Abstract
Iron-sulfur (Fe-S) clusters are ubiquitous cofactors that are crucial for many physiological processes in all organisms. In Escherichia coli, assembly of Fe-S clusters depends on the activity of the iron-sulfur cluster (ISC) assembly and sulfur mobilization (SUF) apparatus. However, the underlying molecular mechanisms and the mechanisms that control Fe-S cluster biogenesis and iron homeostasis are still poorly defined. In this study, we performed a global screen to identify the factors affecting Fe-S cluster biogenesis and iron homeostasis using the Keio collection, which is a library of 3,815 single-gene E. coli knockout mutants. The approach was based on radiolabeling of the cells with [2-14C]dihydrouracil, which entirely depends on the activity of an Fe-S enzyme, dihydropyrimidine dehydrogenase. We identified 49 genes affecting Fe-S cluster biogenesis and/or iron homeostasis, including 23 genes important only under microaerobic/anaerobic conditions. This study defines key proteins associated with Fe-S cluster biogenesis and iron homeostasis, which will aid further understanding of the cellular mechanisms that coordinate the processes. In addition, we applied the [2-14C]dihydrouracil-labeling method to analyze the role of amino acid residues of an Fe-S cluster assembly scaffold (IscU) as a model of the Fe-S cluster assembly apparatus. The analysis showed that Cys37, Cys63, His105, and Cys106 are essential for the function of IscU in vivo, demonstrating the potential of the method to investigate in vivo function of proteins involved in Fe-S cluster assembly.
INTRODUCTION
Iron-sulfur (Fe-S) clusters, which are crucial for all organisms and thought to be among the earliest catalysts in the evolution of biomolecules, serve as electron carriers in redox reactions, substrate binding and activation, redox catalysis, DNA replication and repair, regulation of gene expression, and tRNA modification (1–3). The most abundant types of Fe-S clusters are rhombic [2Fe-2S] and cubane [4Fe-4S] (2, 4), in which the iron atoms are generally bound to the sulfur atoms in the cysteine residues of the peptide backbone and to inorganic sulfurs in the prosthetic group.
The assembly of Fe-S clusters is a very complex process mediated by multiple protein apparatus. In Escherichia coli, Fe-S clusters are primarily synthesized with the help of the iron-sulfur cluster (ISC) assembly components, IscUSA, HscBA, and Fdx, encoded by the isc operon. This system includes a Fe-S cluster assembly scaffold (IscU), a sulfur-trafficking cysteine desulfurase (IscS), molecular chaperones (HscB and HscA), a ferredoxin (Fdx), and a possible iron donor or auxiliary Fe-S cluster assembly protein (IscA) (5–7). The second Fe-S cluster assembly system, the sulfur mobilization (SUF) machinery consisting of six proteins encoded by the sufABCDSE operon, operates during stress conditions such as iron starvation, oxidative damage, or heavy metal exposure (8–10). In addition, several other proteins have potential roles in Fe-S cluster biogenesis: (i) proposed iron donors CyaY (11, 12) and ferritin (13); (ii) the putative intermediate Fe-S cluster carrier/scaffold proteins NfuA (14, 15), GrxD (16), and Mrp (ApbC) (17); (iii) the IscA/SufA-homologous A-type carrier protein ErpA (18, 19); (iv) the putative Fe-S cluster repair protein YtfE (20). Despite extensive studies on Fe-S cluster biosynthesis, defining the molecular mechanism of Fe-S cluster assembly is still a challenge due to the intrinsic chemical nature of Fe-S cluster species. For instance, Fe-S clusters can be spontaneously formed by a nonenzymatic reaction in vitro, making it difficult to assess the specific functions of the individual components of this assembly process.
During our investigation of Fe-S proteins that depend on the iscS gene in E. coli, we identified dihydropyrimidine dehydrogenase (DPD) as one of the iscS-dependent proteins (21). DPD, a heterotetrameric protein composed of two PreT subunits and two PreA subunits, contains a flavin adenine dinucleotide (FAD), a flavin mononucleotide (FMN), and four [4Fe-4S] clusters and catalyzes the reduction of uracil (and also thymine) to its 5,6-dihydro-derivatives using NADH as a specific cosubstrate (22). The enzyme also catalyzes the reverse reaction, i.e., the NAD+-dependent oxidation of 5,6-dihydrouracil to uracil. Interestingly, we found that a DPD-deficient strain cultured in a medium containing radiolabeled dihydrouracil (DHU) was markedly less radioactive than the wild-type strain. This is because radiolabeled uracil produced from radiolabeled DHU by DPD is converted to nucleobases by the pyrimidine metabolic pathway, and the radiolabeled nucleobases are eventually incorporated into both RNA and DNA in the wild-type strain (22) (Fig. 1). The maturation of the Fe-S cluster in DPD appears to be highly sensitive to the function of the ISC system (21). Our previous study suggests that apo-DPD is highly susceptible to degradation since neither PreT nor PreA was detectable in a Western blot analysis of the extract of an iscS mutant strain (21). Therefore, there is a good correlation between activity levels of DPD, which requires an Fe-S cluster biosynthetic system, and the radioactivity of E. coli cells cultured with radiolabeled DHU. This interesting correlation led us to the idea that dysfunctions in Fe-S cluster biogenesis and/or iron homeostasis in an E. coli mutant strain could readily be identified by a low level of radioactivity due to a reduced incorporation of the radiolabeled DHU.
FIG 1.
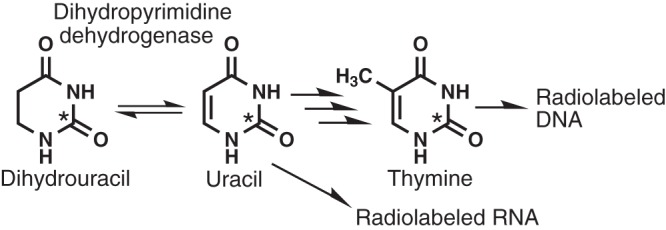
Dihydrouracil metabolism in E. coli K-12. Asterisks indicate the radiolabeled carbon atoms.
In the present work, we have developed a novel strategy using [2-14C]DHU to analyze the in vivo function of Fe-S cluster biogenesis factors. This novel approach has been applied to screen the 3,815 E. coli knockout strains (the Keio collection) for defects in Fe-S cluster biogenesis and iron homeostasis. The Keio collection is a set of systematically created, single-gene knockouts with in-frame (nonpolar) deletions in all nonessential genes in E. coli K-12 (23). Open reading frame coding regions were replaced with a kanamycin cassette, which was further excised to create the in-frame deletions. We have identified several novel factors affecting Fe-S cluster biogenesis and iron homeostasis. Based on the results, we here provide a more comprehensive view of Fe-S cluster biogenesis and iron homeostasis in E. coli. Moreover, we also applied our method to identify amino acid residues of IscU that are essential for in vivo Fe-S cluster assembly. The results suggest that our novel method is a useful and powerful tool for clarifying Fe-S cluster biosynthetic mechanisms in E. coli.
MATERIALS AND METHODS
Strains and materials.
The Keio collection and the parental strain E. coli K-12 BW25113 [genotype, rrnB3 ΔlacZ4787 hsdR514 Δ(araBAD)567 Δ(rhaBAD)568 rph-1] were provided by the National BioResource Project (NBRP), National Institute of Genetics (NIG), Japan. The Keio collection (23) is a systematic single-gene knockout library of all nonessential genes in E. coli, which allows rapid screening and detection of genes (http://www.shigen.nig.ac.jp/ecoli/strain/nbrp/keioCollectionList.jsp). A library of 3,815 nonlethal single-gene knockout strains was used for screening in this study (see Table S1 in the supplemental material). For the static culture, E. coli cells were cultivated in a 1.2-ml 96-well polystyrene plate with a gas-permeable adhesive seal (Thermo Scientific ABgene, Epsom, United Kingdom). [2-14C]dihydrouracil and [2-14C]uracil were from Moravek Biochemicals (Brea, CA). All chemicals used were analytical-grade reagents.
Screening procedure.
For the first screen, each E. coli knockout mutant from the Keio collection was inoculated into 1 ml of Luria-Bertani (LB) medium containing 0.5 μM [2-14C]DHU (962 Bq · ml−1) in a 96-well plate and statically incubated at 37°C for 24 h. The cells were then harvested, washed with 0.8% saline, and added to 3 ml of a liquid scintillation cocktail (Clear-sol II; (Nacalai Tesque, Kyoto, Japan). The radioactivity of the cells was measured using a liquid scintillation counter (Beckman Coulter, Brea, CA). In the second screen, the radioactivity was determined in the same manner as in the first screen and was normalized to the growth (cpm per optical density unit at 600 nm [cpm · OD600−1]) of cells. The third screen was performed similarly, except that [2-14C]uracil (1,028 Bq · ml−1) was used in place of [2-14C]DHU. Further analysis was performed using aerobic cultivation of mutant strains in 1.5 ml of LB medium containing 0.5 μM [2-14C]DHU (962 Bq · ml−1) at 37°C for 24 h. The cells were harvested, and their radioactivity was measured as described above.
Gene cloning and site-directed mutagenesis.
The iscU gene was PCR amplified using E. coli K-12 BW25113 genomic DNA as a template and the two primers EcIscUF and EcIscUR (Table 1). The amplified DNA was inserted into the EcoRI/XbaI sites in pUC118 to yield pIscU, which was used for the expression of IscU under the regulation of a lac promoter. To produce four mutants of IscU (C37H, C63H, H105C, and C106H), the QuikChange site-directed mutagenesis method following the manufacturer's protocol (Qiagen, Venlo, Netherlands) was applied with appropriate mutagenic primers as listed in Table 1.
