Abstract
β-Alanine is a precursor for coenzyme A (CoA) biosynthesis and is a substrate for the bacterial/eukaryotic pantothenate synthetase and archaeal phosphopantothenate synthetase. β-Alanine is synthesized through various enzymes/pathways in bacteria and eukaryotes, including the direct decarboxylation of Asp by aspartate 1-decarboxylase (ADC), the degradation of pyrimidine, or the oxidation of polyamines. However, in most archaea, homologs of these enzymes are not present; thus, the mechanisms of β-alanine biosynthesis remain unclear. Here, we performed a biochemical and genetic study on a glutamate decarboxylase (GAD) homolog encoded by TK1814 from the hyperthermophilic archaeon Thermococcus kodakarensis. GADs are distributed in all three domains of life, generally catalyzing the decarboxylation of Glu to γ-aminobutyrate (GABA). The recombinant TK1814 protein displayed not only GAD activity but also ADC activity using pyridoxal 5′-phosphate as a cofactor. Kinetic studies revealed that the TK1814 protein prefers Asp as its substrate rather than Glu, with nearly a 20-fold difference in catalytic efficiency. Gene disruption of TK1814 resulted in a strain that could not grow in standard medium. Addition of β-alanine, 4′-phosphopantothenate, or CoA complemented the growth defect, whereas GABA could not. Our results provide genetic evidence that TK1814 functions as an ADC in T. kodakarensis, providing the β-alanine necessary for CoA biosynthesis. The results also suggest that the GAD activity of TK1814 is not necessary for growth, at least under the conditions applied in this study. TK1814 homologs are distributed in a wide range of archaea and may be responsible for β-alanine biosynthesis in these organisms.
INTRODUCTION
Coenzyme A (CoA) is an important cofactor found in all three domains of life (1–3). The mechanisms of CoA biosynthesis in bacteria and eukaryotes have been well studied (2, 3). Many microorganisms and plants can synthesize CoA de novo from pyruvate via 2-oxoisovalerate. The conversion of 2-oxoisovalerate to CoA involves eight enzyme reactions. The first three, catalyzed by ketopantoate hydroxymethyltransferase (KPHMT), ketopantoate reductase (KPR), and pantothenate synthetase (PS), lead to the generation of pantothenate. Following this, reactions catalyzed by pantothenate kinase (PanK), phosphopantothenoylcysteine synthetase (PPCS), phosphopantothenoylcysteine decarboxylase (PPCDC), phosphopantetheine adenylyltransferase (PPAT), and dephospho-CoA kinase (DPCK) complete the pathway. Animals do not harbor the first three enzymes and must thus rely on exogenous pantothenate for CoA synthesis.
In comparison, the mechanisms of CoA biosynthesis in archaea are still not completely understood. The PPCS/PPCDC fusion protein from Methanocaldococcus jannaschii (4) and PPAT from Pyrococcus abyssi (5, 6) have been biochemically examined. We have previously clarified that in the hyperthermophilic archaeon Thermococcus kodakarensis, two novel enzymes, pantoate kinase (PoK) and phosphopantothenate synthetase (PPS), are responsible for the conversion of pantoate to 4′-phosphopantothenate, replacing the classical PS/PanK system utilized in bacteria and eukaryotes (7–10). The homologs of PoK and PPS are present on the majority of archaeal genomes, with exceptions limited to Nanoarchaeum equitans, Korarchaeum cryptofilum, and members of the Thermoplasmatales. The presence of a PoK/PPS system has also been indicated in the methanogenic archaeon Methanospirillum hungatei (11). In Thermoplasmatales, the PanK homolog from Picrophilus torridus has been shown to display PanK activity (12). We have recently characterized the KPHMT and KPR proteins from T. kodakarensis (13). KPR from this archaeon is an NADH-dependent enzyme, distinct from the bacterial/eukaryotic counterparts that are NADPH dependent (14–16). Importantly, KPR activity is dramatically inhibited in the presence of CoA, suggesting that CoA biosynthesis in T. kodakarensis is regulated via feedback inhibition toward KPR (13). This regulation mechanism differs from those in many bacteria and eukaryotes, in which PanK is inhibited by CoA and its derivatives (2).
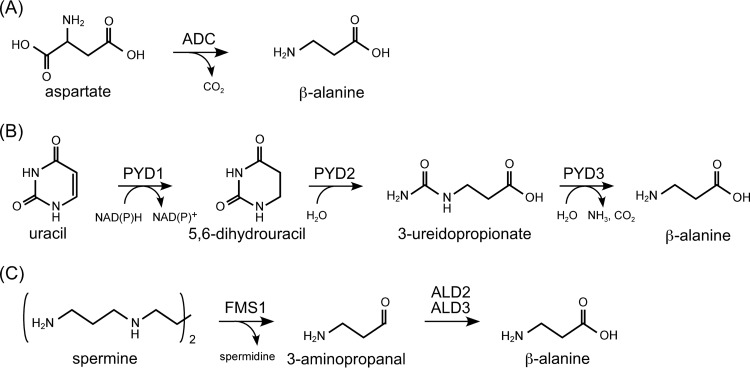
β-Alanine is a substrate of PS (bacteria and eukaryotes) and PPS (archaea) and is a building block of the CoA molecule. In bacteria and eukaryotes, β-alanine is synthesized through various mechanisms. In many bacteria, including Escherichia coli, it is generated by the direct decarboxylation of the C1 carboxylate group of Asp (Fig. 1A). The reaction is catalyzed by aspartate 1-decarboxylase (ADC), encoded by the panD gene (17, 18). ADC from E. coli is a pyruvoyl-dependent enzyme and does not utilize PLP (17, 18). Insects also harbor ADC proteins to generate β-alanine, which is utilized for N-β-alanyl dopamine and carnosine (β-alanyl-l-histidine) biosynthesis, but the insect ADC proteins are not related to the bacterial ADCs in terms of structure and are PLP dependent (19). In plants such as Arabidopsis thaliana, β-alanine is synthesized from uracil via three enzymatic reactions in the pyrimidine degradation pathway (20) (Fig. 1B). Uracil is converted to dihydrouracil by dihydropyrimidine dehydrogenase (PYD1) and then to 3-ureidopropionate by dihydropyrimidinase (PYD2) and finally to β-alanine, CO2, and NH3 by β-ureidopropionase (PYD3). In Saccharomyces cerevisiae, β-alanine is derived from the polyamine degradation pathway by amine oxidase (FMS1) and aldehyde dehydrogenases (ALD2 and ALD3) via spermine and 3-aminopropanal (21, 22) (Fig. 1C).
FIG 1.
Previously reported mechanisms of β-alanine biosynthesis in bacteria and eukaryotes. (A) Direct decarboxylation of Asp by aspartate decarboxylase (ADC) utilized in bacteria and insects. (B) Conversion from uracil via three enzymatic reactions catalyzed by dihydropyrimidine dehydrogenase (PYD1), dihydropyrimidinase (PYD2), and β-ureidopropionase (PYD3) utilized in plants. (C) Oxidation of spermine by polyamine oxidase (FMS1) and aldehyde dehydrogenases (ALD2 and ALD3) utilized in Saccharomyces cerevisiae.
The majority of archaea do not harbor homologs of E. coli ADC, with only a few exceptions, such as Thermoproteus tenax and Pyrobaculum islandicum. The homologs of FMS1 from S. cerevisiae are also absent in archaea. In terms of the pyrimidine degradation pathway, there are several candidate proteins that show low similarities to PYD1 and PYD2 from A. thaliana, but there are no homologs of PYD3, making it unlikely that this pathway functions in the synthesis of β-alanine in archaea.
