Abstract
Many bacteria possess cell density-dependent quorum-sensing (QS) systems that often regulate cooperative secretions involved in host-microbe or microbe-microbe interactions. These secretions, or “public goods,” are frequently coregulated by stress and starvation responses. Here we provide a physiological rationale for such regulatory complexity in the opportunistic pathogen Pseudomonas aeruginosa. Using minimal-medium batch and chemostat cultures, we comprehensively characterized specific growth rate-limiting macronutrients as key triggers for the expression of extracellular enzymes and metabolites directly controlled by the las and rhl QS systems. Expression was unrelated to cell density, depended on the secreted product's elemental composition, and was induced only when the limiting nutrient was not also a building block of the product; rhl-dependent products showed the strongest response, caused by the largely las-independent induction of the regulator RhlR and its cognate signal. In agreement with the prominent role of the rhl system, slow growth inverted the las-to-rhl signal ratio, previously considered a characteristic distinguishing between planktonic and biofilm lifestyles. Our results highlight a supply-driven, metabolically prudent regulation of public goods that minimizes production costs and thereby helps stabilize cooperative behavior. Such regulation would be beneficial for QS-dependent public goods that act broadly and nonspecifically, and whose need cannot always be accurately assessed by the producing cell. Clear differences in the capacities of the las and rhl systems to integrate starvation signals help explain the existence of multiple QS systems in one cell.
INTRODUCTION
In a process termed quorum sensing (QS), bacteria regulate gene expression in response to self-produced diffusible signal molecules (1, 2). As bacterial cell density increases, the accumulation of signals to a critical threshold triggers the activation of target genes. These genes often encode so-called “public goods,” secreted factors implicated in virulence, biofilm formation, nutrient acquisition, and microbial competition. Public goods represent a form of cooperation; they are costly to produce, provide a density-dependent benefit, and are shared among the population (3, 4). In many Gram-negative proteobacteria, QS is mediated by acyl-homoserine lactone (acyl-HSL) signals (1, 2, 4). These signals are produced by a synthase enzyme of the LuxI family and are bound by a cognate receptor of the LuxR family that regulates transcription.
The opportunistic pathogen Pseudomonas aeruginosa serves as a model for the study of acyl-HSL QS (5–7). This organism has the metabolic versatility and regulatory capacity to adapt to diverse environments, including soil, freshwater, and the human host. There are two complete acyl-HSL systems in P. aeruginosa, LasR-LasI (las) and RhlR-RhlI (rhl). LasI and RhlI produce the autoinducers 3-oxododecanoyl (3OC12)-HSL and butanoyl (C4)-HSL, respectively. LasR and RhlR bind their cognate signals to activate the transcription of overlapping regulons. The las system also activates the rhl system (8, 9), although this regulatory hierarchy is conditional (10, 11). There is also an orphan LuxR-type regulator, QscR, and a 2-alkyl-4-quinolone-based QS system (PQS system) (12, 13). A large fraction of the QS-controlled genes in P. aeruginosa encode secreted factors such as exoproteases (e.g., LasA and LasB elastases, P. aeruginosa aminopeptidase [PaAP]) and secondary metabolites (e.g., hydrogen cyanide, pyocyanin, rhamnolipid) implicated in virulence and other group behaviors (7, 14). Most of these secreted factors are at least partially regulated by the rhl system (7). A potential function for multiple QS systems within one organism is to produce a temporally ordered sequence of gene expression (15–17).
While signal accumulation and cell density are important triggers for QS, a threshold concentration is necessary, but not sufficient, to induce most QS-regulated genes. During growth in rich-medium batch culture, most QS-controlled genes are not expressed until stationary phase, even with the addition of exogenous acyl-HSL signals (15, 16, 18). QS is embedded in a network of global regulation, and specific environmental conditions can coregulate QS (6, 19). Starvation has an important role in the induction of QS. For example, the stringent response, the alternative sigma factors RpoS and RpoN, phosphate signaling via PhoRB, and iron regulation via Fur all contribute directly or indirectly to the expression of QS-controlled genes (20–24). Such signal integration has also been observed in the QS systems of other bacterial species (19).
Early biotechnological studies conducted to optimize the production of certain P. aeruginosa-secreted factors, such as exoproteases and rhamnolipid, that later turned out to be quorum controlled showed that the limitation of specific nutrients enhances expression (25, 26). To date, investigations on quorum-controlled gene expression have almost exclusively involved batch cultures, in which the effects of cell density, nutrient starvation, and slow growth cannot be clearly separated. Here we employed a defined minimal medium and a continuous-culture system to identify specific macronutrient limitation and slow growth as key conditions that induce QS-controlled public-good genes. We found that rhl-dependent genes are more strongly affected than las-dependent genes and that the limiting nutrient induces the expression of the secreted product only when it is not a building block of that product, thus minimizing the burden on individual fitness. Our data suggest a functional split between the las and rhl QS systems, with the rhl system integrating stress and starvation signals. Taken together, our results provide a physiological framework for the complex regulation of QS-controlled secretions by stress and starvation responses.
MATERIALS AND METHODS
Bacterial strains, plasmids, and routine culture conditions.
The P. aeruginosa and Escherichia coli strains used in this study are listed in Table 1. Transcriptional fusion strains of P. aeruginosa were constructed by PCR amplification of promoter regions (see Table S1 in the supplemental material), cloning into mini-CTX-lacZ, and integration into the PAO1 chromosome at the attB site, as described previously by Hoang et al. (27). Routine liquid cultures were grown at 37°C in Lennox Luria-Bertani (LB) broth buffered with 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), pH 7.0.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PAO1 | Wild type | 52 |
| PAO1-pepB′-lacZ | Markerless transcriptional fusion of pepB promoter to lacZ in chromosomal attB site; derived from PAO1 | This study |
| PAO1-lasB′-lacZ | Markerless transcriptional fusion of lasB promoter to lacZ in chromosomal attB site; derived from PAO1 | This study |
| PAO1-phzA1′-lacZ | Markerless transcriptional fusion of phzA1 promoter to lacZ in chromosomal attB site; derived from PAO1 | This study |
| PAO1-rhlA′-lacZ | Markerless transcriptional fusion of rhlA promoter to lacZ in chromosomal attB site; derived from PAO1 | This study |
| PAO1 ΔlasR::Tetr | lasR mutant via insertion of tetracycline resistance cassette; derived from PAO1 | 53 |
| PAO1 ΔrhlR::Gmr | rhlR mutant via insertion of gentamicin resistance cassette; derived from PAO1 | 53 |
| PAO1 ΔlasR::Tetr ΔrhlR::Gmr | lasR rhlR double mutant via insertion of tetracycline and gentamicin resistance cassettes, respectively; derived from PAO1 | 53 |
| E. coli DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 | Life Technologies |
| Plasmids | ||
| Mini-CTX-lacZ | Tetr; integration-proficient vector for chromosomal insertion at the attB site | 54 |
| pFLP2 | Ampr; flp recombinase plasmid | 27 |
Batch culture experiments.
