Abstract
For transmission to new hosts, Yersinia pestis, the causative agent of plague, replicates as biofilm in the foregut of fleas that feed on plague-infected animals or humans. Y. pestis biofilm formation has been studied in the rat flea; however, little is known about the cat flea, a species that may bridge zoonotic and anthroponotic plague cycles. Here, we show that Y. pestis infects and replicates as a biofilm in the foregut of cat fleas in a manner requiring hmsFR, two determinants for extracellular biofilm matrix. Examining a library of transposon insertion mutants, we identified the LysR-type transcriptional regulator YfbA, which is essential for Y. pestis colonization and biofilm formation in cat fleas.
INTRODUCTION
Yersinia pestis is the causative agent of plague (1), a frequently lethal disease that affects many mammalian species, including humans (2). Plague is transmitted by fleas, which are infected during blood meals and may be colonized via a gastrointestinal biofilm (3, 4). Transmission of Y. pestis occurs when newly infected (early-phase transmission) or colonized (regurgitative transmission) insects feed on new hosts (5, 6). Several different rodents are enzootic reservoir hosts for the plague pathogen (2). Historically, epizootic outbreaks in certain rodent species have triggered flea-borne transmission of plague into human populations with devastating consequences (7). Examples are the Justinian plague (541 to 767), the Black Death (1346 to the 18th century), and the Asian pandemics (1850 to 1935) (8).
Genetic analysis revealed that Y. pestis is a monomorphic clone of its more diverse parental species, the human gastrointestinal pathogen Yersinia pseudotuberculosis (9). Evolution of Y. pestis occurred >2,600 years ago in the rodent population of China; here, Y. pestis isolates are still scattered over four phylogenetic branches, biovars Orientalis, Medievalis, Antiqua, and Pestoides (10, 11). The Y. pestis genome encompasses 4,012 chromosomal genes, including 149 pseudogenes and three plasmids (12). The pCD1 plasmid encodes the type III secretion machine for delivery of effector proteins (Yops) from the bacterium into immune cells of mammalian hosts (13, 14). pFra harbors genes for the expression and assembly of capsular fraction antigen F1, a surface (pilus) organelle, and the murine toxin (Ymt), which contribute to regurgitative transmission and Y. pestis persistence in the flea vector (15, 16). pPCP1 encodes the Pla surface protease, which is dispensable for Y. pestis survival in the flea but contributes to its escape from mammalian innate immune responses (17, 18). Although the functions of many genes on the three virulence plasmids have been revealed, little is known about chromosomal determinants for the unique life cycle of Y. pestis (19).
The relevance of the oriental rat flea, Xenopsylla cheopis, to the transmission of plague is well established (6). After feeding on infected blood, the flea digestive tract is eventually blocked by massive Y. pestis replication at the proventriculus and midgut (4). Since X. cheopis and several other flea species (for example, Ctenocephalides felis) are intermittent feeders, starvation causes these species to increase their feeding behavior, which is associated with transmission of Y. pestis to new hosts (6). The Y. pestis hms locus is required for rat flea colonization, biofilm formation, intestinal blockade, and regurgitative transmission (6); the locus comprises a four-gene operon (hmsHFRS) for the synthesis of poly-(β1-6)-N-acetylglucosamine (PNAG), an extracellular matrix polymer of Y. pestis biofilms (20, 21). Acquisition of the hms locus predates the evolutionary divergence of Y. pestis from Y. pseudotuberculosis (22). However, Y. pseudotuberculosis, but not Y. pestis, expresses nghA, which encodes a PNAG hydrolase that is thought to interfere with biofilm formation in the flea gut (21, 23). Of note, Y. pseudotuberculosis infection of fleas triggers intestinal toxicity and diarrhea in X. cheopis (24). Further, the rcsA gene, which encodes a negative regulator for the diguanylate cyclase (HmsT) of Y. pseudotuberculosis, contains a 30-bp internal duplication in Y. pestis, rendering rcsA nonfunctional (25, 26). Nevertheless, the signaling molecule cyclic di-GMP is required for Y. pestis PNAG synthesis and biofilm formation in the flea foregut, but not for the pathogenesis of plague in mammalian hosts (27, 28). A positive transcriptional regulator for Y. pestis flea colonization and biofilm formation in the foregut has thus far not been identified (6).
C. felis, the cat flea, has been investigated as a vector for plague transmission in Africa, China, and the United States (7, 29). The host range of C. felis encompasses domesticated animals, household pets, squirrels, rats, and mice, as well as humans (30). This flea species has been studied for the attribute of bridging zoonotic and anthroponotic plague cycles (30, 31). C. felis is a common infestation of dogs, which are infected by Y. pestis and seroconvert to produce F1-specific antibodies but rarely develop fatal disease (32, 33). Dog infestation with infected C. felis is associated with flea contamination of human dwellings and transmission, as documented in Uganda, China, and the United States (34–36). Earlier work revealed the ability of C. felis to produce early-phase transmission of Y. pestis to mammalian hosts (30). This work left unresolved whether Y. pestis forms persistent biofilms in C. felis and by what mechanism such colonization may occur.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Y. pestis CO92(ΔpCD1) and its mutants were grown on heart infusion agar (HIA) at 26°C and stored frozen in 5% monosodium glutamate-5% bovine serum albumin (BSA). When necessary, mutant strains were grown on selective medium containing ampicillin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (30 μg/ml). Where indicated, Y. pestis CO92(ΔpCD1) and its mutants were electroporated with plasmid pEGFP (Clontech, Palo Alto, CA).
Y. pestis transposon mutagenesis.
To generate mini-Tn5 transposon mutants, 50 ml of heart infusion broth (HIB) was inoculated with Y. pestis CO92(ΔpCD1) and grown at 26°C overnight. The following day, the cells were chilled on ice, sedimented by centrifugation, washed twice with ice-cold sterile water, and suspended at a density of 9 × 109 CFU ml−1 for electroporation. One microliter of purified transposase complex of the EZ-Tn5 (Kan-2) transposon kit (Epicentre, Madison, WI) was added to 100 μl bacterial suspension and pulsed at 1,800 V, 100 Ω, 25 μF. Y. pestis organisms were diluted in 1 ml HIB, incubated for 4 h at 26°C, spread on HIA with kanamycin, and finally grown at 26°C for 48 h. Individual colonies were picked, propagated in HIB by overnight growth, and subjected to freezing and isolation of genomic DNA. To identify the positions of Tn5 insertions, the genomic DNA was digested with HhaI and circularized by ligation with T4 DNA ligase. Transposon insertions were PCR amplified with primers provided with the transposon kit and submitted for DNA sequencing. DNA sequences were BLAST analyzed against the Y. pestis CO92 genome sequence to identify the insertion site, and gene-specific primers were designed to confirm insertion in the open reading frame (12, 37).
Infection of cat fleas with Y. pestis.
Mixed-sex adult C. felis fleas were obtained from the Elward II Laboratory (Soquel, CA), separated into 50 to 100 fleas per acrylic cage, and housed in an artificial feeding system at 25°C and 75% humidity (38). The fleas were starved for 48 h prior to infection and fed on defibrinated sheep's blood (Hemostat, Dixon, CA) containing Y. pestis CO92(ΔpCD1) or its variants for 5 h at 37°C. After the initial infectious feeding period, the unfed fleas (identifiable by lack of fresh blood in the midgut and corresponding increase in body size) were removed and the remaining fleas were sustained on daily blood meals with 5 ml sheep's blood for the remainder of the study.
For infection, Y. pestis strains were spread on a Congo Red-heart infusion agar plate and grown at 26°C for 48 h to confirm pigmentation status. A single colony was picked, inoculated into HIB, and grown for 12 h at 26°C. The following day, the optical density was measured and the bacterial load was calculated using a conversion factor of 2.7 × 108 CFU for absorbance at 600 nm (A600). Bacterial cultures were sedimented by centrifugation at 17,090 × g for 10 min, suspended in 1 ml of PBS, and added to 4 ml of prewarmed 37°C sterile sheep's blood (1 × 108 to 1 × 1010 CFU ml−1). Control cohorts of uninfected fleas were fed on 4 ml of sterile sheep's blood diluted with 1 ml PBS.
Enumeration of Y. pestis organisms in infected fleas.
