was used to evaluate virulence of the R. solanacearum strains. Soil soak inoculations were performed by gently pouring a 50-ml bacterial suspension at a final concentration of 108 CFU/g of potting mix into the pots of unwounded 3-week-old tomato plants (24). To measure strain competitive fitness, plants were inoculated with a 1:1 suspension of two strains at a final total concentration of 108 CFU/g of potting mix. Wilt symptom development was assessed daily on a scale from 0 to 4 (0 = 0%, 1 = 1 to 25%, 2 = 26 to 50%, 3 = 51 to 75%, and 4 = 76 to 100% total leaves wilted). Bacteria were inoculated directly into stem xylem tissue by petiole inoculation as previously described (4). Briefly, 3-week-old Bonny Best tomato plants were inoculated with 2,000 CFU of the specified R. solanacearum strain(s) through the cut petiole of the first true leaf and monitored for symptoms as described above. For colonization assays, plants were inoculated by either soil soak or cut petiole inoculations and sampled 2 to 5 days postinoculation or at the first appearance of symptoms, as indicated by the term “Disease Index 1.” A 100-mg segment of midstem tissue was suspended in water to a 1:10 dilution (0.1 g of stem tissue plus 0.9 ml of water), ground, and dilution plated on CPG medium supplied with appropriate antibiotics to measure the CFU per 100 mg of stem tissue. For competitive fitness assays, the relative competitive index (RCI) was calculated as the ratio of mutant to WT CFU per 1 g of stem tissue recovered normalized to the ratio of mutant to WT CFU in the inoculum. A value of 1 indicates the two compared strains behaved similarly. A value of <1 indicates the mutant was less fit than WT, while a value of >1 indicates the mutant was more fit than WT under the conditions tested. Because the coinoculated strains were marked with antibiotic resistance genes, it should be noted that rifampin and gentamicin resistance do not affect virulence or growth in R. solanacearum (25; A. Milling and C. Allen, unpublished data).
To assess the contribution of nitrate assimilation to the ability of R. solanacearum to persist between plant infections, we soil soak inoculated bacterial wilt-resistant H7996 tomato plants with either nasA mutant or wild-type R. solanacearum as described above. Twenty days after the initial inoculation, plants were uprooted and removed from each pot, and the pots continued to be watered daily. Ten days later, a new 3-week-old Bonny Best plant was transplanted into each pot, and disease development was monitored.
Soil survival assessments.
One tomato plant per pot containing 80 g of potting mix was added to each pot (n = 10 per treatment). After 3 weeks of incubation, the plants were removed, and the soil was soaked with 50 ml of either wild type, nasA mutant, or a 1:1 mix of wild-type and nasA mutant R. solanacearum to a final density of 108 CFU/g of soil. Previously used pots were used in order to replicate the conditions used in the original between-plant virulence assay.
Pots were watered with Hoagland's fertilizer every other day, and populations were quantified in the pots immediately after inoculation, at 6 h postinoculation, at 5 days postinoculation, and at 15 days postinoculation. For each time point, a total pooled mass of 1 g of soil was taken from three different locations within each pot. This soil was suspended in 9 ml of sterile water, vortexed, serially diluted, and plated on selective medium to quantify the CFU/g of soil. The RCI was calculated as described above.
EPS analyses.
EPS production was observed by plating 102 CFU of wild-type, nasA mutant, epsB mutant, and nasA + pNascomp R. solanacearum on either CPG or CPG plus 40 mM nitrate. Colony morphology of each strain was visually assessed and imaged after 4 days of incubation at 28°C.
Quantitative analyses of EPS production were conducted using enzyme-linked immunosorbent assay (ELISA), the biochemical Elson-Morgan assay, and viscosity measurements. R. solanacearum strains were grown in CPG medium overnight, rinsed, and diluted to a cell density of 109 CFU/ml. Portions (100 μl) of these cell suspensions were spread on plates of MES-buffered CPG agar or CPG–40 mM nitrate agar and incubated for 4 days at 28°C. The cells were then resuspended in water to a uniform optical density at 600 nm (OD600) of 1 (i.e., 109 CFU/ml). The cell density was confirmed through dilution plating. EPS was quantified by using anti-R. solanacearum EPS antibodies with ELISA (Agdia, Inc., Elkhart, IN) according to the manufacturer's instructions per 10-μl volume (107 CFU) of cell suspension (10 technical replicates were assessed). To measure viscosity of EPS produced by comparable cell numbers (109 CFU/ml) of different strains, bacteria were collected from plates as described above, whole cells were removed from the suspensions (at 109 CFU/ml determined by spectrophotometry [OD600 = 1.0] and confirmed via dilution plating) via centrifugation (as described in reference 14), and the viscosity of the extracellular milieu (mainly consisting of EPS1) was measured. Then, 1 ml of cell-free extracellular milieu was placed in the measurement cup of a Brookfield DV-1 digital viscometer, and the viscosity was measured at 28°C at 60 rpm. Viscosity was determined relative to water, which was given an arbitrary value of 1.
For biochemical analysis, crude EPS was purified, and hexosamine content was quantified per 109 CFU using the Elson-Morgan method as described previously (14). Briefly, EPS was prepared as for the ELISAs, and whole cells were removed as described above for the viscosity assays. Cell-free EPS was lyophilized, dissolved in water, precipitated overnight at −20°C in 4 volumes acetone–20 mM NaCl, and redissolved in water. N-Acetylgalactosamine (Sigma-Aldrich) was used as the standard, and calculated concentrations were normalized to cell density (as determined by the OD600 and dilution plating prior to removing cells via centrifugation).
Root attachment assay.
Tomato seeds (cv. Bonny Best) were surface sterilized, germinated, and grown on Murashige & Skoog (MS) medium plus agar with nitrate (MS Medium Mod 4; PlantMedia) as the sole nitrogen source for 2 days. Bacterial suspensions were generated from overnight cultures grown in CPG plus 10 mM nitrate. Inocula included single strains (wild type, nasA mutant, and epsB mutant R. solanacearum alone), coinoculations (nasA mutant plus wild-type cells [1:1], epsB mutant plus wild-type cells [1:1]), and cell-free EPS-supplemented single strain inoculations containing 2 μg of EPS per cm of root (nasA mutant of EPS produced from wild-type R. solanacearum grown on nitrate-containing plates and epsB mutant plus EPS produced from wild-type R. solanacearum grown on nitrate-containing plates). Then, 10 μl (5 × 105 CFU/ml) of each inoculum was placed on 2-cm sections of individual seedling roots to deliver 2.5 × 103 CFU/cm of root. After incubation for 2 h, inoculated root segments were excised, gently mixed for 10 s in 15-ml conical tubes of sterile water, and blotted dry. Batches of three to five 2-cm root segments were pooled, ground, serially diluted, and plated to quantify CFU of the strain of interest attached per cm of root. To account for small variations in inoculum cell density, the resulting CFU/cm numbers were divided by the initial inoculum, as determined by dilution plating. This number was used to generate a directly comparable percentage of CFU attached from each inoculation. This assay was repeated three times with a total of n = 10 to 16 replicates (each replicate containing 3 to 5 roots) for each treatment.
RESULTS
Nitrate assimilation in R. solanacearum.