TABLE 1.
Primers used in this study
| Primer function and name | Sequence (5′–3′)a |
|---|---|
| Cloning of the E. coli iscU gene | |
| EcIscUF | GGGAATTCAGGAGGTGCCATATGGCTTACAGCGAAAAAGTTAT |
| EcIscUR | GCTCTAGATTATTTTGCTTCACGTTTGCT |
| Site-directed mutagenesis of IscU | |
| IscU C37H Fw | GCACCGGCCCACGGCGACGTG |
| IscU C37H Rv | CACGTCGCCGTGGGCCGGTGC |
| IscU C63H Fw | ACTTACGGCCACGGTTCCGCT |
| IscU C63H Rv | AGCGGAACCGTGGCCGTAAGT |
| IscU H105C Fw | CGGTGAAAATTTGCTGTTCTATTCT |
| IscU H105C Rv | AGAATAGAACAGCAAATTTTCACCG |
| IscU C106H Fw | AAAATTCACCACTCTATTCTG |
| IscU C106H Rv | CAGAATAGAGTGGTGAATTTT |
Underlining indicates the recognition sites of restriction enzymes.
Genetic complementation analysis.
E. coli K-12 iscU-null mutant [genotype, BW25113 ΔiscU] cells harboring a plasmid coding for wild-type or mutated IscU were cultured aerobically in 4 ml of LB medium containing 0.5 μM [2-14C]DHU (962 Bq · ml−1), 100 μg · ml−1 ampicillin, and 1% glycerol at 37°C until stationary phase. The cells were harvested from a 1.5-ml culture and washed once, and their radioactivity was measured.
Enzyme assays.
Cells were grown in LB medium for the DPD assay, in LB medium containing 0.2% glucose for glutamate synthase (GS) (24) and glucose-6-phosphate dehydrogenase assays or in LB medium containing 0.1% gluconate for a 6-phosphogluconate dehydratase (6-PG) assay. Cells were harvested at late logarithmic phase. The crude extract obtained from each strain by sonication was used for the enzyme assays.
6-PG activity was assayed by the production of pyruvate, which was determined using lactate dehydrogenase (25). GS activity was determined by the oxidation of NADPH (26). Isocitrate dehydrogenase (27) and glucose 6-phosphate dehydrogenase (28) activities were assayed by the production of NADPH. DPD activity was determined by the production of DHU from uracil by using high-performance liquid chromatography (HPLC). Protein concentration was determined by the Bradford method (29).
Western blot analysis.
The crude extracts were subjected to SDS-PAGE, and the proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P membrane; Merck Millipore, Billerica, MA). Immunodetection was performed with antibodies raised against E. coli IscU (30) and E. coli GroEL as a loading control (Sigma-Aldrich, Saint Louis, MO) and visualized with an ECL detection reagent (GE Healthcare, Uppsala, Sweden).
RESULTS
Using [2-14C]DHU to examine the in vivo functions of Fe-S cluster biogenesis factors.
Previously, we reported that the radioactivity in the E. coli K-12 cells cultivated in LB medium containing [2-14C]DHU markedly increases as soon as the growth enters stationary phase (22). Radiolabeled uracil and thymine formed from [2-14C]DHU by DPD are incorporated into nucleic acids (i.e., DNA and RNA), resulting in high levels of radioactivity in the wild-type strain (ca. 30,000 cpm · OD600−1) (22). In contrast, the radioactivity in the DPD-deficient strain (ca. 100 cpm · OD600−1) is about 300-fold lower than that in the wild-type strain because the increase in radioactivity is dependent on DPD activity (22). Our site-directed mutagenesis study revealed that each of Cys26, Cys77, Cys344, and Cys378 is required for DPD activity as a ligand for each of the four Fe-S clusters, suggesting that each of four [4Fe-4S] clusters is essential for DPD function (data not shown). Thus, we assumed that defects in the ISC system that is essential for Fe-S cluster biosynthesis lead to DPD inactivation, eventually resulting in low radioactivity in an isc mutant strain compared to that in the wild type. We then aerobically cultivated each of the null mutant ΔiscS, ΔiscU, ΔiscA, ΔhscA, ΔhscB, and Δfdx strains with [2-14C]DHU and examined their radioactivity. As expected, the radioactivity of all the mutant strains was markedly lower than that of the wild-type strain (Fig. 2). The radioactivity levels of ΔiscU and ΔiscA mutants (40% and 30%, respectively, of the wild-type 14C cpm level) were relatively higher than those of the other null mutants (1.3%, 5.1%, 17%, and 27% of the wild-type 14C cpm level for the ΔiscS, ΔhscA, ΔhscB, and Δfdx strains, respectively), consistent with a previous observation that Fe-S enzyme activities are higher in the iscU and iscA mutants than in the iscS, hscA, and fdx strains (31). The results indicate that our new approach is sufficiently sensitive and useful for evaluating the roles of the proteins involved in Fe-S cluster biosynthesis in vivo.
FIG 2.
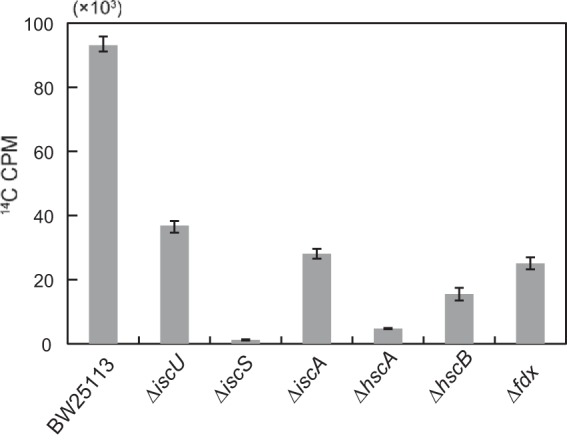
[2-14C]DHU radiolabeling of the wild-type (BW25113) and isc mutant strains that lack the genes essential for Fe-S cluster biosynthesis. Radioactivity (14C cpm) of the wild-type strain (BW25113) and the isc mutant strains, which were aerobically grown in 5 ml of LB medium containing 0.5 μM [2-14C]DHU (962 Bq · ml−1) to stationary phase, was measured with a liquid scintillation counter. The means ± standard deviation of three independent measurements is shown.
Global identification of genes affecting Fe-S enzyme activity.
We employed our radiolabeling method for the screening of the E. coli Keio collection to identify factors affecting Fe-S cluster biogenesis and iron homeostasis. If a strain has a deletion genotype in a gene essential for DPD activity, the radioactivity in the knockout strain would be lower than that in the wild-type strain. Such knockout strains are expected to include those deficient in Fe-S cluster homeostasis (including assembly, disassembly, and stability) and iron homeostasis.
The screening procedure and the results are illustrated in Fig. 3. Under the anaerobic growth conditions used in the first screening experiment, the radioactivity level of the wild-type strain was 1,236 cpm, and the basal radioactivity levels of preT and preA mutants were 4.4% (55 cpm) and 2.4% (30 cpm), respectively, of the wild-type level (see Table S1 in the supplemental material). Based on the observation that the radioactivity levels of the ISC component mutant strains were not more than 40% (for the ΔiscU strain) of that of the wild-type strain (Fig. 2), we applied a threshold of 40% (494 cpm) of the wild-type cpm level (1,236 cpm) to filter out noncandidate mutants for the first screening step. The first step provided 78 gene knockout strains (excluding preT and preA strains) whose radioactivity level was less than 40% of the wild-type cpm level (see Table S1). At this point, the decrease in radioactivity could be attributed either to the inactivation of DPD or to low cell numbers caused by the growth retardation of each mutant. Therefore, in the second screen, the radioactivity and the growth (OD600) of the 78 knockout mutants were measured under the same cultivation conditions as the first screen in order to normalize the net radioactivity to cell number, thereby excluding the possibility that the low radioactivity in a mutant is simply due to a growth defect phenotype. For the second screen, a threshold of 50% of the wild-type 14C cpm · OD600−1 value was employed because the cpm · OD600−1 value of the ΔiscU strain (14,447) was 48% of that of the wild-type strain (30,123) due to the moderate growth retardation phenotype of the ΔiscU strain. This additional screen filtered the results down to 65 knockout strains that carried deletions in candidate genes affecting Fe-S cluster biogenesis and iron homeostasis (see Table S2 in the supplemental material).
FIG 3.