Glutamate decarboxylase (GAD) is an extensively studied enzyme that decarboxylates Glu to form γ-aminobutyrate (GABA). Glu and GABA play central roles in neurotransmission in mammals (23, 24). In bacteria and eukaryotes, GABA can also be utilized as a carbon or energy source (25, 26). In E. coli, GABA is converted to succinate semialdehyde by GABA aminotransferase (GABA-AT) and is further oxidized by succinate semialdehyde dehydrogenase (SSADH) to succinate, entering the tricarboxylic acid (TCA) cycle (27–29). Furthermore, GAD and GABA have been reported to play important roles in acid or stress resistance in bacteria, including E. coli (30), and S. cerevisiae (31, 32).
The GAD proteins from E. coli display specificity for Glu and do not show high levels of activity with Asp (33). On the other hand, in mammals, there are GAD-like proteins such as GADL1 (34). Human GADL1, which is >50% identical to human GADs, does not catalyze the GAD reaction but exhibits ADC activity (34). Similarly, although the mosquito ADC proteins display a high level of homology with the GAD proteins, they exhibit ADC activity (19). Furthermore, a GAD homolog from a hyperthermophilic archaeon, Pyrococcus horikoshii, also shows ADC activity with higher catalytic efficiency toward Asp than toward Glu (35). These findings suggest that it is difficult to distinguish GAD and ADC functions by their primary structures.
Here, we describe a detailed characterization of an archaeal GAD homolog encoded by TK1814 in T. kodakarensis. Although many archaea have GAD homologs on their genomes, their physiological roles have not been reported. Genetic studies clearly suggest that TK1814 plays a predominant role in generating β-alanine for CoA biosynthesis in this archaeon.
MATERIALS AND METHODS
Phylogenetic analysis.
Sequences of GAD and ADC homologs from various organisms were collected and aligned by using the ClustalW program provided by the DNA Databank of Japan (DDBJ). Multiple-sequence alignment was performed with the following default parameters: protein weight matrix, Gonnet; gap open, 10; gap extension, 0.20; gap distances, 5; no end gaps, no; iteration, none; numiter, 1; clustering, NJ. The phylogenetic tree was constructed by the neighbor-joining method (36). Bootstrap resampling was performed 1,000 times.
Strains, media, and culture conditions.
Cultivation of Thermococcus kodakarensis KOD1 (37–39) and its derivative strains was performed under anaerobic conditions at 85°C in a nutrient-rich medium (ASW-YT) or a synthetic medium (ASW-AA) (40). ASW-YT medium consists of 0.8× artificial seawater (ASW) (41), 5.0 g liter−1 yeast extract, 5.0 g liter−1 tryptone, and 0.8 mg liter−1 resazurin. Prior to inoculation, 2.0 g liter−1 elemental sulfur (ASW-YT-S0 medium) or 5.0 g liter−1 sodium pyruvate (ASW-YT-Pyr medium) and Na2S were added to the medium until it became colorless. ASW-AA medium consists of 0.8× ASW, a mixture of 20 amino acids, modified Wolfe's trace minerals, a vitamin mixture, and 2.0 g liter−1 elemental sulfur (ASW-AA-S0 medium) (40, 41). In the case of plate culture used to isolate transformants, elemental sulfur and Na2S 9H2O were replaced with 2 ml of a polysulfide solution (10 g Na2S 9H2O and 3 g sulfur flowers in 15 ml H2O) per liter, and 10 g liter−1 Gelrite was added to solidify the medium.
In order to examine the phenotypes of the TK1814 gene disruption strain, cells were cultivated in ASW-AA-S0 medium (with pantothenate excluded from the vitamin mixture), in the presence of 0.1 mM β-alanine (Sigma-Aldrich, St. Louis, MO), 1 mM GABA, 1 mM CoA, 0.1 mM d-pantoate, d-pantothenate (Sigma-Aldrich), d-4-phosphopantoate, or d-4′-phosphopantothenate. Pantoate was prepared by hydrolyzing d-pantolactone (Sigma-Aldrich) according to previously described methods (9, 13). 4-Phosphopantoate was chemically synthesized in a previous study (8). 4′-Phosphopantothenate was enzymatically synthesized by phosphorylating commercially available pantothenate using recombinant PanK from E. coli, as reported previously (8). Specific modifications of the medium to select and examine the auxotrophy of mutant strains are described below. Escherichia coli strains DH5α and BL21-CodonPlus(DE3)-RIL, used for plasmid construction and heterologous gene expression, respectively, were cultivated at 37°C in Luria-Bertani (LB) medium containing ampicillin (100 mg liter−1). Unless mentioned otherwise, all chemicals were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
Overexpression and purification of the recombinant TK1814 protein.
The TK1814 gene was overexpressed in E. coli. The coding region of the TK1814 gene was amplified from the genomic DNA of T. kodakarensis KOD1 by PCR using primer set 1814F1 (5′-AAAACATATGTTTCCAGAGAGGGGAGC-3′)/1814R1 (5′-AAAAGAATTCTTAAAGCCTTTTTGCAATCTC-3′). Using the NdeI-EcoRI restriction enzyme sites (indicated by underlining) incorporated during PCR, the amplified fragment was inserted into the pET21a(+) expression vector (EMD Millipore, Billerica, MA). After confirming the absence of unintended mutations, the plasmid was introduced into E. coli strain BL21-CodonPlus(DE3)-RIL. The transformant was cultivated at 37°C in LB medium with ampicillin until the optical density at 660 nm reached ∼0.5. Isopropyl 1-thio-β-d-galactopyranoside was added to a final concentration of 0.1 mM to induce expression, and cells were cultivated for a further 4 h. Cells were harvested, resuspended in 50 mM Tris-HCl buffer (pH 7.5), and disrupted by sonication. After centrifugation (20,000 × g at 4°C for 20 min), the soluble cell extracts were incubated at 80°C for 10 min. After removal of thermolabile proteins derived from the host by centrifugation (20,000 × g at 4°C for 20 min), the supernatants were applied for anion-exchange chromatography (HiTrap Q HP; GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom), and proteins were eluted with a linear gradient of NaCl (0 to 1.0 M) in 10 mM Tris-HCl (pH 7.5) and 0.25 mM pyridoxal 5′-phosphate (PLP) at a flow rate of 2.5 ml min−1. After concentrating the sample and exchanging the buffer to 10 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and 0.25 mM PLP using Amicon Ultra-4 10 K (EMD Millipore), the samples were applied onto a Superdex 200 10/300 gel filtration column (GE Healthcare) with a mobile phase of 10 mM Tris-HCl (pH 7.5) containing 150 mM NaCl and 0.25 mM PLP at a flow rate of 0.8 ml min−1. The same column and conditions were used to examine the molecular mass of the recombinant TK1814 protein with the size markers aldolase (158 kDa), conalbumin (75 kDa), albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and RNase A (13.7 kDa) from the High Molecular Weight Gel Filtration Calibration kit, Low Molecular Weight Gel Filtration Calibration kit, and Gel Filtration Low Molecular Weight kit (GE Healthcare). The protein concentration was determined with the Protein Assay system (Bio-Rad, Hercules, CA), using bovine serum albumin as a standard.
