Abstract
SUMMARY
Bacterial genomes are remarkably stable from one generation to the next but are plastic on an evolutionary time scale, substantially shaped by horizontal gene transfer, genome rearrangement, and the activities of mobile DNA elements. This implies the existence of a delicate balance between the maintenance of genome stability and the tolerance of genome instability. In this review, we describe the specialized genetic elements and the endogenous processes that contribute to genome instability. We then discuss the consequences of genome instability at the physiological level, where cells have harnessed instability to mediate phase and antigenic variation, and at the evolutionary level, where horizontal gene transfer has played an important role. Indeed, this ability to share DNA sequences has played a major part in the evolution of life on Earth. The evolutionary plasticity of bacterial genomes, coupled with the vast numbers of bacteria on the planet, substantially limits our ability to control disease.
INTRODUCTION
Bacteria are ubiquitous, extremely numerous, and essential planetary life-forms. It has been estimated that there are about 5 × 1030 bacteria on earth, with the majority residing in oceanic and terrestrial subsurfaces, the open ocean, and soil (1). Bacteria can also be found within and on the surfaces of other organisms, as symbionts or pathogens. Bacteria play important roles in the environment and the ecology of the planet as well as in the evolution of living organisms (by their physical interactions with these organisms and by distributing genetic information by horizontal gene transfer [HGT]).
Maintaining the right balance between genome integrity and instability is essential for the survival of organisms and their offspring. Bacterial chromosomes are complex and dynamic, characteristics that give flexibility to the genome (2, 3). Genome instability can result from point mutations or from genome rearrangements such as deletions, duplications, amplifications, insertions, inversions, or translocations. This review concentrates on instabilities brought about by genome rearrangements and does not discuss the acquisition of genetic information on whole replicons (such as plasmids). Some of these mutations can be silent, while others can lead to phenotypic variation, evolution, and speciation. Deletions, duplications, insertions, and amplifications change the amount of information contained in the genome. Inversions, deletions, insertions, and translocations can disrupt genes. Most forms of genome rearrangement also result in the appearance of new sequences at the sites of the events. These new junctions have the potential to alter the function or expression of proteins. Finally, rearrangements can influence the structure of the chromosome, with indirect effects on phenotype (4). For example, a large inversion in the chromosome of Escherichia coli can have dramatic effects on cell viability (5, 6). Bacteria possess various mechanisms to repair DNA damage. Some of these repair mechanisms, such as nonhomologous end joining (NHEJ) and translesion bypass replication, are mutagenic. Mutagenesis is necessary for adaptation to changing environments and for bacterial evolution, which is significantly dependent on the potential for genetic instability and horizontal gene transfer. Additionally, specific genome instability can be at the origin of regulatory pathways, as in the case of phase and antigenic variation. Restriction-modification (RM) systems and the CRISPR-Cas system (comprising clustered regularly interspaced short palindromic repeats [CRISPR] and CRISPR-associated [Cas] proteins) also use genome instability to protect bacteria against invasion by phages and mobile elements. Genome instability is also used by pathogenic bacteria to facilitate host infection without being attacked by immune systems. Some instabilities are programmed, whereas others are random. They can be the result of specialized genetic elements and/or of the action of endogenous pathways of DNA metabolism. This review focuses mainly on natural chromosomal instability in bacteria. We first describe specialized genetic elements that mediate genome instability. We then report how endogenous processes themselves can create genetic instability by homologous or illegitimate recombination. Finally, we analyze two remarkable consequences of genetic instability in bacteria: phase and antigenic variation and horizontal gene transfer.
INSTABILITY MEDIATED BY SPECIALIZED GENETIC ELEMENTS
There are several kinds of specialized genetic elements playing a role in genomic instability. We first describe a number of mobile elements (insertion sequences [ISs], miniature inverted-repeat transposable elements [MITEs], repetitive extragenic palindromic [REP] sequences, bacterial interspersed mosaic elements [BIMEs], transposons, transposable bacteriophages, and genomic islands), inteins, introns, retroelements, and integrons. We then present two genetic elements that control the stability of mobile elements: postsegregation killing systems and the CRISPR-Cas system.
Mobile Elements
Studies on spontaneous mutations led to the discovery of transposable elements (7). They are found in every kingdom of life, but in bacteria, they are often more abundant in cells living in extreme environments (8–11). Transposable elements are DNA sequences with defined ends that can move locus within and between genomes by means of excision and integration reactions that are independent of homologous recombination. To be mobile, most transposable elements have short terminal inverted repeats (IRs) and use transposases that recognize and process the ends of the elements (Fig. 1). Transposable elements often duplicate the target sequence in which they integrate, creating a short direct-repeat sequence called a target site duplication. Selection of the target site is a function of the transposase and differs at the level of sequence specificity and stringency. The target sites of some elements are very specific (as is the case for Tn7 [12]), whereas other elements display little target specificity (e.g., Tn5 [13]).
FIG 1.
