Abstract
Lipoteichoic acids (LTA) are polymers of alternating units of a polyhydroxy alkane, including glycerol and ribitol, and phosphoric acid, joined to form phosphodiester units that are found in the envelope of Gram-positive bacteria. Here we review four different types of LTA that can be distinguished on the basis of their chemical structure and describe recent advances in the biosynthesis pathway for type I LTA, d-alanylated polyglycerol-phosphate linked to di-glucosyl-diacylglycerol. The physiological functions of type I LTA are discussed in the context of inhibitors that block their synthesis and of mutants with discrete synthesis defects. Research on LTA structure and function represents a large frontier that has been investigated in only few Gram-positive bacteria.
INTRODUCTION
Aiming to reconstitute coenzyme A biosynthesis from extracts of Lactobacillus arabinocus, James Baddiley detected large amounts of soluble nucleotides that were identified as CDP-glycerol and CDP-ribitol (1, 2). Earlier work had revealed that nucleotide-linked sugars, for example, UDP-glucose and UDP-galactose, contribute to metabolism and are polymerized into polysaccharide (3–5). Baddiley proposed that CDP-glycerol and CDP-ribitol contribute to the synthesis of polyglycerol-phosphate [poly(Gro-P)] and polyribitol-phosphate. Large amounts of these polymers could indeed be isolated from the cell walls of Gram-positive bacteria but not from Gram-negative microbes (6). These phosphate-containing polymers harbored glycosyl and d-alanine ester substituents and were eventually designated teichoic acids (7).
Due to their supramolecular structure, the murein sacculi (cell wall peptidoglycan) in extracts of mechanically broken Gram-positive bacteria sediment and teichoic acids associated with such preparations were named wall teichoic acid (WTA) (8). Poly(Gro-P) in the supernatant, when centrifuged, associates with membranes, which explains the designations intracellular teichoic acid (9), membrane teichoic acid (10, 11), and, once the linkage of poly(Gro-P) to glycolipid was revealed, lipoteichoic acid (LTA) (12, 13). The characterization of the structure of teichoic acids and their nucleotide precursors represents a significant achievement at a time when sophisticated technologies such as mass spectrometry or nuclear magnetic resonance (NMR) were not available.
Although initially not appreciated as WTA and LTA, these molecules were identified independently in extracts of Streptococcus pneumoniae as immune-stimulatory compounds. Immunochemical studies had distinguished the somatic antigen fraction C (pneumococcal C-polysaccharide) (14) and a species-specific lipocarbohydrate, also designated heterophile antigen or pneumococcal F-antigen because of its Forssman antigenicity and fatty acid content (15). In contrast to highly variable capsular polysaccharides, the C-polysaccharide and lipocarbohydrate are conserved in strains of S. pneumoniae (15–17). Chemical studies eventually characterized C-polysaccharide as the WTA (15, 18–20) and lipocarbohydrate as the LTA (21) of pneumococci. Unlike other bacterial species, S. pneumoniae incorporates polymeric repeats with identical chemical compositions in both WTA and LTA (17, 22). While a biosynthesis pathway for pneumococcal WTA has been proposed, the synthesis of pneumococcal LTA has not yet been elucidated (see reference 23 for a review).
STRUCTURAL BASIS OF LTA TYPES
Werner Fischer used the generic term “amphiphile” to define polymeric chains associated with bacterial membranes and described five structural types (I to V) (24). The basic structure of LTA consists of a soluble polymer that is tethered to a membrane anchor and faces the outer leaflet of the plasma membrane. The polymer is formed of alternating units of a polyhydroxy alkane, including glycerol and ribitol, joined via phosphodiester linkages. The repeating units are further modified, providing chemical diversity between various bacterial species. It should be noted that methods for the extraction of intact LTA and for the elucidation of its structure have evolved since the early discovery of LTA (12, 13, 25, 26). These advances revised the structural models for some LTA molecules (27–30).
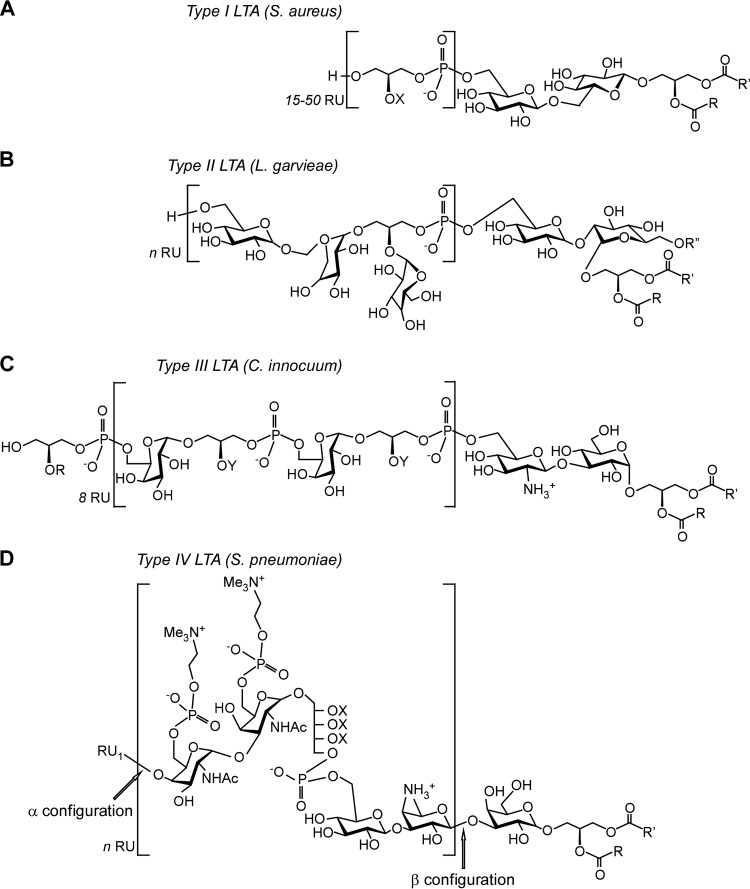
Type I LTA, the most frequently encountered polymer, displays the simple structure poly(Gro-P). Chemical diversity in type I LTA is based on (i) the chemical nature of substituents decorating Gro-P subunits, (ii) the length of the polymer, and (iii) the nature of the glycolipid anchor in the membrane. Type I LTA of Firmicutes (including Staphylococcus aureus, Bacillus subtilis, Enterococcus faecalis, Listeria monocytogenes, Streptococcus agalactiae, and Streptococcus pyogenes) is composed of 1,3-polyglycerolphosphate [poly(Gro-P)]. In S. aureus and B. subitlis, it is linked to the C-6 of the nonreducing glycosyl within the glycolipid anchor gentiobiosyldiacylglycerol Glc(β1–6)Glc(β 1–3)-diacylglycerol [Glc2-DAG] (Fig. 1A) (27, 31, 32). The poly(Gro-P) chain encompasses between 15 and 50 Gro-P units (26). When staphylococci are grown in laboratory media, roughly 70% and 15% of the Gro-P moieties are substituted at position 2 with d-alanine esters and N-acetylglucosamine (GlcNAc), respectively (Fig. 1A) (27, 33, 34). Streptococcus sp. strain DSM 8747, although closely related to S. pneumoniae (type IV LTA), produces type I LTA with the rare glycolipid anchor 3-O-(β-d-galactofuranosyl)-1,2-DAG (35). In group B Streptococcus type III strains, the polyglycerophosphate chain of LTA is substituted only with d-alanine and a kojibiose links the chain to the membrane anchor, resulting in a Glc-α-1,2Glc-α-1-3-DAG (36). The dihexosyl-DAG (Hex2-DAG) appears to be the most common glycolipid anchor of type I LTA, although here too variations exist. For example, Lactobacillus gasseri JCM 1131 carries a tetrahexosylglycerol with either two or three fatty acid chains (37). Some species, including lactobacilli, streptococci, and listeria, may produce two variants of LTA where the glycolipid anchor may be substituted with acyl or phosphatidyl (25, 29, 38). For example, in lactobacilli the structure of the glycolipid anchor may be Glc(β1–6)Gal(α1–2)Glc(β1–3) DAG or Glc(β1–6)Gal(α1–2)6-O-acyl-6Glc(α1–3) DAG (25). The type I LTA of bacilli has been divided into two groups based on their side-chain substituents, α-GlcNAc in group A and α-Gal in group B (39). The fine structure of both type I LTA molecules of L. monocytogenes has recently been resolved by NMR and gas chromatography mass spectrometry, revealing variations in the specific fatty acid distributions between the two LTA molecules (29). Changes in temperature during bacterial growth presumably affect the production of one LTA molecule more than the other (29).
FIG 1.
Structure of LTA molecules from various bacteria. The repeating unit, RU, of each LTA type is indicated within the brackets; it is always linked to a glycolipid. Where known, n (the number of RUs) is indicated. (A) Structure of type I LTA from Staphylococcus aureus. The 1,3-polyglycerol-phosphate RUs are substituted at the C2 position (X) with hydrogen proton (∼15%), d-alanyl ester (∼70%), or N-acetylglucosamine (∼15%). (B) Structure of type II LTA from Lactococcus garvieae. (C) Structure of type III LTA from Clostridium innocuum. Gro-P in the RU can be substituted at the C2 position (Y) with hydrogen proton (∼25%), glucosamine (∼50%), or N-acetylglucosamine (∼50%). (D) Structure of type IV LTA of Streptococcus pneumoniae. The RU may be substituted with hydrogen, d-alanyl, or N-acetylglucosamine (X). Substituents R, R′, and R″ in the glycolipids may be alkyl or branched alkyl chains.