Examination of decarboxylase activity of the TK1814 protein.
Decarboxylase activity of the TK1814 protein toward Glu and Asp was measured by derivatization of GABA and β-alanine using fluorescamine, followed by detection and quantification by high-performance liquid chromatography (HPLC). Unless mentioned otherwise, the Glu/Asp decarboxylase reaction mixture contained 1 mM Glu/Asp, 0.25 mM PLP, 4 μg ml−1 recombinant TK1814 protein, and 50 mM N,N-bis(2-hydroxyethyl)glycine (Bicine)-NaOH (pH 8.0 at 85°C). The reaction was performed at 85°C for 10, 20, and 30 min and stopped by cooling the mixture on ice. The TK1814 proteins were removed by ultrafiltration with Amicon Ultra-0.5 10 K (EMD Millipore), and aliquots were applied for derivatization. The derivatization mixture (1 ml) contained 200-μl aliquots of the decarboxylase reaction mixture, 20 mM NaOH, 0.3 M borate (pH 10.0), and 60 μg fluorescamine in acetonitrile. The mixture was vortexed and applied onto a Cosmosil 5C18-PAQ column (Nacalai Tesque). Compounds were separated with 20 mM sodium acetate (pH 5.9) containing 15% (vol/vol) acetonitrile at a flow rate of 1.0 ml min−1, and the fluorescamine-derivatized compounds were detected by measuring fluorescence (excitation at 390 nm and emission at 460 nm). In order to examine PLP dependency, 0.25 mM PLP in the reaction mixture was replaced with water or 20 μM hydroxylamine (Sigma-Aldrich), and the reactions were carried out at 50°C due to the instability of hydroxylamine at high temperatures. When we examined the effects of pH, the reaction mixture contained 0.5 mM Asp, 0.25 mM PLP, 4 μg ml−1 recombinant dimeric TK1814 protein, and 50 mM various buffers. When we performed an assay to determine the effects of reaction temperature, the reaction mixture contained 0.5 mM Asp, 0.25 mM PLP, 4 μg ml−1 recombinant dimeric TK1814 protein, and 50 mM piperazine-1,4-bis(2-ethanesulfonic acid) (PIPES)-NaOH (pH 7.8 at 85°C).
Thermostability and effects of pH and temperature.
For examining thermostability, the purified dimeric TK1814 protein (0.50 mg ml−1) in 50 mM sodium phosphate (pH 7.5) and 0.25 mM PLP was incubated for various periods of time at 80°C or 90°C. After the protein solutions were cooled on ice for 30 min, aspartate decarboxylase activity was measured. In order to examine the effects of pH, the aspartate decarboxylase reaction was performed at various pH values by using the following buffers at 50 mM: 2-morpholineethanesulfonic acid (MES) (pH 5.5 to 7.0), PIPES (pH 6.5 to 7.5), HEPES (pH 7.0 to 8.0), Bicine (pH 8.0 to 9.0), N-[Tris(hydroxymethyl)methyl]glycine (Tricine) (pH 8.0 to 9.0), and N-cyclohexyl-2-aminoethanesulfonic acid (CHES) (pH 9.0 to 10.0). The reactions were performed at 85°C, and all buffers were prepared so that their pH would reflect accurate values at 85°C. In order to examine the effects of temperature, the aspartate decarboxylase reaction was carried out at various temperatures. The data obtained were used to make an Arrhenius plot.
Construction of the plasmid for gene disruption of TK1814.
The gene disruption plasmid for TK1814 was constructed by first amplifying the gene along with 1 kbp of its 5′- and 3′-flanking regions using the primer set 1814F2 (5′-AAGGATCCCCAGCAACTTCTCTGCAAT-3′)/1814R2 (5′-AAGGATCCCTTCAGAAGCTCGGCTTCA-3′). Using the BamHI restriction enzymes sites (indicated by underlining) incorporated during PCR, the fragment was inserted into pUD3, which contains the pyrF marker gene cassette inserted into the ApaI site of pUC118. Inverse PCR was performed with primer set 1814F3 (5′-AGCTGTCCGACAAATTGACCCATTG-3′)/1814R3 (5′-TTTCTCACCGCCTTGCGGTACTAGC-3′) to remove the coding region, and the amplified fragment was self-ligated. Sequences of the 5′- and 3′-flanking regions were confirmed.
Gene disruption of TK1814.
T. kodakarensis strain KUW1 (ΔpyrF ΔtrpE) (40), which shows uracil and tryptophan auxotrophy, was used as the host strain for TK1814 gene disruption. KUW1 was cultivated in ASW-YT-S0 medium for 12 h at 85°C. Cells were harvested, resuspended in 200 μl of 0.8× ASW, and incubated on ice for 30 min. After addition of 3.0 μg of the gene disruption plasmid and further incubation on ice for an hour, cells were cultivated in ASW-AA-S0 medium without uracil for 24 h at 85°C. Cells were then harvested, diluted with 0.8× ASW, and spread onto solid ASW-YT-S0 medium supplemented with 10 g liter−1 of 5-fluoroorotic acid (5-FOA) and 60 mM NaOH. Only cells that have undergone a pop-out recombination that removes the pyrF gene can grow in the presence of 5-FOA. After cultivation for 2 days at 85°C, transformants displaying 5-FOA resistance were isolated and cultivated in ASW-YT-S0 medium. After cultivation, the genotypes of the isolated transformants were analyzed by PCR with the primer sets 1814F4 (5′-GACGGTAAACGAGTTCTGGATTG-3′)/1814R4 (5′-TCTCAGTGCTCAAGCTGGTCGTTG-3′) and 1814F1/1814R1, which anneal at the extragenic region and intragenic region, respectively. Transformants whose amplified DNA products displayed the expected size were chosen, and relevant sequences were confirmed to have no unintended mutations.
RESULTS
Glutamate decarboxylase homologs in archaea.
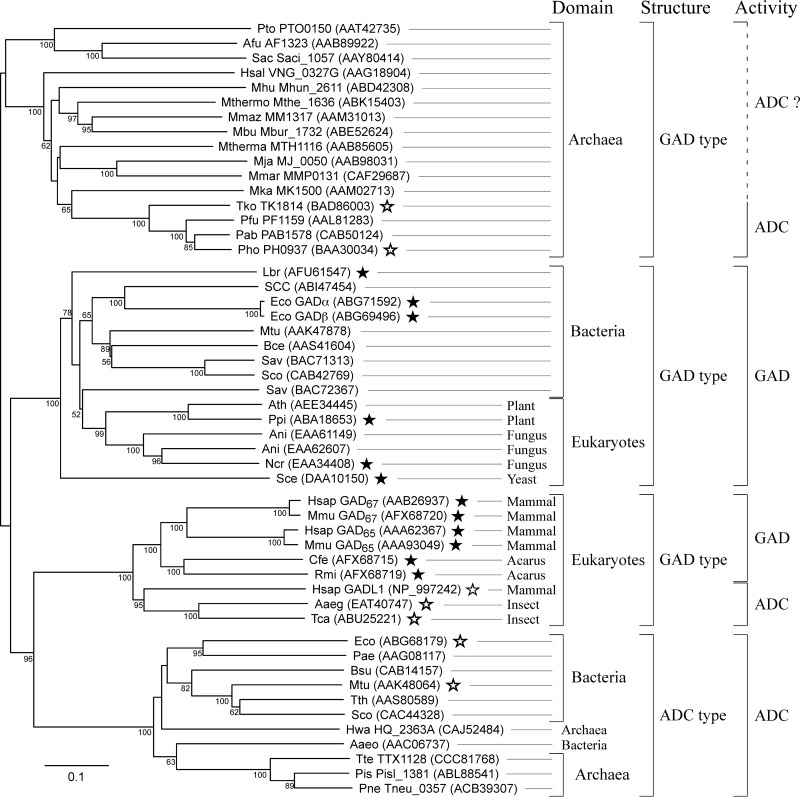
The TK1814 gene from T. kodakarensis encodes a protein that shows similarity to the glutamate decarboxylase (GAD) proteins from human and E. coli. TK1814 is 20% identical to both E. coli GADα and GADβ and is 19% and 24% identical to GAD65 and GAD67 from human, respectively. TK1814 harbors His121/Ala196, Asn54, and Arg358, which correspond to His282/Ala366, Asn203, and Arg558 in human GAD65, which participate in binding with the pyridine ring of PLP and γ-carboxylate and α-carboxylate of Glu, respectively (42) (see Fig. S1 in the supplemental material). TK1814 does not show any similarity to the aspartate decarboxylase (ADC) from E. coli. A phylogenetic tree of the GAD and ADC proteins from a wide range of organisms is shown in Fig. 2.