Infected fleas were frozen at −80°C, surface sterilized with 70% ethanol, and placed into 2-ml vials with 6 to 8 sterile 2.3-mm steel beads (Biospec Products) and 200 μl of HIB. The fleas were mechanically disrupted using agitation in a Bead Beater (Biospec Products) for 45 s at 4.0 m/s. The vials were briefly centrifuged, and the liquid supernatant was removed, serially diluted in PBS, spread on HIA plates, and grown at 26°C for 48 to 72 h prior to enumeration of colonies.
Flea dissection and microscopy.
Fleas were anesthetized by chilling at −20°C for 5 min and then placed into a drop of PBS for dissection. The fleas were decapitated by prying the upper thorax and head off using two U-100 insulin syringes. The digestive tract was withdrawn from the abdomen by gently pulling on the proventriculus with fine tweezers. For microscopic imaging of the digestive tract, the flea guts were placed on a glass carrier with a drop of phosphate-buffered saline (PBS), and coverslips were placed on top. Bright-field microscopic images were captured with a charge-coupled-device (CCD) camera using an Olympus IX81 microscope. Fluorescent images were acquired using a Leica SP5 Tandem Scanner Spectral 2-Photon confocal microscope. Images were acquired with the red Texas Red (594-nm) and green green fluorescent protein (GFP) (488-nm) fluorescence channels. Images were merged using ImageJ software (NIH).
Scanning electron microscopy.
Flea digestive tracts were isolated and fixed in 2% glutaraldehyde in PBS overnight, and then the proventriculus was cut open using a surgical scalpel. Samples were serially dehydrated by consecutive incubations in 25% and 50% ethanol-PBS, 75% and 90% ethanol-water, and 100% ethanol (twice, each step for 5 min), followed by 50% ethanol-hexamethyldisilazane (HMDS) (15 min) and 100% HMDS (20 min), which was allowed to evaporate in the final wash. After overnight evaporation of HMDS at room temperature, the samples were mounted onto specimen mounts (Ted Pella, Inc., Redding, CA) and coated with 80% Pt-20% Pd to 11 nm using a Cressington 208HR Sputter Coater at 20 mA prior to examination with a Fei Nova NanoSEM 200 scanning electron microscope (FEI Co., Hillsboro, OR) at a distance of 5 mm.
Construction of complementation plasmids.
The Y. pestis CO92(ΔpCD1) transposon mutants were complemented with plasmids expressing a wild-type copy of each gene. To generate pyfbA, a 1,097-bp DNA fragment containing the 191-bp upstream and 906-bp coding regions of yfbA from wild-type CO92(ΔpCD1) was amplified by PCR using the specific primers 5′AAACATATGCTCCTGATGCCATCATTAATCAATTGCAGTACAG and 3′AAAGGATCCTCACCGTTCATCCAATTGGCTGAAG. This PCR fragment was subcloned into the 5′ NdeI and 3′ BamHI sites of pMCSG7 (Amp), a high-copy-number pET vector. p2458 was generated by PCR amplification of a 1,182-bp DNA fragment from wild-type CO92(ΔpCD1) of ypo2458 consisting of the 201-bp upstream and 981-bp coding regions of ypo2458 using the specific primers 5′AAACATATGATTTTCATCACCGGGTCCTGTGAGG and 3′AAAGGATCCCTAAAGCACAGCCGGTAGAGGTTTG. This DNA was subcloned into the 5′ NdeI and 3′ BamHI sites of pMCSG7.
To generate pPtac-yfbA, yfbA was amplified using the specific primers 5′AAAACATATGCACGATCTCAATGATCTCTATTACTACGCAGAAGTTGTAG and 3′AAAGGATCCTCACCGTTCATCCAATTGGCTGAAG. This DNA fragment was cloned into the 5′ NdeI and 3′ BamHI sites of a pHSG575 derivative (39, 40) to express yfbA under the control of the tac promoter.
Each complementing plasmid was confirmed by DNA sequencing. pyfbA or pPtac-yfbA was introduced into CO92(ΔpCD1) yfbA::Tn5 by electroporation. CO92(ΔpCD1) ypo2458::Tn5 and wild-type CO92(ΔpCD1) were transformed with p2458 and the pMSCG7 empty vector, respectively, via electroporation. The presence of the complementing plasmids in the mutant strains was confirmed by PCR.
In vitro biofilm analysis.
Strains were grown overnight in HIB supplemented with the relevant antibiotics, 4 mM CaCl2, and 4 mM MgCl2. The following day, the cultures were subcultured to an A600 of 0.02, and 10 replicates of 200 μl each were plated in treated Costar polystyrene 96-well plates in HIB supplemented with 4 mM CaCl2 and 4 mM MgCl2 with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) added to the pPtac-yfbA culture. A row of medium-only wells was used as a control for background absorbance. The plates were incubated at 26°C with shaking at 250 rpm for 24 h. Planktonic cells were removed, and the wells were washed two times with H2O and stained for 15 min with crystal violet (0.02%). The stain was removed, the wells were washed four times with H2O, and the bound crystal violet was solubilized with a 80% ethanol-20% acetone solution. Absorbance was measured at 600 nm using a Synergy HT plate reader (Biotek), and the average absorbance from the medium-only control wells was subtracted from the recorded absorbances to obtain the final measurements.
Fluorescence microscopy of Y. pestis biofilms.
Y. pestis biofilms were grown on 18-mm round glass coverslips pretreated with 1 μg/ml human fibronectin (Sigma) in Costar 12-well polystyrene plates. The wells were washed two times with PBS before adding 400 μl of Y. pestis CO92(ΔpCD1) or its yfbA::Tn5 variant to an A600 of 0.02. The plates were incubated at 26°C for 24 h with shaking at 250 rpm. Glass coverslips were washed twice with PBS and stained with 5 μM Syto9 (Invitrogen) at room temperature for 20 min. The coverslips were washed twice and fixed with 4% formalin before washing, drying, and mounting on glass slides with 1 μl SlowFade Gold (Invitrogen). Fluorescent images of biofilms were acquired in the green GFP channel (488 nm) on an Olympus IX81 microscope using a 40× objective and captured with a CCD camera.