Type II and III LTA molecules contain repeat units of glycosylalditol-phosphate. This definition includes compounds where the repeating units encompass glycosyl residues (24). For example, Lactococcus garvieae and Clostridium innocuum elaborate type II and III LTA molecules with the repeating units (Gal-Gal-Gro-P)n and (Gal-Gro-P)n, respectively (Fig. 1B and C) (24, 40). In L. garvieae, the repeating unit is added to the disaccharide kojibiose linked to DAG. A third fatty acid chain modifies the 6-hydroxy group of the kojibiose. The glycerol moiety in the repeating unit is modified with Gal, and d-alanylation is not observed (40).
Type IV LTA refers to S. pneumoniae WTA and LTA which are substituted with choline (Cho). The structure of pneumococcal teichoic acid has been studied extensively (22, 41) and was recently revised (30). Its repeating unit consists of the pseudopentasaccharide 2-acetamido-4-amino-2,4,6-trideoxygalactose (AATGal), glucose (Glc), and ribitol-phosphate (Rib-P) followed by two N-acetylgalactosamine (GalNAc) moieties, both substituted in position O-6 with P-Cho (Fig. 1D). The terminal repeating unit can occur with or without 6-O-P-Cho substitution, and the hydroxyl groups of Rib-P can be substituted in nonstoichiometric amounts by d-Ala. The first repeating unit is β-linked to the sugar to which it is attached, in this case, the lipid anchor Glc(β 1–3)-diacylglycerol [Glc-DAG] (30). Additional repeating units in the polymer are α-linked to the precedent unit (30). The presence of the nonreducing terminal GalNAc-GalNAc and the common monoglucosyl-DAG provides the molecular basis for the Forssman antigenicity and the identity of the lipid anchor (41).
Type V LTA includes macroamphophiles such as lipoglycans, Gro-P-lipoglucogalactofuranan, and succinyl lipomannan from Bifidobacterium bifidum and Microccocus luteus (42, 43). According to Fischer's definition for LTA structure, these compounds are not composed of repeating phosphodiester-linked units (42, 43). Type V lipoglycans therefore represent polysaccharides attached to lipid (glycolipid or phosphatidylinositol). Fischer excluded the lipopolysaccharide of Gram-negative bacteria from his classification (24). Some Actinobacteria and Tenericutes contain type V lipoglycans as well as type I LTA (44, 45). Nevertheless, the majority of Firmicutes are thought to synthesize only a single type of LTA (24, 45). Lipoglycans are found abundantly in the envelope of acid-fast bacteria, for example, Mycobacterium tuberculosis, which otherwise lack canonical teichoic acids (46, 47).
EARLY INSIGHTS INTO THE SYNTHESIS OF TYPE I LTA
The relatively simple structure of type I LTA invited biochemical approaches to study its biosynthesis in several Gram-positive organisms. Steps involved in the synthesis of the two main building blocks, the glycolipid anchor and the poly(Gro-P) chain, could be distinguished, and modifications of the assembled polymer, i.e., d-alanylation, were attributed to a third group of catalysts. Early attempts at identifying genes involved in LTA synthesis proved confusing, as mutants did not distinguish between specific requirements for WTA and LTA synthesis.
Genetic approaches focused initially on characterizing temperature-sensitive B. subtilis mutants that were also resistant to phage infection or displayed alterations in bacterial shape (48–51). Young used the nomenclature gta and rod to describe mutations in genes regulating the glycosylation of teichoic acid and rod shape (48). Inability to maintain the rod shape under nonpermissive conditions was correlated with cell wall assembly defects, in particular, a decrease in envelope phosphate content and loss of TA enzyme activity in “particulate enzyme preparations” (see reference 52 for a review). Eventually, the tar and tag (teichoic acid ribose and glucose) genes, which are involved in WTA synthesis, were discovered (53). WTA synthesis is initiated on the murein linkage unit tethered to undecaprenol-phosphate, which is also the lipid carrier for peptidoglycan synthesis (54, 55). Nonetheless, several phage resistance alleles in gtaA, gtaB, and gtaC could not completely account for the loss of WTA glycosylation (48, 49, 56). Several decades later, it was discovered that gtaA encodes the enzyme TagE, which is responsible for transferring α-glucose from UDP-glucose to the C-2 position along the poly(Gro-P) polymer backbone of B. subtilis (57, 58). In contrast, pgcA (originally named gtaC or gtaE) and gtaB were shown to be involved in the synthesis of UDP-glucose (56, 59). UDP-glucose was shown to serve as a precursor for the synthesis of WTA in B. licheniformis (60) and as a precursor for the glycolipid anchor of B. subtilis LTA (61). Genetic studies in B. subtilis led Karamata and colleagues to propose that mutations in gtaB and gtaC (pgcA) affect the synthesis of both WTA and LTA (62) (Fig. 2); this model was later confirmed in S. aureus (63).
FIG 2.
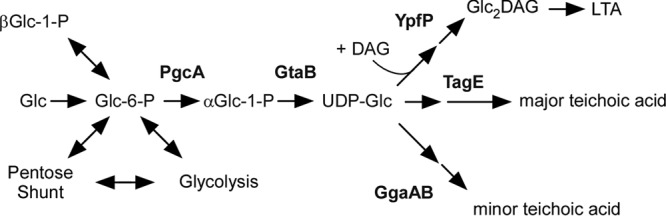
UDP-glucose contributes to wall teichoic acid and lipoteichoic acid synthesis. The diagram shows the flow of the UDP-glucose (UDP-Glc) pool in Bacillus subtilis and the key enzymes involved in synthesis of lipoteichoic acid (LTA) as well as minor and major wall teichoic acids (WTA) (adapted from reference 62). The major WTA from B. subtilis 168 is d-alanyl-[α-d-glycosylated poly(Gro-P)]. It is replaced with a phosphorus-free polysaccharide containing uronic acid residues (minor WTA) when B. subtilis is grown in phosphate-limited medium (89, 137). See the text for details.
The enzyme responsible for the synthesis of poly(Gro-P) was discovered only recently (64) even though its biochemical activity, substrate properties, and catalytic mechanism had been proposed for decades (65). It was initially assumed that type I LTA synthesis involves CDP-glycerol; however, in vivo pulse-labeling experiments revealed phosphatidylglycerol (PG) as a precursor for poly(Gro-P) synthesis (66–68). Careful characterization of lipid extracts from enterococci suggested that sn-glycerol 1-phosphate (Gro-P)-containing phosphoglycolipids contribute to LTA synthesis (69). Using membrane extracts from enterococci, Ganfield and Pieringer demonstrated that phosphatidylkojibiosyl DAG, not CDP-glycerol, functions as the acceptor of Gro-P moieties from PG (65). This study also demonstrated that the responsible enzymatic activity is membrane associated and that in vitro-synthesized poly(Gro-P) is not substituted with a glycolipid anchor (65). Elongation of the poly(Gro-P) moiety was examined in crude membrane preparations containing lipoteichoic acid-synthesizing activity and differential radioisotope labeling (70, 71). These experiments revealed that [14C]acetate appears successively in glucosyl-diacylglycerol (Glc-DAG), Glc2-DAG, and LTA, whereas [2-3H]glycerol is first incorporated into PG and then into glycolipid-anchored LTA (70–72). Stepwise degradation of pulse-labeled LTA with phosphodiesterase and phosphomonoesterase from the glycerol terminus suggested that the polymer grows in a manner distal to the lipid anchor (70, 71). These observations were incorporated into a unifying model whereby transfer of sn-glycerol 1-phosphate from PG to Glc2-DAG and subsequent stepwise addition of Gro-P at the distal end of polymerizing chains lead to LTA assembly (Fig. 3A). This model correctly predicted that LTA synthesis would require enzymes for poly(Gro-P) synthesis and for the transfer of Gro-P from PG to Glc2-DAG (73).
FIG 3.