FIG 2.
Phylogenetic analysis of Glu/Asp decarboxylase homologs. GAD and ADC homologs from various organisms were aligned, and a phylogenetic tree was constructed with the neighbor-joining method (36). Archaeal species names are abbreviated as follows: Afu, Archaeoglobus fulgidus; Hsal, Halobacterium salinarum; Hwa, Haloquadratum walsbyi; Mja, Methanocaldococcus jannaschii; Mbu, Methanococcoides burtonii; MMar, Methanococcus maripaludis; Mka, Methanopyrus kandleri; Mthermo, Methanosaeta thermophila; Mmaz, Methanosarcina mazei; Mhu, Methanospirillum hungatei; Mthema, Methanothermobacter thermautotrophicus; Pto, Picrophilus torridus; Pis, Pyrobaculum islandicum; Pne, Pyrobaculum neutrophilum; Pab, Pyrococcus abyssi; Pfu, Pyrococcus furiosus; Pho, Pyrococcus horikoshii; Sac, Sulfolobus acidocaldarius; Tko, Thermococcus kodakarensis. Bacterial species names are abbreviated as follows: Aaeo, Aquifex aeolicus; Bce, Bacillus cereus; Bsu, Bacillus subtilis; Eco, Escherichia coli; Lbr, Lactobacillus brevis; Mtu, Mycobacterium tuberculosis; Pae, Pseudomonas aeruginosa; Sav, Streptomyces avermitilis; Sco, Streptomyces coelicolor; SCC, Synechococcus sp. strain CC9311; Tth, Thermus thermophilus. Eukaryote species names are abbreviated as follows: Aaeg, Aedes aegypti; Ath, Arabidopsis thaliana; Ani, Aspergillus nidulans; Cfe, Ctenocephalides felis; Hsap, Homo sapiens; Mmu, Mus musculus; Ncr, Neurospora crassa; Ppi, Pinus pinaster; Rmi, Rhipicephalus microplus, Sce, Saccharomyces cerevisiae; Tca, Tribolium castaneum. GenBank accession numbers are shown for all proteins, and locus tags are also shown for archaeal proteins. Bootstrap values above 50 are shown. Closed/open stars indicate GAD/ADC proteins that have been examined experimentally to display GAD/ADC activity, respectively.
Four large clades can be found in the tree. Those whose members display similarity with GADs from human (GAD65 and GAD67) and E. coli are designated GAD-type proteins in the tree, whereas those similar to the ADC from E. coli are designated ADC-type proteins. It seems likely that the GAD-type proteins from bacteria and those from plants, fungi, and yeast are actually GADs, as diverse members of this clade have been experimentally shown to display GAD activity. The six members of GAD-type proteins from mammals and acari (an arachnid subclass) are proven to be authentic GADs, but GADL1 from mammals and GAD-type proteins from insects within this clade have been shown to be ADCs and do not harbor the corresponding Asn residues that are important for recognition of γ-carboxylate of Glu in human GAD65 (Asn203 in human GAD65) (42). The ADC-type proteins from bacteria can be expected to function as ADCs, but none of the ADC-type proteins from hyperthermophilic bacteria (from Aquifex aeolicus) (Fig. 2) or from archaea have been examined. A large number of archaea harbor GAD-type proteins (see Discussion), and one from P. horikoshii has been demonstrated to display ADC activity in addition to GAD activity (35).
Production and purification of the recombinant TK1814 protein.
The TK1814 gene was overexpressed in E. coli, and a soluble protein was obtained. The protein was purified by heat treatment, followed by anion-exchange chromatography and gel filtration chromatography. After gel filtration, we observed two peaks that each contained a single band with a molecular mass corresponding to that of the TK1814 protein calculated from its primary structure (42,543 Da) (see Fig. S2 in the supplemental material). Gel filtration revealed that the two peaks corresponded to a dimeric and a hexameric form of the enzyme at a ratio of 7:3. When we performed gel filtration without PLP, this ratio did not change. When we subjected a dimeric or hexameric fraction of the enzyme to a second gel filtration step, we did not observe a change in the oligomeric state. The oligomeric forms of the enzyme seem to be relatively stable and do not change their state at room temperature for at least 1 h. The two fractions were individually examined for GAD and ADC activity.
Basic enzymatic properties of the TK1814 protein.
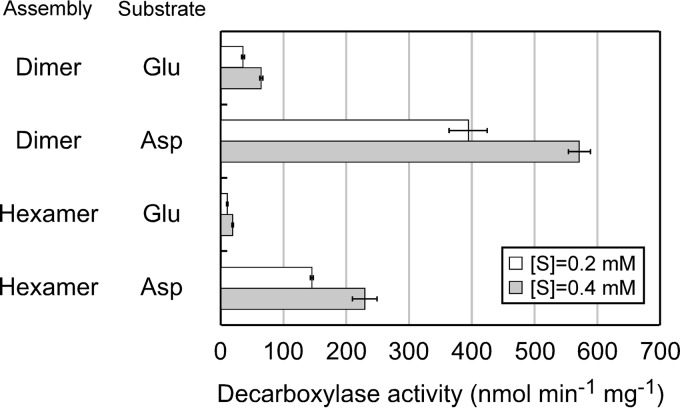
GAD and ADC activity was measured at 85°C. The expected products, GABA and β-alanine, were derivatized with fluorescamine and quantified by HPLC. Both the dimeric and the hexameric enzymes showed GAD and ADC activity, with higher activity levels observed with the dimeric protein (Fig. 3). Both forms displayed higher activity levels with Asp than with Glu. The enzyme reaction was dependent on PLP. When PLP was not added to the reaction mixtures, enzyme activities of both the dimeric and the hexameric enzymes decreased to approximately 30% compared to those in the presence of PLP. The retained activity is most likely due to the incorporation of PLP into the enzyme during its production in E. coli and/or the purification steps, as the cell extract already displayed a yellow color, and PLP was added to the buffers throughout the purification process. Furthermore, when hydroxylamine, a compound known to inhibit PLP-dependent enzymes, was added, activity was completely abolished (data not shown).
FIG 3.