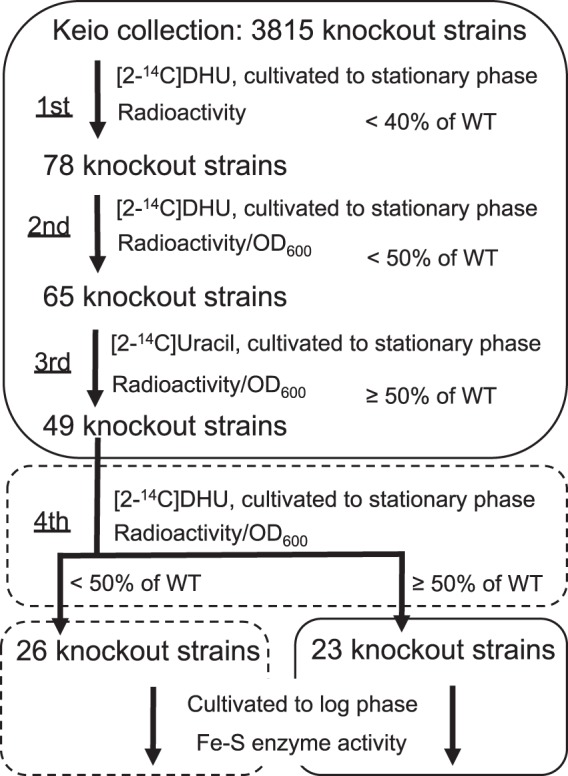
The screening procedure and summary of identification of the genes affecting Fe-S biogenesis and iron homeostasis. In the first screening step, knockout strains whose radioactivity (14C cpm) was less than 40% of that of the wild-type (WT) strain were selected. In the second screen, knockout strains whose radioactivity (14C cpm · OD600−1) was less than 50% of the wild-type level were selected. In the third screening step, knockout strains exhibiting radioactivity less than half of that of the wild-type strain were excluded. In the fourth step, each strain was aerobically cultivated with shaking in LB medium containing [2-14C]DHU. After the fourth step, each of the 26 knockout strains whose radioactivity was less than half of that of the wild-type strain was aerobically cultivated in LB broth with shaking, while each of the 23 knockout strains exhibiting radioactivity more than half of the wild-type strain was statically cultivated in LB medium for Fe-S enzyme assays. The boxes with the solid and broken lines indicate that the cultivations were carried out microaerobically with static culture and aerobically with shaking, respectively.
We assumed that the 65 knockout strains selected by the first and second screens included those with a deletion in nucleobase metabolism and RNA/DNA synthesis because lower radioactivity could also result simply from insufficient incorporation of radiolabeled uracil and its metabolites into RNA and DNA. To exclude such mutant strains, we repeated the radiolabeling method with [2-14C]uracil instead of [2-14C]DHU. Of the 65 strains, 16 knockout strains exhibited less than half the level of radioactivity seen in the wild-type strain (10,024 cpm · OD600−1) (see Table S3 in the supplemental material). These 16 strains include mutants with a deletion in the upp gene encoding uracil phosphoribosyl transferase and the dnaK gene encoding the HSP70 molecular chaperone. This result suggests that these genes are not important for either DPD activation or Fe-S cluster biosynthesis but, instead, play a role in other processes, such as nucleobase metabolism and protein folding. Thus, these 16 strains were excluded, and the remaining 49 strains were chosen for further analysis.
The screens described above were carried out using static cultivation, which is considered to recapitulate microaerobic/anaerobic conditions. In order to clarify whether the genes we identified are also important for Fe-S cluster biogenesis and/or iron homeostasis under fully aerobic conditions, we next examined the 49 knockout strains using the radiolabeling method in cultures that were shaken extensively. A threshold of 50% of the wild-type 14C cpm · OD600−1 value was again used in this step for the above-mentioned reason. This experiment identified 23 strains with radioactivity more than half of that of the wild-type strain (12,427 cpm · OD600−1) under aerobic conditions (see Table S4 in the supplemental material). This result suggests that the genes disrupted in these strains are essential for the activity of DPD only under microaerobic/anaerobic conditions. On the other hand, the remaining 26 strains with reduced radioactivity under both microaerobic/anaerobic and highly aerobic conditions likely have deletions in genes essential for the DPD activity, irrespective of aeration conditions during cultivation (see Table S4).
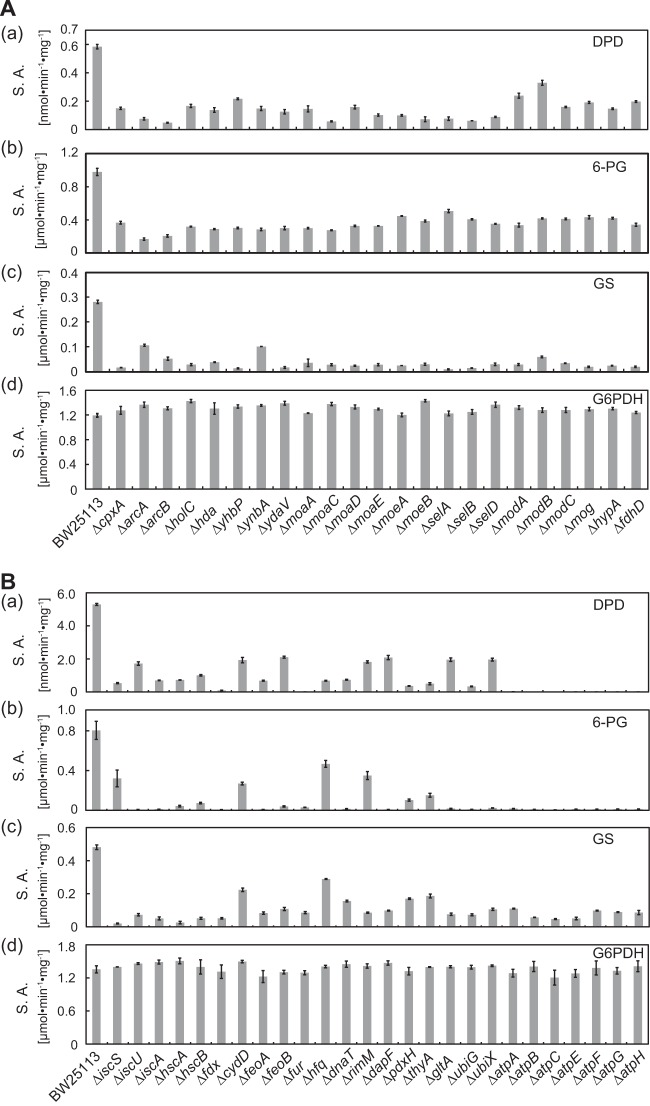
To validate the role of the 49 candidate genes identified above in Fe-S cluster biogenesis and iron homeostasis, the activities of Fe-S enzymes (DPD, 6-PG, and GS) in crude extracts of the mutant strains grown to the late logarithmic phase by static cultivation (23 strains) or fully aerobic cultivation (26 strains) were evaluated. We found that the Fe-S enzyme activities in all 49 knockout strains were near half or less than half of those of the wild-type strain (Fig. 4). Although both 6-PG and GS contain one Fe-S cluster apiece, 6-PG activity was less affected than GS activity by those mutations, with only a few exceptions, under static cultivation conditions (Fig. 4A). DPD, which contains four Fe-S clusters per heterodimer, was more significantly impaired by the disruption of arcAB, moaCE, moeAB, and selABD than other genes (Fig. 4A). There was apparently little or no correlation between the Fe-S cluster content in the enzymes and the effect of the gene disruption on the enzyme activities. Under aerobic conditions, 6-PG activity was less affected in the iscS, cydD, hfq, and rimM strains than in others (Fig. 4B). The disruption in cydD and hfq also caused a relatively moderate deficit of the GS activity. The mutation in each of the atpABCEFGH genes encoding the ATPase subunits resulted in the complete inactivation of DPD and 6-PG, suggesting that these enzymes especially require a large amount of ATP for maintaining their Fe-S clusters. This may be related to the fact that both DPD and the 6-PG have labile Fe-S clusters highly susceptible to inactivation by oxidants (31). In contrast, the activities of the non-Fe-S-containing enzyme glucose-6-phosphate dehydrogenase were similar in the mutant and the wild-type strains. These results suggest that each of the deleted genes in the 49 mutants indeed affects Fe-S cluster biogenesis and/or iron homeostasis.
FIG 4.
Fe-S enzyme activity of the knockouts identified by the radiolabeling screen under microaerobic or anaerobic conditions. The crude extracts were prepared from the strains cultivated under a microaerobic (A) or an aerobic (B) conditions. The specific activities (S.A.) of the Fe-S enzymes, (a) dihydropyrimidine dehydrogenase (DPD), (b) 6-phosphogluconate dehydratase (6-PG), and (c) glutamate synthase (GS) from the wild-type and mutant strains were determined. The activity of glucose 6-phosphate dehydrogenase (G6PDH), which does not contain an Fe-S cluster, was measured as a control (d). The means ± standard errors from two independent measurements are shown.
The 23 strains that exhibited Fe-S enzyme deficiency only under microaerobic/anaerobic conditions harbored disruption in genes related to molybdenum cofactor biogenesis (moaACDE, moeAB, mog, and modABC) (32, 33), selenocysteine incorporation (selABD) (34), formate dehydrogenase maturation (fdhD) (35), hydrogenase maturation (hypA) (36), gene regulation (cpxA and arcAB) (37, 38), DNA replication (hda and holC) (39, 40), and unknown functions (ynbA, yhbP, and ydaV). Interestingly, the processes of molybdenum cofactor biogenesis, selenocysteine incorporation, and FdhD-mediated sulfur transfer are essential for the activity of the three formate dehydrogenase isozymes required for anaerobic respiration (41–43).
On the other hand, genes disrupted in the 26 strains that are deficient in the Fe-S enzyme activities under both aerobic and anaerobic conditions can be classified into seven function-based categories: Fe-S cluster formation (iscSUA, hscAB, and fdx), transport (cydD and feoAB), gene regulation (fur and hfq), DNA replication (dnaT), ribosome maturation (rimM), metabolism (dapF, pdxH, thyA, gltA, and ubiGX), and ATP biosynthesis (atpABCEFGH).
Analysis of the in vivo role of IscU residues in Fe-S cluster maturation by the [2-14C]DHU-labeling method.