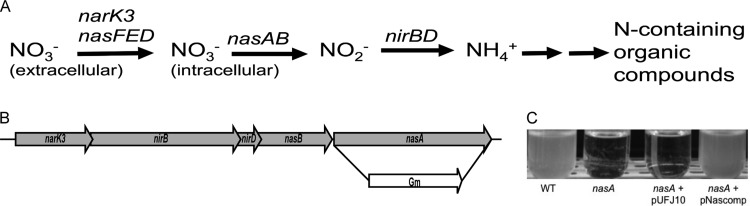
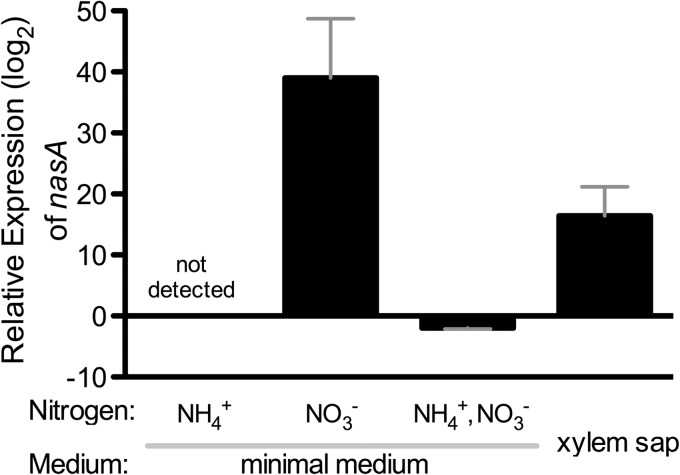
The genomic structure of R. solanacearum's nitrate assimilation network is unique to a subset of Betaproteobacteria. Genomic analysis suggests R. solanacearum has two assimilatory nitrate uptake systems, encoded by nasFED and narK3 (26, 27). The nasFED operon, predicted to encode the typical assimilatory nitrate transporter, is atypically distant from the metabolic operon, suggesting a relatively recent genomic rearrangement (27). The metabolic operon is predicted to encode enzymes responsible for the reduction of nitrate and nitrite. It begins with narK3, predicted to encode an ATP-independent high-affinity nitrate transporter similar to those that uptake nitrate for respiration (Fig. 1) (26, 28). narK3 is directly upstream of nirB and nirD, which encode the assimilatory nitrite reductase, and nasB and nasA, which encode the assimilatory nitrate reductase. This arrangement (narK3-nirBD-nasBA) is limited to R. solanacearum, R. pickettii, R. eutropha, Cupriavidus taiwanensis, and Herbaspirillum seropedicae. Entirely unique to R. solanacearum is the positioning of this operon between aer1 (encoding an energy taxis receptor [29]) and a cluster of predicted filamentous hemagglutinin genes. This nas operon has no nearby regulator, which is uncommon (27). However, a predicted NasR regulator is encoded immediately upstream of the nasFED operon. Although nasR and narK3-nirBD-nasBA are genomically distant and the leader sequence of the metabolic operon has no clearly identifiable stem-loop secondary structure motif (see below), we nonetheless observed the typical nitrate-induced regulatory patterns (Fig. 2) (30). In organisms such as Klebsiella oxytoca, a stem-loop structure forms in the leader region of the metabolic operon. This leads to transcriptional termination. When nitrate is present, however, NasR aids in the formation of an antiterminator in this region, allowing transcription of the nitrate assimilatory genes (31). Using qRT-PCR to measure transcription of nasA, we determined that R. solanacearum nitrate assimilation is induced by nitrate (Fig. 2). The expression of nasA was highly induced (39-fold) in minimal medium with 5 mM nitrate relative to in medium without nitrogen. There was no detectable nasA transcript in minimal medium with 5 mM ammonium, and growth in 5 mM nitrate and 5 mM ammonium decreased nasA expression 2-fold compared to in minimal medium with no nitrogen. This, together with the failure to detect any nasA transcript in the 5 mM ammonium condition, indicates that ammonium represses transcription of nitrate assimilation genes, as is typical in most nitrate-assimilating bacteria (27). Growth in diluted tomato xylem sap, a biologically relevant condition, increased R. solanacearum nasA expression 16-fold (Fig. 2).
FIG 1.
Organization of the R. solanacearum nitrate assimilation pathway and its function in culture. (A) The nitrate assimilation pathway allows for the uptake of nitrate and reduction of both nitrate and nitrite for use in the synthesis of organic compounds. The relevant genes are listed above each enzymatic step. Annotations: narK3 and nasFED, nitrate uptake transporters; nasA, large catalytic subunit of the assimilatory nitrate reductase; nasB, small subunit of the assimilatory nitrate reductase; nirB, large catalytic subunit of the assimilatory nitrite reductase; nirD, small subunit of the assimilatory nitrite reductase. (B) The R. solanacearum nitrate assimilation operon and construction of the nasA mutant strain. The complete nasA ORF was replaced with a gentamicin resistance cassette, as shown. (C) Growth of R. solanacearum strains in minimal medium with 5 mM nitrate as the sole nitrogen source, imaged after 72 h. WT, wild-type R. solanacearum UW551; nasA, nasA deletion mutant; nasA + pUFJ10, nasA mutant plus empty vector; nasA + pNascomp, complemented nasA mutant.
FIG 2.
Expression of the nasA gene was induced by nitrate and repressed by ammonium. RNA was extracted from wild-type R. solanacearum cells incubated for 3 h at a density of 2 × 108 CFU/ml in either minimal medium (with no nitrogen, ammonium, nitrate, or both ammonium and nitrate) or in 1:10-diluted xylem sap. After cDNA synthesis, gene expression levels were determined with relative value quantitative real-time PCR using rplM for normalization. The data are presented relative to nasA expression in wild-type R. solanacearum cells in minimal medium with no nitrogen source. Bars indicate the standard errors; n = 9 biological replicates for each treatment.
The nas cluster is expressed by R. solanacearum during plant pathogenesis.
We predicted that if R. solanacearum uses nitrate assimilation during pathogenesis, the genes of the nas operon would be expressed during plant infection. The data from a previous transcriptional analysis indicate that the nas operon is indeed expressed at 5 × 108 CFU/ml both in culture (data not shown) and during tomato pathogenesis (Table 3) (4). R. solanacearum also expresses genes encoding components of the ammonium-deficient response, such as ntrBC, in the plant during disease. This confirms that under the conditions tested, ammonium, the preferred nitrogen source of most bacteria and the one that requires the least energy to channel into assimilation, is absent. This suggests that R. solanacearum is not subject to ammonium-mediated repression of nasA during plant pathogenesis and may actively assimilate nitrate.
TABLE 3.
Expression of R. solanacearum genes involved in nitrate assimilation and scavenging during tomato pathogenesisa
| Gene | Annotation (NCBI) |
R. solanacearum UW551 |
R. solanacearum GMI1000 |
||
|---|---|---|---|---|---|
| Locus tag | In planta absolute expression | Locus tag | In planta absolute expression | ||
| narK3 | High-affinity nitrate transporter transmembrane protein | RRSL_01120 | 9.66 | RSp1223 | 8.95 |
| nirB | Nitrite reductase NADPH large subunit | RRSL_01119 | 9.72 | RSp1222 | 8.94 |
| nirD | Nitrite reductase NADPH small subunit | RRSL_01118 | 7.97 | RSp1221 | 8.29 |
| nasB | Flavoprotein FAD oxidoreductase | RRSL_01117 | 8.26 | RSp1220 | 7.94 |
| nasA | Nitrate reductase large subunit oxidoreductase | RRSL_01116 | 7.08 | RSp1219 | 7.95 |
| nasF | Nitrate transporter protein | RRSL_03280 | 9.01 | RSc0381 | 9.83 |
| nasE | Nitrate transmembrane ABC transporter protein | RRSL_03281 | 7.80 | RSc0380 | 9.17 |
| nasD | Nitrate binding ABC transporter protein | RRSL_03282 | 8.71 | RSc0379 | 7.97 |
| glnA1 | Glutamine synthetase | RRSL_01421 | 11.71 | RSc1258 | 14.03 |
| ntrB | Nitrogen regulation (sensor protein kinase transcription regulator) | RRSL_01419 | 9.68 | RSc1260 | 11.31 |
| ntrC | Nitrogen assimilation regulatory response regulator transcription regulator protein | RRSL_01418 | 9.84 | RSc1261 | 9.95 |
| glnA2 | Glutamine synthetase | RRSL_03114 | 8.51 | RSp0886 | 8.41 |
| glnB | Nitrogen regulatory P-II transcription regulator protein | RRSL_00967 | 12.05 | RSc2345 | 12.91 |
| amtB | Ammonium transporter | RRSL_03319 | 10.07 | RSc0343 | 10.88 |
| glnK | Nitrogen regulatory P-II transcription regulator protein | RRSL_03320 | 10.68 | RSc0342 | 11.51 |
To determine the in planta absolute expression, R. solanacearum cells were centrifuged from the stems of symptomatic tomato plants inoculated with the indicated strain, and absolute gene expression levels were determined using strain-specific microarrays as described previously (4).