RESULTS
Y. pestis infection and colonization of C. felis.
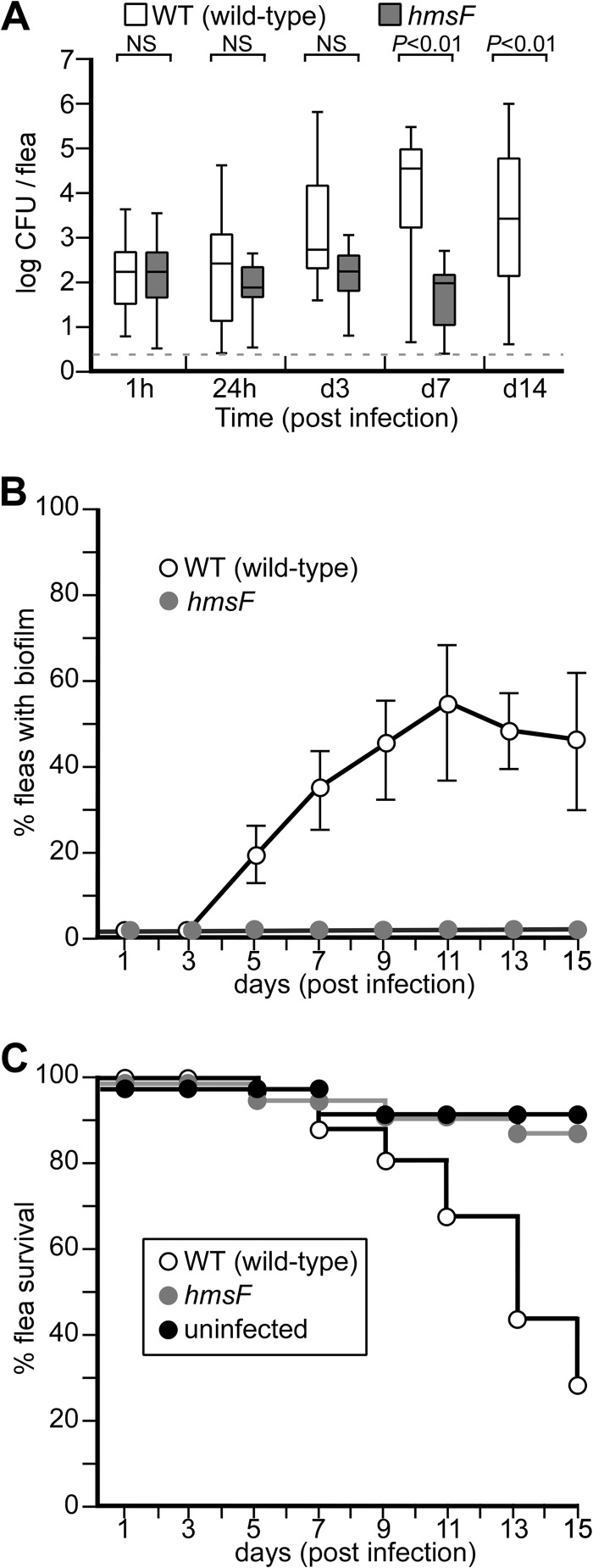
Mixed-sex adult C. felis fleas (n = 50 to 100 fleas) were isolated in acrylic cages at 25°C and starved for 48 h prior to feeding on defibrinated sheep's blood mixed with 1 × 108 CFU ml−1 Y. pestis CO92(ΔpCD1) or its variants. Blood samples were maintained for 5 h at 37°C in a humidified feeding chamber (artificial dog) during feeding (41). The fleas were subsequently fed daily on defibrinated sheep's blood without added bacteria and analyzed for Y. pestis colonization. To determine the Y. pestis load in the gut, flea homogenates were spread on agar plates and the bacteria were enumerated. These studies revealed that the flea guts harbored 1.47 × 102 CFU Y. pestis CO92 1 h after the end of feeding (Fig. 1A). Between 24 h and 3 days postinfection, Y. pestis was able to replicate inside the fleas (Fig. 1A). At day 7 after feeding, the bacterial load increased to 2.89 × 104 CFU (Fig. 1A). Increases in the bacterial load were correlated with decreased flea survival. By day 14, more than 60% of the infected flea population succumbed to Y. pestis infection (Fig. 1C). Approximately half of all infected cat fleas (49%) were not colonized by wild-type Y. pestis CO92(ΔpCD1) (see below). The Y. pestis(ΔpCD1, ΔhmsF) mutant lacks the (β1-6)-N-acetylglucosamine deacetylase (HmsF) required for functional assembly of PNAG and biofilm formation (42). When the Y. pestis hmsF mutant was used for infection of C. felis, 1.5 × 102 CFU were isolated 1 h after feeding, similar to wild-type Y. pestis (Fig. 1A). This observation indicates that both wild-type and hmsF mutant Y. pestis CO92 infect cat fleas, in agreement with the recent report that C. felis is capable of early-phase plague transmission between mammalian hosts (5). Nevertheless, the hmsF mutant did not replicate in the flea gut and did not affect the survival of infected C. felis compared to mock-infected insects (Fig. 1C).
FIG 1.
Y. pestis colonization of cat fleas requires the hmsF locus. (A) Cat fleas (n = 100) were infected by one-time feeding on defibrinated sheep's blood inoculated with 1 × 108 CFU wild-type Y. pestis CO92(ΔpCD1) or its hmsF mutant and were subsequently fed sterile blood. At various time intervals of hours (h) or days (d), the infected fleas were homogenized and plated on HIA, and CFU were enumerated. The box plot represents Y. pestis flea colonization data in log CFU, where the horizontal line denotes the mean and the whiskers indicate variability outside the upper and lower quartiles. The dashed line identifies the limit of detection. NS, not significant. (B) Infected-flea cohorts (n = 100) were monitored by dissection of the gut and microscopy to detect biofilm formation. Fleas with a visible mass spreading from the proventriculus into the midgut were scored positive. The data shown represent averages of five independent experiments. The error bars indicate standard deviations. (C) Survival of cat fleas (n = 100) that were either left uninfected or infected with wild-type or hmsF mutant Y. pestis CO92(ΔpCD1) and monitored for survival. The data are representative of more than five independent experiments.
Y. pestis forms a biofilm in the proventriculus of C. felis.
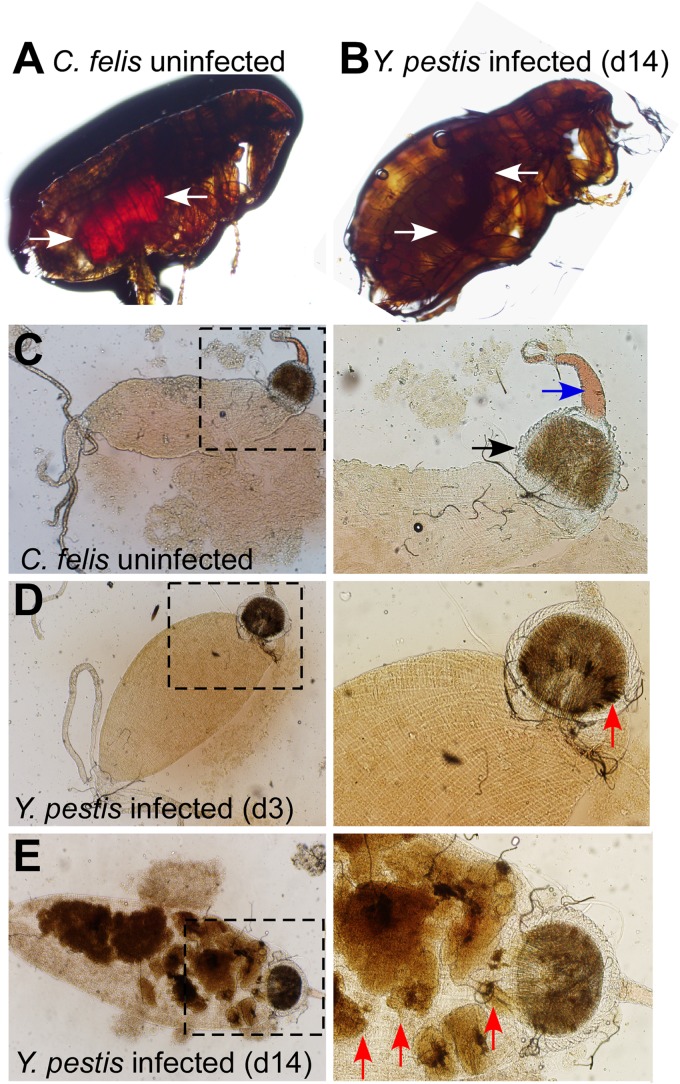
Y. pestis-infected or mock-infected cat fleas were immobilized and viewed by light microscopy (Fig. 2). While the guts of mock-infected fleas could be discerned as a red (hemin-colored) contour within the flea body, the gut of Y. pestis CO92(ΔpCD1) infected fleas appeared as dark-brown-stained material (Fig. 2A and B). The latter was difficult to discern against the pigmented chitin bodies of the insects. To analyze the flea gut, insects were decapitated, the intestines were removed, and the proventriculus and midgut were viewed by microscopy (Fig. 2C to E). In uninfected fleas, the esophagus and proventriculus could be discerned by their characteristic shape and dense structure, respectively (Fig. 2C). The midgut of cat fleas presented as an amorphous, bag-like structure connected to the proventriculus, which was filled with granular, light-brown-pigmented material (Fig. 2C). Immediately following Y. pestis infection, the esophagus, proventriculus, and midgut of cat fleas were not affected compared to uninfected C. felis. On day 3, dark-brown pigment was detected on the proventriculus of Y. pestis-infected fleas (Fig. 2D). By day 14, most of the midgut was filled with the dark-brown pigmented material (Fig. 2E). In X. cheopis, this pigmented material was previously shown to represent Y. pestis biofilm (43).
FIG 2.
Y. pestis induced aggregates and gut blockade in cat fleas. (A and B) Micrographs of immobilized live adult uninfected (A) and Y. pestis CO92(ΔpCD1)-infected (B) cat fleas maintained in an artificial feeder for 14 days. Evidence of a recent blood meal is indicated by a red-stained midgut (arrows in panel A). In infected fleas, the digestive tract appears smaller and is filled with a dark mass extending from the proventriculus along the entire gut (arrows in panel B). (C) Gut of a dissected uninfected cat flea (boxed area magnified on the right) identifying the esophagus (blue arrow) and proventriculus (black arrow). (D) Gut of a dissected cat flea 3 days (d3) after infection with Y. pestis CO92(ΔpCD1) (boxed area magnified on the right) identifying dark-pigmented material deposited in the proventriculus (red arrow). (E) Gut of a dissected cat flea 14 days after infection with Y. pestis CO92(ΔpCD1) (boxed area magnified on the right) identifying large amounts of dark-pigmented material in the proventriculus, midgut, and hindgut (red arrows). The images are representative of more than 50 analyzed cat fleas in each cohort.