During the ISC assembly apparatus-mediated Fe-S cluster synthesis, the cysteine desulfurase IscS decomposes l-cysteine to l-alanine and sulfane sulfur via the formation of an enzyme-bound persulfide intermediate (44, 45). The scaffold protein IscU receives sulfur directly from the sulfane sulfur on IscS (46, 47) to form a [2Fe-2S]/[4Fe-4S] cluster in vitro (48, 49). It is proposed that the Fe-S cluster assembled on the IscU scaffold can be incorporated into target proteins with the help of the DnaKJ-like proteins HscA and HscB (50–52). Analysis of the solution structure has revealed that apo-IscU exists in two different conformations, one structured and the other disordered, which can interconvert (53, 54). The disordered conformational state of IscU interacts with IscS and then converts to a structured state to stabilize the cluster when it is assembled (55). Based on in vitro analyses, Cys37, Cys63, His105, and Cys106 of IscU are putative ligands for the Fe-S cluster (56). Although this is supported by a study using a genetic approach for the functional analysis of the conserved amino acid residues within Azotobacter vinelandii IscU (57) and by structural analysis of an IscS-IscU complex from Archaeoglobus fulgidus (58), direct evidence demonstrating in vivo relevance of the potential amino acid ligands of E. coli IscU is limited.
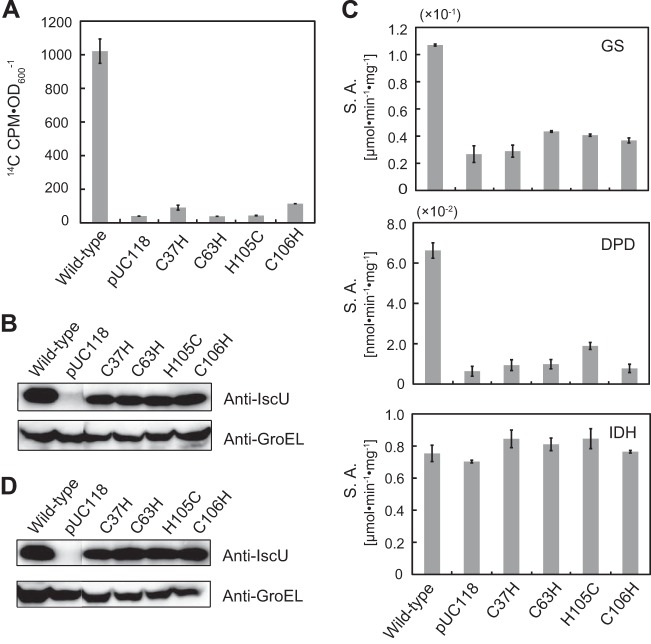
We applied the [2-14C]DHU-labeling method to the iscU-deficient E. coli strains, each of which harbored a plasmid encoding different IscU mutants (C37H, C63H, H105C, and C106H). The radioactivity of each of the mutant strains was less than 1/10 of that of the wild-type strain (Fig. 5A); this was not due to differences in protein expression levels (Fig. 5B). This result suggests that C37, C63, H105, and C106 are indeed essential for the in vivo function of IscU. We also measured the activity of the Fe-S enzymes DPD and glutamate synthase in each of the strains in order to validate our approach for analyzing an Fe-S cluster biosynthetic mechanism. The Fe-S enzyme activities of the mutant strains were less than half of those of the wild-type strain (Fig. 5C), and, again, the expression of wild-type IscU did not significantly differ from that of its variants (Fig. 5D). In contrast, the control enzyme isocitrate dehydrogenase, which does not contain an Fe-S cluster, showed similar activity levels in all of the strains tested. These data are highly consistent with the result of the [2-14C]DHU-labeling experiment described above. Taken together, the results show that our radiolabeling method is very useful in identifying the genes involved in Fe-S biosynthesis.
FIG 5.
In vivo functions of IscU and its mutants evaluated by the [2-14C]DHU-labeling method. (A) The cells were grown in LB medium containing 0.5 μM [2-14C]DHU (962 Bq · ml−1) and 1% glycerol with shaking to late stationary phase. The radioactivity of the cells was measured with a liquid scintillation counter. The means ± standard errors from two independent measurements are shown. (B) Crude extracts (25 μg protein) obtained from the cells used in the experiment shown in panel A were subjected to SDS-PAGE and analyzed by Western blotting using antibodies raised against E. coli IscU (upper) and E. coli GroEL (lower) as a loading control. (C) The specific activity (S.A.) of the Fe-S enzymes, dihydropyrimidine dehydrogenase (DPD), glutamate synthase (GS), and isocitrate dehydrogenase (IDH), in the crude extract from each strain was determined. The data represent the means ± standard errors from two independent measurements. The cells were grown in LB medium containing 0.5 μM DHU and 1% glycerol with shaking to late logarithmic phase. (D) Crude extracts (25 μg protein) from the cells used in the experiment shown in panel C were subjected to SDS-PAGE and analyzed by Western blotting using antibodies against E. coli IscU and E. coli GroEL as a loading control.
DISCUSSION
Although a great deal is now known regarding Fe-S cluster maturation, difficulties in elucidating the precise steps governing its maturation arise from the plasticity of Fe-S cluster species, i.e., the capacity of free iron and sulfide to activate apo-forms of Fe-S proteins in vitro and the inherent lability of Fe-S cluster species assembled on scaffold proteins (59). In addition, only limited progress toward understanding molecular links between Fe-S cluster biogenesis and iron homeostasis has been achieved. In this study, we have identified 49 genes affecting in vivo Fe-S cluster biogenesis and iron homeostasis under two different aeration conditions. Of these genes, 23 were important for Fe-S cluster biogenesis and/or iron homeostasis under microaerobic/anaerobic conditions, and the remaining 26 genes were identified to be crucial under both highly aerobic and microaerobic/anaerobic conditions. All known ISC machinery factors, IscUSA, HscAB, and Fdx, were identified by our screening method with no exception, indicating the reliability and validity of the screening method.
It must be noted that genes essential for cell viability were not tested in our screen because the Keio collection consists of only nonessential gene knockout strains (23). Therefore, a subset of potential genes required for Fe-S cluster biogenesis and iron homeostasis could not be identified in the present study. For example, ErpA, which is an A-type carrier protein essential for cell growth during aerobic and anaerobic respiration (60), was not identified in our screen. Genes important for Fe-S cluster biogenesis and iron homeostasis under stress conditions, such as exposure to reactive oxygen species and iron scarcity, are expected to be different from those essential under normal conditions. In fact, the SUF components, which are expressed upon stress (9), were not identified in our screens. Alternative Fe-S cluster scaffold proteins NfuA (14, 15) and a monothiol glutaredoxin, GrxD (16), function as important Fe-S cluster carriers under iron scarcity or oxidative stress conditions, and an iron donor protein, CyaY, modulates the quality of the Fe-S cluster in the presence of excessive amounts of iron (11, 12). None of these proteins was identified in our screens, suggesting that these factors are not essential under normal growth conditions. In addition, IscR, a master regulator for Fe-S cluster homeostasis, which controls the expression of more than 40 genes including ISC and SUF operons (61), was not identified in our screen. As a deletion of the iscR gene results in high expression of the ISC operon irrespective of the aeration conditions (62), Fe-S cluster synthesis in DPD may not be impaired due to increased levels of the ISC component in the iscR mutant strain, suggesting that our screen is not necessarily sensitive enough to identify a subset of genes involved in the complex homeostatic regulation of Fe-S cluster biogenesis. Interestingly, the genes identified in our screen do not overlap those identified in a global transcriptional profiling study of an E. coli iscR mutant strain (61).
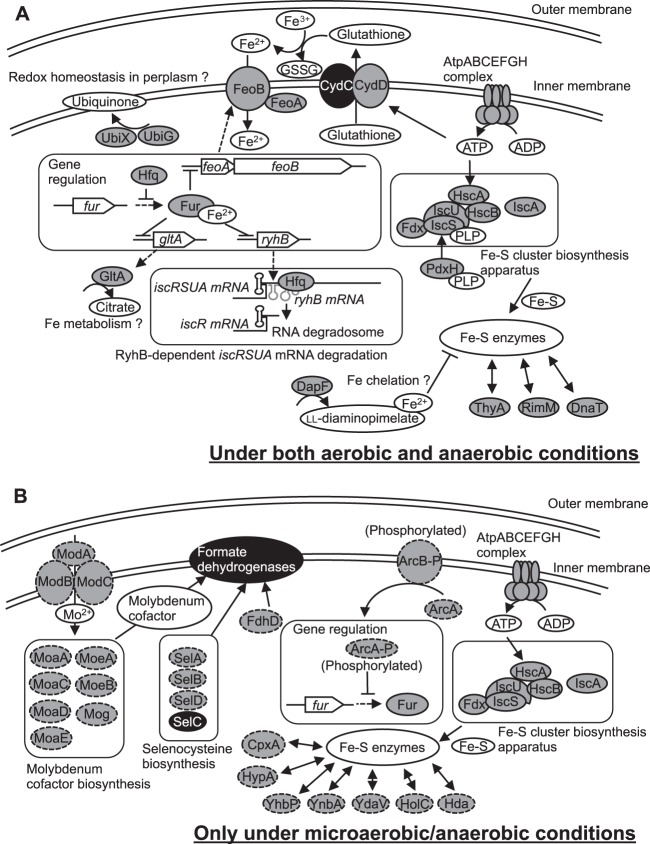
The functions of the 49 genes identified in this study are already known to some extent, but here we define new and vital roles for them in the control of Fe-S cluster biogenesis and iron homeostasis in vivo. Notably, the identification of the Fe-regulator fur and of the ferrous iron transporter feoAB genes involved in iron homeostasis enables us to predict a major iron transport pathway for Fe-S cluster biogenesis. It remains unclear whether links between the identified genes and Fe-S cluster biogenesis and/or iron homeostasis are direct or indirect. Nevertheless, possible roles for several of them during Fe-S cluster biogenesis and/or iron homeostasis are illustrated in Fig. 6 and discussed based on the known function of each gene, as described below.