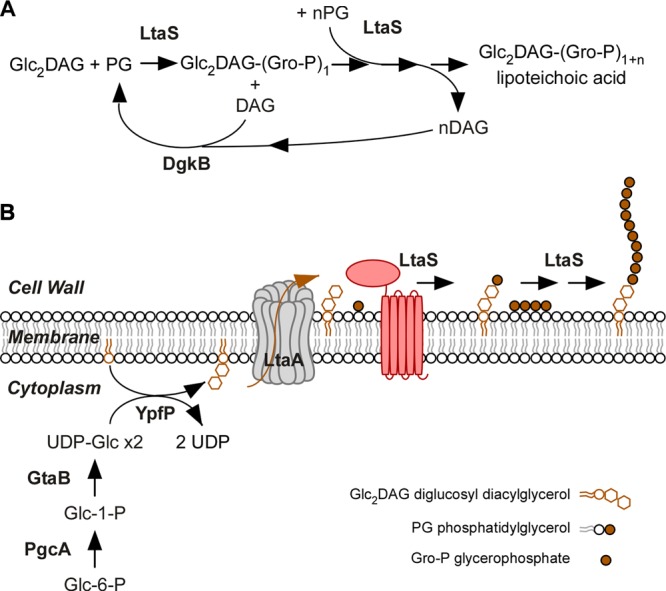
Synthesis of lipoteichoic acid in Staphylococcus aureus. (A) Diagram showing the reaction catalyzed by LtaS. The first glycerophosphate (Gro-P) subunit is cleaved from phosphatidyl glycerol (PG) and attached to the glycolipid anchor diglycosyl-diacylglycerol (Glc2DAG). This reaction leads to the release of DAG. Polymerization of Gro-P by LtaS occurs at the distal end of Glc2DAG-(Gro-P)1, utilizing additional PG molecules (n = ∼50). For some bacterial species, the first reaction is thought to require a specific primase (see the text for details). The DgkB enzyme is responsible for the recycling of DAG (see the text for details). (B) Diagram showing the biosynthesis of LTA in S. aureus. The enzymes PgcA, GtaB, and YpfP synthesize the Glc2DAG glycolipid anchor. LtaA, a 12-transmembrane domain protein (in gray), flips Glc2DAG across the plasma membrane. LtaS (red) spans the plasma membrane five times, and its C-terminal domain polymerizes the poly(Gro-P) chain on Glc2DAG on the trans side of the membrane.
LTA SYNTHESIS IN S. AUREUS
It is now appreciated that the synthesis of the type I LTA glycolipid anchor, Glc2-DAG, occurs via three enzymatic steps. First, α-phosphoglucomutase (PgcA) converts glucose-6-phosphate to glucose-1-phosphate (60, 62). Next, UTP:α-glucose-1-phosphate uridyl transferase (GtaB) converts glucose-1-phosphate to UDP glucose (UDP-Glc) (56, 59). Finally, the processive glycosyl-transferase YpfP strings two UDP-Glc molecules onto DAG to generate Glc2-DAG (Fig. 2 and 3) (61, 74, 75). Analysis of pgcA, gtaB, and ypfP mutations in S. aureus corroborated this model (63). Importantly, pgcA, gtaB, and ypfP mutants continue to synthesize poly(Gro-P); however, LTA is tethered to the membrane via a terminal DAG, instead of Glc2-DAG (63, 75, 76). In staphylococci, ypfP is located in an operon with a second gene, ltaA, which is conserved in several Firmicutes species (63). Mutants lacking ltaA synthesize (Gro-P) chains attached to DAG instead of Glc2-DAG, although Glc2-DAG continues to be synthesized and accumulates in the membrane (63). Bioinformatic predictions identify LtaA as a member of the major facilitator superfamily (77); this polytopic membrane protein has been proposed to translocate Glc2-DAG (63) (Fig. 3).
The enzyme responsible for the synthesis of poly(Gro-P) was discovered based on the model whereby PG is used as a substrate for stepwise addition of sn-glycerol 1-phosphate into poly(Gro-P)-polymerizing chains and transfer onto Glc2-DAG (Fig. 3A) (73). As the membranes of Escherichia coli harbor PG and Glc2-DAG but lack poly(Gro-P), it seemed plausible that expression of the gene for LTA synthesis in this organism could lead to the formation of poly(Gro-P). A plasmid library of staphylococcal genomic DNA fragments was introduced into E. coli, and several hundred clones were screened by immunoblotting of bacterial extracts with LTA [poly(Gro-P)]-specific monoclonal antibody (64). This approach identified ltaS (64), which encodes a polytopic membrane protein with a large C-terminal domain on the outer surface of the bacterial membrane, annotated in the Pfam database as a sulfatase domain (www.sanger.ac.uk/Software/Pfam; pfam00884) (64) (Fig. 3B). The protein was named LTA synthase (LtaS); mutations in the corresponding S. aureus ltaS gene cause severe cell division defects and loss of viability (64, 78).
LtaS proteins contain hydrophobic domains predicted to span the plasma membrane five times with short intervening loops (64) (Fig. 3B). The fifth hydrophobic segment is followed by a predicted signal peptidase I recognition motif that has been experimentally validated for S. aureus LtaS (79). The three-dimensional structures of the extracellular catalytic domain of S. aureus and B. subtilis LtaS (eLtaS) have been determined (80, 81). Overall, eLtaS assumes a sulfatase-like fold but with discrete changes in the active site (80). In S. aureus eLtaS, Thr300 (T300), together with E255, D475, and H476, coordinates a manganese ion and forms the binding pocket for Gro-P (80). Catalysis has been proposed to involve PG docking in the active site of eLtaS to enable the nucleophilic attack from the deprotonated hydroxyl of T300, thereby generating the Gro-P-threonine intermediate and releasing DAG. The Gro-P-threonine intermediate may subsequently be resolved by the nucleophilic attack of the terminal OH group of PG to extend the LTA chain (80).
LTA SYNTHESIS IN OTHER GRAM-POSITIVE BACTERIA
The chemical structure of type I LTA differs for various bacterial species, and the enzymes responsible for their synthesis and translocation and for polymerization of both the glycolipid anchor and the hydrophilic polymer must therefore also differ. In S. aureus, synthesis of Glc2-DAG-linked LTA requires PgcA, GtaB, LtaA, and YpfP (32, 82); however, ltaA is not found in the genome of B. subtilis (61, 83). In L. monocytogenes, YpfP is replaced with two glycosyltransferases, LafA and LafB (LTA anchor formation protein), for the synthesis of the more complex GalGlc-DAG anchor and the transmembrane LafC protein is required to flip this glycolipid across the membrane (84). Pathways similar to the glycolipid anchor synthesis pathway in L. monocytogenes are thought to exist in E. faecalis, S. agalactiae, and S. pneumoniae (83). The function of LtaS homologues has been characterized in several Gram-positive bacteria, including B. subtilis, L. monocytogenes, and B. anthracis. These investigations confirmed that proteins with a predicted sulfatase domain (pfam00884 family) are indeed responsible for the polymerization of poly(Gro-P) and the anchoring of LTA to the membrane (81, 84–86). B. subtilis and B. anthracis encode four LtaS homologues, and genetic deletion of the corresponding genes suggested an overlap in activity or even redundancy (see below) (64, 81, 85, 86). L. monocytogenes was found to encode two LtaS enzymes, and it was proposed that one of the enzymes, designated LTA primase, adds a single Gro-P moiety to the glycolipid anchor to initiate chain polymerization (84). It has been suggested that B. subtilis YvgJ may catalyze a similar reaction (85). Nevertheless, LTA primase is not required for the synthesis of glycolipid-linked LTA in either B. subtilis or L. monocytogenes (84, 85) and its molecular function may not yet be fully appreciated. These data can be incorporated into a predictive model whereby microbes expressing an ltaS-like gene are likely to synthesize a polyglycerol-containing LTA. S. pneumoniae with its more complex polymer lacks an ltaS orthologue. Here, synthesis of WTA and type IV LTA has been proposed to involve cytoplasmic CDP-ribitol polymerized on a lipid carrier that is subsequently flipped across the membrane for delivery to a peptidoglycan ligase or glycolipid transferase (41, 87, 88).
LTA d-ALANYLATION
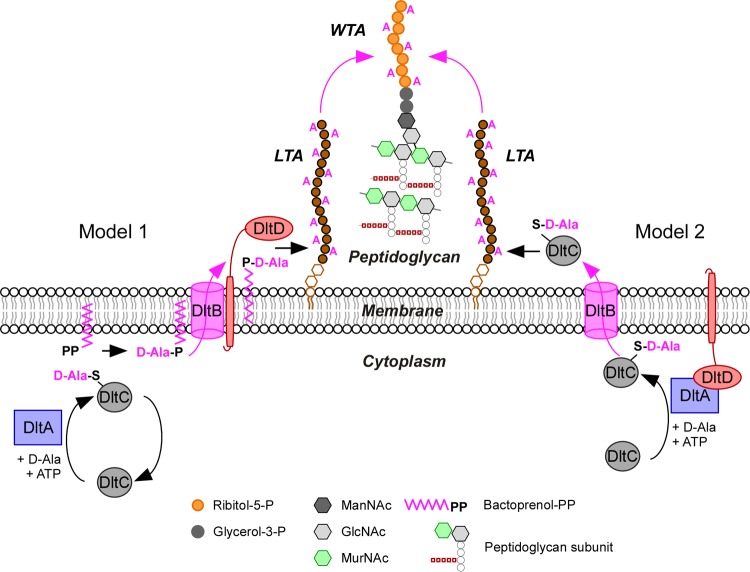
Type I LTA is typically substituted with d-alanine esters, which confer positive charges onto an otherwise negatively charged polymer (89). Studying extracts of enterococci, Baddiley and Neuhaus detected an enzyme activity that activated d-alanine in the presence of ATP (90). In addition to this 56-kDa activating enzyme, a 6-kDa heat-stable protein, membranes, ATP, and d-alanine were required for the d-alanylation of LTA (89). d-Alanine activating ligase (Dcl) is a member of the family of proteins that activates amino acids or fatty acids and which functions here to ligate activated d-alanine to the 4′-phosphopantetheine prosthetic group of Dcp, the 6-kDa d-alanyl carrier protein (Fig. 4) (91, 92). Francis Neuhaus and colleagues identified the genetic determinants for Dcl and Dcp, which were designated dltA and dltC, respectively. Both genes are located in an operon along with two other determinants, dltB and dltD (93). In S. aureus, this operon consists of five genes, dltXABCD (94). Although also found in other species, the function of dltX is still unknown. In B. subtilis, each gene of the dltABCD operon is required for the d-alanylation of LTA (94, 95). Two models have been proposed to account for the contributions of DltB and DltD. DltB spans the plasma membrane several times, and DltD appears to be anchored to the membrane via an N-terminal hydrophobic sequence. In the first model, DltB transfers d-alanine from DltC to undecaprenol-phosphate and flips the lipid-linked intermediate across the membrane, whereas DltD, acting on the trans side of the membrane, transfers d-alanine to LTA (Fig. 4; model 1) (95). In the second model, DltD promotes transfer of d-alanine between DltA and DltC in the cytoplasm. Alanylated DltC is translocated across the membrane by DltB and may then transfer d-alanine directly onto LTA (Fig. 4; model 2) (89). An analysis of DltD membrane topology suggests that the protein may reside outside the cytoplasm, on the trans side of the membrane (96), as predicted by the first model (Fig. 4; model 1). Nevertheless, experimental support for this model lacks a demonstration that undecaprenol-phosphate or its synthetic intermediates indeed contribute to LTA d-alanylation and transport of d-alanyl across the membrane. Of note, the Dlt system appears to d-alanylate LTA but not WTA. Both in vitro and in vivo pulse-chase experiments suggest that d-alanyl esters are transferred from LTA to WTA (97, 98). If so, one wonders whether d-alanylation of WTA requires a dedicated catalyst or whether catalysis is provided by d-alanylated LTA (Fig. 4).