R. solanacearum nasA encodes a necessary component of a functional assimilatory nitrate reductase.
To explore the role of nitrate assimilation in R. solanacearum biology, we created a deletion mutant lacking nasA (Fig. 1B). As predicted by annotation, nasA was required for R. solanacearum growth on nitrate as a sole nitrogen source but was dispensable for growth on ammonium (Fig. 1C; data not shown). Measuring continuous growth of the nasA mutant and wild-type R. solanacearum strains over 96 h confirmed that nitrate assimilation was suppressed by the presence of ammonium, as is typical of bacterial nitrate assimilation networks (data not shown) (32–34). The two strains grew indistinguishably in minimal medium supplemented with ammonium or ammonium + nitrate. However, the wild type grew significantly slower on nitrate than on ammonium when nitrate was being assimilated, suggesting again that ammonium is a more efficiently used nitrogen source than nitrate and is preferred over nitrate when available. The nasA mutant nitrate assimilation defect was complemented by addition of the wild-type narK3-nirBD-nasBA operon in trans, but the empty vector did not affect nitrate assimilation (Fig. 1C).
nasA contributes to bacterial wilt virulence early in infection.
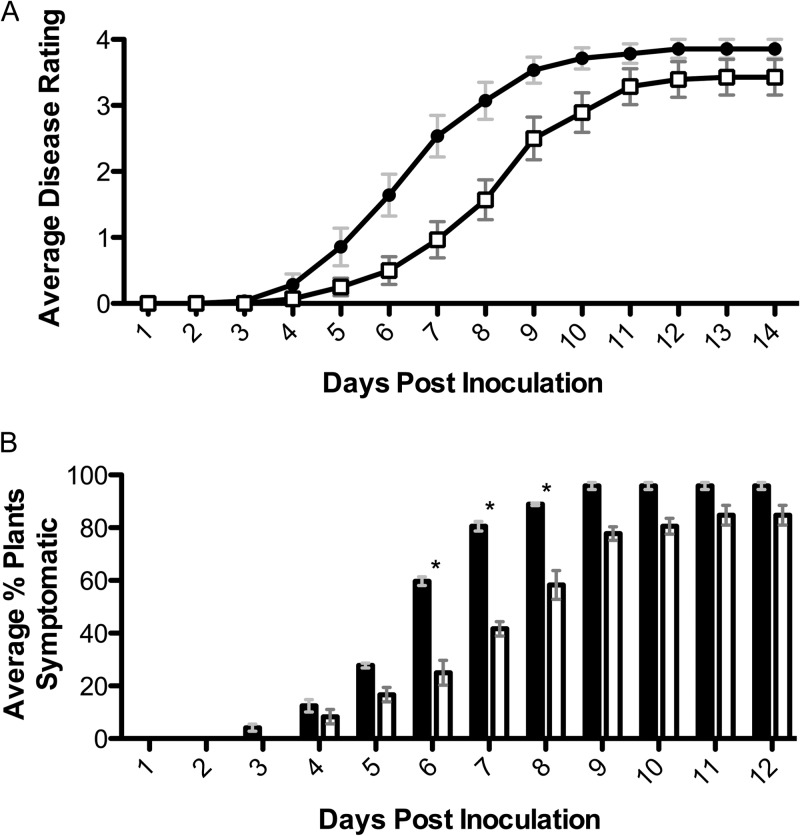
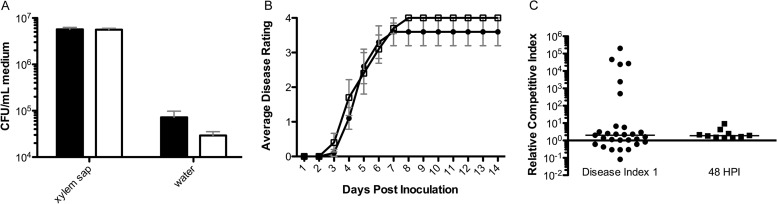
The nasA mutant was significantly delayed in virulence on tomato following naturalistic soil soak inoculation (P = 0.002, repeated-measures analysis of variance [ANOVA]; Fig. 3A). Soil soak inoculation of unwounded plants forces the bacterium to survive in soil, chemotax to root exudates, attach, form microcolonies, enter the root, and move through the cortex to colonize the xylem. Based on the gene expression data, the abundance of nitrate in the agricultural environment and the virulence defect of the nasA mutant, we hypothesized that nitrate is an important nitrogen source for R. solanacearum in the xylem environment. However, the nasA and wild-type strains grew equally well in filter-sterilized tomato xylem sap (P > 0.05, Mann-Whitney t test) (Fig. 4A). To determine whether during colonization, the plant provides nitrate or other nitrate assimilation-promoting factors that are not present in ex vivo xylem sap from healthy plants, we inoculated nasA mutant and wild-type strains directly into xylem tissue via a cut leaf petiole and measured symptom development. With this inoculation method, the nasA mutant and wild-type strains were equally virulent (P > 0.05, repeated-measures ANOVA) (Fig. 4B). To determine whether nitrate assimilation confers a competitive fitness benefit on R. solanacearum cells growing in xylem, we measured population sizes of marked nasA mutant and wild-type bacteria after coinoculation via cut petiole at either 48 h postinoculation or when wilt symptoms first appeared. Under these conditions the nasA mutant grew better than the wild type at both sampling points (P = 0.0026 and 0.0059, respectively) (Fig. 4C). This indicates that NasA provides no detectable fitness benefit to the pathogen during stem xylem colonization. Overall, these experiments demonstrate that nitrate assimilation was not required for bacterial growth or competitive fitness in host xylem. This suggests that nitrate assimilation makes its contribution to R. solanacearum virulence during the early stages of pathogenesis.
FIG 3.
Nitrate assimilation is required for full R. solanacearum virulence on tomato. (A) Disease caused by wild-type strain UW551 (●) or assimilatory nitrate reductase mutant nasA (□) following naturalistic soil soak inoculations of susceptible tomato plants (cv. Bonny Best), as illustrated in Fig. 5A. The data shown reflect mean daily ratings on a disease index scale of 0 to 4 of three biological replicates, each containing 8 to 16 plants/treatment for a total n = 28 plants. P = 0.002 (two-way repeated-measures ANOVA). (B) Percentage of plants showing any wilt symptoms each day for the experiment described above. Black bars, wild-type strain UW551; white bars, nasA mutant. Bars indicate the standard errors of the mean; asterisks indicate days when percent symptomatic plants were significantly different between wild-type and nasA treatments (P < 0.05 [unpaired t test]).
FIG 4.
R. solanacearum does not require nitrate assimilation for wild-type growth, virulence, or competitive fitness in tomato xylem. (A) Growth of wild-type strain UW551 (■) or nasA mutant (□) in either filter-sterilized xylem sap or water inoculated with 104 CFU of bacteria. Growth was assessed 72 h later by dilution plating. The data shown are means of nine replicates; P > 0.05 (Mann-Whitney t test). (B) Disease progress of tomato plants inoculated directly into xylem tissue via a cut leaf petiole with 2,000 CFU of either R. solanacearum wild-type UW551 (black) or nasA mutant (white). Plants were rated daily on a 0 to 4 disease index scale; n = 10; P > 0.05 (repeated-measures ANOVA). (C) Competitive indices of marked R. solanacearum strains following cut-petiole inoculation of tomato plants with a 1:1 mixture of wild-type and nasA mutant bacteria (2,000 cells total). Portions (100 mg) of plant tissue were ground and dilution plated to quantify each strain at either the first sign of disease (disease index 1; n = 29) or at 48 h postinoculation (n = 10). The relative competitive index was calculated as the ratio of CFU of nas mutant recovered/g of stem to the CFU of UW551 wild-type recovered/g of stem, adjusted to inoculum levels. Horizontal lines represent the medians. The nasA mutant significantly outcompeted wild-type bacteria (P = 0.0026 and 0.0059 [Wilcoxon signed-rank test] for disease index 1 and 48 h postinfection [HPI], respectively).
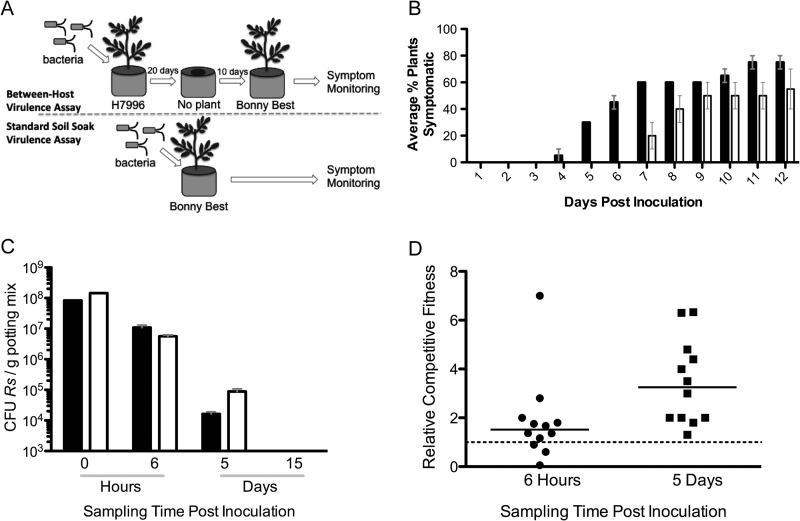
Nitrate assimilation contributes to pathogen success over the disease cycle.