Nutrient-limiting batch culture experiments were carried out in MOPS minimal medium as described by Neidhardt et al. (28), with minor modifications. The medium contains 50 mM MOPS (pH 7.0), 4 mM Tricine (pH 7.0), 50 mM NaCl, 15 mM NH4Cl, 4 mM K2HPO4, 1 mM K2SO4, 0.05 mM MgCl2, 0.01 mM CaCl2, 0.005 mM FeCl2, trace elements, and a carbon source (25 mM monosodium glutamate, 25 mM disodium succinate, or 25 mM d-glucose). Specific nutrients were diluted to yield a C-, N-, P-, S-, or Fe-limiting medium. Nutrient dilution cultures were inoculated to an optical density at 600 nm (OD600) of 0.005 from mid-exponential-phase cultures containing full-strength medium, and they were grown for 16 h in 96 deep-well plates (VWR) at 37°C with agitation (250 rpm). Samples were taken for the measurement of the OD600 and β-galactosidase activity. For these experiments, OD600 values were read by a microplate reader (Infiniti M200; Tecan) and are reported without conversion to a 1-cm path length.
To measure gene expression in different growth phases by quantitative real-time PCR (qPCR), batch cultures, inoculated as described above for the nutrient dilution experiments, were grown in glass tubes at 37°C with agitation. The minimal medium was used at full strength. Exponential-phase, early-stationary-phase, and late-stationary-phase samples were taken at OD600 values (1-cm path length) of 0.15, 1.5, and 2.5, respectively.
Chemostat experiments.
Continuous-culture experiments were carried out in glass vessels containing 100 ml of MOPS minimal medium, as described by Whiteley et al. (29) with minor modifications. The medium contained 75 mM MOPS, 93.5 mM NH4Cl, 42.8 mM NaCl, 4 mM KH2PO4, 1 mM MgSO4, 0.0075 mM FeSO4, and trace elements. P. aeruginosa strains were grown at 37°C with aeration, in either a C-limited medium (4 mM KH2PO4 and 3 mM monosodium glutamate) or a P-limited medium (0.11 mM KH2PO4 and 10 mM monosodium glutamate). Chemostat vessels were routinely treated with Sigmacote (Sigma-Aldrich) to minimize biofilm formation. Chemostat cultures were inoculated to an OD600 of 0.01 from an overnight culture and were grown to an OD600 of approximately 0.1 before the flow of medium was initiated. All chemostat experiments began at the highest dilution rate (D) of 0.5 h−1 and were incrementally reduced to the lowest D of 0.125 h−1. Bacteria were allowed to adjust to steady state for at least three doublings before samples were taken to measure the OD600 and gene expression, acyl-HSL, pyocyanin, and protein levels. At steady state, D is equal to the specific growth rate (μ).
β-Galactosidase assay.
β-Galactosidase activity in batch and chemostat cultures was measured in a Tecan microplate reader by using a Galacto-Light Plus kit (Applied Biosystems) as described previously (15). Batch culture results are expressed as the fold difference in the number of photons per OD600 unit between undiluted and diluted medium. To visualize these data in a heat map, the pheatmap package in R, version 2.15.3, was used (http://CRAN.R-project.org/package=pheatmap).
Quantitative real-time PCR.
Transcript levels in batch and chemostat cultures were analyzed by a two-step reverse transcription qPCR procedure by using the Applied Biosystems 7300 sequencing system as described previously (30, 31). Total-RNA isolation and cDNA synthesis with semirandom primers were performed as described previously (16). An Agilent 2100 Bioanalyzer was used to determine RNA quality. The 25-μl qPCR mixture contained forward and reverse primers (300 nM each; listed in Table S1 in the supplemental material), 1 ng of purified cDNA, and Power SYBR green PCR master mix reagents (Applied Biosystems). Equal amounts of cDNA were used for all qPCRs. Relative expression levels were determined using a genomic DNA standard curve.
Acyl-HSL detection.
E. coli reporter strain bioassays were used to quantify 3OC12-HSL and C4-HSL levels from ethyl acetate extracts of chemostat culture supernatants as described previously (32, 33).
Pyocyanin production.
Pyocyanin was extracted from chemostat culture supernatants and measured as described by Essar et al. (34). Briefly, pyocyanin was extracted from 5 ml of the culture supernatant with 3 ml of chloroform. One milliliter of 0.2 M HCl was used to extract the pyocyanin from chloroform. Absorbance was measured at 520 nm, and values were converted to micrograms per milliliter via a previously reported conversion factor of 17.072 (34).
Western blot analysis.
Western blotting was performed as described previously (31). Chemostat culture aliquots were harvested by centrifugation, and pellets were resuspended in lysis buffer. Cells were lysed by sonication, and the lysates were centrifuged to remove insoluble material. The protein concentration of the soluble fraction was determined by a Bradford assay. Equal amounts of soluble protein were separated by polyacrylamide gel electrophoresis. Separated proteins were blotted onto a nitrocellulose membrane and were probed with polyclonal anti-LasR and anti-RhlR antibodies (31).
RESULTS
Serial dilution of macronutrients in batch culture yields distinct expression profiles for QS-controlled genes.
We began our study by investigating the responses of individual QS-controlled genes to the limitation of specific macronutrients during batch culture growth in defined medium. We selected four QS-controlled genes, pepB, lasB, phzA1, and rhlA, associated with public-good production (Table 2). Our previous transcriptome analysis in rich medium showed that these genes exhibit differing specificities for the las and rhl QS systems and that their expression increases at the transition to stationary phase, regardless of signal concentration (see Fig. S1 in the supplemental material) (16). Expression is controlled directly by LasR, RhlR, or both regulators (16, 35). To quantify the expression of these genes, we fused a lacZ reporter, encoding β-galactosidase, to the respective promoter regions and inserted the resulting constructs into a neutral site on the P. aeruginosa chromosome. We grew the reporter strains in a minimal medium with the synthetic organic buffer MOPS (28). In this medium, each macronutrient is present at a sufficiently high level to meet the nutritional needs of the bacterial cell. We separately and progressively diluted five macronutrients: carbon (C), nitrogen (N), phosphorus (P), sulfur (S), and iron (Fe). We also tested three sources of C and energy: succinate, glutamate, and glucose. The preferences of P. aeruginosa for these C sources differ and can influence QS-controlled phenotypes (36). Organic acids such as tricarboxylic acid (TCA) cycle intermediates are generally preferred, since they repress the utilization of sugars and other substrates through carbon catabolite repression (37).