FIG 4.
Models for d-alanylation of teichoic acids. DltA (Dcl), DltB, DltC (Dcp), and DltD are required for the d-alanylation of LTA, and two models have been proposed to explain the molecular basis. Model 1 proposes that the combined activity of DltA and DltC in the cytoplasm results in the transfer of d-Ala onto the lipid carrier bactoprenol pyrophosphate (PP) (95). Next, DltB flips d-Ala-bactoprenol-PP across the plasma membrane. It is unclear whether DltD assists the DltB flippase or contributes to d-Ala transfer onto LTA (96). In model 2, DltD facing the cytoplasm assists DltA for the loading of d-Ala onto the carrier protein DltC. Next, DltC-S-d-Ala is translocated across the membrane by DltB and transfers d-Ala onto LTA (89). d-Alanylation of WTA is not thought to require catalysis. Presumably, d-Ala moieties of LTA are transferred onto WTA. MurNAc, N-acetylmuramic acid; GlcNAc, N-acetylglucosamine; ManNAc, N-acetylmannosamine.
TEICHOIC ACIDS AND BACTERIAL GROWTH
It had long been presumed that WTA and LTA synthesis is essential for bacterial growth (89, 99, 100). However, genes for enzymes that catalyze the first two steps in the WTA synthesis, tagO and tagA, can be deleted in B. subtilis or S. aureus without abolishing growth (101–103). In contrast, genes for enzymes that function downstream in the WTA synthesis cannot be deleted unless tagA or tagO is also mutated (102). A model has been developed to explain this synthetic viability phenotype: the accumulation of WTA synthesis intermediates limits the availability of undecaprenyl-phosphate for peptidoglycan synthesis, which is essential for bacterial growth (101). Thus, strictly speaking, WTA is not required for bacterial growth albeit S. aureus cells lacking WTA are enlarged and do not divide in the same manner as wild-type cells (see below). The genetic requirements for LTA are best discussed for each of its building blocks. For example, glycolipid synthesis enzymes such as those encoded by pgcA, gtaB, and ypfP are dispensable for S. aureus growth even though the mutants display morphological alterations such as increased size and aberrant cell shape (63, 75). These pleiotropic phenotypes could be attributed to the possibility that UDP-glucose may be perceived as a metabolic signal, coupling cell division with increased cell mass (104, 105). In B. subtilis, UgtP (YpfP) has been shown to inhibit FtsZ assembly (106). ypfP is dispensable for LTA synthesis, as S. aureus ypfP mutants anchor the polymer onto DAG, instead of Glc2-DAG; nevertheless, LTA production is greatly reduced in ltaS mutants (63, 75, 76).
Reduced ltaS expression or chemical inhibition of the ltaS gene product results in severe cell division defects, a phenotype that is exacerbated when staphylococci are grown under conditions above 30°C (64, 78, 107). Some organisms carry more than one ltaS-like gene. For example, B. subtilis and B. anthracis carry four ltaS genes. The B. subtilis yflE mutant grows very slowly, which earned YflE the designation of a housekeeping LTA synthase (81, 85). Two housekeeping LtaS enzymes were identified in B. anthracis, and deletion of their corresponding genes, ltaS1 and ltaS2, resulted in a three-log reduction in plating efficiency and in an inability to sporulate (86). Deletion of all four ltaS genes in B. anthracis could not be achieved (86). A mutant lacking all four paralogues of ltaS in B. subtilis remained viable albeit with severe morphological defects in cell structure and filament formation (81, 85). Further, B. subtilis does not tolerate the simultaneous deletion of tagO (WTA synthesis) and ltaS (LTA synthesis) (81). Deletion of the two ltaS genes in L. monocytogenes reduced bacterial plating efficiency by more than 6-log (81, 84–86). By screening a library of 167,405 compounds for inhibition of S. aureus growth at 42°C, compound 1771 was identified as an inhibitor of LTA synthesis (107). The spectrum of antibiotic activity for compound 1771 was analyzed against several Gram-positive bacteria harboring poly(Gro-P) LTA as well as LtaS homologues. Compound 1771 inhibited the growth of antibiotic-resistant methicillin-resistant S. aureus (MRSA), including USA300 LAC, and VRE, i.e., vancomycin-resistant Enterococcus faecalis and E. faecium, whose genomes harbor two ltaS homologues. Gram-positive bacteria with three ltaS homologues (Clostridium perfringens) or four ltaS homologues (B. cereus and B. anthracis) were also susceptible to compound 1771-mediated growth inhibition (107). Scanning and thin-section transmission electron microscopy of bacterial cultures treated with subinhibitory concentrations of 1771 revealed dispersal of bacterial cluster or chain formation, increases in cell size, deformation of cell surface and shape, thickening, and structural disorganization of the cell wall envelope (107). Thus, LTA synthesis, but not WTA, may be essential for growth in several different Gram-positive bacteria and chemical inhibition may be used to assess the presence of LtaS enzymes and type I LTA in less-well-characterized organisms.
Genetic suppression analyses also suggest a role of LTA and its building blocks in metabolism. For example, the B. subtilis housekeeping ltaS gene yflE was identified in a screen for suppressors of the Mg2+-dependent growth defect of mbl mutants, a gene otherwise required for elongation of rod-shaped bacteria (81). Another investigation revealed that disruption of B. subtilis yflE or yfnI suppresses the lethality caused by dgkB repression (108). DAG is phosphorylated by diacylglycerol (diglyceride) kinase (DgkB) and thereby shunted into phosphatidylglycerol synthesis (Fig. 3A) (71). LtaS enzymes hydrolyze PG, a process that results in polymerization of poly(Gro-P) and formation of the byproduct DAG. The loss of viability of dgkB mutants has been attributed to accumulation of DAG, and it seems plausible that mutations in ltaS can alleviate this phenotype (108). In S. aureus, the growth defect caused by loss of LTA in an ltaS mutant could be rescued by inactivation of gdpP (109), a phosphodiesterase that hydrolyzes the essential signaling molecule cyclic-di-AMP (110). Increased cyclic-di-AMP in the gdpP mutant was shown to be associated with increased peptidoglycan cross-linking, which could somehow compensate for the lack of LTA (109). A receptor(s) for cyclic-di-AMP, i.e., the potassium transporters Kdp and Ktr, was identified, which suggests a role for the second messenger in ion transport, maintenance of cytoplasmic pH, and osmotic homeostasis in S. aureus (111, 112).
OTHER FUNCTIONS OF TEICHOIC ACIDS
Teichoic acids are zwitterionic molecules, and LTA d-alanylation is known to modulate ion hemostasis (89). d-Alanylation of LTA has been proposed to promote Mg2+ ion scavenging and to target autolysins to discrete locations in the bacterial envelope, which is essential for the separation of peptidoglycan in dividing cells (113, 114). d-Alanylated LTA may restrict the activity of autolysins and cell wall active antibiotics, thereby maintaining the integrity of the bacterial envelope (115, 116) (also reviewed in references 54 and 89). Modifications of TA polymers with d-alanyl esters or N-acetylglucosamine are also important for escape from innate immune defenses such as host antimicrobial peptides (117). In B. subtilis, d-alanylation-deficient mutants restore the protein secretion defect associated with mutations in prsA, which encodes a secretion chaperone (118). Presumably, an increased anionic charge through the increased binding of Ca2+ and Mg2+ to teichoic acids can stabilize and fold proteins in the absence of PrsA (118).