Virulence assays typically ignore key steps in the biological disease cycle such as survival and production of virulence factors between crop plantings. To test the hypothesis that nitrate assimilation contributes to the ability of R. solanacearum to cause disease following a period of soil incubation, we compared symptom development in tomato plants grown in soil previously colonized by either nasA mutant or wild-type strains (Fig. 5A). This assay required bacteria to both survive in soil without a living plant, and to successfully infect a new host. To generate infested soil, the moderately wilt-resistant tomato line H7996 was soil soak inoculated with nasA mutant or wild-type bacteria. Importantly, H7996 is equally susceptible to nasA mutant and wild-type strains (data not shown). After 20 days, the plants were uprooted and discarded, leaving the bacteria to survive under conditions analogous to those in a harvested field. A 3-week-old susceptible tomato plant was transplanted into each pot, and wilt symptoms were assessed over time. Plants growing in pots that previously contained nasA mutant-infected plants were less likely to wilt and also developed symptoms several days later than plants growing in pots that formerly contained wild-type infected plants (P = 0.0009; repeated-measures ANOVA) (Fig. 5B). This suggests that the ability to assimilate nitrate contributes to the ability of R. solanacearum to infect new plants brought into an infested environment.
FIG 5.
nasA contributes to tomato plant bacterial wilt following a period of soil survival. (A) Virulence assay comparison. In a between-host virulence assay (see panel B), unwounded plants of moderately wilt-resistant tomato H7996 were inoculated with 108 CFU of R. solanacearum/g soil. After 20 days, the plants were removed from each pot, and 10 days later a susceptible Bonny Best plant was transplanted into each pot. This is in contrast to the standard soil soak virulence assay (see the results presented in Fig. 2), where 108 CFU of R. solanacearum were added to pots containing a 3-week-old Bonny Best plant. (B) Between-host virulence assay. The percentages of Bonny Best tomato plants showing any wilt symptoms each day after transplanting into pots previously containing a H7996 plant inoculated with either wild-type UW551 (■) or nasA mutant (□) strains are shown. This assay was repeated twice, with 20 plants/treatment in total (P = 0.0009 [repeated-measures ANOVA]). (C) Soil survival assessments. Pots were inoculated via soil soak as described above with 108 CFU of either nasA mutant or wild-type R. solanacearum/g of soil. At 0 h, 6 h, 5 days, and 15 days postinoculation, 1-g soil samples were dilution plated to determine the population sizes of R. solanacearum in each pot. (D) Soil survival competitive index assessments. Pots were inoculated via soil soak with a 1:1 ratio of nasA mutant and wild-type R. solanacearum cells. Population sizes of each strain were determined by dilution plating on selective media at 6 h and 5 days postinoculation. The relative competitive index was calculated as the ratio of CFU nasA mutant to CFU wild-type recovered/g of stem, adjusted to inoculum levels. Horizontal bars represent the medians.
The nasA mutant was not defective in soil survival.
To test the hypothesis that the nasA mutant's delayed symptom development was due to reduced ability to survive in soil, we measured bacterial populations sizes over time under the conditions described above. Pots that previously contained 3-week-old tomato plants were inoculated with 108 CFU of either the nasA mutant, the wild type, or a 1:1 mix of nasA mutant plus wild type/g of soil, and the population sizes in soil were measured over time. The population sizes of both strains declined rapidly and at similar rates over the experiment (Fig. 5C). In single-strain treatments, the nasA mutant population sizes were slightly higher than those of the wild type, and in coinoculation treatments the nasA mutant strain was slightly more fit than the wild-type strain (P = 0.0025; Wilcoxon signed-rank test) at 5 days postinoculation (Fig. 5D). By 15 days postinoculation, all populations were below the detection limit of 103 CFU/g of soil. These data indicate that the virulence defect of the nasA mutant is not due to reduced ability to survive in soil. In addition, the nasA mutant exhibited wild-type levels of motility and chemotaxis in a plate assay (35), indicating that loss of nitrate assimilation did not affect these known early-disease virulence factors (data not shown).
Nitrate assimilation influences EPS production in a nasA-dependent fashion.
Because soil survival did not explain the nasA mutant virulence defect (Fig. 5D), we hypothesized that the nasA mutant produces qualitatively or quantitatively different virulence factors. Since EPS, the major virulence factor of this organism, is influenced by environmental conditions, we investigated the possibility that EPS production was linked to nitrate assimilation. Qualitatively, we assessed EPS production and the influence of nitrate assimilation on it, We assessed colony morphologies of nasA mutant and wild-type R. solanacearum cells growing on CPG plates with or without added nitrate (Fig. 6A). On CPG, R. solanacearum colonies are mucoid and irregular, reflecting the organism's abundant production of EPS. Adding nitrate to CPG reduced the mucoidy of wild-type colonies, but the nasA mutant colony morphology was not affected by the addition of nitrate (Fig. 6A).
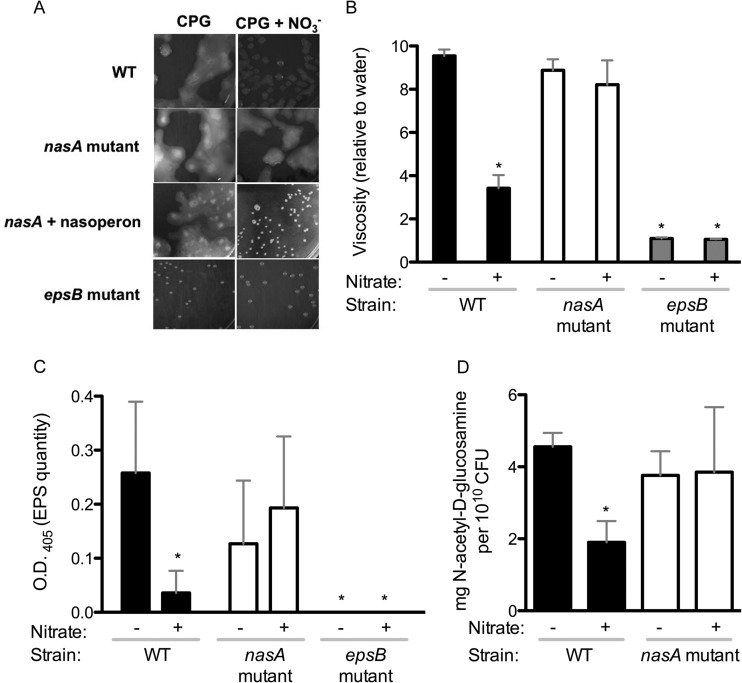
FIG 6.
Nitrate influences the quantity and viscosity of extracellular polysaccharide (EPS) via nasA. (A) Colony morphology of R. solanacearum wild-type UW551, nasA mutant, nasA mutant complemented with pNascomp, or epsB mutant, grown on either CPG medium plates (left) or on CPG plus 40 mM nitrate (right) and imaged after 4 days of growth at 28°C. (B) Effect of nitrate supplementation and nasA mutation on viscosity of a crude EPS suspension. Colonies grown for 4 days on CPG with (+) or without (−) 40 mM nitrate were suspended in water to 109 CFU/ml and centrifuged, and the viscosity of the cell-free supernatant (from 109 CFU) was measured with a Brookfield model DV-1 viscometer. The viscosity shown is normalized to that of water, arbitrarily set at 1. Bars: ■, WT UW551; □, nasA mutant;  , epsB mutant. Bars indicate the standard errors. Asterisks indicate significant differences from wild-type cells growing without nitrate (P < 0.05, t test). (C) Immunological quantification of EPS in supernatants using ELISA with anti-R. solanacearum EPS antibodies (OD405). Colonies grown for 4 days on CPG with (+) or without (−) 40 mM nitrate were suspended in water to 109 CFU/ml. EPS was quantified in 107 CFU (10 μl) by ELISA. Asterisks indicate significant differences in OD405 values from wild-type cells growing without nitrate (P < 0.05, t test). (D) Biochemical quantification of EPS hexosamines. Purified EPS from colonies grown for 4 days on CPG with (+) or without (−) 40 mM nitrate was quantified by using the Elson-Morgan assay with known concentrations of N-acetylgalactosamine as a standard. The total hexosamine was quantified and normalized to the amount of CFU in the samples. An asterisk indicates a significant difference from wild-type cells growing without nitrate (P = 0.0014; t test).