FIG 6.
An overview of proposed roles of the identified genes in Fe-S cluster biogenesis and iron homeostasis. Proposed roles of the genes and/or their products in Fe-S biogenesis and iron homeostasis under both aerobic and anaerobic conditions (A) and under only microaerobic/anaerobic conditions (B) are shown. The proteins indicated by the shaded circles with solid lines represent those important for Fe-S biogenesis and/or iron homeostasis under both aerobic and anaerobic conditions, while those indicated by the shaded circles with broken lines are essential under only microaerobic/anaerobic conditions. The proteins indicated by the black circles were shown for better representation although their genes were not identified in our screen. The dashed arrows represent processes of transcription and translation. The curved and straight arrows indicate metabolic pathways. The double-headed arrows indicate possible correlations between Fe-S enzymes and each protein. The AtpABCEFGH complex, the Fe-S cluster biosynthesis apparatus, and Fur were shown in the both panels to facilitate the model representation.
Essential transcriptional regulation factors for Fe-S cluster biogenesis and iron homeostasis.
The genes identified in this study include fur and hfq. Fur is a transcriptional regulator that plays a key role in regulating iron homeostasis (63, 64). When intracellular iron levels become sufficiently high, expression of certain iron acquisition-related genes is repressed by a Fur-Fe2+ complex. In contrast, when iron concentrations fall below a certain threshold, Fur becomes inactive, and the repression of iron acquisition genes is relieved. Under low-iron conditions, the small regulatory RNA RyhB, which is regulated by Fur, causes degradation of mRNAs that encode Fe-S proteins, including succinate dehydrogenase (65). RyhB is expressed and pairs with the 5′ untranslated region of iscS. This promotes the Hfq-mediated recruitment of the RNA degradosome and degradation of the mRNAs encoding iscS, iscU, and iscA. In a fur-null mutant strain, RyhB expression triggers such mRNA degradation (66, 67). Therefore, activities of Fe-S enzymes dependent on the ISC machinery are lower in a fur-null mutant strain than in a wild-type strain, as demonstrated in this study (Fig. 6A).
The role of Hfq in RyhB regulation involves both the recruitment of the major endo-RNase, RNase E, for RyhB-dependent target mRNA cleavage and protection from endonucleolytic cleavage of RyhB (68). Therefore, loss of Hfq in the cell is expected to result in insufficient RyhB-dependent degradation of iscS, iscU, and iscA mRNAs. However, our data show that Fe-S enzyme activity in the hfq-deficient strain were lower than that in the wild type. This discrepancy can be explained by the finding that the fur mRNA is also the target of negative posttranscriptional control by Hfq (69). Upregulation of Fur in the hfq-null mutant strain reduces the expression of iron acquisition genes, including that of the FeoAB iron transporter (70). Therefore, we speculate that disruption of the hfq gene reduces the abundance of intracellular iron, thereby impairing maturation of Fe-S clusters. Thus, maintaining appropriate expression levels of the fur and hfq genes may be important for Fe-S cluster homeostasis. Further studies using a double-knockout mutant of the fur and hfq genes may be required to address the relationship between these two genes in Fe-S cluster homeostasis.
The 23 knockout strains included those with a deletion in arcAB and cpxA genes that are important for signal transduction (Fig. 6B). ArcA and ArcB comprise a two-component transduction system, which negatively regulates the fur gene under anaerobic conditions (71), suggesting that ArcAB may be involved in anaerobic iron homeostasis. The CpxA protein senses envelope stress, the causes of which include misfolded or mislocalized proteins in the periplasm, and regulates the phosphorylation of the regulator CpxR. This phosphorylated CpxR upregulates adaptive response genes encoding protein folding and degradation factors (38, 72). Because the cpxR gene was not identified in the first screen, the exact functional correlation between cpxA and Fe-S cluster biogenesis remains to be resolved.
Activation of Fe-S cluster biogenesis factors.
We identified the pdxH gene coding for pyridoxine 5′-phosphate oxidase (73), which catalyzes the FMN-dependent oxidation of pyridoxine 5′-phosphate, the terminal step in the biosynthesis of pyridoxal 5′-phosphate (PLP). This enzyme contains a PLP molecule bound at noncatalytic site, which is readily transferred to a PLP-requiring apo-enzyme by a possible channeling mechanism (74). Thus, pdxH is important for Fe-S cluster biogenesis, most likely because it is required to transfer PLP to apo-IscS to form the active holoenzyme (Fig. 6A).
We also identified a set of genes, atpABCEFGH (not atpD), each of which encodes an ATP synthase subunit which, along with an electrochemical proton gradient generated by using a number of alternative electron acceptors (75), is involved in ATP synthesis. AtpD was deemed nonessential for unknown reasons. Obviously, ATP is needed for many cellular processes, as indicated by the reduced growth of the mutants lacking any of the atpABCEFGH components (see Table S4 in the supplemental material). Nevertheless, the effect of the deficiency of atpABCEFGH on Fe-S cluster biogenesis and iron homeostasis appears more significant than its effect on growth. ATP is required to generate a driving force for the CydD glutathione transporter (76, 77) and for HscA to transfer a preformed Fe-S cluster to various target apo-proteins (50–52), consistent with the result that impaired ATP biosynthesis in the atp mutants results in decreased Fe-S enzyme activities. Thus, Fe-S cluster maturation and/or iron homeostasis may be strictly influenced by ATP availability (Fig. 6A).
The genes for molybdenum cofactor (moaACDE, moeAB, mog, and modABC) biogenesis (32, 33), selenocysteine incorporation (selABD) (34), and a sulfur transferase, FdhD (35), are required for production of mature forms of the formate dehydrogenase isozymes, all of which contain molybdenum cofactors and selenocysteine residues. Formate dehydrogenases are involved in alternative electron transfer reactions allowing cellular energy conservation and ATP generation via the proton-translocating ATPase (41–43). Therefore, the ATP generation supported by formate dehydrogenase activity may be associated with Fe-S cluster biogenesis and iron homeostasis under microaerobic/anaerobic conditions (Fig. 6B). The genes for the three formate dehydrogenase isozymes were not identified in our screens, probably due to their functional compensation.
Iron uptake.
Since iron is insoluble in the ferric form present at neutral pH under aerobic conditions, E. coli has developed ferric iron-solubilizing and uptake strategies using high-affinity molecules known as siderophores (78, 79). FepA (80), FhuA (81), FhuE (82), and FecA (83) are the transporters for the distinct siderophores enterobactin, ferrichrome, ferric coprogen, and ferric dicitrate, respectively. E. coli also express the EfeUOB transporter system that is responsible for uptake of ferrous iron under conditions of iron limitation and low pH (84). In addition, there are several ferric iron-reducing enzymes in the cytosol (85), such as NfnB (86), Fre (87), Fpr (86), and YqjH (88). Interestingly however, none of these transporters or ferric iron-reducing enzymes was identified in our screen as essential factors for Fe-S cluster maturation (see Table S1 in the supplemental material). On the other hand, we found that deletion of either feoA or feoB resulted in a decrease in Fe-S enzyme activities, irrespective of the aeration conditions during cultivation. FeoAB, a ferrous iron transport system localized in the cytoplasmic membrane, likely plays a key role in controlling the iron supply to cells (70, 89). We speculate that Fur may promote derepression of RyhB under conditions when intracellular ferrous iron sources are low since the protein-ferrous iron complex represses the expression of RyhB. Another possibility is that the small amount of intracellular ferrous iron may reduce the amount of iron incorporated into the scaffold protein during Fe-S cluster maturation. Collectively, these results strongly indicate that iron uptake by FeoAB may be the main pathway connecting the periplasmic space to the cytosol for intracellular iron supply (Fig. 6A).
Our results suggest that cydD is associated with Fe-S biogenesis and/or iron homeostasis. CydD forms a complex with CydC to transport cysteine and glutathione from the cytosol to the periplasm (76, 77). The cydC gene was not identified in our screen because the genetic knockout was not included in the Keio collection (23). Glutathione is a major thiol-disulfide redox buffer, reaching ∼10 mM in E. coli cytoplasm (90). A defect in the cydD gene may result in the disturbance of periplasmic redox balance since glutathione plays a key role in periplasmic redox homeostasis. In addition, the cydD defect may cause an insufficiency in iron(II) supply because glutathione has a buffering effect on ferric iron; that is, glutathione-chelated ferric iron is rapidly reduced back to a glutathione chelated-ferrous iron form (91). Therefore, it is likely that the glutathione transported from the cytosol by CydCD may fulfill a trafficking role of an iron(II) to FeoAB transporter in the periplasm (Fig. 6A).
Iron chelation.