Other LTA types are not thought to contain d-alanine modifications. Instead, type IV LTA of S. pneumoniae is extensively modified with choline, a highly unusual component for bacteria (119). LTA choline substituents are essential for the deposition of LytB glucosaminidase and LytA autolysin at the septum of dividing pneumococci, indicating that S. pneumoniae teichoic acids contribute to cell separation and that choline may play a role similar to that of d-alanylation (120).
LTA has been shown to bind the GW modules of internalin B of L. monocytogenes (121). Each GW module is approximately 80 amino acids long, with a highly conserved glycine-tryptophan dipeptide (121). The GW module is also present in staphylococcal autolysins AtlA and AtlE from S. aureus and S. epidermidis (122, 123). A structural model for their interaction with LTA was recently proposed (124). Because autolysins contribute to cell division and biofilm formation (123), it is not surprising that bacteria with reduced amounts of LTA display pleiotropic phenotypes (76).
LTA may play an important role in bacterial infectious diseases, as the polymer has been proposed to activate the immune system of infected hosts and promote inflammation (125–127). It was first proposed that LTA triggers innate immune responses by activating CD14 and Toll-like receptor (TLR) ligands (reviewed in references 128, 129, and 130). However, this model has been recently challenged with the argument that LTA extracted from bacteria may be contaminated with lipoproteins and peptidoglycan, compounds that are well known for their immune-modulatory activities (131, 132). The total chemical synthesis of various LTA types has provided valuable information for assessing their immunostimulatory attributes, as this technology can eliminate the concern of contamination with other cell wall compounds. Further, chemical synthesis of LTA building blocks affords the opportunity of assessing individual contributions of the glycolipid anchor, polymer repeat, and polymer substituents for immune system stimulation (36, 40, 132). These approaches have confirmed that LTA from several different bacterial species may be endowed with inflammatory activity, albeit Toll-like receptors (TLRs), in particular, TLR2, may not be involved in LTA recognition (40). Nevertheless, it seems unlikely that type I LTA displays an immune-stimulatory function that is as potent as that observed for lipoproteins (131, 133) or lipopolysaccharides (132).
CONCLUSIONS AND PERSPECTIVES
In their landmark review, Neuhaus and Baddiley detailed the discovery, synthesis, and functions of teichoic acids, emphasizing the importance of LTA as a polyelectrolyte that forms a “continuum of anionic charges” between the plasma membrane and the extracellular milieu (89). To date, the molecular mechanisms whereby teichoic acids can contribute to the integrity of the bacterial cell wall envelope or to bacterial growth have remained poorly understood. Nonetheless, we have learned that type I LTA is positioned at the crossroads of several biosynthesis pathways that are perturbed by the accumulation or depletion of metabolites such as DAG, lipids, UDP-Glc, and cyclic-di-AMP (105, 108, 109). Only few of the enzymes that are involved in the synthesis of type I LTA synthesis have been examined in detail, and this gap needs to be addressed. Further, while we begin to appreciate the function of LtaS, it is not clear why some microbes synthesize LTA with only one type of synthesis enzyme whereas others employ multiple different LtaS enzymes. Both type I LTA and WTA contribute positional information for murein hydrolases that must act on peptidoglycan during bacterial growth and cell division (116, 124, 134). This cell biological function, i.e., the markings of new and old peptidoglycan or of discrete subcellular sites, may explain the essential contributions of teichoic acids to bacterial growth. Technological advances such as NMR and mass spectrometry have provided the means of identifying and characterizing the structures of new amphiphiles from Gram-positive bacteria (44, 135). Yet we remain unable to build models for the synthesis of many different types of LTA molecules. To quote Neuhaus and Baddiley, it seems that indeed “there may be no clear beginning and certainly there is no clear end to the cell surface…” of Gram-positive bacteria (89, 136).
ACKNOWLEDGMENTS
Research on LTA synthesis and inhibition in the laboratories of O.S. and D.M. is supported by the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIH Award 1-U54-AI-057153).
Footnotes
Published ahead of print 10 January 2014
REFERENCES
- 1.Baddiley J, Buchanan JG, Carss B, Mathias AP. 1956. Cytidine diphosphate ribitol. Biochim. Biophys. Acta 21:191–192. 10.1016/0006-3002(56)90123-8 [DOI] [PubMed] [Google Scholar]
- 2.Baddiley J, Buchanan JG, Carss B, Mathias AP, Sanderson AR. 1956. The isolation of cytidine diphosphate glycerol, cytidine diphosphate ribitol and mannitol 1-phosphate from Lactobacillus arabinosus. Biochem. J. 64:599–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leloir LF. 1951. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch. Biochem. Biophys. 33:186–190. 10.1016/0003-9861(51)90096-3 [DOI] [PubMed] [Google Scholar]
- 4.Glaser L. 1957. The enzymic synthesis of cellulose by Acetobacter xylinum. Biochim. Biophys. Acta 25:436. 10.1016/0006-3002(57)90501-2 [DOI] [PubMed] [Google Scholar]
- 5.Leloir LF, Cardini CE. 1957. Biosynthesis of glycogen from uridine diphosphate glucose. J. Am. Chem. Soc. 79:6340–6341. 10.1021/ja01580a061 [DOI] [Google Scholar]
- 6.Baddiley J. 1989. Bacterial cell walls and membranes. Discovery of the teichoic acids. Bioessays 10:207–210 [DOI] [PubMed] [Google Scholar]
- 7.Armstrong JJ, Baddiley J, Buchanan JG, Carss B, Greenberg GR. 1958. Isolation and structure of ribitol phosphate derivatives (teichoic acids) from bacterial cell walls. J. Chem. Soc. 1958:4344–4354. 10.1039/JR9580004344 [DOI] [Google Scholar]
- 8.Salton MR, Horne RW. 1951. Studies of the bacterial cell wall. II. Methods of preparation and some properties of cell walls. Biochim. Biophys. Acta 7:177–197 [DOI] [PubMed] [Google Scholar]
- 9.Kelemen MV, Baddiley J. 1961. Structure of the intracellular glycerol teichoic acid from Lactobacillus casei A.T.C.C. 7469. Biochem. J. 80:246–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hay JB, Wicken AJ, Baddiley J. 1963. The location of intracellular teichoic acids. Biochim. Biophys. Acta 71:188–190. 10.1016/0006-3002(63)90999-5 [DOI] [PubMed] [Google Scholar]
- 11.Shockman GD, Slade HD. 1964. The cellular location of the streptococcal group D antigen. J. Gen. Microbiol. 37:297–305. 10.1099/00221287-37-3-297 [DOI] [PubMed] [Google Scholar]
- 12.Wicken AJ, Knox KW. 1970. Studies on the group F antigen of lactobacilli: isolation of a teichoic acid-lipid complex from Lactobacillus fermenti NCTC 6991. J. Gen. Microbiol. 60:293–301. 10.1099/00221287-60-3-293 [DOI] [PubMed] [Google Scholar]
- 13.Toon P, Brown PE, Baddiley J. 1972. The lipid-teichoic acid complex in the cytoplasmic membrane of Streptococcus faecalis N.C.I.B. 8191. Biochem. J. 127:399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tillett WS, Francis T. 1930. Serological reactions in pneumonia with a non-protein somatic fraction of pneumococcus. J. Exp. Med. 52:561–571. 10.1084/jem.52.4.561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goebel WF, Adams MH. 1943. The immunological properties of the heterophile antigen and somatic polysaccharide of pneumococcus. J. Exp. Med. 77:435–449. 10.1084/jem.77.5.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goebel WF, Shedlovsky T, Lavin GI, Adams MH. 1943. The heterophile antigen of pneumococcus. J. Biol. Chem. 148:1–15 http://www.jbc.org/content/148/1/1.citation [Google Scholar]
- 17.Sørensen UB, Henrichsen J. 1987. Cross-reactions between pneumococci and other streptococci due to C polysaccharide and F antigen. J. Clin. Microbiol. 25:1854–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotschlich EC, Liu TY. 1967. Structural and immunological studies on the pneumococcal C polysaccharide. J. Biol. Chem. 242:463–470 [PubMed] [Google Scholar]
- 19.Brundish DE, Baddiley J. 1967. The characterization of pneumococcal C-polysaccharide as a ribitol teichoic acid. Biochem. J. 105:30C–31C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poxton IR, Tarelli E, Baddiley J. 1978. The structure of C-polysaccharide from the walls of Streptococcus pneumoniae. Biochem. J. 175:1033–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Briles EB, Tomasz A. 1973. Pneumococcal Forssman antigen. A choline-containing lipoteichoic acid. J. Biol. Chem. 248:6394–6397 [PubMed] [Google Scholar]