, epsB mutant. Bars indicate the standard errors. Asterisks indicate significant differences from wild-type cells growing without nitrate (P < 0.05, t test). (C) Immunological quantification of EPS in supernatants using ELISA with anti-R. solanacearum EPS antibodies (OD405). Colonies grown for 4 days on CPG with (+) or without (−) 40 mM nitrate were suspended in water to 109 CFU/ml. EPS was quantified in 107 CFU (10 μl) by ELISA. Asterisks indicate significant differences in OD405 values from wild-type cells growing without nitrate (P < 0.05, t test). (D) Biochemical quantification of EPS hexosamines. Purified EPS from colonies grown for 4 days on CPG with (+) or without (−) 40 mM nitrate was quantified by using the Elson-Morgan assay with known concentrations of N-acetylgalactosamine as a standard. The total hexosamine was quantified and normalized to the amount of CFU in the samples. An asterisk indicates a significant difference from wild-type cells growing without nitrate (P = 0.0014; t test).
We further qualitatively assessed the EPS by measuring viscosity of cell-free supernatants of bacteria harvested from plates (Fig. 6B). The EPS from the wild-type strain was significantly less viscous (P < 0.05, t test) when cells grew with supplemental nitrate, but nitrate levels did not affect the viscosity of EPS from the nasA mutant (Fig. 6B). Viscosity measurements are shown in values relative to water (which was given an arbitrary value of 1).
We used ELISA to quantify total EPS produced by comparable numbers (107 CFU) of R. solanacearum cells growing on plates with or without nitrate. In the absence of supplemental nitrate, the nasA mutant and the wild-type strain produced similar quantities of EPS. However, in the presence of nitrate, the wild-type strain produced significantly less EPS than the nasA mutant and less than the wild-type strain without added nitrate (P < 0.05, t test) (Fig. 6C). An epsB mutant, which lacks a key EPS biosynthetic enzyme and forms small round dry colonies, produced no detectable EPS in this assay.
We quantified a major component of R. solanacearum EPS, hexosamines, using the Elson-Morgan biochemical assay, using cell-free preparations from 109 CFU (Fig. 6D). This method also indicated that nitrate reduced the amount of EPS produced by the wild-type strain (P < 0.05, t test) but did not affect EPS levels in the nasA mutant. Together, these results indicate that high environmental nitrate levels reduce EPS production in R. solanacearum and that nasA is required for this response.
The nasA mutant was delayed in host plant colonization; this phenotype was rescued by coinoculation with wild-type, but not EPS-deficient, bacteria.
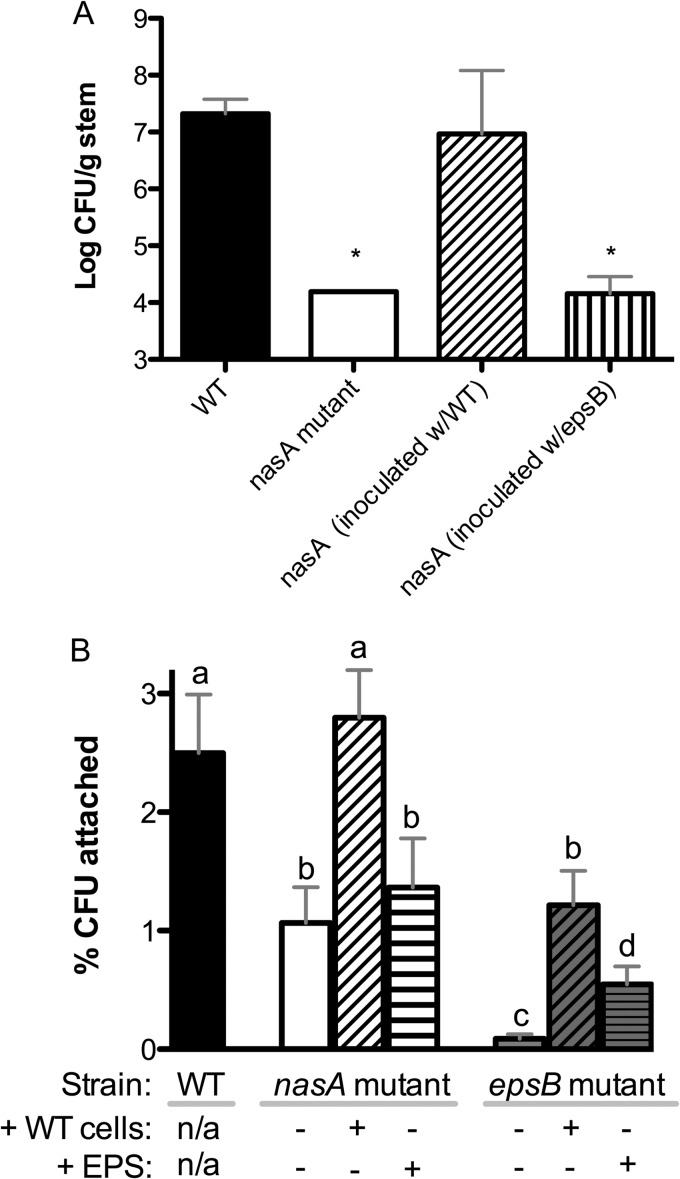
Because the nasA mutant caused slower symptom development than wild-type following soil soak inoculation, it seemed likely that this mutant would also be delayed in plant colonization. To test this hypothesis, we compared bacterial population sizes in tomato stems 5 days after soil soak inoculation. As predicted, the nasA mutant was significantly delayed in stem colonization (P < 0.05, t test) (Fig. 6A). By 8 days postinoculation, the nasA mutant reached stem populations comparable to those of wild type (data not shown). Therefore, the lower population sizes at 5 days postinoculation represent a delay and not an inability to reach wild-type population sizes during infection.
To test the hypothesis that aberrant EPS is responsible for the nasA mutant's slower stem colonization, we coinoculated plants with the nasA mutant and an equal number of either wild-type or epsB mutant bacteria. Interestingly, coinoculation with the wild-type strain externally complemented the nasA mutant's colonization defect (P < 0.05, t test) (Fig. 7A). In contrast, coinoculation with an epsB mutant had no effect on the nasA mutant colonization delay. This result suggested that altered EPS production explains the nasA mutant's colonization defect.
FIG 7.
Loss of nitrate assimilation or EPS production reduced R. solanacearum tomato stem colonization and attachment to tomato roots. (A) Colonization of tomato stems following soil soak inoculation of tomato plants with wild-type UW551 (WT, ■), nasA mutant (□), nasA mutant coinoculated with UW551 wild type (▨), and nasA mutant coinoculated with epsB mutant ( ). Bacteria were quantified by dilution plating of ground midstem tissue 5 days after inoculation. Asterisks indicate significant differences from the WT (P < 0.05, t test; n = 9 plants/treatment; bars indicate the standard errors [nasA mutant error bars are too small to see]). (B) R. solanacearum attachment to tomato roots after 2 h of incubation with 2.5 × 103 CFU/cm of root of wild-type UW551 (■), nasA mutant (□), or epsB mutant (
). Bacteria were quantified by dilution plating of ground midstem tissue 5 days after inoculation. Asterisks indicate significant differences from the WT (P < 0.05, t test; n = 9 plants/treatment; bars indicate the standard errors [nasA mutant error bars are too small to see]). (B) R. solanacearum attachment to tomato roots after 2 h of incubation with 2.5 × 103 CFU/cm of root of wild-type UW551 (■), nasA mutant (□), or epsB mutant ( ). Three rinsed and blotted roots were pooled per biological replicate (n = 10 to 16 replicates), and each replicate was ground and dilution plated to quantify CFU/cm of root. As indicated below the columns, some treatments were combined with either an equal number of wild-type (WT) cells/root or 2.5 mg of cell-free EPS/cm of root (purified from wild-type cells grown in the presence of NO3−). Bars represent the standard errors. Columns with different letters above them are significantly different from each other (P < 0.05, t test).