The dapF gene, which encodes diaminopimelate epimerase (DapF) involved in lysine biosynthesis, was also identified in this study. It is shown that blocking the expression of dapB, which encodes dihydrodipicolinate reductase in the same pathway, leads to intracellular accumulation of dihydrodipicolinate, which is an excellent iron chelator (92). Its cellular accumulation triggers a large increase in the intracellular pool of chelated iron and a decrease of free iron in the cell, which in turn causes derepression of the Fur regulon. The dapF-null mutant accumulated large amounts of ll-diaminopimelate (93), which possibly serves as an iron chelator, and may also trigger a large decrease in the intracellular pool of free iron and derepression of the Fur regulon. This leads to RyhB induction, iscRSUA mRNA degradation, and a decrease in the Fe-S enzyme activity (Fig. 6A).
One of the most interesting results of this study is that citrate synthase (94) encoded by the gltA gene was found to be important for Fe-S enzyme activities. This gene is repressed by the Fur-Fe2+ complex, suggesting that citrate may have a role in iron uptake and metabolism (95). However, ferric citrate is not likely to be the predominant iron source for Fe-S clusters as the fecA ferric citrate transporter gene was not identified in our screen (see Table S1 in the supplemental material). Thus, we hypothesize that citrate contributes to cytosolic iron storage for Fe-S cluster maturation (Fig. 6A).
Cellular redox homeostasis.
UbiX and UbiG are members of ubiquinone biosynthetic enzyme family (96). Ubiquinone acts as the final electron acceptor in the periplasmic disulfide bond redox machinery (97). Under low-ubiquinone conditions, a reduced form of the Dsb protein, which is the disulfide catalyst in the periplasm, is increased and inhibits the appropriate folding and integration of outer membrane proteins (98, 99). Therefore, it is likely that the lack of the UbiX or UbiG causes a functional defect in iron transporters (e.g., FeoAB) or other proteins important for Fe-S cluster maturation (Fig. 6A). Another possibility is that Fe-S cluster biogenesis was inhibited by a pleiotropic effect of ubiquinone deficiency, as reported previously (100). This may be due to increased sensitivity to reactive oxygen species in the mutant strains. In fact, the level of [2-14C]DHU-derived radioactivity in the hpx strain (101) (a kind gift from J. A. Imlay) that lacks a gene essential for eliminating reactive oxygen species was lower than that in the wild-type strain because [4Fe-4S] in DPD is susceptible to hydrogen peroxide (data not shown).
Other functions.
The dnaT, hda, and holC genes play key roles in adequate DNA replication (39, 40, 102). Unlike the hda and holC genes, the dnaT gene was important for Fe-S cluster biogenesis even under aerobic conditions. The rimM gene is involved in the assembly of the 30S ribosomal subunit (103). The thyA gene encodes thymidylate synthase catalyzing the 5,10-methylene tetrahydrofolate-dependent conversion of deoxyuridine monophosphate to deoxythymidine monophosphate (104). The correlations between these genes and Fe-S cluster biogenesis and/or iron homeostasis are still puzzling. The hypA gene required for anaerobic respiration via nickel incorporation into hydrogenase 3 (36) was identified in our screen, but other known hydrogenase maturation factors (105) were not, suggesting that the decreased Fe-S enzyme activities in the hypA strain are not due to the inactivation of hydrogenase. The roles of yhbP, ynbA, and ydaV in Fe-S biogenesis/homeostasis also remain interesting open questions. Future studies will address the relationship between these proteins and Fe-S cluster biogenesis and/or iron homeostasis.
Analyzing Fe-S cluster biogenesis with the [2-14C]DHU-radiolabeling method.
Using the radiolabeling method, we clarified that 4 amino acid residues of IscU (Cys37, Cys63, His105, and Cys106) were essential for its in vivo function in Fe-S cluster biosynthesis. The radiolabeling method we have developed will be useful in shedding light on yet-unknown molecular mechanisms involved in Fe-S cluster biogenesis. The method relies on the observation that the levels of radioactivity derived from [2-14C]DHU in E. coli cells depends on DPD, which requires Fe-S clusters for its activity. As such, it provides a facile and sensitive method to analyze the essential amino acid composition of Fe-S cluster biosynthesis factors, as well as a high-throughput format to screen for genes affecting Fe-S cluster homeostasis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by a Grant-in-Aid for Scientific Research (B) 23380060 (to H.M.) from JSPS and by a Grant-in-Aid for Challenging Exploratory Research 24658086 (to H.M.) from JSPS.
We thank Machiko Utsunomiya for valuable technical assistance for cell cultivation.
Footnotes
Published ahead of print 10 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01160-13.
REFERENCES
- 1.Py B, Barras F. 2010. Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8:436–446. 10.1038/nrmicro2356 [DOI] [PubMed] [Google Scholar]
- 2.Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247–281. 10.1146/annurev.biochem.74.082803.133518 [DOI] [PubMed] [Google Scholar]
- 3.Beinert H. 2000. Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5:2–15. 10.1007/s007750050002 [DOI] [PubMed] [Google Scholar]
- 4.Drennan CL, Peters JW. 2003. Surprising cofactors in metalloenzymes. Curr. Opin. Struct. Biol. 13:220–226. 10.1016/S0959-440X(03)00038-1 [DOI] [PubMed] [Google Scholar]
- 5.Schwartz CJ, Djaman O, Imlay JA, Kiley PJ. 2000. The cysteine desulfurase, IscS, has a major role in in vivo Fe-S cluster formation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:9009–9014. 10.1073/pnas.160261497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tokumoto U, Takahashi Y. 2001. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J. Biochem. 130:63–71. 10.1093/oxfordjournals.jbchem.a002963 [DOI] [PubMed] [Google Scholar]
- 7.Hidese R, Mihara H, Esaki N. 2011. Bacterial cysteine desulfurases: versatile key players in biosynthetic pathways of sulfur-containing biofactors. Appl. Microbiol. Biotechnol. 91:47–61. 10.1007/s00253-011-3336-x [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, Yeo WS, Roe JH. 2004. Induction of the sufA operon encoding Fe-S assembly proteins by superoxide generators and hydrogen peroxide: involvement of OxyR, IHF and an unidentified oxidant-responsive factor. Mol. Microbiol. 51:1745–1755. 10.1111/j.1365-2958.2003.03946.x [DOI] [PubMed] [Google Scholar]
- 9.Yeo WS, Lee JH, Lee KC, Roe JH. 2006. IscR acts as an activator in response to oxidative stress for the suf operon encoding Fe-S assembly proteins. Mol. Microbiol. 61:206–218. 10.1111/j.1365-2958.2006.05220.x [DOI] [PubMed] [Google Scholar]
- 10.Jang S, Imlay JA. 2010. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol. Microbiol. 78:1448–1467. 10.1111/j.1365-2958.2010.07418.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adinolfi S, Iannuzzi C, Prischi F, Pastore C, Iametti S, Martin SR, Bonomi F, Pastore A. 2009. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 16:390–396. 10.1038/nsmb.1579 [DOI] [PubMed] [Google Scholar]
- 12.Iannuzzi C, Adinolfi S, Howes BD, Garcia-Serres R, Clémancey M, Latour JM, Smulevich G, Pastore A. 2011. The role of CyaY in iron sulfur cluster assembly on the E. coli IscU scaffold protein. PLoS One 6:e21992. 10.1371/journal.pone.0021992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Velayudhan J, Castor M, Richardson A, Main-Hester KL, Fang FC. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63:1495–1507. 10.1111/j.1365-2958.2007.05600.x [DOI] [PubMed] [Google Scholar]
- 14.Py B, Gerez C, Angelini S, Planel R, Vinella D, Loiseau L, Talla E, Brochier-Armanet C, Garcia Serres R, Latour JM, Ollagnier-de Choudens S, Fontecave M, Barras F. 2012. Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol. Microbiol. 86:155–171. 10.1111/j.1365-2958.2012.08181.x [DOI] [PubMed] [Google Scholar]
- 15.Angelini S, Gerez C, Ollagnier-de Choudens S, Sanakis Y, Fontecave M, Barras F, Py B. 2008. NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283:14084–14091. 10.1074/jbc.M709405200 [DOI] [PubMed] [Google Scholar]
- 16.Yeung N, Gold B, Liu NL, Prathapam R, Sterling HJ, Willams ER, Butland G. 2011. The E. coli monothiol glutaredoxin GrxD forms homodimeric and heterodimeric FeS cluster containing complexes. Biochemistry 50:8957–8969. 10.1021/bi2008883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyd JM, Sondelski JL, Downs DM. 2009. Bacterial ApbC protein has two biochemical activities that are required for in vivo function. J. Biol. Chem. 284:110–118. 10.1074/jbc.M807003200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loiseau L, Gerez C, Bekker M, Ollagnier-de Choudens S, Py B, Sanakis Y, Teixeira de Mattos J, Fontecave M, Barras F. 2007. ErpA, an iron sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104:13626–13631. 10.1073/pnas.0705829104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vinella D, Loiseau L, de Choudens SO, Fontecave M, Barras F. 2013. In vivo [Fe-S] cluster acquisition by IscR and NsrR, two stress regulators in Escherichia coli. Mol. Microbiol. 87:493–508. 10.1111/mmi.12135 [DOI] [PubMed] [Google Scholar]