TABLE 2.
QS-controlled genes and their propertiesa
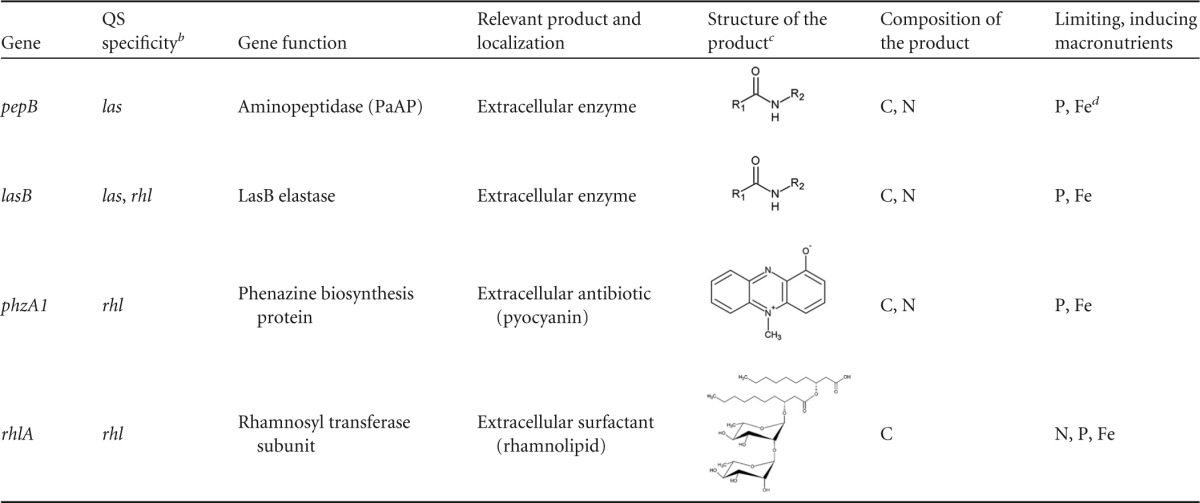
This table includes data from previous studies (cited in the text) and from this work.
In the text, we distinguish primarily between a “las-specific” gene, pepB, which responds only to the las system and the “rhl-dependent” genes lasB, phzA1, and rhlA, which respond only to the rhl system or to both systems.
The peptide bonds shown symbolize proteins.
The effect on gene expression was small.
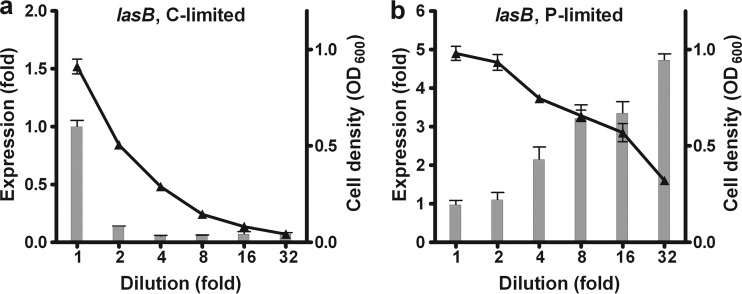
First, we used the lasB-lacZ reporter strain to test progressive 2-fold dilutions of essential nutrients (examples of C and P limitation are shown in Fig. 1; see Fig. S2 to S4 in the supplemental material, lasB, for complete data). Limitation of all macronutrients except S substantially decreased the growth yields of the cultures. Despite this reduction in cell density, P and Fe limitation stimulated lasB expression. C and N limitation, on the other hand, reduced lasB expression. The expression of lasB was highest when succinate was the sole C source. The C source glutamate can also serve as an N source, so N is not limiting in glutamate medium. S dilution in these experiments had only a small effect on growth yield, with no appreciable impact on gene expression. It is possible that P. aeruginosa can utilize MOPS as an S source, as has been shown for some strains of E. coli (28, 38).
FIG 1.
Examples of nutrient dilution series with the P. aeruginosa lasB reporter strain. Progressive 2-fold dilutions of C (a) and P (b) were carried out in minimal medium batch cultures with glutamate as the sole C source. Bars indicate fold changes from β-galactosidase expression in undiluted medium (left y axis). Triangles indicate the OD600 (right y axis). Fold change values were normalized to OD600 values. Values are means for three independent biological replicates. Error bars indicate standard deviations of the means.
For the analysis of the remaining reporter fusions, those with pepB, phzA1, and rhlA, we limited ourselves to the 8- and 32-fold nutrient dilutions (see Fig. S2 to S4 in the supplemental material, pepB, phzA1, and rhlA; Fig. 2 presents a summary of all gene expression data). We found that pepB, phzA1, and rhlA, like lasB, are induced by P and Fe limitation and suppressed by C limitation. In contrast, rhlA was induced by N limitation, whereas the other genes either were suppressed or showed no response. Also, pepB was not as highly induced (≤2-fold) as lasB, phzA1, and rhlA. In some cases, expression decreased after further dilution of an inducing macronutrient. Induction patterns were generally qualitatively similar for all three C sources (excluding N dilutions in glutamate medium).
FIG 2.
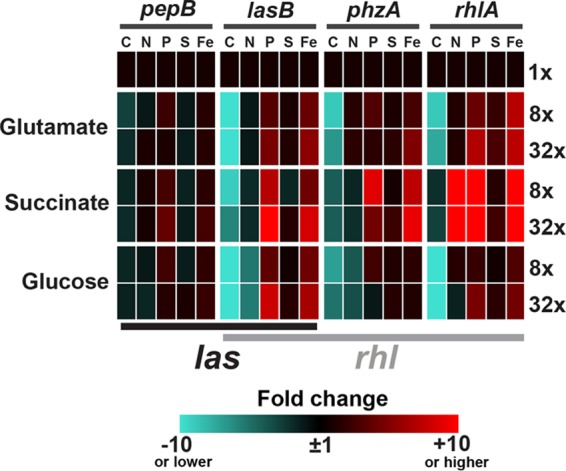
QS-controlled gene expression in P. aeruginosa during specific macronutrient limitation in batch culture. To compare the expression of QS-controlled genes (pepB, lasB, phzA1, and rhlA) across many nutrient conditions, fold change data from Fig. S2 to S4 in the supplemental material are visualized in a heat map. Numbers on the right indicate fold dilutions of the limiting macronutrient (either C, N, P, S, or Fe). Carbon sources are given on the left, and QS-controlled genes and limiting nutrients are given above the heat map. lasR (black) or rhlR (gray) promoter specificity is indicated on the bottom. In the text, genes regulated exclusively by the las system are referred to as las specific (pepB), and genes dependent on regulation by the rhl system are referred to as rhl dependent (lasB, phzA1, and rhlA).