Examination of decarboxylase activity toward Glu and Asp. Shown are activities of the dimeric and hexameric proteins toward Glu and Asp. Reactions were performed at 85°C in the presence of 0.2 mM or 0.4 mM Glu or Asp and 0.25 mM PLP. All measurements were carried out in triplicate.
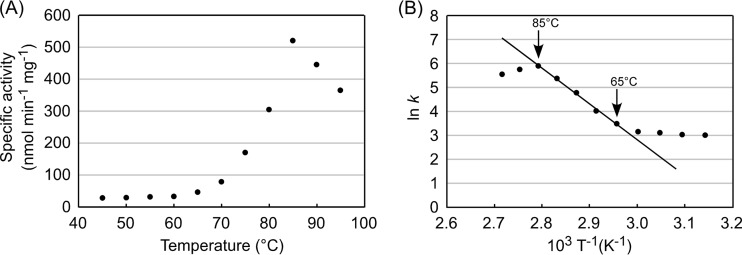
As activity levels were higher, further examination of the enzyme was carried out on the dimeric protein. We next examined the effect of pH on the reaction and observed high activity at neutral pH, with a maximum at pH 8.0 in 50 mM Bicine-NaOH buffer (see Fig. S3 in the supplemental material). In terms of temperature, the recombinant TK1814 protein showed the highest level of activity at 85°C (Fig. 4A). An Arrhenius plot of the data showed linearity between 65°C and 85°C (Fig. 4B), and the activation energy of the reaction was calculated to be 124 kJ mol−1. Thermostability of the TK1814 protein was examined by incubating the protein at 80°C and 90°C for various periods of time and by measuring residual activity. Half-lives of the protein at 80°C and 90°C were 10 h and 5.5 h, respectively.
FIG 4.
Effects of temperature on the aspartate decarboxylase activity of the TK1814 protein. (A) Effects of temperature on aspartate decarboxylase activity. (B) Arrhenius plot of the data shown in panel A.
Kinetic examinations.
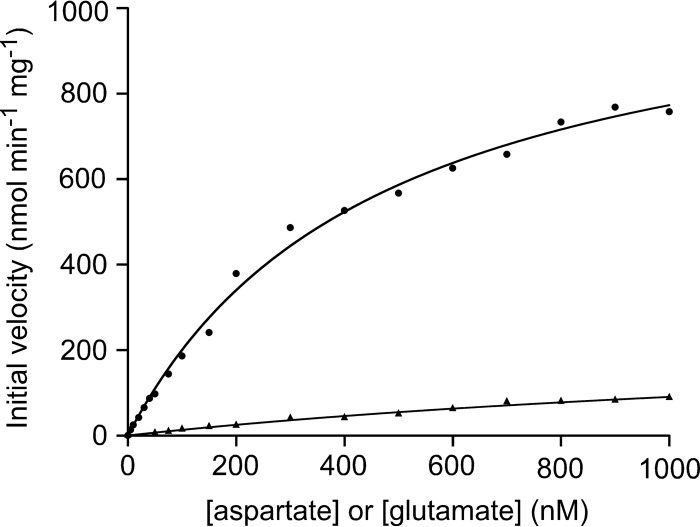
Kinetic studies of GAD and ADC activities were performed by measuring initial velocities in the presence of various concentrations of Glu or Asp. The kinetics toward both amino acids followed Michaelis-Menten kinetics (Fig. 5). The obtained parameters are shown in Table 1. The kcat/Km value with Asp was much higher than that with Glu, suggesting that Asp is the preferred substrate of the TK1814 protein.
FIG 5.
Kinetic studies of the TK1814 protein. Shown are initial velocities of the glutamate/aspartate decarboxylase reactions with various concentrations of Glu/Asp and 0.25 mM PLP. Symbols: circles, Asp; triangles, Glu.
TABLE 1.
Kinetic parameters of TK1814a
| Substrate | Mean Vmax (μmol min−1 mg−1) ± SD | Mean Km (mM) ± SD | Mean kcat (s−1) ± SD | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| Aspartate | 1.13 ± 0.04 | 0.47 ± 0.04 | 0.80 ± 0.03 | 1.70 |
| Glutamate | 0.26 ± 0.05 | 1.92 ± 0.50 | 0.18 ± 0.04 | 0.09 |
ADC and GAD activities with various concentrations of Asp or Glu were measured in the presence of 0.25 mM PLP. Values are means ± standard deviations or ratios.
Gene disruption of TK1814.
In order to examine the physiological roles of the ADC and GAD activities of TK1814 in T. kodakarensis, we constructed a gene disruption strain and compared its phenotype with that of the host strain, T. kodakarensis KUW1. The disruption plasmid was designed so that gene disruption would occur via single-crossover insertion of the plasmid, followed by pop-out recombination. Cells that had undergone single-crossover insertion were enriched by cultivating the transformants in a uracil-free medium, and cells that had further undergone pop-out recombination were selected on solid, nutrient-rich media that included 5-FOA (see Fig. S4A in the supplemental material). PCR analysis and DNA sequencing of the genomic DNA confirmed that the transformant contained a disrupted TK1814 gene (see Fig. S4B in the supplemental material).
Growth characteristics of the gene disruption strain of TK1814.
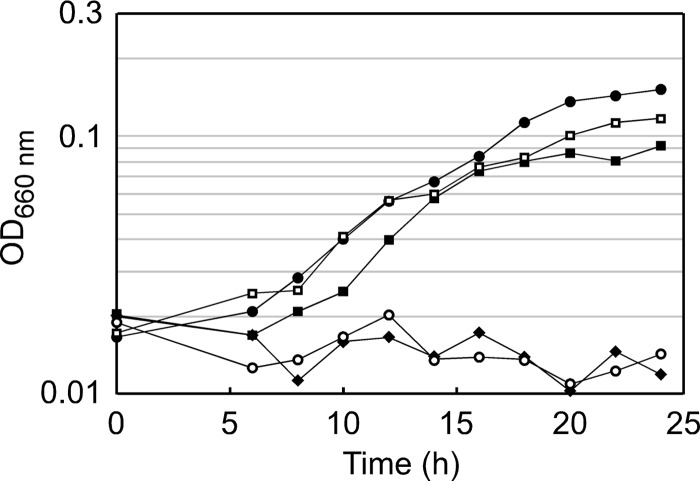
The TK1814 gene disruption strain (ΔTK1814) and its host strain, KUW1, were grown in ASW-AA-S0 medium (Fig. 6). We found that the ΔTK1814 strain did not show growth for 24 h, suggesting that the ADC and/or GAD activities of TK1814 were essential for growth in this medium. When exogenous β-alanine, the product of ADC activity, was added to the medium, the growth defects were almost fully recovered. In contrast, the addition of GABA, the product of GAD activity, did not complement TK1814 disruption at all. These results strongly suggest that, at least under the growth conditions applied in this study, the function of TK1814 that is important for T. kodakarensis is its ADC activity and not its GAD activity. We also found that the addition of CoA restores growth of the TK1814 gene disruption strain. This suggests that the β-alanine generated by TK1814 is utilized mainly for CoA biosynthesis. Furthermore, we examined the effects of adding pantoate, 4-phosphopantoate, 4′-phosphopantothenate, and pantothenate to the medium. As expected, addition of pantoate and 4-phosphopantoate, metabolites upstream of the β-alanine-utilizing PPS reaction, did not restore the growth of the ΔTK1814 strain in ASW-AA-S0 medium (optical density at 660 nm [OD660] values were 0.011 and 0.009, respectively, at 24 h). On the other hand, the addition of 4′-phosphopantothenate, the product of the PPS reaction, restored growth of the ΔTK1814 strain (OD660 = 0.083 at 24 h). To our surprise, the addition of pantothenate also supported a low but detectable level of growth (OD660 = 0.037). This raises the possibility that pantothenate may be hydrolyzed in T. kodakarensis, providing low levels of β-alanine for use in the PPS reaction. The growth recovery with β-alanine, 4′-phosphopantothenate, and CoA, but not with pantoate and 4-phosphopantoate, clearly indicates the involvement of TK1814 in β-alanine/CoA biosynthesis.