To discern whether the dark-brown-pigmented material in the proventriculus and midgut of cat fleas indeed represented Y. pestis biofilms, wild-type and hmsF mutant strains were transformed with pEGFP, a plasmid providing for constitutive expression of GFP. Following infection of fleas with wild-type or hmsF mutant Y. pestis CO92(ΔpCD1, pEGFP), the fleas were dissected, and their intestines were examined by fluorescence microscopy. C. felis displayed GFP fluorescence in the proventriculus on day 3 for wild-type- but not hmsF mutant-infected insects (Fig. 3A to E). On day 7, GFP fluorescence had expanded into large aggregates in the proventriculus and midgut of fleas infected with wild-type Y. pestis (Fig. 3G to I). C. felis infected with the hmsF mutant harbored only weak GFP fluorescence signals in the flea midgut (Fig. 3A to C).
FIG 3.
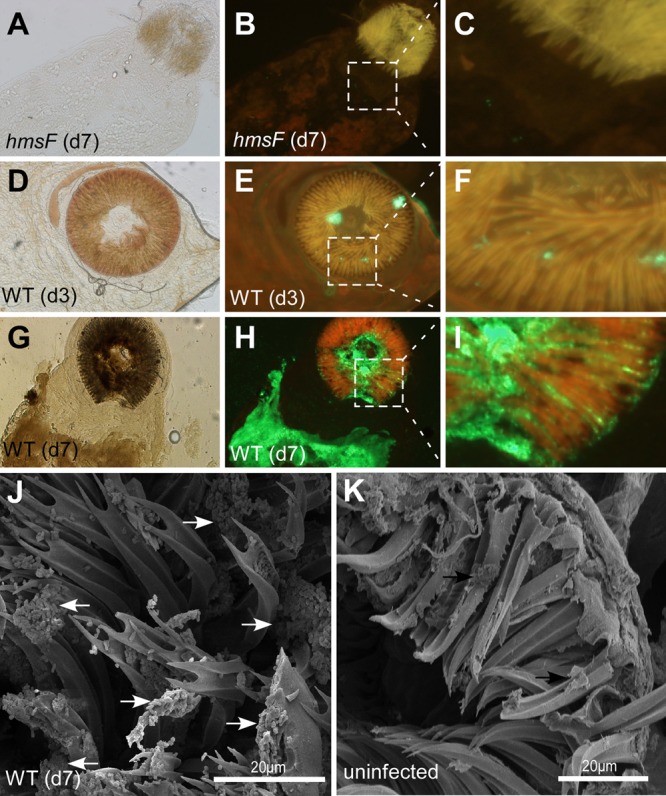
Y. pestis forms biofilms in the proventriculus and in the gut of infected cat fleas. (A to C) Representative bright-field (A) and fluorescence microscopy (B) images (the boxed area is magnified in panel C) of the dissected digestive tracts of fleas (n = 30) 7 days (d7) after infection with hmsF mutant Y. pestis CO92(ΔpCD1, pGFP). The fluorescence images were acquired in red Texas Red (594-nm) and green GFP (488-nm) fluorescence channels and merged, which, due to autofluorescence, reveals the contours of the proventriculus as orange signals and GFP-expressing Y. pestis in green. (D to F) Representative bright-field (D) and fluorescence microscopy (E) images (the boxed area is magnified in panel F) of dissected digestive tracts of fleas (n = 30) 3 days after infection with wild-type Y. pestis CO92(ΔpCD1, pGFP). Note the areas of green fluorescence (F) derived from GFP-expressing wild-type Y. pestis in the proventriculus. (G to I) Representative bright-field (G) and fluorescence microscopy (H) images (the boxed area is magnified in panel I) of dissected digestive tracts of fleas (n = 30) 7 days after infection with wild-type Y. pestis CO92(ΔpCD1, pGFP). Note the massive biofilm of GFP-expressing Y. pestis in the midgut and proventriculus. (J and K) The dissected proventriculus of fleas infected with wild-type Y. pestis CO92(ΔpCD1) (J) or uninfected fleas (K) was fixed and viewed by scanning electron microscopy (SEM). The arrows indicate Y. pestis CO92(ΔpCD1) aggregates on the spines of the cat flea proventriculus.
To obtain further evidence for the formation of biofilms in the proventriculus of cat fleas, we analyzed the foregut of mock- or Y. pestis-nfected C. felis by scanning electron microscopy. Individual bacteria, as well as large clumps of bacterial masses, could be detected at the tip and along the spines of the proventriculus of infected C. felis (Fig. 3J). The spines in the proventriculus of uninfected C. felis were not associated with bacterial deposits (Fig. 3K). Taken together, these data indicate that wild-type Y. pestis forms a biofilm on the proventriculus and in the midgut of infected C. felis. Biofilm formation likely requires the production of PNAG, as hmsF mutant Y. pestis cannot persistently colonize cat fleas or assemble as bacterial aggregates within an extracellular matrix (Fig. 1B). In contrast, wild-type Y. pestis formed massive intestinal biofilms in up to 60% of infected fleas (Fig. 1B).
The Y. pestis mutant is defective for in vitro biofilm formation.
We sought to identify other genes required for Y. pestis replication in C. felis. Assuming that genes required for bacterial biofilm growth in vitro may also be required for flea colonization, a library of mutants with insertional mini-Tn5 lesions in the genome of Y. pestis(ΔpCD1) was generated and screened for defects in in vitro biofilm formation. As reported earlier, Y. pestis(ΔpCD1) forms biofilms during stationary growth at 26°C, which can be detected and quantified by crystal violet staining and A600 (44). As controls, Y. pestis CO92(ΔpCD1) formed a robust biofilm, whereas the hmsR::Tn5 mutant, which catalyzes the polymerization of poly-β(1-6)-N-acetylglucosamine in the cytoplasmic membrane (42, 45), did not (Fig. 4A). hms mutants, such as an hmsR::Tn5 mutant, were also detected via a nonpigmentation phenotype when grown at 26°C on Congo Red agar plates (Fig. 4E); Congo Red staining identifies PNAG, the Y. pestis exopolysaccharide (44) (Fig. 4D). The Y. pestis mutant with an insertional lesion in ypo2150, here designated yfbA (Yersinia pestis flea biofilm regulator A), failed to form biofilms when grown as static in vitro cultures, similar to the hmsR variant (Fig. 4A). This defect could also be visualized with fluorescence microscopy of Syto9-stained static cultures, comparing wild-type and yfbA mutant biofilms (Fig. 4B and C). The biofilm defect of the yfbA mutant could be complemented in trans with plasmid-borne expression of wild-type yfbA(pyfbA) or IPTG-induced expression from the Ptac promoter (pPtac-yfbA) (Fig. 4A). Of note, the Y. pestis yfbA::Tn5 mutant formed red colonies on Congo Red agar plates, indicating that the gene and its product are not required for the biosynthesis of PNAG (Fig. 4E and F). Tn5 insertions in two other genes, ypo2458 and ypo3682, also caused a reduction of in vitro biofilm formation, although the phenotype was not as pronounced as in yfbA and hmsR mutant Y. pestis (Fig. 4A).
FIG 4.