Our results suggest that nutrient stress is a major trigger of QS-controlled expression of public goods and that individual QS-controlled genes respond differently to the limitation of specific nutrients. The extent of regulation appears to correlate with las versus rhl promoter specificity (Table 2; Fig. 2); the rhl-dependent genes are more responsive to nutrient starvation than the las-specific gene. Most intriguingly, as we discuss in more detail below, responses to individual nutrients appear to be a function of the molecular composition of the regulated secreted product (Table 2). Limitation of a particular macronutrient enhances expression only if it is not a building block of the product.
Slow growth induces QS-controlled gene expression in continuous culture when specific nutrients are limiting.
The batch culture experiments discussed above indicate that QS-controlled expression of public goods is triggered by specific nutrient limitation rather than by cell density. To further investigate the role of nutrient-limited slow growth independently of cell density, we employed a chemostat culture system. In a chemostat, the cell density is determined by the concentration of the growth-limiting nutrient in the medium reservoir. The specific growth rate (μ), on the other hand, is determined by the flow rate and hence by the dilution rate (D) of the medium. As D decreases, the actual concentration of the growth-limiting substrate in the chemostat decreases and consequently reduces μ without affecting density. As cells reach a steady state, μ equals D. This way, cells can be maintained in balanced growth at rates similar to those transiently experienced by batch cultures during entry into stationary phase, when many QS genes are induced. We measured QS-controlled gene expression at four different μ values, 0.5 h−1, 0.33 h−1, 0.25 h−1, and 0.13 h−1, by using qPCR. The range of μ values was dictated, at least in part, by culturing constraints. A μ of >0.5 h−1 approaches the maximum possible growth rate in the medium (μmax, 0.83 ± 0.15 h−1), resulting in a decrease in culture density and eventual washout. A μ of <0.13 h−1 causes biofilm formation on the walls of the chemostat vessel. The cell density, measured as the OD600, was kept constant at approximately 0.15. This density is below the quorum threshold in standard batch culture (see Fig. S1 in the supplemental material; also see below). We used P as an example of a nutrient that induces QS when limiting and C as a nutrient that does not induce QS when limiting. We chose glutamate as the C source.
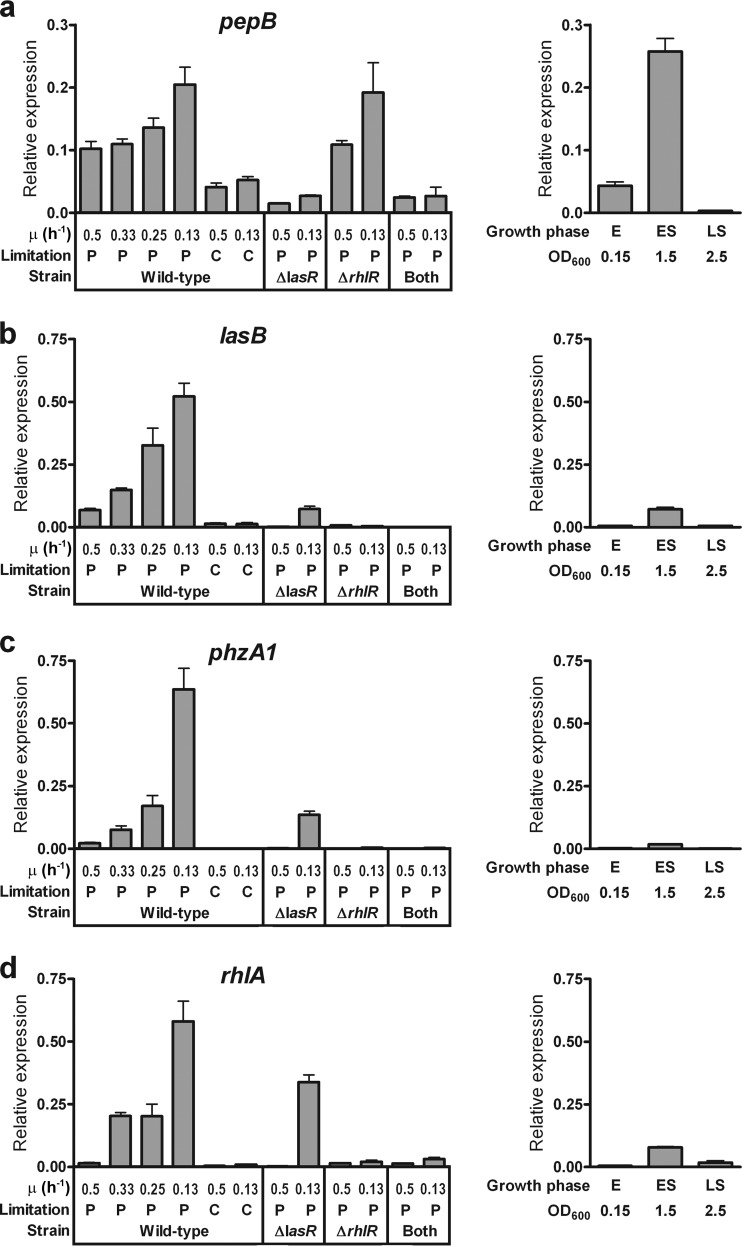
We found that P-limited, but not C-limited, slow growth induces QS-controlled gene expression and that rhl-dependent genes are more strongly induced than a las-specific gene (Fig. 3, left, wild type). Between a μ of 0.5 h−1 and a μ of 0.13 h−1, the expression of the las-specific gene pepB increased by ≤2-fold, whereas the expression of the rhl-dependent genes lasB, phzA1, and rhlA increased by 7.5-, 28-, and 36-fold, respectively (Fig. 3, left, wild type). The induction of phzA1 correlated with a striking blue-green pigmentation of the chemostat culture, indicative of high-level pyocyanin secretion (see Fig. S5 in the supplemental material), demonstrating that transcription correlates with actual public-good production.
FIG 3.
Transcript levels of QS-controlled genes in chemostat and batch cultures. Relative abundances of pepB (a), lasB (b), phzA1 (c), and rhlA (d) mRNAs were measured by qPCR. Wild-type P. aeruginosa was cultured in glutamate minimal medium in a P- or C-limited chemostat (left) and in batch culture (right). (Left) A lasR mutant, an rhlR mutant, and a lasR rhlR double mutant (indicated as “both”) were cultured in a P-limited chemostat for comparison with the wild type. Chemostat cultures were at a steady-state OD600 of 0.15. (Right) Batch cultures were sampled during the exponential (E), early-stationary (ES), and late-stationary (LS) phases, at the indicated densities. Values are means for three independent biological replicates. Error bars indicate standard deviations of the means.