FIG 6.
Growth characteristics of T. kodakarensis KUW1 and the ΔTK1814 strain. Cells were cultivated in ASW-AA-S0 medium with uracil at 85°C. Symbols: closed circles, KUW1; open circles, the ΔTK1814 strain; closed squares, the ΔTK1814 strain with 1 mM CoA; open squares, the ΔTK1814 strain with 0.1 mM β-alanine; diamonds, the ΔTK1814 strain with 1 mM GABA.
DISCUSSION
In this study, we have carried out detailed biochemical and genetic analyses of an archaeal GAD homolog encoded by TK1814 in T. kodakarensis. Kinetic analyses revealed that the kcat/Km value with Asp was much higher than that with Glu. This indicates that GAD from T. kodakarensis preferentially utilizes Asp. This was not surprising, as ADC activity, along with GAD activity, was also found in the GAD homolog of the closely related organism P. horikoshii (35). However, the differences in kcat/Km were much more striking in the case of the TK1814 protein. The ratios of the kcat/Km for Asp to the kcat/Km for Glu were 18.9 for the enzyme from T. kodakarensis and 4.2 for the enzyme from P. horikoshii (35). The most important findings of this study were obtained from genetic studies: (i) the ΔTK1814 strain displayed β-alanine auxotrophy, and (ii) the addition of GABA could not restore growth, whereas (iii) the addition of CoA relieved the growth defects. The results indicate that (i) the important function of TK1814 is its ADC activity in supplying β-alanine, (ii) GAD activity is most likely irrelevant in vivo or at least is not important for growth in the media applied here, and (iii) the requirement for β-alanine formation is mainly for CoA biosynthesis. We cannot exclude the possibility that exogenous CoA is degraded to supply β-alanine for other purposes in T. kodakarensis, but this is unlikely, as we do not find any other metabolic pathways predicted to utilize β-alanine.
As described above, the majority of archaea do not possess homologs of bacterial ADC, and genes with notable similarity to those encoding enzymes of the pyrimidine degradation pathway and the polyamine oxidation pathway do not seem to be present in archaea. As the TK1814 disruption strain could not grow in the absence of exogenous β-alanine, this suggests that TK1814 is the major supplier of β-alanine in T. kodakarensis and that other routes are not present or at least do not have the capacity to support growth.
Homologs of TK1814, although with various degrees of identity, can be found throughout most of the Euryarchaeota (see Table S1 in the supplemental material). Those from members of the Thermococcales can be expected to exhibit ADC activity, as even the most distantly related protein, from Pyrococcus furiosus, is 71% identical to the TK1814 protein. Homologs from Methanocaldococcus jannaschii (42% identical), Methanothermobacter thermautotrophicus (46%), Methanosarcina acetivorans (41%), Halobacterium salinarum (37%), and Haloferax volcanii (39%) also display considerable similarity, and taking into account the absence of alternative routes, this suggests that these proteins may also function as ADCs in supplying β-alanine. Several intriguing points in the distribution of the TK1814 homologs indicate that they do function as ADCs in Euryarchaeota. There is only one genome from halophilic archaea, that from Haloquadratum walsbyi, that does not harbor a TK1814 homolog (see Table S1 in the supplemental material). Coincidentally, this genome is the only one among the halophiles (and Euryarchaeota) that harbors an ADC homolog. Another point is that among the Thermoplasmatales, which do not utilize the PoK/PPS system for conversion of pantoate to 4′-phosphopantothenate, two species, Ferroplasma acidarmanus and P. torridus, harbor a PanK homolog, suggesting that they utilize a PS/PanK system, similarly to bacteria and eukaryotes (12). Thermoplasma acidophilum and Thermoplasma volcanium do not harbor any genes structurally related to PS, PanK, PoK, or PPS, raising the possibility that these two organisms cannot synthesize 4′-phosphopantothenate. Correlating with this, TK1814 homologs are found only on the genomes F. acidarmanus and P. torridus (see Table S1 in the supplemental material), suggesting a relationship of these homologs to CoA biosynthesis.
We aligned the sequences of 16 representative archaeal GAD homologs and the mammalian GADs and were able to identify highly conserved residues. There were 27 residues conserved among the archaeal homologs, among which 14 had reactive side chains (Cys, Asp, Glu, His, Lys, Arg, Ser, and Thr). Among these 14 residues, 9 residues (Glu68, His121, Lys126, Asp175, His192, Asp194, His230, Lys231, and Thr273 in the TK1814 protein) were also found in mammalian GADs. These residues correspond to Glu217, His282, Lys287, Asp345, His362, Asp364, His395, Lys396, and Thr439 in human GAD65. His282 is known to participate in PLP binding (42). All residues conserved in both archaeal GAD homologs and mammalian GADs were also conserved in GADL1. The 5 residues found only in the archaeal homologs are Asp57, Thr93, Glu94, Asp228, and His364 in the TK1814 protein and may be involved in its ADC activity. However, residues specifically conserved in the archaeal sequences were not found in GADL1, making it difficult to identify residues involved in the distinct substrate specificities of these enzymes.
One point that requires further consideration is the metabolic fate of GABA, generated by the GAD activity of the TK1814 protein. This is more so the case in P. horikoshii, as the GAD homolog displays relatively higher levels of activity toward Glu. As the addition of β-alanine fully restored the growth of the TK1814 disruption strain, the GAD activity of TK1814 does not seem to be necessary in T. kodakarensis. Asp concentrations have been reported to be much higher than those of Glu in T. kodakarensis cells (43), and considering the catalytic efficiencies of the TK1814 protein, GABA production can be considered limited. In bacteria and eukaryotes, GABA is converted to succinate semialdehyde and succinate via GABA-AT and SSADH, respectively (26–28, 44). T. kodakarensis harbors two homologs of GABA-AT, while P. horikoshii harbors four. However, the only gene that displays homology with SSADH in T. kodakarensis has been shown not to display SSADH activity (45), and no homolog is found in P. horikoshii. There may be an unidentified enzyme in these organisms that converts succinate semialdehyde to succinyl-CoA, which can be utilized for ATP synthesis (46), or an oxidoreductase that would extract reducing equivalents through the conversion of succinate semialdehyde to succinate. For Sulfolobus solfataricus, it has been reported that SSO1629 and SSO1842 show SSADH activities (47). As two GABA-AT homologs are also present in this archaeon, there may be a route to convert GABA to succinate, which can then be utilized by the TCA cycle that is present in this archaeon.