). Three rinsed and blotted roots were pooled per biological replicate (n = 10 to 16 replicates), and each replicate was ground and dilution plated to quantify CFU/cm of root. As indicated below the columns, some treatments were combined with either an equal number of wild-type (WT) cells/root or 2.5 mg of cell-free EPS/cm of root (purified from wild-type cells grown in the presence of NO3−). Bars represent the standard errors. Columns with different letters above them are significantly different from each other (P < 0.05, t test).
EPS and nasA are needed for R. solanacearum attachment to roots.
Although it has not been explored in R. solanacearum, EPS is involved in root attachment in other bacteria (36, 37). Both EPS production and early stages of pathogenesis were compromised in a strain lacking the ability to assimilate nitrate, so we hypothesized that under high nitrate conditions a nasA mutant strain would be affected in root attachment, one of the first events in R. solanacearum pathogenesis. To test this hypothesis, we compared attachment to intact sterile tomato roots of wild-type UW551, the nasA mutant, and an EPS-deficient epsB mutant. After 2 h of incubation with roots, more than twice as many wild-type bacteria were attached as the nasA mutant (P < 0.05, t test), while very few cells of the epsB mutant were attached (Fig. 7B). Similar results were obtained with a 30-min incubation (data not shown). To rule out the possibility that the reduced recovery of nasA mutant cells from roots was due to an inability to survive oxidative plant-produced defenses, we measured the survival of the nasA mutant and wild-type R. solanacearum after exposure to various concentrations of nitric oxide and hydrogen peroxide. There was no difference between the strains in survival (data not shown).
To further define the contribution of EPS to R. solanacearum root attachment, we measured the root attachment of various strains in the presence of either wild-type cells or purified EPS from wild-type cells. Coinoculation with wild-type cells at a 1:1 ratio significantly enhanced attachment of the nasA mutant and epsB mutant strains to tomato roots (P < 0.05, t test) (Fig. 7B). The addition of 2 μg of EPS/cm of root significantly enhanced the ability of the epsB mutant to attach (P < 0.05, t test) and also slightly increased nasA attachment, although not significantly (Fig. 7B). These results suggest that EPS increases the ability of R. solanacearum to attach to host roots and that the reduced attachment of the nasA mutant may be at least in part due to its aberrant EPS production.
DISCUSSION
R. solanacearum uses nitrate assimilation during pathogenesis.
The form of inorganic nitrogen in fertilizer is known to affect plant disease development by altering host susceptibility. However, the pathogen's contribution to this phenomenon is poorly understood (38). When the R. solanacearum nasA mutant entered plants through its natural infection court in the root, it was significantly less virulent than its wild-type parent; this defect was exacerbated if the mutant was incubated in soil for 10 days between hosts. To the best of our knowledge, this is the first time nitrate assimilation has been directly implicated in the virulence of any bacterial pathogen, although nitrogen starvation is known to induce the expression of type 3 secretion systems and secreted effector genes in several plant pathogens (39). Once the nasA mutant reached the xylem, it grew and caused disease as well as the wild-type strain. Interestingly, a nitrate assimilation-deficient Pseudomonas syringae mutant likewise grows as well as the wild type on nitrate-rich leaf surfaces (3). Our results suggest that nitrate assimilation contributes to early-stage virulence. This defect could reflect only the effect of nasA on EPS that we observed, but nitrate assimilation may be involved in additional early-pathogenesis events. Nitrate is abundant and highly mobile in soil. Nitrate is also relatively abundant in xylem sap, but other accessible nitrogen sources such as amino acids are present as well, though not at comparable concentrations (40–43). Our finding that nasA is dispensable for R. solanacearum growth and competitive fitness in tomato xylem suggests strongly that this environment offers the bacterium sufficient amounts of non-nitrate nitrogen sources.
R. solanacearum scavenges nitrogen in the xylem environment.
The transcriptomic data suggest that during growth in tomato xylem R. solanacearum expresses several nitrogen assimilatory systems (ammonium uptake, amino acid uptake and metabolism, and nitrate uptake and metabolism). This affirms that ammonium, an easily metabolized nitrogen source that represses uptake and metabolism of other nitrogen sources, is not present in this environment (4). Although this nitrogen scavenging behavior is consistent with the long-held idea that plant xylem is a relatively nutrient-poor environment, this description should be used carefully (44). The nitrogen-scavenging behavior revealed by gene expression analysis does not necessarily indicate that the organism is nitrogen starved. Although tomato xylem sap does not contain ammonium, it does contain substantial amounts of nitrate, some amino acids, and potentially other forms of nitrogen.
Over the course of its life cycle R. solanacearum moves between diverse environments such as water, soil, rhizosphere, root cortex, and host plant xylem, so the ability to quickly access any nitrogen source that becomes available may be advantageous. We speculate that metabolic flexibility contributes to this bacterium's ability to survive in nutrient poor environments such as surface water (45, 46). In several symbiotic relationships, including the squid-Vibrio fischeri interaction, the microbe relies on host-supplied amino acids for growth (47). R. solanacearum may likewise primarily use host-derived amino acids for growth. In culture, R. solanacearum can use single amino acids as sole sources of both carbon and nitrogen (35; B. L. Dalsing, unpublished data), and our finding that a nasA mutant grew like the wild type in xylem sap indicates that xylem sap contains enough non-nitrate nitrogen to support R. solanacearum colonization and pathogenesis. Further, this result suggests that nitrogen is not quantitatively limited in xylem.
Nitrate assimilation alters EPS.
EPS production in R. solanacearum is tightly controlled by a complicated network of virulence regulators and quorum sensing (16). Although our results indicate that nitrate assimilation influences EPS production, the mechanism remains to be determined, and it is unclear exactly how nitrate assimilation fits into the broader context of EPS regulation. Nitrogen source and nutrient abundance influence EPS-like substances produced by Burkholderia cenocepacia, Staphylococcus aureus, Cryptococcus neoformans, Bradyrhizobium japonicum, Aureobasidium pullulans, and Azotobacter vinelandii, although no regulatory mechanism has been identified (48–53). It is possible that low levels of nitrite within the cell trigger reduced EPS production in the wild-type strain. This could be caused by changes in intracellular pH or directly by nitrite-responsive signaling. Aerobically supplied nitrate differentially induced expression of 18% of the Pseudomonas aeruginosa genome, perhaps in part due to nitrate assimilation-dependent signaling (54). Alternatively, nitrate assimilation may affect EPS production by changing the intracellular C/N ratio. Nitrate assimilation requires higher carbon input as the process involves more ATP than, for example, ammonium assimilation. This, in turn, could affect the overall intracellular C/N ratio, a core metabolic signal that can affect production of many secondary metabolites, including nitrogen-containing virulence factors such as EPS (55, 56).
It remains to be determined how EPS production changes when cells are assimilating nitrate. Our immunological, biochemical, and viscometric measurements indicate that wild-type, but not nasA mutant, bacteria produce less EPS in the presence of nitrate. Our data do not indicate whether this EPS is qualitatively different because of, for example, altered patterns of cross-linking, branching, or N acetylation. This question could be addressed with structural analyses and assessments of physical properties of EPS produced under conditions with or without nitrate assimilation.
Nitrate, EPS, and root attachment.
The almost complete failure of an EPS-deficient mutant to attach to tomato roots offers direct evidence that EPS is necessary for R. solanacearum root attachment. The role of nitrate assimilation in this process is less obvious, since the nasA mutant actually produces more EPS than the wild-type strain in the presence of agriculturally relevant quantities of nitrate. It remains to be determined how nitrate assimilation regulates EPS production in R. solanacearum: does it simply reduce the total amount of EPS, or does it alter the EPS structure in a way that affects binding efficiency? It should be noted the root attachment assay used here may not accurately reflect the physiochemical or biological conditions of roots growing under more natural conditions. Further studies are needed to determine whether high soil nitrate alters R. solanacearum EPS in ways that promote pathogen binding to host roots in agricultural environments.
EPSs of diverse biochemistries are key for root attachment by several plant pathogens and symbionts (36, 37), but EPS has not previously been implicated in attachment of R. solanacearum. To date, only type 4 pili are known to be required for normal attachment (57). Here, we demonstrate that EPS is also important for attachment to roots. However, the mechanism by which EPS aids attachment remains to be determined.