To relate the chemostat data to those from more-familiar batch cultures, we also measured gene expression during batch culture growth (Fig. 3, right), acknowledging that direct comparison is somewhat limited by the fact that chemostat cultures are at steady state, whereas batch cultures are not. The minimal medium for the batch cultures was used at full strength, with P and C concentrations higher than those used for chemostat cultures. This allowed us to establish baseline gene expression at μmax during exponential growth, at a density identical to that of the chemostat. In batch cultures, the expression of all QS-controlled genes was lowest during exponential phase, increased in early-stationary phase, and decreased again in late-stationary phase (Fig. 3, right), an expression pattern similar to that in rich medium (see Fig. S1 in the supplemental material). When glutamate is the C source, the growth yield in the full-strength minimal medium is C limited, although other macronutrients are also approaching depletion (see Fig. S2 in the supplemental material). Interestingly, the expression of rhl-dependent genes in P-limited chemostats was higher than that in batch cultures at a comparatively higher cell density. Taken together, these data show that a reduction in the growth rate under specific nutrient limitation induces the expression of secreted products, independently of cell density.
To confirm the chemostat expression data obtained by qPCR, we independently assessed QS gene expression via our transcriptional lacZ reporter fusions (see Fig. S6 in the supplemental material). These two approaches yielded very similar results. The reporter data also confirm the notion that the starvation-dependent regulation of QS transcripts is at the level of transcription initiation.
Slow growth primarily induces the rhl system in continuous culture when specific nutrients are limiting.
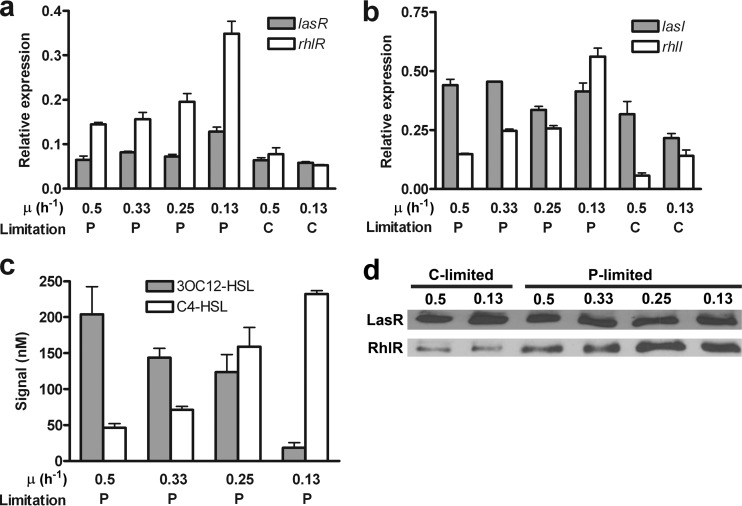
We hypothesized that the preferential induction of rhl-dependent genes during P-limited slow growth in chemostat culture is reflected in the differential activation of the las versus the rhl system. To test this, we compared the transcript levels of lasR, rhlR, lasI, and rhlI by qPCR (Fig. 4a and b). We further examined acyl-HSL levels by bioassay and LasR and RhlR protein levels by Western blotting (Fig. 4c and d, respectively). Growth rate reduction had a much more pronounced effect under P-limiting than under C-limiting conditions and affected the rhl system more than the las system, in support of our hypothesis. The increase in rhlR and rhlI transcript levels under P-limited slow growth correlated with the increase in RhlR protein and C4-HSL levels, respectively (Fig. 4). On the other hand, while lasR transcript levels showed a modest increase, lasI transcript levels showed little to no increase during P-limited slow growth (Fig. 4a and b). This corresponded to virtually unchanged LasR protein levels and an actual decrease in 3OC12-HSL levels (Fig. 4c and d). Interestingly, the ratios of the two acyl-HSL signals were dramatically affected by the growth rate (Fig. 4c). During faster growth (μ, 0.5 h−1), the concentration of 3OC12-HSL was much higher than that of C4-HSL. During slow growth (μ, 0.13 h−1), in contrast, the concentration of C4-HSL was much higher than that of 3OC12-HSL.
FIG 4.
Expression levels of las and rhl system components in chemostat culture. Wild-type P. aeruginosa was cultured in P- or C-limited glutamate minimal medium as indicated. Chemostat cultures were at a steady-state OD600 of 0.15. (a and b) Relative transcript abundances of lasR and rhlR (a) and of lasI and rhlI (b) as measured by qPCR. (c) Acyl-HSL concentrations measured by a bioassay. Values in panels a, b, and c are means for three independent biological replicates. Error bars indicate standard deviations of the means. (d) LasR and RhlR protein levels determined by Western blotting. Equal amounts of total protein were loaded for each condition.
To finally demonstrate that preferential induction of the rhl system by P-limited slow growth actually causes the activation of target genes, we measured the expression of QS-controlled public-good genes in a lasR mutant, an rhlR mutant, and a lasR rhlR double mutant. We found that lasB, phzA1, and rhlA expression increases substantially during slow growth in a lasR mutant but not in an rhlR mutant (Fig. 3, left). However, expression in the lasR mutant was significantly lower than that in the wild type (P, ≤0.049 at a μ value of 0.5 h−1 or 0.13 h−1 by a two-tailed t test), indicating that lasR is still required for full induction, presumably as an epistatic regulator of the rhl system, and also as a coregulator in the case of lasB. The expression of pepB was greatly reduced in a lasR mutant but was not significantly different from that of the wild type in an rhlR mutant (P, ≥0.60 at a μ value of 0.5 h−1 or 0.13 h−1 by a two-tailed t test), confirming that this gene is las specific (Fig. 3, left). As expected, all genes exhibited little to no expression in the lasR rhlR double mutant control (Fig. 3, left). Taken together, these data suggest that the rhl system is mainly responsible for increased QS-controlled gene expression during P-limited slow growth and that the las system plays a minor role.
DISCUSSION
Our work consolidates a large body of literature on the complex regulation of secreted products in P. aeruginosa and provides insights into the evolutionary and ecological purpose of such regulation. While research shows that the regulation of public goods by QS optimizes the costs and benefits of their production (39, 40), the purpose of their coregulation by numerous other pathways and environmental conditions has been less apparent. Our approach was to comprehensively identify specific nutrient conditions that induce QS-controlled public-good genes and to determine whether these genes are controlled in a growth rate-dependent manner. We utilized a defined minimal medium that permits the limitation of each macronutrient and a continuous-culture system that separates the effects of growth rate and cell density. We found that QS-controlled genes are differentially regulated under different nutrient conditions and during slow growth. Successive reductions in the growth rate through specific nutrient limitation augmented induction (Fig. 5 presents a schematic model, additional details of which are discussed below).
FIG 5.