- 20.Justino MC, Almeida CC, Teixeira M, Saraiva LM. 2007. Escherichia coli di-iron YtfE protein is necessary for the repair of stress-damaged iron-sulfur clusters. J. Biol. Chem. 282:10352–10359. 10.1074/jbc.M610656200 [DOI] [PubMed] [Google Scholar]
- 21.Mihara H, Hidese R, Yamane M, Kurihara T, Esaki N. 2008. The iscS gene deficiency affects the expression of pyrimidine metabolism genes. Biochem. Biophys. Res. Commun. 372:407–411. 10.1016/j.bbrc.2008.05.019 [DOI] [PubMed] [Google Scholar]
- 22.Hidese R, Mihara H, Kurihara T, Esaki N. 2011. Escherichia coli dihydropyrimidine dehydrogenase is a novel NAD-dependent heterotetramer essential for the production of 5,6-dihydrouracil. J. Bacteriol. 193:989–993. 10.1128/JB.01178-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergstrom DE, Leonard NJ. 1972. Structure of the borohydride reduction product of photolinked 4-thiouracil and cytosine. Fluorescent probe of transfer ribonucleic acid tertiary structure. J. Am. Chem. Soc. 94:6178–6182 [DOI] [PubMed] [Google Scholar]
- 25.Fraenkel DG, Horecker BL. 1964. Pathways of d-glucose metabolism in Salmonella typhimurium. A study of a mutant lacking phosphoglucose isomerase. J. Biol. Chem. 239:2765–2771 [PubMed] [Google Scholar]
- 26.Miller RE, Stadtman ER. 1972. Glutamate synthase from Escherichia coli. An iron-sulfide flavoprotein. J. Biol. Chem. 247:7407–7419 [PubMed] [Google Scholar]
- 27.Cribbs R, Englesberg E. 1964. l-Arabinose negative mutants of the l-ribulokinase structural gene affecting the levels of l-arabinose isomerase in Escherichia coli. Genetics 49:95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee S, Fraenkel DG. 1972. Glucose-6-phosphate dehydrogenase from Escherichia coli and from a “high-level” mutant. J. Bacteriol. 110:155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254. 10.1016/0003-2697(76)90527-3 [DOI] [PubMed] [Google Scholar]
- 30.Kato S, Mihara H, Kurihara T, Takahashi Y, Tokumoto U, Yoshimura T, Esaki N. 2002. Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: implications for the mechanism of iron-sulfur cluster assembly. Proc. Natl. Acad. Sci. U. S. A. 99:5948–5952. 10.1073/pnas.082123599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Djaman O, Outten FW, Imlay JA. 2004. Repair of oxidized iron-sulfur clusters in Escherichia coli. J. Biol. Chem. 279:44590–44599. 10.1074/jbc.M406487200 [DOI] [PubMed] [Google Scholar]
- 32.Zhang W, Urban A, Mihara H, Leimkühler S, Kurihara T, Esaki N. 2010. IscS functions as a primary sulfur-donating enzyme by interacting specifically with MoeB and MoaD in the biosynthesis of molybdopterin in Escherichia coli. J. Biol. Chem. 285:2302–2308. 10.1074/jbc.M109.082172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iobbi-Nivol C, Leimkühler S. 2013. Molybdenum enzymes, their maturation and molybdenum cofactor biosynthesis in Escherichia coli. Biochim. Biophys. Acta 1827:1086–1101. 10.1016/j.bbabio.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 34.Yoshizawa S, Böck A. 2009. The many levels of control on bacterial selenoprotein synthesis. Biochim. Biophys. Acta 1790:1404–1414. 10.1016/j.bbagen.2009.03.010 [DOI] [PubMed] [Google Scholar]
- 35.Thomé R, Gust A, Toci R, Mendel R, Bittner F, Magalon A, Walburger A. 2012. A sulfur transferase is essential for activity of formate dehydrogenases in Escherichia coli. J. Biol. Chem. 287:4671–4678. 10.1074/jbc.M111.327122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan Chung KC, Zamble DB. 2011. Protein interactions and localization of the Escherichia coli accessory protein HypA during nickel insertion to [NiFe] hydrogenase. J. Biol. Chem. 286:43081–43090. 10.1074/jbc.M111.290726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malpica R, Sandoval GR, Rodríguez C, Franco B, Georgellis D. 2006. Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8:781–795. 10.1089/ars.2006.8.781 [DOI] [PubMed] [Google Scholar]
- 38.Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326:2–11. 10.1111/j.1574-6968.2011.02406.x [DOI] [PubMed] [Google Scholar]
- 39.Kato J, Katayama T. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253–4262. 10.1093/emboj/20.15.4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witte G, Urbanke C, Curth U. 2003. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 31:4434–4440. 10.1093/nar/gkg498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berg BL, Li J, Heider J, Stewart V. 1991. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J. Biol. Chem. 266:22380–22385 [PubMed] [Google Scholar]
- 42.Abaibou H, Pommier J, Benoit S, Giordano G, Mandrand-Berthelot MA. 1995. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 177:7141–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. 1997. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275:1305–1308. 10.1126/science.275.5304.1305 [DOI] [PubMed] [Google Scholar]
- 44.Mihara H, Kurihara T, Yoshimura T, Esaki N. 2000. Kinetic and mutational studies of three NifS homologs from Escherichia coli: mechanistic difference between l-cysteine desulfurase and l-selenocysteine lyase reactions. J. Biochem. 127:559–567. 10.1093/oxfordjournals.jbchem.a022641 [DOI] [PubMed] [Google Scholar]
- 45.Flint DH. 1996. Escherichia coli contains a protein that is homologous in function and N-terminal sequence to the protein encoded by the nifS gene of Azotobacter vinelandii and that can participate in the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase. J. Biol. Chem. 271:16068–16074 [PubMed] [Google Scholar]
- 46.Urbina HD, Silberg JJ, Hoff KG, Vickery LE. 2001. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem. 276:44521–44526. 10.1074/jbc.M106907200 [DOI] [PubMed] [Google Scholar]
- 47.Smith AD, Agar JN, Johnson KA, Frazzon J, Amster IJ, Dean DR, Johnson MK. 2001. Sulfur transfer from IscS to IscU: the first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123:11103–11104. 10.1021/ja016757n [DOI] [PubMed] [Google Scholar]
- 48.Chandramouli K, Unciuleac MC, Naik S, Dean DR, Huynh BH, Johnson MK. 2007. Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry 46:6804–6811. 10.1021/bi6026659 [DOI] [PubMed] [Google Scholar]
- 49.Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. 2007. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry 46:6812–6821. 10.1021/bi6026665 [DOI] [PubMed] [Google Scholar]
- 50.Chandramouli K, Johnson MK. 2006. HscA and HscB stimulate [2Fe-2S] cluster transfer from IscU to apoferredoxin in an ATP-dependent reaction. Biochemistry 45:11087–11095. 10.1021/bi061237w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. 2008. Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry 47:12795–12801. 10.1021/bi801565j [DOI] [PubMed] [Google Scholar]
- 52.Kim JH, Tonelli M, Frederick RO, Chow DC, Markley JL. 2012. Specialized Hsp70 chaperone (HscA) binds preferentially to the disordered form, whereas J-protein (HscB) binds preferentially to the structured form of the iron-sulfur cluster scaffold protein (IscU). J. Biol. Chem. 287:31406–31413. 10.1074/jbc.M112.352617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai Z, Tonelli M, Markley JL. 2012. Metamorphic protein IscU changes conformation by cis-trans isomerizations of two peptidyl-prolyl peptide bonds. Biochemistry 51:9595–9602. 10.1021/bi301413y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JH, Füzéry AK, Tonelli M, Ta DT, Westler WM, Vickery LE, Markley JL. 2009. Structure and dynamics of the iron-sulfur cluster assembly scaffold protein IscU and its interaction with the cochaperone HscB. Biochemistry 48:6062–6071. 10.1021/bi9002277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JH, Tonelli M, Markley JL. 2012. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc. Natl. Acad. Sci. U. S. A. 109:454–459. 10.1073/pnas.1114372109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. 2011. Facilitated transfer of IscU-[2Fe2S] clusters by chaperone-mediated ligand exchange. Biochemistry 50:9641–9650. 10.1021/bi201123z [DOI] [PubMed] [Google Scholar]
- 57.Johnson DC, Unciuleac MC, Dean DR. 2006. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J. Bacteriol. 188:7551–7561. 10.1128/JB.00596-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marinoni EN, de Oliveira JS, Nicolet Y, Raulfs EC, Amara P, Dean DR, Fontecilla-Camps JC. 2012. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 51:5439–5442. 10.1002/anie.201201708 [DOI] [PubMed] [Google Scholar]
- 59.Raulfs EC, O'Carroll IP, Dos Santos PC, Unciuleac MC, Dean DR. 2008. In vivo iron-sulfur cluster formation. Proc. Natl. Acad. Sci. U. S. A. 105:8591–8596. 10.1073/pnas.0803173105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pinske C, Sawers RG. 2012. A-type carrier protein ErpA is essential for formation of an active formate-nitrate respiratory pathway in Escherichia coli K-12. J. Bacteriol. 194:346–353. 10.1128/JB.06024-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 60:1058–1075. 10.1111/j.1365-2958.2006.05160.x [DOI] [PubMed] [Google Scholar]
- 62.Giel JL, Nesbit AD, Mettert EL, Fleischhacker AS, Wanta BT, Kiley PJ. 2013. Regulation of iron-sulphur cluster homeostasis through transcriptional control of the Isc pathway by [2Fe-2S]-IscR in Escherichia coli. Mol. Microbiol. 87:478–492. 10.1111/mmi.12052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee JW, Helmann JD. 2007. Functional specialization within the Fur family of metalloregulators. Biometals 20:485–499. 10.1007/s10534-006-9070-7 [DOI] [PubMed] [Google Scholar]