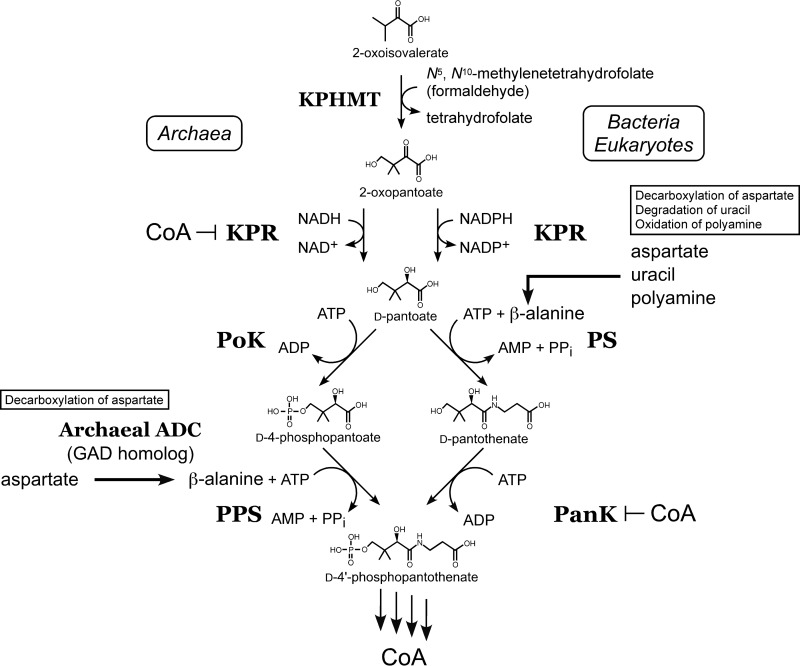
In Fig. 7, we summarize what is known regarding CoA biosynthesis in T. kodakarensis. PoK and PPS function to convert pantoate to 4′-phosphopantothenate instead of the classical PS and PanK utilized in bacteria and eukaryotes (10). The first four enzymes (KPHMT, KPR, PoK, and PPS) have been biochemically examined (8, 9, 13). KPHMT, PoK, and PPS are not inhibited by CoA/acetyl-CoA, whereas KPR is dramatically inhibited, indicating that KPR is the target of feedback inhibition and is involved in the regulation of CoA biosynthesis in T. kodakarensis (8, 9, 13). In this study, we have further clarified that TK1814 is responsible for β-alanine biosynthesis.
FIG 7.
Diagram illustrating the first four enzyme reactions in CoA biosynthesis in T. kodakarensis in comparison with those in bacteria and eukaryotes. The first four enzyme reactions of the archaeal CoA biosynthesis pathway in T. kodakarensis, catalyzed by KPHMT, KPR, PoK, and PPS, are shown on the left. Inhibition of archaeal KPR and bacterial/eukaryotic PanK by CoA is indicated. In bacteria and eukaryotes, β-alanine is synthesized by either the direct decarboxylation of Asp, the degradation of uracil, or the oxidation of polyamines. In archaea, β-alanine is synthesized by the archaeal ADC identified in this study, which is a GAD homolog.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by a grant-in-aid for the Japan Society for the Promotion of Science (JSPS) fellows (grant number [24.2716] to H.T.).
Footnotes
Published ahead of print 10 January 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01327-13.
REFERENCES
- 1.Genschel U. 2004. Coenzyme A biosynthesis: reconstruction of the pathway in archaea and an evolutionary scenario based on comparative genomics. Mol. Biol. Evol. 21:1242–1251. 10.1093/molbev/msh119 [DOI] [PubMed] [Google Scholar]
- 2.Leonardi R, Zhang YM, Rock CO, Jackowski S. 2005. Coenzyme A: back in action. Prog. Lipid Res. 44:125–153. 10.1016/j.plipres.2005.04.001 [DOI] [PubMed] [Google Scholar]
- 3.Spry C, Kirk K, Saliba KJ. 2008. Coenzyme A biosynthesis: an antimicrobial drug target. FEMS Microbiol. Rev. 32:56–106. 10.1111/j.1574-6976.2007.00093.x [DOI] [PubMed] [Google Scholar]
- 4.Kupke T, Schwarz W. 2006. 4′-Phosphopantetheine biosynthesis in Archaea. J. Biol. Chem. 281:5435–5444. 10.1074/jbc.M510056200 [DOI] [PubMed] [Google Scholar]
- 5.Armengaud J, Fernandez B, Chaumont V, Rollin-Genetet F, Finet S, Marchetti C, Myllykallio H, Vidaud C, Pellequer JL, Gribaldo S, Forterre P, Gans P. 2003. Identification, purification, and characterization of an eukaryotic-like phosphopantetheine adenylyltransferase (coenzyme A biosynthetic pathway) in the hyperthermophilic archaeon Pyrococcus abyssi. J. Biol. Chem. 278:31078–31087. 10.1074/jbc.M301891200 [DOI] [PubMed] [Google Scholar]
- 6.Nálezkova M, de Groot A, Graf M, Gans P, Blanchard L. 2005. Overexpression and purification of Pyrococcus abyssi phosphopantetheine adenylyltransferase from an optimized synthetic gene for NMR studies. Protein Expr. Purif. 39:296–306. 10.1016/j.pep.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 7.Atomi H, Tomita H, Ishibashi T, Yokooji Y, Imanaka T. 2013. CoA biosynthesis in archaea. Biochem. Soc. Trans. 41:427–431. 10.1042/BST20120311 [DOI] [PubMed] [Google Scholar]
- 8.Ishibashi T, Tomita H, Yokooji Y, Morikita T, Watanabe B, Hiratake J, Kishimoto A, Kita A, Miki K, Imanaka T, Atomi H. 2012. A detailed biochemical characterization of phosphopantothenate synthetase, a novel enzyme involved in coenzyme A biosynthesis in the Archaea. Extremophiles 16:819–828. 10.1007/s00792-012-0477-5 [DOI] [PubMed] [Google Scholar]
- 9.Tomita H, Yokooji Y, Ishibashi T, Imanaka T, Atomi H. 2012. Biochemical characterization of pantoate kinase, a novel enzyme necessary for coenzyme A biosynthesis in the Archaea. J. Bacteriol. 194:5434–5443. 10.1128/JB.06624-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokooji Y, Tomita H, Atomi H, Imanaka T. 2009. Pantoate kinase and phosphopantothenate synthetase, two novel enzymes necessary for CoA biosynthesis in the Archaea. J. Biol. Chem. 284:28137–28145. 10.1074/jbc.M109.009696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh H, Tamaki H, Tokutake Y, Hanada S, Chohnan S. 2013. Identification of pantoate kinase and phosphopantothenate synthetase from Methanospirillum hungatei. J. Biosci. Bioeng. 115:372–376. 10.1016/j.jbiosc.2012.10.019 [DOI] [PubMed] [Google Scholar]
- 12.Takagi M, Tamaki H, Miyamoto Y, Leonardi R, Hanada S, Jackowski S, Chohnan S. 2010. Pantothenate kinase from the thermoacidophilic archaeon Picrophilus torridus. J. Bacteriol. 192:233–241. 10.1128/JB.01021-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomita H, Imanaka T, Atomi H. 2013. Identification and characterization of an archaeal ketopantoate reductase and its involvement in regulation of coenzyme A biosynthesis. Mol. Microbiol. 90:307–321. 10.1111/mmi.12363 [DOI] [PubMed] [Google Scholar]
- 14.King HL, Jr, Dyar RE, Wilken DR. 1974. Ketopantoyl lactone and ketopantoic acid reductases. Characterization of the reactions and purification of two forms of ketopantoyl lactone reductase. J. Biol. Chem. 249:4689–4695 [PubMed] [Google Scholar]
- 15.Shimizu S, Kataoka M, Chung MC, Yamada H. 1988. Ketopantoic acid reductase of Pseudomonas maltophilia 845. Purification, characterization, and role in pantothenate biosynthesis. J. Biol. Chem. 263:12077–12084 [PubMed] [Google Scholar]
- 16.Zheng R, Blanchard JS. 2003. Substrate specificity and kinetic isotope effect analysis of the Escherichia coli ketopantoate reductase. Biochemistry 42:11289–11296. 10.1021/bi030101k [DOI] [PubMed] [Google Scholar]
- 17.Cronan JE., Jr 1980. β-Alanine synthesis in Escherichia coli. J. Bacteriol. 141:1291–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williamson JM, Brown GM. 1979. Purification and properties of l-aspartate-α-decarboxylase, an enzyme that catalyzes the formation of β-alanine in Escherichia coli. J. Biol. Chem. 254:8074–8082 [PubMed] [Google Scholar]