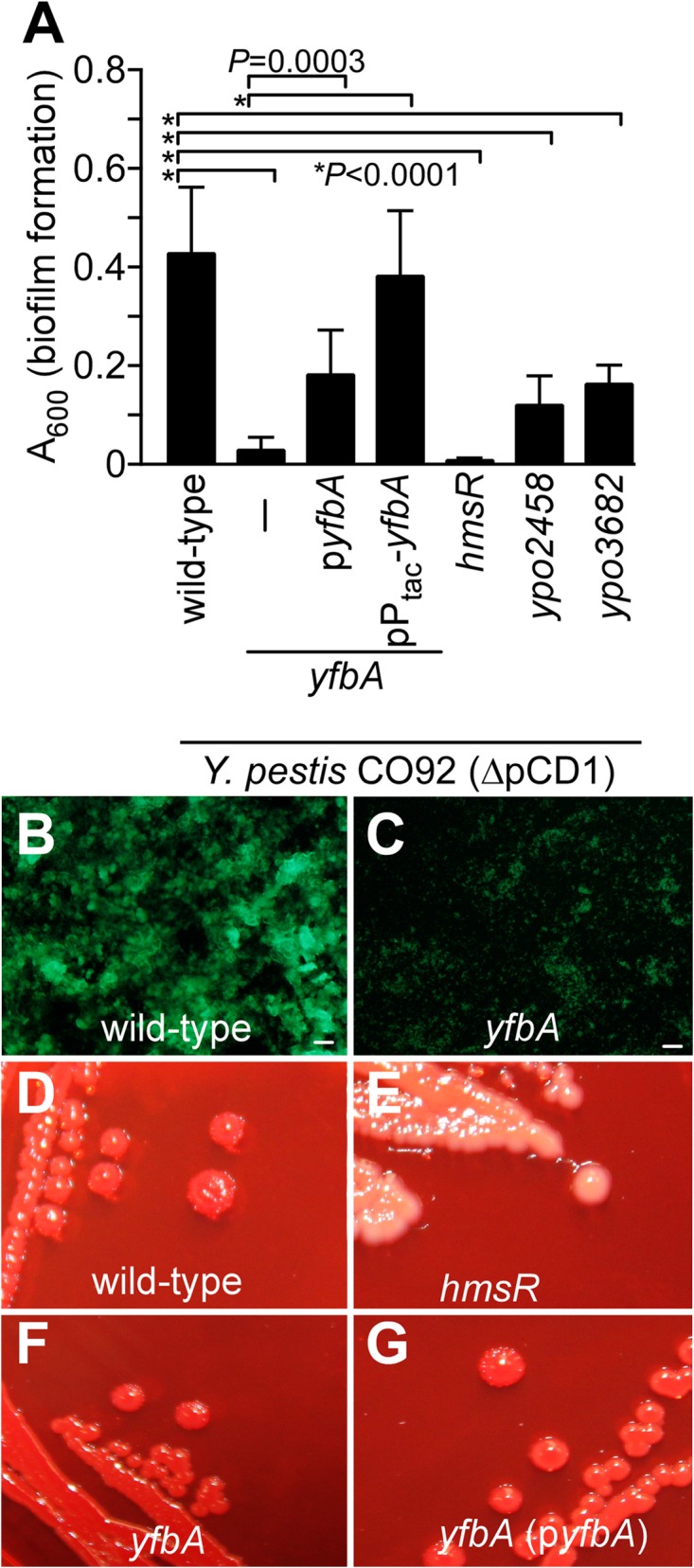
Three LysR-type transcriptional regulators contribute to Y. pestis in vitro biofilm formation. (A) A library of Y. pestis CO92(ΔpCD1) mutants with mini-Tn5 insertional lesions was screened for in vitro biofilm defects by incubating static cultures for 24 h at 26°C and crystal violet staining. This approach identified variants with insertions in ypo2150 (yfbA), ypo2458, and ypo3682, each of which encodes an LTTR. As a control, a Y. pestis mutant with an insertional lesion in hmsR was also defective for biofilm formation and plasmids pyfbA (yfbA under the control of its native promoter) and pPtac-yfbA (yfbA under the control of the IPTG-inducible Ptac promoter) increased biofilm formation of the yfbA mutant. The data represent the means and standard errors of the mean from 10 independent experimental determinations. (B and C) Biofilm cultures of wild-type Y. pestis CO92(ΔpCD1) (B) and its yfbA mutant (C) were stained with Syto9, and fluorescence microcopy images were acquired (bars, 20 μm). (D and E) A Congo Red agar plate was inoculated with Y. pestis strains and incubated at 26°C for 48 h. The microscopy images reveal the PNAG (hms)-dependent Congo Red pigmentation phenotype of wild-type Y. pestis CO92(ΔpCD1) (D) compared to the nonpigmented staining phenotype of the hmsR mutant (E). (F and G) Y. pestis CO92(ΔpCD1) yfbA (F) and yfbA(pyfbA) (G) strains form pigmented colonies on Congo Red agar plates.
yfbA is required for Y. pestis colonization of C. felis.
yfbA, ypo2458, and ypo3682 encode LysR-type transcriptional regulators (LTTRs) (12, 46). Earlier work by Vadyvaloo and colleagues and Sebbane and colleagues examined the expression of Y. pestis KIM (Medievalis) genes with microarray studies using in vitro growth at 26°C and 37°C, as well as in the gut of X. cheopis fleas or rat bubos (47, 48). This work revealed the expression of yfbA (y2171), ypo2458 (y1731), and ypo3682 (y0181) in the flea gut but not in rat bubos (47). LTTRs encompass an N-terminal DNA-binding helix-turn-helix motif and a C-terminal coinducer binding domain (49). LTRRs dimerize and bind DNA, thereby increasing the affinity of RNA polymerase for specific promoters. Their transcriptional activity can be restricted to either individual genes or many dozens of different genes (49).
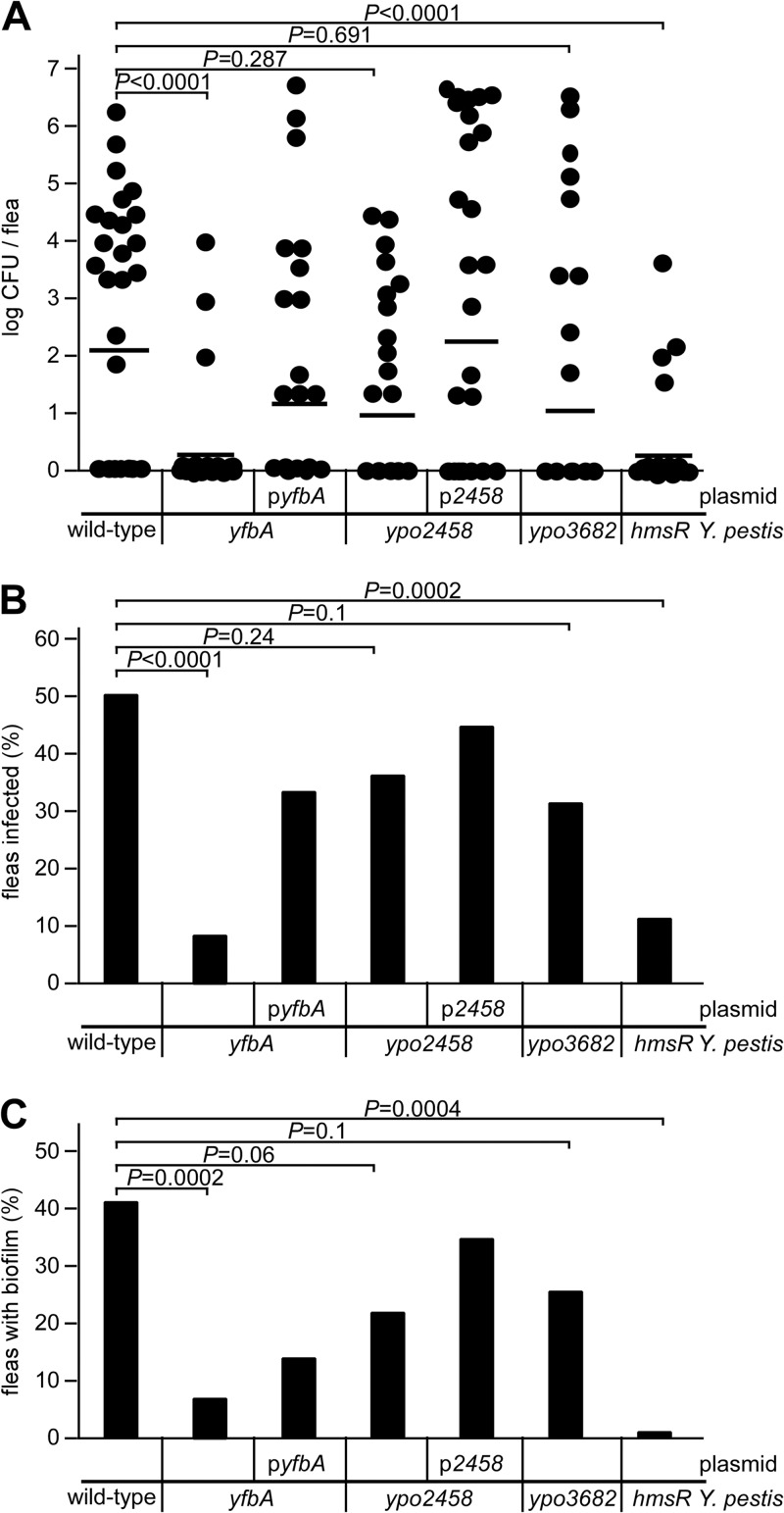
We wondered whether yfbA, ypo2458, or ypo3682 is required for Y. pestis colonization of C. felis. Fourteen days after infection, cat fleas were homogenized and analyzed for their bacterial loads. Compared to fleas infected with wild-type Y. pestis, the yfbA mutant displayed a reduced bacterial load (P < 0.0001) in the flea gut (Fig. 5A). This defect was complemented by plasmid-borne expression of yfbA(pyfbA) (Fig. 5A). Whereas 51% of infected cat fleas were colonized with wild-type Y. pestis, only 8% of infected fleas were colonized with the yfbA mutant (Fig. 5B). This colonization defect was in part restored by the plasmid-borne expression of yfbA(pyfbA) (Fig. 5B). As expected, the hmsR mutant was also defective in colonizing cat fleas (Fig. 5A B). Y. pestis ypo2458 or ypo3682 encodes other members of the LTTR family of transcriptional regulators (12). Similar to yfbA (ypo2150), Y. pestis ypo2458 or ypo3682 is preferentially expressed in the gut of Y. pestis KIM-infected X. cheopis (47). Though Y. pestis ypo2458 and ypo3682 mutants showed a reduction in biofilm formation in an in vitro assay, these mutants did not display defects in the colonization of the intestines of infected cat fleas (Fig. 5A and B).