- 64.Chen Z, Lewis KA, Shultzaberger RK, Lyakhov IG, Zheng M, Doan B, Storz G, Schneider TD. 2007. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 35:6762–6777. 10.1093/nar/gkm631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620–4625. 10.1073/pnas.032066599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jacques JF, Jang S, Prévost K, Desnoyers G, Desmarais M, Imlay J, Massé E. 2006. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 62:1181–1190. 10.1111/j.1365-2958.2006.05439.x [DOI] [PubMed] [Google Scholar]
- 67.Desnoyers G, Morissette A, Prévost K, Massé E. 2009. Small RNA-induced differential degradation of the polycistronic mRNA iscRSUA. EMBO J. 28:1551–1561. 10.1038/emboj.2009.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geissmann TA, Touati D. 2004. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 23:396–405. 10.1038/sj.emboj.7600058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Večerek B, Moll I, Afonyushkin T, Kaberdin V, Blasi U. 2003. Interaction of the RNA chaperone Hfq with mRNAs: direct and indirect roles of Hfq in iron metabolism of Escherichia coli. Mol. Microbiol. 50:897–909. 10.1046/j.1365-2958.2003.03727.x [DOI] [PubMed] [Google Scholar]
- 70.Kammler M, Schön C, Hantke K. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu X, De Wulf P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588–12597. 10.1074/jbc.M313454200 [DOI] [PubMed] [Google Scholar]
- 72.Price NL, Raivio TL. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191:1798–1815. 10.1128/JB.00798-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao G, Winkler ME. 1995. Kinetic limitation and cellular amount of pyridoxine (pyridoxamine) 5′-phosphate oxidase of Escherichia coli K-12. J. Bacteriol. 177:883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Safo MK, Musayev FN, di Salvo ML, Schirch V. 2001. X-ray structure of Escherichia coli pyridoxine 5′-phosphate oxidase complexed with pyridoxal 5′-phosphate at 2.0 Å resolution. J. Mol. Biol. 310:817–826. 10.1006/jmbi.2001.4734 [DOI] [PubMed] [Google Scholar]
- 75.Nakamoto RK, Baylis Scanlon JA, Al-Shawi MK. 2008. The rotary mechanism of the ATP synthase. Arch. Biochem. Biophys. 476:43–50. 10.1016/j.abb.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pittman MS, Robinson HC, Poole RK. 2005. A bacterial glutathione transporter (Escherichia coli CydDC) exports reductant to the periplasm. J. Biol. Chem. 280:32254–32261. 10.1074/jbc.M503075200 [DOI] [PubMed] [Google Scholar]
- 77.Pittman MS, Corker H, Wu G, Binet MB, Moir AJ, Poole RK. 2002. Cysteine is exported from the Escherichia coli cytoplasm by CydDC, an ATP-binding cassette-type transporter required for cytochrome assembly. J. Biol. Chem. 277:49841–49849. 10.1074/jbc.M205615200 [DOI] [PubMed] [Google Scholar]
- 78.Andrews SC, Robinson AK, Rodríguez-Quiñones F. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215–237. 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- 79.Braun V, Braun M. 2002. Iron transport and signaling in Escherichia coli. FEBS Lett. 529:78–85. 10.1016/S0014-5793(02)03185-X [DOI] [PubMed] [Google Scholar]
- 80.Buchanan SK, Smith BS, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. 1999. Crystal structure of the outer membrane active transporter FepA from Escherichia coli. Nat. Struct. Biol. 6:56–63. 10.1038/4931 [DOI] [PubMed] [Google Scholar]
- 81.Locher KP, Rees B, Koebnik R, Mitschler A, Moulinier L, Rosenbusch JP, Moras D. 1998. Transmembrane signaling across the ligand-gated FhuA receptor: crystal structures of free and ferrichrome-bound states reveal allosteric changes. Cell 95:771–778. 10.1016/S0092-8674(00)81700-6 [DOI] [PubMed] [Google Scholar]
- 82.Sauer M, Hantke K, Braun V. 1987. Ferric-coprogen receptor FhuE of Escherichia coli: processing and sequence common to all TonB-dependent outer membrane receptor proteins. J. Bacteriol. 169:2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ferguson AD, Chakraborty R, Smith BS, Esser L, van der Helm D, Deisenhofer J. 2002. Structural basis of gating by the outer membrane transporter FecA. Science 295:1715–1719. 10.1126/science.1067313 [DOI] [PubMed] [Google Scholar]
- 84.Cao J, Woodhall MR, Alvarez J, Cartron ML, Andrews SC. 2007. EfeUOB (YcdNOB) is a tripartite, acid-induced and CpxAR-regulated, low-pH Fe2+ transporter that is cryptic in Escherichia coli K-12 but functional in E. coli O157:H7. Mol. Microbiol. 65:857–875. 10.1111/j.1365-2958.2007.05802.x [DOI] [PubMed] [Google Scholar]
- 85.Schröder I, Johnson E, de Vries S. 2003. Microbial ferric iron reductases. FEMS Microbiol. Rev. 27:427–447. 10.1016/S0168-6445(03)00043-3 [DOI] [PubMed] [Google Scholar]
- 86.Takeda K, Sato J, Goto K, Fujita T, Watanabe T, Abo M, Yoshimura E, Nakagawa J, Abe A, Kawasaki S, Niimura Y. 2010. Escherichia coli ferredoxin-NADP+ reductase and oxygen-insensitive nitroreductase are capable of functioning as ferric reductase and of driving the Fenton reaction. Biometals 23:727–737. 10.1007/s10534-010-9339-8 [DOI] [PubMed] [Google Scholar]
- 87.Ingelman M, Ramaswamy S, Niviere V, Fontecave M, Eklund H. 1999. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry 38:7040–7049. 10.1021/bi982849m [DOI] [PubMed] [Google Scholar]
- 88.Miethke M, Hou J, Marahiel MA. 2011. The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 50:10951–10964. 10.1021/bi201517h [DOI] [PubMed] [Google Scholar]
- 89.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157. 10.1007/s10534-006-0003-2 [DOI] [PubMed] [Google Scholar]
- 90.Fahey RC, Brown WC, Adams WB, Worsham MB. 1978. Occurrence of glutathione in bacteria. J. Bacteriol. 133:1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hider RC, Kong XL. 2011. Glutathione: a key component of the cytoplasmic labile iron pool. Biometals 24:1179–1187. 10.1007/s10534-011-9476-8 [DOI] [PubMed] [Google Scholar]
- 92.Maringanti S, Imlay JA. 1999. An intracellular iron chelator pleiotropically suppresses enzymatic and growth defects of superoxide dismutase-deficient Escherichia coli. J. Bacteriol. 181:3792–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Richaud C, Higgins W, Mengin-Lecreulx D, Stragier P. 1987. Molecular cloning, characterization, and chromosomal localization of dapF, the Escherichia coli gene for diaminopimelate epimerase. J. Bacteriol. 169:1454–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Faloona GR, Srere PA. 1969. Escherichia coli citrate synthase. Purification and the effect of potassium on some properties. Biochemistry 8:4497–4503 [DOI] [PubMed] [Google Scholar]
- 95.McHugh JP, Rodríguez-Quiñones F, Abdul-Tehrani H, Svistunenko DA, Poole RK, Cooper CE, Andrews SC. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478–29486. 10.1074/jbc.M303381200 [DOI] [PubMed] [Google Scholar]
- 96.Gulmezian M, Hyman KR, Marbois BN, Clarke CF, Javor GT. 2007. The role of UbiX in Escherichia coli coenzyme Q. biosynthesis. Arch. Biochem. Biophys. 467:144–153. 10.1016/j.abb.2007.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bader M, Muse W, Ballou DP, Gassner C, Bardwell JC. 1999. Oxidative protein folding is driven by the electron transport system. Cell 98:217–227. 10.1016/S0092-8674(00)81016-8 [DOI] [PubMed] [Google Scholar]
- 98.Missiakas D, Raina S. 1997. Protein folding in the bacterial periplasm. J. Bacteriol. 179:2465–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zeng H, Snavely I, Zamorano P, Javor GT. 1998. Low ubiquinone content in Escherichia coli causes thiol hypersensitivity. J. Bacteriol. 180:3681–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Collis CM, Grigg GW. 1989. An Escherichia coli mutant resistant to phleomycin, bleomycin, and heat inactivation is defective in ubiquinone synthesis. J. Bacteriol. 171:4792–4798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Park S, You X, Imlay JA. 2005. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx− mutants of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 102:9317–9322. 10.1073/pnas.0502051102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCool JD, Ford CC, Sandler SJ. 2004. A dnaT mutant with phenotypes similar to those of a priA2::kan mutant in Escherichia coli K-12. Genetics 167:569–578. 10.1534/genetics.103.025296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guo Q, Goto S, Chen Y, Feng B, Xu Y, Muto A, Himeno H, Deng H, Lei J, Gao N. 2013. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 41:2609–2620. 10.1093/nar/gks1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Newby Z, Lee TT, Morse RJ, Liu Y, Liu L, Venkatraman P, Santi DV, Finer-Moore JS, Stroud RM. 2006. The role of protein dynamics in thymidylate synthase catalysis: variants of conserved 2′-deoxyuridine 5′-monophosphate (dUMP)-binding Tyr-261. Biochemistry 45:7415–7428. 10.1021/bi060152s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bürstel I, Siebert E, Winter G, Hummel P, Zebger I, Friedrich B, Lenz O. 2012. A universal scaffold for synthesis of the Fe(CN)2(CO) moiety of [NiFe] hydrogenase. J. Biol. Chem. 287:38845–38853. 10.1074/jbc.M112.376947 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.