Two independent lines of evidence support a link between the reduced virulence of the nasA mutant and the production of EPS. First, nitrate affects the quantity of EPS in a nasA-dependent manner in culture. Second, EPS is required for R. solanacearum attachment to host roots, a critical first step in pathogenesis. Attachment of the nasA mutant was increased by coinoculation with either wild-type (EPS+) cells or purified cell-free EPS. Although these data do not directly prove that nitrate assimilation changes EPS to facilitate root attachment, they do indicate that this hypothesis deserves further exploration.
This work initiated exploration of inorganic nitrogen metabolism, specifically nitrate assimilation, in R. solanacearum pathogenesis. Further work is ongoing to understand additional roles of this primary metabolite in R. solanacearum, in particular functions associated with nitrate respiration and denitrification.
ACKNOWLEDGMENTS
This study was supported by a National Science Foundation Predoctoral Fellowship to B.L.D., National Science Foundation grant IOS-1258082, USDA-Hatch project WIS01502, and the University of Wisconsin-Madison College of Agricultural and Life Sciences.
We thank Anne Alvarez (University of Hawaii-Manoa) for the gift of anti-EPS antibodies, Raka Mitra (Carleton College) for her sterile root protocol and helpful discussions, and Jonathan M. Jacobs for the gentamicin resistance cassette primer sequences, as well as for sharing the transcriptomic data in Table 3 and helpful discussions. We are very grateful to Jean-Claude Davidian (SupAgro INRA, Montpellier, France) for HPLC measurement of tomato xylem nitrate.
Footnotes
Published ahead of print 20 December 2013
REFERENCES
- 1.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. 10.1016/j.chom.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.da Silva ACR, Ferro JA, Relnach FC, Farah CS, Furlan LR, Quaggio RB, Monteiro-Vitorello CB, Van Sluys MA, Almeida NF, Alves LMC, do Amaral AM, Camargo CBMLEA, Camarotte G, Cannavan F, Cardozo J, Chambergo F, Clapina LP, Cicarelli RMB, Coutinho LL, Cursino-Santos JR, El-Dorry H, Faria JB, Ferreira AJS, Ferreira RCC, Ferro MIT, Formighleri EF, Franco MC, Greggio CC, Gruber A, Katsuyama AM, Kishi LT, Lette RP, Lemos EGM, Lemos MVF, Locali EC, Machado MA, Madeira AMBN, Miyaki CY, Moon DH, Moreira LM, Novo MTM, Okura VK, Oliveria MC, Oliveria VR, Pereira HA, Rossi A, Sena JAD, Silva C, de Souza RF, Spinola LAF, Takita MA, Tamura RE, Telxeira EC, Tezza RID, Trindade dos Santos M, Truffi D, Tsai SM, White FF, Setubal JC, Kitajima JP. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459–463. 10.1038/417459a [DOI] [PubMed] [Google Scholar]
- 3.Parangan-Smith A, Lindow S. 2013. Contribution of nitrate assimilation to the fitness of Pseudomonas syringae pv. syringae B728a on plants. Appl. Environ. Microbiol. 79:678–687. 10.1128/AEM.02511-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs JM, Babujee L, Meng F, Milling A, Allen C. 2012. The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. mBio 3(4):e00114-12. 10.1128/mBio.00114-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayward AC. 1994. The hosts of Pseudomonas solanacearum, p 9–24 In Hayward AC, Hartman GL. (ed), Bacterial wilt: the disease and its causative agent, Pseudomonas solanacearum. CAB International, Wallingford, United Kingdom [Google Scholar]
- 6.Genin S, Denny TP. 2012. Pathogenomics of the Ralstonia solanacearum species complex. Annu. Rev. Phytopathol. 50:67–89. 10.1146/annurev-phyto-081211-173000 [DOI] [PubMed] [Google Scholar]
- 7.Vasse J, Frey P, Trigalet A. 1995. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol. Plant-Microbe Interact. 8:241–251. 10.1094/MPMI-8-0241 [DOI] [Google Scholar]
- 8.Denny TP. 2006. Plant pathogenic Ralstonia species, p 573–644 In Gnanamanickam SS. (ed), Plant-associated bacteria. Springer Publishing, Dordrecht, Netherlands [Google Scholar]
- 9.McGarvey JA, Denny TP, Schell MA. 1999. Spatial-temporal and quantitative analysis of growth and EPS I production by Ralstonia solanacearum in resistant and susceptible tomato cultivars. Phytopathology 89:1233–1239. 10.1094/PHYTO.1999.89.12.1233 [DOI] [PubMed] [Google Scholar]
- 10.Orgambide G, Montrozier H, Servin P, Roussel J, Trigalet-Demery D, Trigalet A. 1999. High heterogeneity of the exopolysaccharides of Pseudomonas solanacearum strain GMI 1000 and the complete structure of the major polysaccharide. J. Biol. Chem. 266:8312–8321 [PubMed] [Google Scholar]
- 11.Kao CC, Barlow E, Sequeira L. 1992. Extracellular polysaccharide is required for wild-type virulence of Pseudomonas solanacearum. J. Bacteriol. 174:1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saile E, McGarvey JA, Schell MA, Denny TP. 1997. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology 87:1264–1271. 10.1094/PHYTO.1997.87.12.1264 [DOI] [PubMed] [Google Scholar]
- 13.Coplin DL, Cook D. 1990. Molecular genetics of extracellular polysaccharide biosynthesis in vascular phytopathogenic bacteria. Mol. Plant-Microbe Interact. 3:271–279. 10.1094/MPMI-3-271 [DOI] [PubMed] [Google Scholar]
- 14.Milling A, Babujee L, Allen C. 2011. Ralstonia solanacearum extracellular polysaccharide is a specific elicitor of defense responses in wilt-resistant tomato plants. PLoS One 6:e15853. 10.1371/journal.pone.0015853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garg RP, Huang JY, Denny WTP, Schell MA. 2000. Multicomponent transcriptional regulation at the complex promoter of the exopolysaccharide I biosynthetic operon of Ralstonia solanacearum. J. Bacteriol. 182:6659–6666. 10.1128/JB.182.23.6659-6666.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schell MA. 2000. Control of virulence and pathogenicity genes of Ralstonia solanacearum by an elaborate sensory network. Annu. Rev. Phytopathol. 38:263–292. 10.1146/annurev.phyto.38.1.263 [DOI] [PubMed] [Google Scholar]
- 17.Raven JA, Smith FA. 1976. Nitrogen assimilation and transport in vascular land plants in relation to intracellular pH regulation. New Phytologist 76:415–431. 10.1111/j.1469-8137.1976.tb01477.x [DOI] [Google Scholar]
- 18.Pate JS. 1973. Uptake, assimilation and transport of nitrogen compounds by plants. Soil Biol. Biochem. 5:109–119. 10.1016/0038-0717(73)90097-7 [DOI] [Google Scholar]
- 19.Hendrick CA, Sequeira L. 1984. Lipopolysaccharide-defective mutants of the wilt pathogen Pseudomonas solanacearum. Appl. Environ. Microbiol. 48:94–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boucher C, Barberis P, Trigalet A, Demery D. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. J. Gen. Microbiol. 131:2449–2457 [Google Scholar]
- 21.Ge L, Rudolph P. 1997. Simultaneous introduction of multiple mutations using overlap extension PCR. Biotechniques 22:28–30 [DOI] [PubMed] [Google Scholar]
- 22.Brown DG, Swanson JK, Allen C. 2007. Two host-induced Ralstonia solanacearum genes, acrA and dinF, encode multidrug efflux pumps and contribute to bacterial wilt virulence. Appl. Environ. Microbiol. 73:2777–2786. 10.1128/AEM.00984-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel DW, Allen C, Schell M, Denny TP, Greenberg JT, Duan YP, Flores-Cruz Z, Huang Q, Clifford JM, Presting G, Gonzalez ET, Reddy J, Elphinstone J, Swanson J, Yao J, Mulholland V, Liu L, Farmerie W, Patnaikuni M, Balogh B, Norman D, Alvarez A, Castillo JA, Jones J, Saddler G, Walunas T, Zhukov A, Mikhailova N. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol. Plant-Microbe Interact. 19:69–79. 10.1094/MPMI-19-0069 [DOI] [PubMed] [Google Scholar]