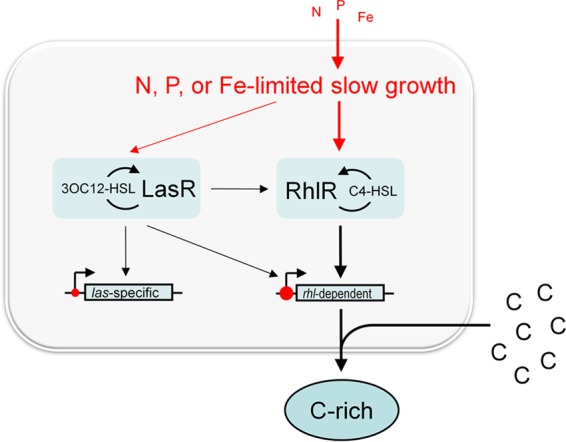
Supply-driven regulation of QS-dependent public goods in P. aeruginosa. Specific macronutrient limitation and concomitant reduction in the growth rate induce the expression of secreted products, primarily those under the direct control of the rhl system (indicated by thicker arrows). Nutrient limitation induces RhlR and C4-HSL expression but likely also directly regulates target gene expression at the promoter level (indicated as red circles; a larger circle indicates a stronger effect). In the example shown, the secretion of a C-rich molecule is induced by N, P, or Fe starvation in the presence of excess C. In another example from this study, the secretion of C- and N-rich molecules (e.g., extracellular enzymes) is induced by P or Fe starvation in the presence of excess C and N.
Our data indicate that central factors in governing QS gene expression are regulation by either the las or the rhl system and the molecular composition of the relevant secreted product. First, a las-controlled gene showed little response to nutrient starvation compared to genes directly controlled by the rhl system, and the observed expression pattern correlated with, and was caused by, the preferential induction of the rhl system. Second, products were generally highly expressed when their molecular building blocks were in excess and other nutrients were limiting, but their expression was suppressed when their molecular building blocks became limiting (Fig. 2; Table 2). A simple statistical thought experiment indicates that this association between molecular composition and macronutrient regulation is strong. The probability of the particular expression pattern observed for all genes, namely, that they are induced by P and Fe (not building blocks) but not by C (a building block), is 1 in 23 possible permutations, and the probability that only rhlA, and none of the three other genes, is induced by N (rhamnolipids are the only product devoid of N) is 1 in 24 possible permutations. Thus, the combined probability for the expression pattern observed, which is the only pattern consistent with our conclusion, is 0.0078 (1/8 × 1/16).
Our results attribute general validity to the “metabolically prudent” regulation of secretions that minimizes the costs of their production (41). The term “metabolic prudence” was introduced by Xavier et al. to describe the nutritionally conditional regulation of rhamnolipid secretion in P. aeruginosa (41). Expression of this C-rich molecule is induced only when another nutrient, N, is limiting, when there is excess carbon, and when the cells approach the stationary phase of growth. Metabolic prudence minimizes the fitness costs of cooperation through efficient use of available resources, because cells do not secrete molecular building blocks that could instead be used for growth. This regulatory strategy, therefore, has the potential to stabilize cooperative behavior according to inclusive fitness theory (3, 42) and to limit the spread of noncooperating mutants, or “cheaters,” as demonstrated previously (41).
What might be the ecological significance of such supply-driven regulation, seemingly unrelated to the function of the public goods? As we have pointed out previously (43), QS-dependent public goods are secreted irrespective of whether they provide a direct fitness benefit (i.e., promote the growth of clonal populations) or not. P. aeruginosa and other opportunistic pathogens frequently reside in nonparasitic environments, and it is possible that secreted products primarily provide an indirect fitness benefit in this context. Many of the QS-controlled secretions have the potential to harm other cells. In light of the recently proposed “competition-sensing” hypothesis (44), such secretions can be viewed as a broad, nonspecific microbial counterattack against ecological competition inferred from nutrient starvation. If the presence of competition cannot be sensed directly and reliably, then the production of secretions appears prudent whenever the cost of their production is low.
In our work, successive limitation of C, and in some cases N, during batch culture growth reduced the expression of public goods. Even though none of the macronutrients in the full-strength minimal medium were in great excess, we can infer from the individual nutrient dilution experiments that the medium was C and N limited when succinate and glucose were the C sources and was C limited when glutamate was the C source (see Fig. S2 to S4 in the supplemental material). Even under C-limited conditions, there was some induction of QS-controlled public-good genes in glutamate medium batch culture in stationary phase (Fig. 3). This is consistent with previous work. Individual QS-controlled genes are induced during C-limited slow growth in casein medium (30), and exoprotease activity is induced during C-limited chemostat culture at very low growth rates (μ, ≥0.05 h−1, attained at reduced temperatures with a different P. aeruginosa strain and an antifoaming agent) (25). However, we also showed that the level of QS gene expression decreases in batch cultures when C limitation becomes more pronounced through successive dilutions. This decrease was specific to certain macronutrients and target genes and is thus unlikely to be caused solely by a concomitant reduction in cell density. These data point to the ratio of macronutrients as a key determinant in regulating secretions (26). For example, gene expression decreases with decreasing C-to-N or C-to-P ratios. The inclusion of C levels as a regulatory cue is equally “prudent,” since all QS-controlled secreted products in P. aeruginosa are rich in C (31).
Additionally, we found that QS gene expression in batch cultures decreases when cells enter late-stationary phase in C-limited medium (Fig. 3) and, in some cases, when an inducing macronutrient is highly diluted (see Fig. S2 to S4 in the supplemental material). This pattern may be interpreted as a complete “shutdown” of cells in response to severe starvation.
To address the mechanism of the preferential regulation of rhl-dependent public goods, we measured the expression of central QS components in the wild type, and of QS target genes in signal receptor mutants, during P- and C-limited chemostat growth. Quantitation of transcript, acyl-HSL, and regulator protein levels indicates that the rhl system is greatly induced by P-limited slow growth and that this induction is largely responsible for the expression pattern observed for QS target genes. In agreement with the predominant role of the rhl system, we found that progressively lower growth rates invert the 3OC12-HSL/C4-HSL ratio in favor of C4-HSL. Lower dilution rates could conceivably have an impact on the steady-state concentrations of each signal due to their different chemical stabilities. However, if this were the case, the effect would be the opposite of what we observed, because long-chain acyl-HSL signals are more resistant to lactonolysis than short-chain signals (45). Thus, signal ratios likely represent real physiological changes rather than an artifact of the culturing method. Signal ratios have been proposed as a diagnostic marker for the growth of P. aeruginosa as a biofilm in cystic fibrosis (CF) lung infections (46). The signal ratio, as measured by radiometry, was the same in laboratory biofilms and in CF sputum but was opposite that in planktonic culture (46). Our results suggest more broadly that the signal ratio is primarily an indicator of the growth rate and nutrient status, irrespective of the mode of growth.