- 19.Richardson G, Ding H, Rocheleau T, Mayhew G, Reddy E, Han Q, Christensen BM, Li J. 2010. An examination of aspartate decarboxylase and glutamate decarboxylase activity in mosquitoes. Mol. Biol. Rep. 37:3199–3205. 10.1007/s11033-009-9902-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zrenner R, Riegler H, Marquard CR, Lange PR, Geserick C, Bartosz CE, Chen CT, Slocum RD. 2009. A functional analysis of the pyrimidine catabolic pathway in Arabidopsis. New Phytol. 183:117–132. 10.1111/j.1469-8137.2009.02843.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White WH, Gunyuzlu PL, Toyn JH. 2001. Saccharomyces cerevisiae is capable of de novo pantothenic acid biosynthesis involving a novel pathway of β-alanine production from spermine. J. Biol. Chem. 276:10794–10800. 10.1074/jbc.M009804200 [DOI] [PubMed] [Google Scholar]
- 22.White WH, Skatrud PL, Xue Z, Toyn JH. 2003. Specialization of function among aldehyde dehydrogenases: the ALD2 and ALD3 genes are required for β-alanine biosynthesis in Saccharomyces cerevisiae. Genetics 163:69–77 http://www.genetics.org/content/163/1/69.full [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mody I, De Koninck Y, Otis TS, Soltesz I. 1994. Bridging the cleft at GABA synapses in the brain. Trends Neurosci. 17:517–525. 10.1016/0166-2236(94)90155-4 [DOI] [PubMed] [Google Scholar]
- 24.Roberts E, Frankel S. 1950. γ-Aminobutyric acid in brain: its formation from glutamic acid. J. Biol. Chem. 187:55–63 [PubMed] [Google Scholar]
- 25.Kumar S, Punekar NS. 1997. The metabolism of 4-aminobutyrate (GABA) in fungi. Mycol. Res. 101:403–409. 10.1017/S0953756296002742 [DOI] [Google Scholar]
- 26.Shelp BJ, Bown AW, McLean MD. 1999. Metabolism and functions of gamma-aminobutyric acid. Trends Plant Sci. 4:446–452. 10.1016/S1360-1385(99)01486-7 [DOI] [PubMed] [Google Scholar]
- 27.Bartsch K, von Johnn-Marteville A, Schulz A. 1990. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD). J. Bacteriol. 172:7035–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dover S, Halpern YS. 1972. Utilization of γ-aminobutyric acid as the sole carbon and nitrogen source by Escherichia coli K-12 mutants. J. Bacteriol. 109:835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuhrer T, Chen L, Sauer U, Vitkup D. 2007. Computational prediction and experimental verification of the gene encoding the NAD+/NADP+-dependent succinate semialdehyde dehydrogenase in Escherichia coli. J. Bacteriol. 189:8073–8078. 10.1128/JB.01027-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Biase D, Pennacchietti E. 2012. Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86:770–786. 10.1111/mmi.12020 [DOI] [PubMed] [Google Scholar]
- 31.Cao J, Barbosa JM, Singh NK, Locy RD. 2013. GABA shunt mediates thermotolerance in Saccharomyces cerevisiae by reducing reactive oxygen production. Yeast 30:129–144. 10.1002/yea.2948 [DOI] [PubMed] [Google Scholar]
- 32.Coleman ST, Fang TK, Rovinsky SA, Turano FJ, Moye-Rowley WS. 2001. Expression of a glutamate decarboxylase homologue is required for normal oxidative stress tolerance in Saccharomyces cerevisiae. J. Biol. Chem. 276:244–250. 10.1074/jbc.M007103200 [DOI] [PubMed] [Google Scholar]
- 33.Fonda ML. 1972. Glutamate decarboxylase. Substrate specificity and inhibition by carboxylic acids. Biochemistry 11:1304–1309 [DOI] [PubMed] [Google Scholar]
- 34.Liu P, Ge X, Ding H, Jiang H, Christensen BM, Li J. 2012. Role of glutamate decarboxylase-like protein 1 (GADL1) in taurine biosynthesis. J. Biol. Chem. 287:40898–40906. 10.1074/jbc.M112.393728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim HW, Kashima Y, Ishikawa K, Yamano N. 2009. Purification and characterization of the first archaeal glutamate decarboxylase from Pyrococcus horikoshii. Biosci. Biotechnol. Biochem. 73:224–227. 10.1271/bbb.80583 [DOI] [PubMed] [Google Scholar]
- 36.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 37.Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267. 10.1155/2004/204953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352–363. 10.1101/gr.3003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morikawa M, Izawa Y, Rashid N, Hoaki T, Imanaka T. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889–3899. 10.1128/AEM.71.7.3889-3899.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robb FT, Place AR. 1995. Media for thermophiles, p 167–168 In Robb FT, Place AR. (ed), Archaea: a laboratory manual—thermophiles. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42.Capitani G, De Biase D, Gut H, Ahmed S, Grütter MG. 2005. Structural model of human GAD65: prediction and interpretation of biochemical and immunogenic features. Proteins 59:7–14. 10.1002/prot.20372 [DOI] [PubMed] [Google Scholar]
- 43.Borges N, Matsumi R, Imanaka T, Atomi H, Santos H. 2010. Thermococcus kodakarensis mutants deficient in di-myo-inositol phosphate use aspartate to cope with heat stress. J. Bacteriol. 192:191–197. 10.1128/JB.01115-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dover S, Halpern YS. 1972. Control of the pathway of γ-aminobutyrate breakdown in Escherichia coli K-12. J. Bacteriol. 110:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsubara K, Yokooji Y, Atomi H, Imanaka T. 2011. Biochemical and genetic characterization of the three metabolic routes in Thermococcus kodakarensis linking glyceraldehyde 3-phosphate and 3-phosphoglycerate. Mol. Microbiol. 81:1300–1312. 10.1111/j.1365-2958.2011.07762.x [DOI] [PubMed] [Google Scholar]
- 46.Yokooji Y, Sato T, Fujiwara S, Imanaka T, Atomi H. 2013. Genetic examination of initial amino acid oxidation and glutamate catabolism in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 195:1940–1948. 10.1128/JB.01979-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Esser D, Kouril T, Talfournier F, Polkowska J, Schrader T, Bräsen C, Siebers B. 2013. Unraveling the function of paralogs of the aldehyde dehydrogenase super family from Sulfolobus solfataricus. Extremophiles 17:205–216. 10.1007/s00792-012-0507-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.