- 22.Fischer W. 1997. Pneumococcal lipoteichoic and teichoic acid. Microb. Drug Resist. 3:309–325. 10.1089/mdr.1997.3.309 [DOI] [PubMed] [Google Scholar]
- 23.Denapaite D, Bruckner R, Hakenbeck R, Vollmer W. 2012. Biosynthesis of teichoic acids in Streptococcus pneumoniae and closely related species: lessons from genomes. Microb. Drug Resist. 18:344–358. 10.1089/mdr.2012.0026 [DOI] [PubMed] [Google Scholar]
- 24.Fischer W. 1994. Lipoteichoic acids and lipoglycans, p 199–215 In Ghuysen JM, Hakenbeck R. (ed), New comprehensive biochemistry, vol 27 Elsevier Science, Amsterdam, The Netherlands [Google Scholar]
- 25.Fischer W, Koch HU, Haas R. 1983. Improved preparation of lipoteichoic acids. Eur. J. Biochem. 133:523–530. 10.1111/j.1432-1033.1983.tb07495.x [DOI] [PubMed] [Google Scholar]
- 26.Fischer W. 1988. Physiology of lipoteichoic acids in bacteria. Adv. Microb. Physiol. 29:233–302. 10.1016/S0065-2911(08)60349-5 [DOI] [PubMed] [Google Scholar]
- 27.Morath S, Geyer A, Hartung T. 2001. Structure-function relationship of cytokine induction by lipoteichoic acid from Staphylococcus aureus. J. Exp. Med. 193:393–397. 10.1084/jem.193.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehner MD, Morath S, Michelsen KS, Schumann RR, Hartung T. 2001. Induction of cross-tolerance by lipopolysaccharide and highly purified lipoteichoic acid via different Toll-like receptors independent of paracrine mediators. J. Immunol. 166:5161–5167 [DOI] [PubMed] [Google Scholar]
- 29.Dehus O, Pfitzenmaier M, Stuebs G, Fischer N, Schwaeble W, Morath S, Hartung T, Geyer A, Hermann C. 2011. Growth temperature-dependent expression of structural variants of Listeria monocytogenes lipoteichoic acid. Immunobiology 216:24–31. 10.1016/j.imbio.2010.03.008 [DOI] [PubMed] [Google Scholar]
- 30.Gisch N, Kohler T, Ulmer AJ, Muthing J, Pribyl T, Fischer K, Lindner B, Hammerschmidt S, Zahringer U. 2013. Structural reevaluation of Streptococcus pneumoniae lipoteichoic acid and new insights into its immunostimulatory potency. J. Biol. Chem. 288:15654–15667. 10.1074/jbc.M112.446963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morath S, Geyer A, Spreitzer I, Hermann C, Hartung T. 2002. Structural decomposition and heterogeneity of commercial lipoteichoic acid preparations. Infect. Immun. 70:938–944. 10.1128/IAI.70.2.938-944.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duckworth M, Archibald AR, Baddiley J. 1975. Lipoteichoic acid and lipoteichoic acid carrier in Staphylococcus aureus H. FEBS Lett. 53:176–179. 10.1016/0014-5793(75)80013-5 [DOI] [PubMed] [Google Scholar]
- 33.Fischer W, Rosel P. 1980. The alanine ester substitution of lipoteichoic acid (LTA) in Staphylococcus aureus. FEBS Lett. 119:224–226. 10.1016/0014-5793(80)80257-2 [DOI] [PubMed] [Google Scholar]
- 34.Rajbhandary UL, Baddiley J. 1963. The intracellular teichoic acid from Staphylococcus aureus H. Biochem. J. 87:429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roethlisberger P, Iida-Tanaka N, Hollemeyer K, Heinzle E, Ishizuka I, Fischer W. 2000. Unique poly(glycerophosphate) lipoteichoic acid and the glycolipids of a Streptococcus sp. closely related to Streptococcus pneumoniae. Eur. J. Biochem. 267:5520–5530. 10.1046/j.1432-1327.2000.01613.x [DOI] [PubMed] [Google Scholar]
- 36.Henneke P, Morath S, Uematsu S, Weichert S, Pfitzenmaier M, Takeuchi O, Muller A, Poyart C, Akira S, Berner R, Teti G, Geyer A, Hartung T, Trieu-Cuot P, Kasper DL, Golenbock DT. 2005. Role of lipoteichoic acid in the phagocyte response to group B streptococcus. J. Immunol. 174:6449–6455 [DOI] [PubMed] [Google Scholar]
- 37.Shiraishi T, Yokota S, Morita N, Fukiya S, Tomita S, Tanaka N, Okada S, Yokota A. 2013. Characterization of a Lactobacillus gasseri JCM 1131T lipoteichoic acid with a novel glycolipid anchor structure. Appl. Environ. Microbiol. 79:3315–3318. 10.1128/AEM.00243-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uchikawa K, Sekikawa I, Azuma I. 1986. Structural studies on lipoteichoic acids from four Listeria strains. J. Bacteriol. 168:115–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwasaki H, Shimada A, Ito E. 1986. Comparative studies of lipoteichoic acids from several Bacillus strains. J. Bacteriol. 167:508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt RR, Pedersen CM, Qiao Y, Zahringer U. 2011. Chemical synthesis of bacterial lipoteichoic acids: an insight on its biological significance. Org. Biomol. Chem. 9:2040–2052. 10.1039/c0ob00794c [DOI] [PubMed] [Google Scholar]
- 41.Seo HS, Cartee RT, Pritchard DG, Nahm MH. 2008. A new model of pneumococcal lipoteichoic acid structure resolves biochemical, biosynthetic, and serologic inconsistencies of the current model. J. Bacteriol. 190:2379–2387. 10.1128/JB.01795-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer W. 1987. ‘Lipoteichoic acid’ of Bifidobacterium bifidum subspecies pennsylvanicum DSM 20239. A lipoglycan with monoglycerophosphate side chains. Eur. J. Biochem. 165:639–646 [DOI] [PubMed] [Google Scholar]
- 43.Fischer W. 1991. One-step purification of bacterial lipid macroamphiphiles by hydrophobic interaction chromatography. Anal. Biochem. 194:353–358. 10.1016/0003-2697(91)90240-T [DOI] [PubMed] [Google Scholar]
- 44.Rahman O, Pfitzenmaier M, Pester O, Morath S, Cummings SP, Hartung T, Sutcliffe IC. 2009. Macroamphiphilic components of thermophilic actinomycetes: identification of lipoteichoic acid in Thermobifida fusca. J. Bacteriol. 191:152–160. 10.1128/JB.01105-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahman O, Dover LG, Sutcliffe IC. 2009. Lipoteichoic acid biosynthesis: two steps forwards, one step sideways? Trends Microbiol. 17:219–225. 10.1016/j.tim.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 46.Berg S, Kaur D, Jackson M, Brennan PJ. 2007. The glycosyltransferases of Mycobacterium tuberculosis - roles in the synthesis of arabinogalactan, lipoarabinomannan, and other glycoconjugates. Glycobiology 17:35R–56R. 10.1093/glycob/cwm010 [DOI] [PubMed] [Google Scholar]
- 47.Mishra AK, Driessen NN, Appelmelk BJ, Besra GS. 2011. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol. Rev. 35:1126–1157. 10.1111/j.1574-6976.2011.00276.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young FE. 1967. Requirement of glucosylated teichoic acid for adsorption of phage in Bacillus subtilis 168. Proc. Natl. Acad. Sci. U. S. A. 58:2377–2384. 10.1073/pnas.58.6.2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young FE, Smith C, Reilly BE. 1969. Chromosomal location of genes regulating resistance to bacteriophage in Bacillus subtilis. J. Bacteriol. 98:1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boylan RJ, Mendelson NH, Brooks D, Young FE. 1972. Regulation of the bacterial cell wall: analysis of a mutant of Bacillus subtilis defective in biosynthesis of teichoic acid. J. Bacteriol. 110:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karamata D, McConnell M, Rogers HJ. 1972. Mapping of rod mutants of Bacillus subtilis. J. Bacteriol. 111:73–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward JB. 1981. Teichoic and teichuronic acids: biosynthesis, assembly, and location. Microbiol. Rev. 45:211–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karamata D, Pooley HM, Monod M. 1987. Expression of heterologous genes for wall teichoic acid in Bacillus subtilis 168. Mol. Gen. Genet. 207:73–81. 10.1007/BF00331493 [DOI] [PubMed] [Google Scholar]
- 54.Weidenmaier C, Peschel A. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6:276–287. 10.1038/nrmicro1861 [DOI] [PubMed] [Google Scholar]
- 55.Brown S, Santa Maria JP, Jr, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67:313–336. 10.1146/annurev-micro-092412-155620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soldo B, Lazarevic V, Margot P, Karamata D. 1993. Sequencing and analysis of the divergon comprising gtaB, the structural gene of UDP-glucose pyrophosphorylase of Bacillus subtilis 168. J. Gen. Microbiol. 139:3185–3195. 10.1099/00221287-139-12-3185 [DOI] [PubMed] [Google Scholar]
- 57.Mauël C, Young M, Karamata D. 1991. Genes concerned with synthesis of poly(glycerol phosphate), the essential teichoic acid in Bacillus subtilis strain 168, are organized in two divergent transcription units. J. Gen. Microbiol. 137:929–941. 10.1099/00221287-137-4-929 [DOI] [PubMed] [Google Scholar]
- 58.Allison SE, D'Elia MA, Arar S, Monteiro MA, Brown ED. 2011. Studies of the genetics, function, and kinetic mechanism of TagE, the wall teichoic acid glycosyltransferase in Bacillus subtilis 168. J. Biol. Chem. 286:23708–23716. 10.1074/jbc.M111.241265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pooley HM, Paschoud D, Karamata D. 1987. The gtaB marker in Bacillus subtilis 168 is associated with a deficiency in UDPglucose pyrophosphorylase. J. Gen. Microbiol. 133:3481–3493 [DOI] [PubMed] [Google Scholar]