FIG 5.
The yfbA gene is required for Y. pestis CO92(ΔpCD1) cat flea colonization and biofilm formation. (A) Y. pestis loads in the intestinal tracts of cat fleas (n = 36) infected for 14 days with wild-type or yfbA, yfbA(pyfbA), ypo2458, ypo2458(pypo2458), ypo3682, or hmsR mutant strains. The horizontal bar indicates the nonparametric mean of each data set. Significance was calculated using the Mann-Whitney test for nonparametric distributions. (B) Percentages of fleas (n = 36) producing viable CFU 14 days after infection with wild-type or yfbA, yfbA(pyfbA), ypo2458, ypo2458(pypo2458), ypo3682, or hmsR mutant Y. pestis. Significance was calculated using the Student t test. (C) Percentages of fleas harboring biofilm formation, indicative of gut blockage (n = 20 to 30), 14 days after infection with wild-type or yfbA, yfbA(pyfbA), ypo2458, ypo2458(pypo2458), ypo3682, or hmsR mutant Y. pestis. The fleas were dissected in PBS. The proventriculus and midgut were removed and viewed at ×20 magnification using an Olympus IX81 microscope. The data are representative of two independent experiments. Significance was calculated with the Student t test.
yfbA is required for Y. pestis biofilm formation in infected C. felis.
To examine the ability of the Y. pestis yfbA mutant to form biofilms in the proventriculus, infected cat fleas were examined by microscopy (Fig. 6). Compared to fleas infected with wild-type Y. pestis CO92(ΔpCD1) (42% biofilm formation), only 6% of C. felis fleas infected with the yfbA mutant harbored biofilms (Fig. 5C). This defect was in part complemented in Y. pestis yfbA(pyfbA) (Fig. 5C). As a control, the hmsR mutant did not form biofilms in any of the infected fleas (Fig. 5C). Representative images of infected flea guts and biofilm formation are shown in Fig. 6.
FIG 6.
The yfbA gene is required for Y. pestis CO92(ΔpCD1) biofilm formation in cat fleas. Cat fleas (n = 30) were dissected, and their intestinal tracts were analyzed by bright-field microscopy 14 days (d14) after infection with either wild-type Y. pestis CO92(ΔpCD1) (WT) or its hmsR, yfbA, or yfbA(pyfbA) mutant strain. Representative images were acquired and inspected for biofilm formation (arrows). Bars denote 0.1 mm.
DISCUSSION
Y. pestis infects many mammalian species, including humans, and remains endemic as a zoonotic pathogen in African, North and South American, and Asian countries (2). Although Y. pestis causes pandemic disease outbreaks in humans and remains a public health threat, plague is primarily a disease of rodents and their fleas. Rodent species are highly variable in their susceptibilities, ranging from enzootic species with high-level prolonged resistance to plague, e.g., the great gerbil (Rhombomys opimus) in Kazakhstan, to epizootic, highly susceptible species (black-tailed prairie dog) (50, 51). Plague transmission cycles were modeled, and infected off-host fleas, where Y. pestis can survive for more than a year (52), are thought to play important roles in maintaining enzootic cycles (53). Off-host questing fleas may become infected and transmit plague to new hosts, which may precipitate epizootic outbreaks from on-host fleas that transiently maintain high infectious loads of Y. pestis through repeated infectious feeds (53). Early-phase transmission by fleas can drive plague dynamics at the population level; however, late-phase regurgitative transmission also contributes to plague persistence and outbreaks (4, 6, 7, 53, 54).
C. felis, the cat flea, is one of the most abundant and widespread flea species (30). Cat fleas are capable of being infected with Y. pestis and promoting early-phase transmission of plague disease between animals and perhaps also animals and humans (30). C. felis has been investigated as a vector for plague transmission in Africa, China, and the United States (7, 29). Cat fleas feed on domesticated animals, household pets, squirrels, rats, and mice, as well as humans (30). Cat fleas can also be categorized by their behavior in leaving their hosts after feeding and remaining on the floors of animal burrows or human dwellings, where they could serve as an off-host vector for plague transmission (31). Although cat fleas promote early-phase transmission of Y. pestis, a specific mechanism or gene required for cat flea colonization or transmission of plague was heretofore not known (30). Here, we show that Y. pestis colonizes cat fleas and forms biofilms in its proventriculus and midgut, which may contribute to the accelerated mortality of infected fleas. Using a library of Y. pestis CO92(ΔpCD1) mutants with insertional Tn5 lesions and screening for in vitro biofilm defects, we identified three genes whose expression is required for efficient biofilm growth: ypo2150, ypo2458, and ypo3682. All three genes encode LTTRs, and Y. pestis KIM5 microarray studies revealed that these genes are expressed in the gastrointestinal tract of infected X. cheopis fleas (47). However, when examined for their contribution to Y. pestis colonization and biofilm formation in the intestinal tract of cat fleas, only ypo2150, but not ypo2458 and ypo3682, was required. We have designated ypo2150 yfbA (Yersinia pestis flea biofilm A), as the gene appears to contribute to efficient colonization and biofilm formation by Y. pestis in cat fleas. It seems plausible that yfbA may control the expression of a still unidentified biofilm or colonization factor. Similar phenotypes have been reported for two regulatory factors, PhoP (a two-component response regulator) and Hfq (a bacterial RNA binding protein) when studying biofilm formation in the rat flea (55, 56).
Y. pestis biofilm formation in the intestinal tract of X. cheopis is dependent on the hmsHFRS genes and production of PNAG, an extracellular polysaccharide (6). In some fleas, biofilm formation leads to blockage of the gut. Blocked or partially blocked fleas accumulate blood meals in the esophagus and may regurgitate blood mixed with biofilm material during feeding; this may contribute to the transmission of Y. pestis to new hosts (4, 6). Work over the past 2 decades identified several genetic traits of Y. pestis that are required for either colonization or biofilm formation and blockage of the intestinal tract of X. cheopis, including the ymt, hfq, gmhA, and rcs genes, as well as genes involved in the synthesis and regulation of the second messenger cyclic di-GMP. The contributions of two diguanylate cyclase genes (hmsT and hmsD), as well as hmsP, which encodes a cyclic di-GMP phosphodiesterase, and of the Rcs phosphorelay system are to the regulation of PNAG production (25, 27, 28, 44). However, the contributions of ymt, hfq, and gmhA genes cannot be explained as regulators of hmsHFRS. Hfq is a cytoplasmic protein that binds many different small RNAs and controls their stability and regulatory attributes (57, 58). Y. pestis hfq is expressed both in the mammalian host and in flea intestines (47, 48); however, a specific RNA(s) that supports bacterial adaptation to the flea gut is not yet known (56). Y. pestis ymt encodes cytoplasmic phospholipase D, which is not required for laboratory growth or the pathogenesis of plague in mice (16). Y. pestis ymt mutants are eliminated from the flea gut and assume a spheroplast-like morphology, suggesting that phospholipase D may be responsible for neutralizing a toxic compound in flea intestines (59). Finally, gmhA, encoding a phosphoheptose isomerase, is thought to promote lipopolysaccharide (LPS) biosynthesis, and a gmhA mutant cannot effectively block the proventriculus of infected fleas (60).