- 24.Tans-Kersten J, Guan Y, Allen C. 1998. Ralstonia solanacearum pectin methylesterase is required for growth on methylated pectin but not for bacterial wilt virulence. Appl. Environ. Microbiol. 64:4918–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swanson JK, Yao J, Tans-Kersten J, Allen C. 2005. Behavior of Ralstonia solanacearum Race 3 biovar 2 during latent and active infection of geranium. Phytopathology 95:136–143. 10.1094/PHYTO-95-0136 [DOI] [PubMed] [Google Scholar]
- 26.Moir JW, Wood NJ. 2001. Nitrate and nitrite transport in bacteria. Cell. Mol. Life Sci. 58:215–224. 10.1007/PL00000849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luque-Almagro VM, Gates AJ, Moreno-Vivian C, Ferguson SJ, Richardson DJ, Roldan MD. 2011. Bacterial nitrate assimilation: gene distribution and regulation. Biochem. Soc. Trans. 39:1838–1843. 10.1042/BST20110688 [DOI] [PubMed] [Google Scholar]
- 28.Zumft WG. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Allen C. 2007. The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189:6415–6424. 10.1128/JB.00398-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender RA, Friedrich B. 1990. Regulation of assimilatory nitrate reductase formation in Klebsiella aerogenes W70. J. Bacteriol. 172:7256–7259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chai W, Stewart V. 1999. RNA sequence requirements for NasR-mediated, nitrate-responsive transcription antitermination of the Klebsiella oxytoca M5aI nasF operon leader. J. Mol. Biol. 292:203–216. 10.1006/jmbi.1999.3084 [DOI] [PubMed] [Google Scholar]
- 32.Wu SQ, Chai W, Lin JT, Stewart V. 1999. General nitrogen regulation of nitrate assimilation regulatory gene nasR expression in Klebsiella oxytoca M5al. J. Bacteriol. 181:7274–7284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toukdarian A, Kennedy C. 1986. Regulation of nitrogen metabolism in Azotobacter vinelandii: isolation of ntr and glnA genes and construction of ntr mutants. EMBO J. 5:399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Lu CD. 2007. Regulation of carbon and nitrogen utilization by CbrAB and NtrBC two-component systems in Pseudomonas aeruginosa. J. Bacteriol. 189:5413–5420. 10.1128/JB.00432-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yao J, Allen C. 2006. Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 188:3697–3708. 10.1128/JB.188.10.3697-3708.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meneses CH, Rouws LF, Simoes-Araujo JL, Vidal MS, Baldani JI. 2011. Exopolysaccharide production is required for biofilm formation and plant colonization by the nitrogen-fixing endophyte Gluconacetobacter diazotrophicus. Mol. Plant-Microbe Interact. 24:1448–1458. 10.1094/MPMI-05-11-0127 [DOI] [PubMed] [Google Scholar]
- 37.Skorupska A, Janczarek M, Marczak M, Mazur A, Krol J. 2006. Rhizobial exopolysaccharides: genetic control and symbiotic functions. Microb. Cell Factories 5:10.1186/1475-2859-1185-1187. 10.1186/1475-2859-5-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agrios GN. 2003. Plant pathology, 5th ed. Elsevier Academic Press, Amsterdam, Netherlands [Google Scholar]
- 39.Snoeijers SS, Pérez-García A, Joostem MHAJ, De Wit PJGM. 2000. The effect of nitrogen on disease development and gene expression in bacterial and fungal pathogens. Eur. J. Plant Pathol. 106:493–506. 10.1023/A:1008720704105 [DOI] [Google Scholar]
- 40.Pate JS. 1980. Transport and partitioning of nitrogenous solutes. Annu. Rev. Plant Physiol. 31:313–340. 10.1146/annurev.pp.31.060180.001525 [DOI] [Google Scholar]
- 41.Bialczyk J, Lechowski Z. 1995. Chemical composition of xylem sap of tomato grown on bicarbonate containing medium. J. Plant Nutr. 18:2005–2021. 10.1080/01904169509365040 [DOI] [Google Scholar]
- 42.Dixon GR, Pegg GF. 1972. Changes in amino-acid content of tomato xylem sap following infection with strains of Verticillium albo-atrum. Ann. Bot. 36:147–154 [Google Scholar]
- 43.Senden MHMN, Van Der Meer AJGM, Limborgh J, Wolterbeek HT. 1992. Analysis of major tomato xylem organic acids and PITC-derivatives of amino acids by RP-HPLC and UV detection. Plant Soil 142:81–89 [Google Scholar]
- 44.Pegg GF. 1985. Life in a black hole: the microenvironment of the vascular pathogen. Trans. Br. Mycol. Soc. 85:1–20 [Google Scholar]
- 45.Alvarez B, Lopez MM, Biosca EG. 2008. Survival strategies and pathogenicity of Ralstonia solanacearum phylotype II subjected to prolonged starvation in environmental water microcosms. Microbiology 154:3590–3598. 10.1099/mic.0.2008/019448-0 [DOI] [PubMed] [Google Scholar]
- 46.Milling A, Meng F, Denny TP, Allen C. 2009. Interactions with hosts at cool temperatures, not cold tolerance, explain the unique epidemiology of Ralstonia solanacearum race 3 biovar 2. Phytopathology 99:1127–1134. 10.1094/PHYTO-99-10-1127 [DOI] [PubMed] [Google Scholar]
- 47.Graf J, Ruby EG. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc. Natl. Acad. Sci. U. S. A. 95:1818–1822. 10.1073/pnas.95.4.1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wattanaphon HT, Kerdsin A, Thammacharoen C, Sangvanich P, Vangnai AS. 2008. A biosurfactant from Burkholderia cenocepacia BSP3 and its enhancement of pesticide solubilization. J. Appl. Microbiol. 105:416–423. 10.1111/j.1365-2672.2008.03755.x [DOI] [PubMed] [Google Scholar]
- 49.Poutrel B, Gilbert FB, Lebrun M. 1995. Effects of culture conditions on production of type 5 capsular polysaccharide by human and bovine Staphylococcus aureus strains. Clin. Diagn. Lab. Immunol. 2:166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guimaraes AJ, Frases S, Cordero RJ, Nimrichter L, Casadevall A, Nosanchuk JD. 2010. Cryptococcus neoformans responds to mannitol by increasing capsule size in vitro and in vivo. Cell. Microbiol. 12:740–753. 10.1111/j.1462-5822.2010.01430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Quelas JI, Lopez-Garcia SL, Casabuono A, Althabegoiti MJ, Mongiardini EJ, Perez-Gimenez J, Couto A, Lodeiro AR. 2006. Effects of N-starvation and C-source on Bradyrhizobium japonicum exopolysaccharide production and composition, and bacterial infectivity to soybean roots. Arch. Microbiol. 186:119–128. 10.1007/s00203-006-0127-3 [DOI] [PubMed] [Google Scholar]
- 52.Orr D, Zheng W, Campbell BS, McDougall BM, Seviour RJ. 2009. Culture conditions affect the chemical composition of the exopolysaccharide synthesized by the fungus Aureobasidium pullulans. J. Appl. Microbiol. 107:691–698. 10.1111/j.1365-2672.2009.04247.x [DOI] [PubMed] [Google Scholar]
- 53.Vargas-Garcia MC, Lopez MJ, Elorrieta MA, Suarez F, Moreno J. 2001. Influence of nutritional and environmental factors on polysaccharide production by Azotobacter vinelandii cultured on 4-hydroxybenzoic acid. J. Ind. Microbiol. Biotechnol. 27:5–10. 10.1038/sj.jim.7000152 [DOI] [PubMed] [Google Scholar]
- 54.Filiatrault MJ, Wagner VE, Bushnell D, Haidaris CG, Iglewski BH, Passador L. 2005. Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect. Immun. 73:3764–3772. 10.1128/IAI.73.6.3764-3772.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jackson MA, Schisler DA. 1992. The composition and attributes of Colletotrichum spores are altered by the nutritional environment. Appl. Environ. Microbiol. 58:2260–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Santos AS, Sampaio AP, Vasquez GS, Santa Anna LM, Pereira NJ, Freire DM. 2002. Evaluation of different carbon and nitrogen sources in production of rhamnolipids by a strain of Pseudomonas aeruginosa. Appl. Biochem. Biotechnol. 98–100:1025–1035. 10.1385/ABAB:98-100:1-9:1025 [DOI] [PubMed] [Google Scholar]
- 57.Kang Y, Liu H, Genin S, Schell MA, Denny TP. 2002. Ralstonia solanacearum requires type 4 pili to adhere to multiple surfaces and for natural transformation and virulence. Mol. Microbiol. 46:427–437. 10.1046/j.1365-2958.2002.03187.x [DOI] [PubMed] [Google Scholar]