- 60.Forsberg CW, Wyrick PB, Ward JB, Rogers HJ. 1973. Effect of phosphate limitation on the morphology and wall composition of Bacillus licheniformis and its phosphoglucomutase-deficient mutants. J. Bacteriol. 113:969–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorasch P, Wolter FP, Zahringer U, Heinz E. 1998. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol. Microbiol. 29:419–430. 10.1046/j.1365-2958.1998.00930.x [DOI] [PubMed] [Google Scholar]
- 62.Lazarevic V, Soldo B, Medico N, Pooley H, Bron S, Karamata D. 2005. Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl. Environ. Microbiol. 71:39–45. 10.1128/AEM.71.1.39-45.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gründling A, Schneewind O. 2007. Genes required for glycolipid synthesis and lipoteichoic acid anchoring in Staphylococcus aureus. J. Bacteriol. 189:2521–2530. 10.1128/JB.01683-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gründling A, Schneewind O. 2007. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc. Nat. Acad. Sci. U. S. A. 104:8478–8483. 10.1073/pnas.0701821104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ganfield MC, Pieringer RA. 1980. The biosynthesis of nascent membrane lipoteichoic acid of Streptococcus faecium (S. faecalis ATCC 9790) from phosphatidylkojibiosyl diacylglycerol and phosphatidylglycerol. J. Biol. Chem. 255:5164–5169 [PubMed] [Google Scholar]
- 66.Emdur LI, Chiu TH. 1974. Turnover of phosphatidylglycerol in Streptococcus sanguis. Biochem. Biophys. Res. Commun. 59:1137–1144. 10.1016/S0006-291X(74)80097-5 [DOI] [PubMed] [Google Scholar]
- 67.Glaser L, Lindsay B. 1974. The synthesis of lipoteichoic acid carrier. Biochem. Biophys. Res. Commun. 59:1131–1136. 10.1016/S0006-291X(74)80096-3 [DOI] [PubMed] [Google Scholar]
- 68.Emdur L, Chiu T-H. 1975. The role of phosphatidylglycerol in the in vitro biosynthesis of teichoic acid and lipoteichoic acid. FEBS Lett. 55:216–219. 10.1016/0014-5793(75)80995-1 [DOI] [PubMed] [Google Scholar]
- 69.Fischer W, Landgraf HR. 1975. Glycerophosphoryl phosphatidyl kojibiosyl diacylglycerol, a novel phosphoglucolipid from Streptococcus faecalis. Biochim. Biophys. Acta 380:227–244. 10.1016/0005-2760(75)90009-0 [DOI] [PubMed] [Google Scholar]
- 70.Cabacungan E, Pieringer RA. 1981. Mode of elongation of the glycerol phosphate polymer of membrane lipoteichoic acid of Streptococcus faecium ATCC 9790. J. Bacteriol. 147:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taron DJ, Childs WC, III, Neuhaus FC. 1983. Biosynthesis of D-alanyl-lipoteichoic acid: role of diglyceride kinase in the synthesis of phosphatidylglycerol for chain elongation. J. Bacteriol. 154:1110–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koch HU, Haas R, Fischer W. 1984. The role of lipoteichoic acid biosynthesis in membrane lipid metabolism of growing Staphylococcus aureus. Eur. J. Biochem. 138:357–363. 10.1111/j.1432-1033.1984.tb07923.x [DOI] [PubMed] [Google Scholar]
- 73.Fischer W. 1994. Lipoteichoic acid and lipids in the membrane of Staphylococcus aureus. Med. Microbiol. Immunol. 183:61–76. 10.1007/BF00277157 [DOI] [PubMed] [Google Scholar]
- 74.Jorasch P, Warnecke DC, Lindner B, Zahringer U, Heinz E. 2000. Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycolipids, glycophospholipids, glycosphingolipids, and glycosylsterols. Eur. J. Biochem. 267:3770–3783. 10.1046/j.1432-1327.2000.01414.x [DOI] [PubMed] [Google Scholar]
- 75.Kiriukhin MY, Debabov DV, Shinabarger DL, Neuhaus FC. 2001. Biosynthesis of the glycolipid anchor in lipoteichoic acid of Staphylococcus aureus RN4220: role of YpfP, the diglucosyldiacylglycerol synthase. J. Bacteriol. 183:3506–3514. 10.1128/JB.183.11.3506-3514.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fedtke I, Mader D, Kohler T, Moll H, Nicholson G, Biswas R, Henseler K, Gotz F, Zahringer U, Peschel A. 2007. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol. Microbiol. 65:1078–1091. 10.1111/j.1365-2958.2007.05854.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee B, Sekimizu K. 2009. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J. Bacteriol. 191:141–151. 10.1128/JB.01221-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wörmann ME, Reichmann NT, Malone CL, Horswill AR, Gründling A. 2011. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J. Bacteriol. 193:5279–5291. 10.1128/JB.00369-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lu D, Wormann ME, Zhang X, Schneewind O, Grundling A, Freemont PS. 2009. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc. Natl. Acad. Sci. U. S. A. 106:1584–1589. 10.1073/pnas.0809020106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schirner K, Marles-Wright J, Lewis RJ, Errington J. 2009. Distinct and essential morphogenetic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 28:830–842. 10.1038/emboj.2009.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Coley J, Duckworth M, Baddiley J. 1972. The occurrence of lipoteichoic acids in the membranes of Gram-positive bacteria. J. Gen. Microbiol. 73:587–591. 10.1099/00221287-73-3-587 [DOI] [PubMed] [Google Scholar]
- 83.Reichmann NT, Gründling A. 2011. Location, synthesis and function of glycolipids and polyglycerolphosphate lipoteichoic acid in Gram-positive bacteria of the phylum Firmicutes. FEMS Microbiol. Lett. 319:97–105. 10.1111/j.1574-6968.2011.02260.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Webb AJ, Karatsa-Dodgson M, Gründling A. 2009. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol. Microbiol. 74:299–314. 10.1111/j.1365-2958.2009.06829.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wörmann ME, Corrigan RM, Simpson PJ, Matthews SJ, Gründling A. 2011. Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol. Microbiol. 79:566–583. 10.1111/j.1365-2958.2010.07472.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garufi G, Hendrickx AP, Beeri K, Kern JW, Sharma A, Richter SG, Schneewind O, Missiakas D. 8 June 2012. Synthesis of lipoteichoic acids in Bacillus anthracis. J. Bacteriol. 10.1128/JB.00626-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Damjanovic M, Kharat AS, Eberhardt A, Tomasz A, Vollmer W. 2007. The essential tacF gene is responsible for the choline-dependent growth phenotype of Streptococcus pneumoniae. J. Bacteriol. 189:7105–7111. 10.1128/JB.00681-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baur S, Marles-Wright J, Buckenmaier S, Lewis RJ, Vollmer W. 2009. Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae. J. Bacteriol. 191:1200–1210. 10.1128/JB.01120-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686–723. 10.1128/MMBR.67.4.686-723.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baddiley J, Neuhaus FC. 1960. The enzymic activation of D-alanine. Biochem. J. 75:579–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Heaton MP, Neuhaus FC. 1992. Biosynthesis of D-alanyl-lipoteichoic acid: cloning, nucleotide sequence, and expression of the Lactobacillus casei gene for the D-alanine-activating enzyme. J. Bacteriol. 174:4707–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Heaton MP, Neuhaus FC. 1994. Role of the D-alanyl carrier protein in the biosynthesis of D-alanyl-lipoteichoic acid. J. Bacteriol. 176:681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Neuhaus FC, Heaton MP, Debabov DV, Zhang Q. 1996. The dlt operon in the biosynthesis of D-alanyl-lipoteichoic acid in Lactobacillus casei. Microb. Drug Resist. 2:77–84. 10.1089/mdr.1996.2.77 [DOI] [PubMed] [Google Scholar]
- 94.Koprivnjak T, Mlakar V, Swanson L, Fournier B, Peschel A, Weiss JP. 2006. Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J. Bacteriol. 188:3622–3630. 10.1128/JB.188.10.3622-3630.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. 1995. Incorporation of D-alanine into lipoteichoic acid and wall teichoic acid in Bacillus subtilis. Identification of genes and regulation. J. Biol. Chem. 270:15598–15606 [DOI] [PubMed] [Google Scholar]
- 96.Reichmann NT, Cassona CP, Grundling A. 2013. Revised mechanism of D-alanine incorporation into cell wall polymers in Gram-positive bacteria. Microbiology 159:1868–1877. 10.1099/mic.0.069898-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Haas R, Koch HU, Fischer W. 1984. Alanyl turnover from lipoteichoic acid to teichoic acid in Staphylococcus aureus. FEMS Microbiol. Lett. 21:27–31. 10.1111/j.1574-6968.1984.tb00180.x [DOI] [Google Scholar]