Douglas and Wheeler studied the capacity of C. felis to become infected with Y. pestis and to transmit plague from infected guinea pigs or mice to healthy animals (61). This work suggested that cat fleas may become infected but are not blocked following Y. pestis infection (61). While infected C. felis fleas were capable of early-phase transmission with mass-infected fleas, two trials with small numbers of fleas (either 6 or 8 C. felis fleas) did not show transmission of Y. pestis, suggesting that cat fleas have low transmission potential (62). Using an artificial feeding system with defribrinated sheep's blood, we show here that about 50% of infected C. felis fleas are colonized and eventually blocked, which was associated with diminished survival of colonized fleas. We have not examined blocked C. felis for its ability to transmit Y. pestis to mammalian hosts. If one considers the abundance and global distribution of C. felis and its propensity as a questing, off-host species to feed on domesticated animals, rodents, and humans, a more thorough investigation of cat fleas as a vector for early-phase and late-phase transmission of plague seems warranted. The foundation for such studies is provided here, as we demonstrate both early-phase colonization and late-phase biofilm formation in Y. pestis-infected cat fleas.
ACKNOWLEDGMENTS
We thank Lauriane Quenee, Bill Blaylock, and members of our laboratory for experimental assistance, critical comments and discussion.
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under grant/contract no. U19 AI107792 and RO1AI042797. We acknowledge membership of and support from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIH award 1-U54-AI-057153).
Footnotes
Published ahead of print 3 January 2014
REFERENCES
- 1.Yersin A. 1894. La peste bubonique à Hong-Kong. Ann. Inst. Pasteur 2:428–430 [Google Scholar]
- 2.Gage KL, Kosoy MY. 2005. Natural history of plague: perspectives from more than a century of research. Annu. Rev. Entomol. 50:505–528. 10.1146/annurev.ento.50.071803.130337 [DOI] [PubMed] [Google Scholar]
- 3.Simond PL. 1898. La propagation de la peste. Ann. Inst. Pasteur 12:625–687 [Google Scholar]
- 4.Bacot AW, Martin CJ. 1914. Observations on the mechanism of the transmission of plague by fleas. J. Hyg. 13:423–439 [PMC free article] [PubMed] [Google Scholar]
- 5.Vetter SM, Eisen RJ, Schotthoefer AM, Montenieri JA, Holmes JL, Bobrov AG, Bearden SW, Perry RD, Gage KL. 2010. Biofilm formation is not required for early-phase transmission of Yersinia pestis. Microbiology 156:2216–2225. 10.1099/mic.0.037952-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinnebusch BJ. 2012. Biofilm-dependent and biofilm-independent mechanisms of transmission of Yersinia pestis by fleas. Adv. Exp. Med. Biol. 954:237–243. 10.1007/978-1-4614-3561-7_30 [DOI] [PubMed] [Google Scholar]
- 7.Eisen RJ, Gage KL. 2012. Transmission of flea-borne zoonotic agents. Annu. Rev. Entomol. 57:61–82. 10.1146/annurev-ento-120710-100717 [DOI] [PubMed] [Google Scholar]
- 8.Pollitzer R. 1951. Plague studies. 1. A summary of the history and survey of the present distribution of the disease. Bull. World Health Organ. 4:475–533 [PMC free article] [PubMed] [Google Scholar]
- 9.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 96:14043–14048. 10.1073/pnas.96.24.14043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anisimov A, Lindler L, Pier G. 2004. Intraspecific diversity of Yersinia pestis. Clin. Microbiol. Rev. 17:434–464. 10.1128/CMR.17.2.434-464.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, Feldkamp M, Kusecek B, Vogler AJ, Li Y, Cui Y, Thomson NR, Jombart T, Leblois R, Lichtner P, Rahalison L, Petersen JM, Balloux F, Keim P, Wirth T, Ravel J, Yang R, Carniel E, Achtman M. 2010. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat. Genet. 42:1140–1143. 10.1038/ng.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkhill J, Wren BW, Thompson NR, Titball RW, Holden MT, Prentice MB, Sebaihia M, James KD, Churcher C, Mungall KL, Baker S, Dasham D, Bentley SD, Brokks K, Cerdeno-Tarraga AM, Chillingworth T, Cronin A, Davies RM, Davis P, Dougan G, Feltwell T, Hamlin N, Holroyd S, Jagels K, Karlyshev AV, Leather S, Moule S, Oyston PC, Quail M, Rutherford K, Simmonds M, Skelton J, Stevens K, Whitehead S, Barrell BG. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523–527. 10.1038/35097083 [DOI] [PubMed] [Google Scholar]
- 13.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol. Rev. 62:1315–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marketon MM, DePaolo RW, DeBord KL, Jabri B, Schneewind O. 2005. Plague bacteria target immune cells during infection. Science 309:1739–1741. 10.1126/science.1114580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sebbane F, Jarrett C, Gardner D, Long D, Hinnebusch BJ. 2009. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect. Immun. 77:1222–1229. 10.1128/IAI.00950-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733–735. 10.1126/science.1069972 [DOI] [PubMed] [Google Scholar]
- 17.Hinnebusch BJ, Fischer ER, Schwan TG. 1998. Evaluation of the role of the Yersinia pestis plasminogen activator and other plasmid-encoded factors in temperature-dependent blockage of the flea. J. Infect. Dis. 178:1406–1415. 10.1086/314456 [DOI] [PubMed] [Google Scholar]
- 18.Sodeinde O, Subrahmanyam Y, Stark K, Quan T, Bao Y, Goguen J. 1992. A surface protease and the invasive character of plague. Science 258:1004–1007. 10.1126/science.1439793 [DOI] [PubMed] [Google Scholar]
- 19.Perry RD, Fetherston JD. 1997. Yersinia pestis: etiologic agent of plague. Clin. Microbiol. Rev. 10:35–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perry RD, Pendrak ML, Schuetze P. 1990. Identification and cloning of a hemin storage locus involved in the pigmentation phenotype of Yersinia pestis. J. Bacteriol. 172:5929–5937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erickson DL, Jarrett CO, Callison JA, Fischer ER, Hinnebusch BJ. 2008. Loss of biofilm-inhibiting glycosyl hydrolase during the emergence of Yersinia pestis. J. Bacteriol. 190:8163–8170. 10.1128/JB.01181-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chain PS, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. U. S. A. 101:13826–13831. 10.1073/pnas.0404012101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erickson DL, Jarrett CO, Wren BW, Hinnebusch BJ. 2006. Serotype differences and lack of biofilm formation characterize Yersinia pseudotuberculosis infection of the Xenopsylla cheopis flea vector of Yersinia pestis. J. Bacteriol. 188:1113–1119. 10.1128/JB.188.3.1113-1119.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erickson DL, Waterfield NR, Vadyvaloo V, Long D, Fischer ER, Ffrench-Constant R, Hinnebusch BJ. 2007. Acute oral toxicity of Yersinia pseudotuberculosis to fleas: implications for the evolution of vector-borne transmission of plague. Cell Microbiol. 9:2658–2666. 10.1111/j.1462-5822.2007.00986.x [DOI] [PubMed] [Google Scholar]
- 25.Sun YC, Guo XP, Hinnebusch BJ, Darby C. 2012. The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J. Bacteriol. 194:2020–2026. 10.1128/JB.06243-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun YC, Hinnebusch BJ, Darby C. 2008. Experimental evidence for negative selection in the evolution of a Yersinia pestis pseudogene. Proc. Natl. Acad. Sci. U. S. A. 105:8097–8101. 10.1073/pnas.0803525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79:533–551. 10.1111/j.1365-2958.2010.07470.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun YC, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, Darby C, Hinnebusch BJ. 2011. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6:e19267. 10.1371/journal.pone.0019267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gage KL, Dennis DT, Orloski KA, Ettestad P, Brown TL, Reynolds PJ, Pape WJ, Fritz CL, Carter LG, Stein JD. 2000. Cases of cat-associated human plague in the Western US, 1977–1998. Clin. Infect. Dis. 30:893–900. 10.1086/313804 [DOI] [PubMed] [Google Scholar]