Previous studies identified mechanisms that allow P. aeruginosa to respond to low N, P, and Fe levels during batch culture growth, and they provide the link between QS and the physiological cues identified in this study. The two-component regulatory system PhoRB induces rhlR expression when growth becomes P limited (23). Regulatory control appears to be direct, since rhlR possesses a PhoB binding site in its promoter region (23), but may also be indirect, through a newly identified non-acyl-HSL QS signal, termed IQS (for “integrating QS and stress response”) (47). Fe depletion enhances rhlR and rhlI expression via ferric uptake regulator protein and the PQS system (22, 23), whereas N starvation enhances rhlR transcription via the alternative sigma factor RpoN (10). However, regulatory control at the level of rhlR and/or rhlI is unlikely to be sufficient to produce the diverse responses to macronutrient limitation that we observed. While induction by P and Fe limitation was universal, induction by N limitation was specific to only one secreted product, rhamnolipid. Further control must therefore exist at the level of the target promoters of the individual QS-controlled genes. This conclusion is consistent with the finding that overexpression of rhlR and the simultaneous addition of C4-HSL are not sufficient to induce rhlA expression (48).
The stringent response can activate the las and rhl QS systems in P. aeruginosa during amino acid starvation (20). Because the minimal media employed in this study lack amino acids and require de novo synthesis, the amino acid-dependent stringent response likely contributed to a basal level of target gene induction. It is conceivable that N-, P-, or Fe-limited slow growth resulted in further activation of the stringent response, in addition to the activation of specific starvation pathways as described above. For example, in E. coli, the stringent response is also activated upon P limitation in addition to amino acid limitation (49).
Macronutrient ratios may be integrated at the level of intracellular metabolite fluxes. This is perhaps best exemplified by the signal transduction pathway that regulates nitrogen assimilation. It consists of the two-component system NtrBC and the signal protein PII, which senses nitrogen availability via glutamate (50). However, PII is also responsive to cellular carbon levels and energy charge through direct binding of α-ketoglutarate and ATP (50).
Our results help explain why there is a need for multiple QS systems in one organism. While two QS systems may serve to temporally scale gene induction (15–17), the integration of distinct stress signals in P. aeruginosa QS suggests an additional role. The las system appears to respond predominantly to autoinducer accumulation, whereas the rhl system integrates nutritional cues. Thus, multiple QS systems can provide a functional split between gene products that should be prudently regulated during starvation and gene products that should be expressed whenever the cell density is adequate for their efficient use. Interestingly, the rhl system was acquired later in the evolutionary history of P. aeruginosa via horizontal gene transfer (51). It is intriguing to speculate that this event facilitated the integration of quorum and stress responses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rashmi Gupta for cloning the lasB promoter and Herbert P. Schweizer for the mini-CTX-lacZ construct. We also thank Cara Wilder, Rashmi Gupta, D. Joseph Sexton, Kyle Asfahl, and Erin Bredeweg for helpful discussions.
This work was supported by National Institutes of Health grant AI079454 and by National Science Foundation grant 1158553 (both to M.S.). B.M. was supported in part by the P.F. and Nellie Buck Yerex Graduate Fellowship.
Footnotes
Published ahead of print 27 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01223-13.
REFERENCES
- 1.Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346. 10.1146/annurev.cellbio.21.012704.131001 [DOI] [PubMed] [Google Scholar]
- 2.Williams P, Winzer K, Chan WC, Camara M. 2007. Look who's talking: communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 362:1119–1134. 10.1098/rstb.2007.2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West SA, Griffin AS, Gardner A, Diggle SP. 2006. Social evolution theory for microorganisms. Nat. Rev. Microbiol. 4:597–607. 10.1038/nrmicro1461 [DOI] [PubMed] [Google Scholar]
- 4.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu. Rev. Microbiol. 67:43–63. 10.1146/annurev-micro-092412-155635 [DOI] [PubMed] [Google Scholar]
- 5.Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, Quax WJ. 2012. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 76:46–65. 10.1128/MMBR.05007-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams P, Camara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12:182–191. 10.1016/j.mib.2009.01.005 [DOI] [PubMed] [Google Scholar]
- 7.Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296:73–81. 10.1016/j.ijmm.2006.01.036 [DOI] [PubMed] [Google Scholar]
- 8.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137–1146. 10.1046/j.1365-2958.1996.00063.x [DOI] [PubMed] [Google Scholar]
- 9.Pesci EC, Pearson JP, Seed PC, Iglewski BH. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina G, Juarez K, Diaz R, Soberon-Chavez G. 2003. Transcriptional regulation of Pseudomonas aeruginosa rhlR, encoding a quorum-sensing regulatory protein. Microbiology 149:3073–3081. 10.1099/mic.0.26282-0 [DOI] [PubMed] [Google Scholar]
- 11.Dekimpe V, Deziel E. 2009. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155:712–723. 10.1099/mic.0.022764-0 [DOI] [PubMed] [Google Scholar]
- 12.Lee JH, Lequette Y, Greenberg EP. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59:602–609. 10.1111/j.1365-2958.2005.04960.x [DOI] [PubMed] [Google Scholar]
- 13.Dubern JF, Diggle SP. 2008. Quorum sensing by 2-alkyl-4-quinolones in Pseudomonas aeruginosa and other bacterial species. Mol. Biosyst. 4:882–888. 10.1039/b803796p [DOI] [PubMed] [Google Scholar]
- 14.Girard G, Bloemberg GV. 2008. Central role of quorum sensing in regulating the production of pathogenicity factors in Pseudomonas aeruginosa. Future Microbiol. 3:97–106. 10.2217/17460913.3.1.97 [DOI] [PubMed] [Google Scholar]
- 15.Whiteley M, Lee KM, Greenberg EP. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 96:13904–13909. 10.1073/pnas.96.24.13904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuster M, Lohstroh CP, Ogi T, Greenberg EP. 2003. Identification, timing and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066–2079. 10.1128/JB.185.7.2066-2079.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long T, Tu KC, Wang Y, Mehta P, Ong NP, Bassler BL, Wingreen NS. 2009. Quantifying the integration of quorum-sensing signals with single-cell resolution. PLoS Biol. 7:e68. 10.1371/journal.pbio.1000068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diggle SP, Winzer K, Lazdunski A, Williams P, Camara M. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576–2586. 10.1128/JB.184.10.2576-2586.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellbye B, Schuster M. 2011. More than just a quorum: integration of stress and other environmental cues in acyl-homoserine lactone signaling, p 349–363 In Storz G, Hengge R. (ed), Bacterial stress responses, 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 20.van Delden C, Comte R, Bally AM. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376–5384. 10.1128/JB.183.18.5376-5384.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973–985. 10.1046/j.1365-2958.2003.03886.x [DOI] [PubMed] [Google Scholar]