- 98.Koch HU, Doker R, Fischer W. 1985. Maintenance of D-alanine ester substitution of lipoteichoic acid by reesterification in Staphylococcus aureus. J. Bacteriol. 164:1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McCarty M. 1959. The occurrence of polyglycerophosphate as an antigenic component of various gram-positive bacterial species. J. Exp. Med. 109:361–378. 10.1084/jem.109.4.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mauël C, Young M, Margot P, Karamata D. 1989. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol. Gen. Genet. 215:388–394. 10.1007/BF00427034 [DOI] [PubMed] [Google Scholar]
- 101.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J. Bacteriol. 188:8313–8316. 10.1128/JB.01336-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J. Bacteriol. 188:4183–4189. 10.1128/JB.00197-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weidenmaier C, Kokai-Kun JF, Kristian SA, Chanturiya T, Kalbacher H, Gross M, Nicholson G, Neumeister B, Mond JJ, Peschel A. 2004. Role of teichoic acids in Staphylococcus aureus nasal colonization, a major risk factor in nosocomial infections. Nat. Med. 10:243–245. 10.1038/nm991 [DOI] [PubMed] [Google Scholar]
- 104.Shiomi D, Margolin W. 2007. A sweet sensor for size-conscious bacteria. Cell 130:216–218. 10.1016/j.cell.2007.07.011 [DOI] [PubMed] [Google Scholar]
- 105.Wang JD, Levin PA. 2009. Metabolism, cell growth and the bacterial cell cycle. Nat. Rev. Microbiol. 7:822–827. 10.1038/nrmicro2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. 2007. A metabolic sensor governing cell size in bacteria. Cell 130:335–347. 10.1016/j.cell.2007.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Richter SG, Elli D, Kim HK, Hendrickx AP, Sorg JA, Schneewind O, Missiakas D. 2013. Small molecule inhibitor of lipoteichoic acid synthesis is an antibiotic for Gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 110:3531–3536. 10.1073/pnas.1217337110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Matsuoka S, Hashimoto M, Kamiya Y, Miyazawa T, Ishikawa K, Hara H, Matsumoto K. 2011. The Bacillus subtilis essential gene dgkB is dispensable in mutants with defective lipoteichoic acid synthesis. Genes Genet. Syst. 86:365–376. 10.1266/ggs.86.365 [DOI] [PubMed] [Google Scholar]
- 109.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7:e1002217. 10.1371/journal.ppat.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285:473–482. 10.1074/jbc.M109.040238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Corrigan RM, Campeotto I, Jeganathan T, Roelofs KG, Lee VT, Grundling A. 2013. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proc. Natl. Acad. Sci. U. S. A. 110:9084–9089. 10.1073/pnas.1300595110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Price-Whelan A, Poon CK, Benson MA, Eidem TT, Roux CM, Boyd JM, Dunman PM, Torres VJ, Krulwich TA. 2013. Transcriptional profiling of Staphylococcus aureus during growth in 2 M NaCl leads to clarification of physiological roles for Kdp and Ktr K+ uptake systems. mBio. 4:e00407-13. 10.1128/mBio.00407-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lambert PA, Hancock IC, Baddiley J. 1977. Occurrence and function of membrane teichoic acids. Biochim. Biophys. Acta 472:1–12. 10.1016/0304-4157(77)90012-0 [DOI] [PubMed] [Google Scholar]
- 114.Cleveland RF, Höltje JV, Wicken AJ, Tomasz A, Daneo-Moore L, Shockman GD. 1975. Inhibition of bacterial wall lysins by lipoteichoic acids and related compounds. Biochem. Biophys. Res. Commun. 67:1128–1135. 10.1016/0006-291X(75)90791-3 [DOI] [PubMed] [Google Scholar]
- 115.Peschel A, Vuong C, Otto M, Götz F. 2000. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob. Agents Chemother. 44:2845–2847. 10.1128/AAC.44.10.2845-2847.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schlag M, Biswas R, Krismer B, Kohler T, Zoll S, Yu W, Schwarz H, Peschel A, Gotz F. 2010. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol. Microbiol. 75:864–873. 10.1111/j.1365-2958.2009.07007.x [DOI] [PubMed] [Google Scholar]
- 117.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410. 10.1074/jbc.274.13.8405 [DOI] [PubMed] [Google Scholar]
- 118.Hyyrylainen HL, Vitikainen M, Thwaite J, Wu H, Sarvas M, Harwood CR, Kontinen VP, Stephenson K. 2000. D-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J. Biol. Chem. 275:26696–26703. 10.1074/jbc.M003804200 [DOI] [PubMed] [Google Scholar]
- 119.Tomasz A. 1967. Choline in the cell wall of a bacterium: novel type of polymer-linked choline in Pneumococcus. Science 157:694–697. 10.1126/science.157.3789.694 [DOI] [PubMed] [Google Scholar]
- 120.Mosser JL, Tomasz A. 1970. Choline-containing teichoic acid as a structural component of pneumococcal cell wall and its role in sensitivity to lysis by an autolytic enzyme. J. Biol. Chem. 245:287–298 [PubMed] [Google Scholar]
- 121.Jonquières R, Bierne H, Fiedler F, Gounon P, Cossart P. 1999. Interaction between the protein InlB of Listeria monocytogenes and lipoteichoic acid: a novel mechanism of protein association at the surface of gram-positive bacteria. Mol. Microbiol. 34:902–914. 10.1046/j.1365-2958.1999.01652.x [DOI] [PubMed] [Google Scholar]
- 122.Oshida T, Sugai M, Komatsuzawa H, Hong YM, Suginaka H, Tomasz A. 1995. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc. Natl. Acad. Sci. U. S. A. 92:285–289. 10.1073/pnas.92.1.285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Heilmann C, Hussain M, Peters G, Gotz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013–1024. 10.1046/j.1365-2958.1997.4101774.x [DOI] [PubMed] [Google Scholar]
- 124.Zoll S, Schlag M, Shkumatov AV, Rautenberg M, Svergun DI, Gotz F, Stehle T. 2012. Ligand-binding properties and conformational dynamics of autolysin repeat domains in staphylococcal cell wall recognition. J. Bacteriol. 194:3789–3802. 10.1128/JB.00331-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ginsburg I, Fligiel SE, Ward PA, Varani J. 1988. Lipoteichoic acid-antilipoteichoic acid complexes induce superoxide generation by human neutrophils. Inflammation 12:525–548. 10.1007/BF00914316 [DOI] [PubMed] [Google Scholar]
- 126.Riesenfeld-Orn I, Wolpe S, Garcia-Bustos JF, Hoffmann MK, Tuomanen E. 1989. Production of interleukin-1 but not tumor necrosis factor by human monocytes stimulated with pneumococcal cell surface components. Infect. Immun. 57:1890–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cleveland MG, Gorham JD, Murphy TL, Tuomanen E, Murphy KM. 1996. Lipoteichoic acid preparations of gram-positive bacteria induce interleukin-12 through a CD14-dependent pathway. Infect. Immun. 64:1906–1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ginsburg I. 2002. Role of lipoteichoic acid in infection and inflammation. Lancet Infect. Dis. ii:171–179 [DOI] [PubMed] [Google Scholar]
- 129.Morath S, von Aulock S, Hartung T. 2005. Structure/function relationships of lipoteichoic acids. J. Endotoxin Res. 11:348–356. 10.1177/09680519050110061001 [DOI] [PubMed] [Google Scholar]
- 130.Hermann C. 2007. Review: variability of host-pathogen interaction. J. Endotoxin Res. 13:199–218. 10.1177/0968051907082605 [DOI] [PubMed] [Google Scholar]
- 131.Hashimoto M, Tawaratsumida K, Kariya H, Kiyohara A, Suda Y, Krikae F, Kirikae T, Gotz F. 2006. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. Immunol. 177:3162–3169 [DOI] [PubMed] [Google Scholar]
- 132.Rockel C, Hartung T. 2012. Systematic review of membrane components of gram-positive bacteria responsible as pyrogens for inducing human monocyte/macrophage cytokine release. Front. Pharmacol. 3:56. 10.3389/fphar.2012.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bubeck Wardenburg J, Williams WA, Missiakas D. 2006. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 103:13831–13836. 10.1073/pnas.0603072103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Frankel MB, Schneewind O. 2012. Determinants of murein hydrolase targeting to cross-wall of Staphylococcus aureus peptidoglycan. J. Biol. Chem. 287:10460–10471. 10.1074/jbc.M111.336404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Reid CW, Vinogradov E, Li J, Jarrell HC, Logan SM, Brisson JR. 2012. Structural characterization of surface glycans from Clostridium difficile. Carbohydr. Res. 354:65–73. 10.1016/j.carres.2012.02.002 [DOI] [PubMed] [Google Scholar]
- 136.Rogers HJ. 1988. The bacterial surface—where does it begin and end?, p 639–648 In Actor P, Daneo-Moore L, Higgins ML, Salton MR, Schockman GD. (ed), Antibiotic inhibition of bacterial cell surface assembly and function. American Society for Microbiology, Washington, DC [Google Scholar]
- 137.Ellwood DC, Tempest DW. 1972. Influence of culture pH on the content and composition of teichoic acids in the walls of Bacillus subtilis. J. Gen. Microbiol. 73:395–402. 10.1099/00221287-73-2-395 [DOI] [PubMed] [Google Scholar]