- 30.Eisen RJ, Borchert JN, Holmes JL, Amatre G, Van Wyk K, Enscore RE, Babi N, Atiku LA, Wilder AP, Vetter SM, Bearden SW, Montenieri JA, Gage KL. 2008. Early-phase transmission of Yersinia pestis by cat fleas (Ctenocephalides felis) and their potential role as vectors in a plague-endemic region of Uganda. Am. J. Trop. Med. Hyg. 78:949–956 [PubMed] [Google Scholar]
- 31.Graham CB, Borchert JN, Black WC, Atiku LA, Mpanga JT, Boegler KA, Moore SM, Gage KL, Eisen RJ. 2013. Blood meal identification in off-host cat fleas (Ctenocephalides felis) from a plague-endemic region of Uganda. Am. J. Trop. Med. Hyg. 88:381–389. 10.4269/ajtmh.2012.12-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salkeld DJ, Stapp P. 2006. Seroprevalence rates and transmission of plague (Yersinia pestis) in mammalian carnivores. Vector Borne Zoonotic Dis. 6:231–239. 10.1089/vbz.2006.6.231 [DOI] [PubMed] [Google Scholar]
- 33.Orloski KA, Eidson M. 1995. Yersinia pestis infection in three dogs. J. Am. Vet. Med. Assoc. 207:316–318 [PubMed] [Google Scholar]
- 34.Kilonzo BS, Mbise TJ, Mwalimu DC, Kindamba L. 2006. Observations on the endemicity of plague in Karatu and Ngorongo, northern Tanzania. Tanzan. Health Res. Bull. 8:1–6 [DOI] [PubMed] [Google Scholar]
- 35.Gould LH, Pape J, Ettestad P, Griffith KS, Mead PS. 2008. Dog-associated risk factors for human plague. Zoonoses Public Health 55:448–454. 10.1111/j.1863-2378.2008.01132.x [DOI] [PubMed] [Google Scholar]
- 36.Yin JX, Geater A, Chongsuvivatwong V, Dong XQ, Du CH, Zhong YH. 2011. Predictors for abundance of host flea and floor flea in households of villages with endemic commensal rodent plague, Yunnan Province, China. PLoS Negl. Trop. Dis. 5:e997. 10.1371/journal.pntd.0000997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 38.Wade SE, Georgi JR. 1988. Survival and reproduction of artificially fed cat fleas, Ctenocephalides felis Bouché (Siphonaptera: Pulicidae). J. Med. Entomol. 25:186–190 [DOI] [PubMed] [Google Scholar]
- 39.Anderson DM, Schneewind O. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140–1143. 10.1126/science.278.5340.1140 [DOI] [PubMed] [Google Scholar]
- 40.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. 1987. High-copy-number and low-copy-number plasmid vectors for LacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene 61:63–74. 10.1016/0378-1119(87)90365-9 [DOI] [PubMed] [Google Scholar]
- 41.Noden BH, Radulovic S, Higgins JA, Azad AF. 1998. Molecular identification of Rickettsia typhi and R. felis in co-infected Ctenocephalides felis (Siphonaptera: Pulicidae). J. Med. Entomol. 35:410–414 [DOI] [PubMed] [Google Scholar]
- 42.Forman S, Bobrov AG, Kirillina O, Craig SK, Abney J, Fetherston JD, Perry RD. 2006. Identification of critical amino acid residues in the plague biofilm Hms proteins. Microbiology 152:3399–3410. 10.1099/mic.0.29224-0 [DOI] [PubMed] [Google Scholar]
- 43.Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, Hinnebusch BJ. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783–792. 10.1086/422695 [DOI] [PubMed] [Google Scholar]
- 44.Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:75–88. 10.1111/j.1365-2958.2004.04253.x [DOI] [PubMed] [Google Scholar]
- 45.Gerke C, Kraft A, Süssmuth R, Schweitzer O, Götz F. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586–18593. 10.1074/jbc.273.29.18586 [DOI] [PubMed] [Google Scholar]
- 46.Henikoff S, Haughn GW, Calvo JM, Wallace JC. 1988. A large family of bacterial activator proteins. Proc. Natl. Acad. Sci. U. S. A. 85:6602–6606. 10.1073/pnas.85.18.6602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog. 6:e1000783. 10.1371/journal.ppat.1000783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sebbane F, Lemaître N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. 2006. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. U. S. A. 103:11766–11771. 10.1073/pnas.0601182103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maddocks SE, Oyston PC. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609–3623. 10.1099/mic.0.2008/022772-0 [DOI] [PubMed] [Google Scholar]
- 50.Webb CT, Brooks CP, Gage KL, Antolin MF. 2006. Classic flea-borne transmission does not drive plague epizootics in prairie dogs. Proc. Natl. Acad. Sci. U. S. A. 103:6236–6241. 10.1073/pnas.0510090103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis S, Begon M, De Bruyn L, Ageyev VS, Klasovskiy NL, Pole SB, Viljugrein H, Stenseth NC, Leirs H. 2004. Predictive thresholds for plague in Kazakhstan. Science 304:736–738. 10.1126/science.1095854 [DOI] [PubMed] [Google Scholar]
- 52.Bazanova LP, Maevskii MP. 1996. The duration of the persistence of plague microbe in the body of flea Citellophilus tesquorum altaicus. Med. Parasitol. 2:45–48 [PubMed] [Google Scholar]
- 53.Buhnerkempe MG, Eisen RJ, Goodell B, Gage KL, Antolin MF, Webb CT. 2011. Transmission shifts underlie variability in population responses to Yersinia pestis infection. PLoS One 6:e22498. 10.1371/journal.pone.0022498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. 2006. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc. Natl. Acad. Sci. U. S. A. 103:15380–15385. 10.1073/pnas.0606831103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rebeil R, Jarrett CO, Driver JD, Ernst RK, Oyston PCF, Hinnebusch BJ. 2013. Induction of the Yersinia pestis PhoP-PhoQ regulatory system in the flea and its role in producing a transmissible infection. J. Bacteriol. 195:1920–1930. 10.1128/JB.02000-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rempe KA, Hinz AK, Vadyvaloo V. 2012. Hfq regulates biofilm gut blockage that facilitates flea-borne transmission of Yersinia pestis. J. Bacteriol. 194:2036–2040. 10.1128/JB.06568-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koo JT, Alleyne TM, Schiano CA, Jafari N, Lathem WW. 2011. Global discovery of small RNAs in Yersinia pseudotuberculosis identifies Yersinia-specific small, noncoding RNAs required for virulence. Proc. Natl. Acad. Sci. U. S. A. 108:E710–E717. 10.1073/pnas.1101655108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Lay N, Schu DJ, Gottesman S. 2013. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288:7996–8003. 10.1074/jbc.R112.441386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hinnebusch BJ. 2003. Transmission factors: Yersinia pestis genes required to infect the flea vector of plague. Adv. Exp. Med. Biol. 529:55–62. 10.1007/0-306-48416-1_11 [DOI] [PubMed] [Google Scholar]
- 60.Darby C, Ananth SL, Tan L, Hinnebusch BJ. 2005. Identification of gmhA, a Yersinia pestis gene required for flea blockage, by using a Caenorhabditis elegans biofilm system. Infect. Immun. 73:7236–7242. 10.1128/IAI.73.11.7236-7242.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Douglas JR, Wheeler CM. 1943. Sylvatic plague studies. II. The fate of Pasteurella pestis in the flea. J. Infect. Dis. 72:18–30 [Google Scholar]
- 62.Wheeler CM, Douglas JR. 1945. Sylvatic plague studies. V. The determination of vector efficiency. J. Infect. Dis. 77:1–12 [Google Scholar]