- 22.Oglesby AG, Farrow JM, III, Lee JH, Tomaras AP, Greenberg EP, Pesci EC, Vasil ML. 2008. The influence of iron on Pseudomonas aeruginosa physiology: a regulatory link between iron and quorum sensing. J. Biol. Chem. 283:15558–15567. 10.1074/jbc.M707840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen V, Lons D, Zaoui C, Bredenbruch F, Meissner A, Dieterich G, Munch R, Haussler S. 2006. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J. Bacteriol. 188:8601–8606. 10.1128/JB.01378-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medina G, Juarez K, Valderrama B, Soberon-Chavez G. 2003. Mechanism of Pseudomonas aeruginosa RhlR transcriptional regulation of the rhlAB promoter. J. Bacteriol. 185:5976–5983. 10.1128/JB.185.20.5976-5983.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whooley MA, O'Callaghan JA, McLoughlin AJ. 1983. Effect of substrate on the regulation of exoprotease production by Pseudomonas aeruginosa ATCC 10145. J. Gen. Microbiol. 129:981–988 [DOI] [PubMed] [Google Scholar]
- 26.Guerra-Santos L, Kappeli O, Fiechter A. 1984. Pseudomonas aeruginosa biosurfactant production in continuous culture with glucose as carbon source. Appl. Environ. Microbiol. 48:301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–71. 10.1006/plas.1999.1441 [DOI] [PubMed] [Google Scholar]
- 28.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteley M, Bangera MG, Bumgartner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860–864. 10.1038/35101627 [DOI] [PubMed] [Google Scholar]
- 30.Sandoz K, Mitzimberg S, Schuster M. 2007. Social cheating in Pseudomonas aeruginosa quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 104:15876–15881. 10.1073/pnas.0705653104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuster M, Greenberg EP. 2007. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics 8:287. 10.1186/1471-2164-8-287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson JP, Pesci EC, Iglewski BH. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson JP, Gray KM, Passador L, Tucker KD, Eberhard A, Iglewski BH, Greenberg EP. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U. S. A. 91:197–201. 10.1073/pnas.91.1.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Essar DW, Eberly L, Hadero A, Crawford IP. 1990. Identification and characterization of genes for a second anthranilate synthase in Pseudomonas aeruginosa: interchangeability of the two anthranilate synthases and evolutionary implications. J. Bacteriol. 172:884–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert KB, Kim TH, Gupta R, Greenberg EP, Schuster M. 2009. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 73:1072–1085. 10.1111/j.1365-2958.2009.06832.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. 2006. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol. Microbiol. 62:1264–1277. 10.1111/j.1365-2958.2006.05421.x [DOI] [PubMed] [Google Scholar]
- 37.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34:658–684. 10.1111/j.1574-6976.2010.00218.x [DOI] [PubMed] [Google Scholar]
- 38.van Der Ploeg JR, Iwanicka-Nowicka R, Bykowski T, Hryniewicz MM, Leisinger T. 1999. The Escherichia coli ssuEADCB gene cluster is required for the utilization of sulfur from aliphatic sulfonates and is regulated by the transcriptional activator Cbl. J. Biol. Chem. 274:29358–29365. 10.1074/jbc.274.41.29358 [DOI] [PubMed] [Google Scholar]
- 39.Darch SE, West SA, Winzer K, Diggle SP. 2012. Density-dependent fitness benefits in quorum-sensing bacterial populations. Proc. Natl. Acad. Sci. U. S. A. 109:8259–8263. 10.1073/pnas.1118131109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pai A, Tanouchi Y, You L. 2012. Optimality and robustness in quorum sensing (QS)-mediated regulation of a costly public good enzyme. Proc. Natl. Acad. Sci. U. S. A. 109:19810–19815. 10.1073/pnas.1211072109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xavier JB, Kim W, Foster KR. 2011. A molecular mechanism that stabilizes cooperative secretions in Pseudomonas aeruginosa. Mol. Microbiol. 79:166–179. 10.1111/j.1365-2958.2010.07436.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamilton WD. 1964. The genetical evolution of social behaviour, I and II. J. Theor. Biol. 7:1–52. 10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- 43.Gupta R, Schuster M. 2013. Negative regulation of bacterial quorum sensing tunes public goods cooperation. ISME J. 7:2159–2168. 10.1038/ismej.2013.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cornforth DM, Foster KR. 2013. Competition sensing: the social side of bacterial stress responses. Nat. Rev. Microbiol. 11:285–293. 10.1038/nrmicro2977 [DOI] [PubMed] [Google Scholar]
- 45.Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Camara M, Smith H, Williams P. 2002. N-Acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70:5635–5646. 10.1128/IAI.70.10.5635-5646.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh PK, Schaefer AL, Parsek MR, Moninger TO, Welsh MJ, Greenberg EP. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762–764. 10.1038/35037627 [DOI] [PubMed] [Google Scholar]
- 47.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, Chang C, Dong Y, Williams P, Zhang LH. 2013. A cell-cell communication signal integrates quorum sensing and stress response. Nat. Chem. Biol. 9:339–343. 10.1038/nchembio.1225 [DOI] [PubMed] [Google Scholar]
- 48.Medina G, Juarez K, Soberon-Chavez G. 2003. The Pseudomonas aeruginosa rhlAB operon is not expressed during the logarithmic phase of growth even in the presence of its activator RhlR and the autoinducer N-butyryl-homoserine lactone. J. Bacteriol. 185:377–380. 10.1128/JB.185.1.377-380.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spira B, Silberstein N, Yagil E. 1995. Guanosine 3′,5′-bispyrophosphate (ppGpp) synthesis in cells of Escherichia coli starved for Pi. J. Bacteriol. 177:4053–4058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leigh JA, Dodsworth JA. 2007. Nitrogen regulation in bacteria and archaea. Annu. Rev. Microbiol. 61:349–377. 10.1146/annurev.micro.61.080706.093409 [DOI] [PubMed] [Google Scholar]
- 51.Lerat E, Moran NA. 2004. The evolutionary history of quorum-sensing systems in bacteria. Mol. Biol. Evol. 21:903–913. 10.1093/molbev/msh097 [DOI] [PubMed] [Google Scholar]
- 52.Holloway JB, Krishnapillai V, Morgan AF. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rahim R, Ochsner UA, Olvera C, Graninger M, Messner P, Lam JS, Soberon-Chavez G. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40:708–718. 10.1046/j.1365-2958.2001.02420.x [DOI] [PubMed] [Google Scholar]
- 54.Becher A, Schweizer HP. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948–950, 952 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.