Schematic organization of different transposable elements inserted into a genome. (A) Organization of a typical IS (represented as a rectangle). It contains a single open reading frame (sometimes two), encoding the transposase, that extends within the right inverted repeat (IRR). The transposase promoter (P) is partially localized in the left inverted repeat (IRL). DR is the target fragment that has been duplicated to become a direct repeat following the insertion of the IS. (B) Organization of a typical MITE. (C) Organization of a typical transposon. Long terminal inverted repeated (IR) sequences surround function modules of genes. (D) Organization of a typical composite transposon. A chromosomal sequence is transposed together with two IS elements that surround it (here ISs are in a direct orientation, but they can be inverted). At least one of the transposases needs to be active. The internal DR of the IS elements can be absent. (E) Organization of a typical conjugative transposon (or ICE). The element contains inverted repeats surrounding various modules of genes for maintenance (recombination), regulation, and dissemination (conjugation) and some accessory proteins. A conjugal origin of transfer, oriT, is situated in the dissemination module. (F) Schematic organization of Mu, a typical transposable bacteriophage. Mu is delimited by inverted repeats. The element contains various modules of genes for regulation, transposition, lysis, and head and tail proteins. The G region is invertible, enabling the synthesis of different tail fiber proteins.
Despite their ubiquity, transposable elements are not the only forms of mobile DNA in bacteria. The characteristics of these elements can overlap and intertwine, and mobile elements often invade each other. Therefore, some of these elements can be difficult to categorize. In addition to plasmids (which are mobile but are not discussed in this review), there are various main groups of bacterial elements that are potentially mobile (Fig. 1; see also Fig. 2 and 3): ISs, MITEs, REP sequences, BIMEs, transposons (including integrative conjugative elements [ICEs]), transposable bacteriophages, and, to some extent, inteins, introns, homing endonucleases, and retroelements (see “Inteins, Introns, and Retroelements,” below). All mobile elements need to regulate their mobility to avoid excessive mutagenesis, which would be detrimental for the cell.
FIG 2.
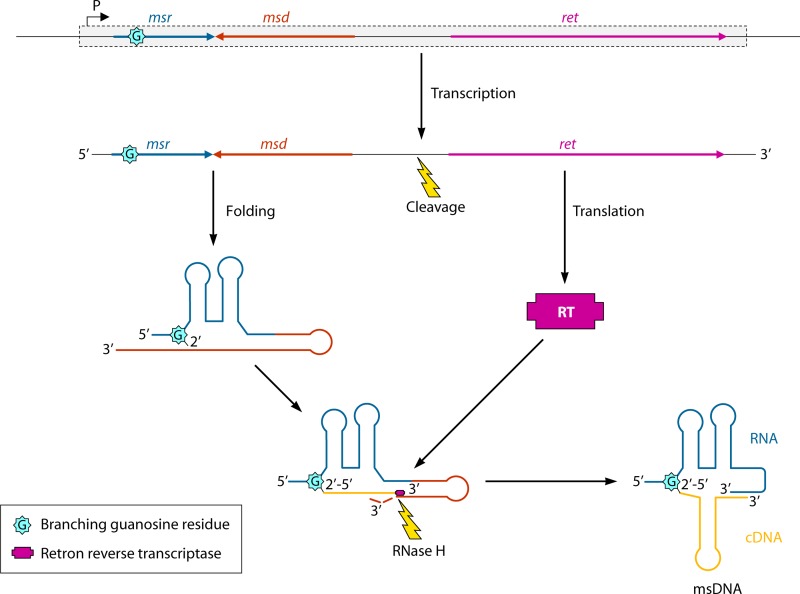
Schematic diagram describing the synthesis of msDNA. The retron element (represented as a rectangle) is transcribed, and the retron-specific reverse transcriptase (RT) is produced, whereas the part of the mRNA containing the transcription product of the msr and msd genes folds into a particular secondary structure. Thanks to this structure, the 2′-OH group of a specific branching guanosine residue (G) becomes the primer permitting the reverse transcription of the msd gene, while RNase H cleaves the mRNA template. Transcription stops at a fixed point, resulting in the msDNA molecule: an RNA and a cDNA molecule covalently linked. (Based on references 167–169.)
FIG 3.
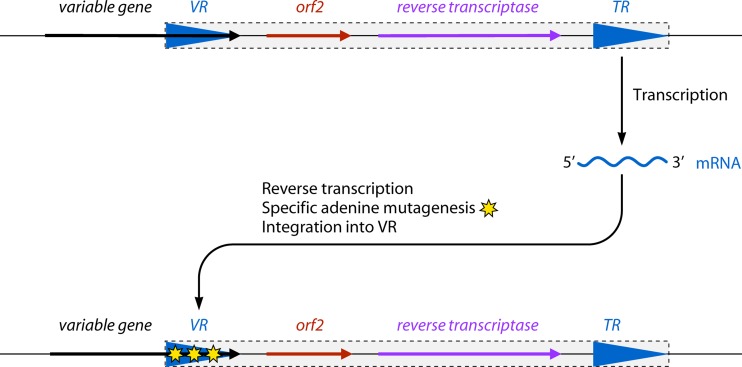
Schematic organization of a diversity-generating retroelement (DGR). A DGR (represented as a rectangle) is generally composed of two repeated sequences and one or two ORFs. VR is the variable repeat, which is at the 3′ end of a variable gene. TR is the template repeat, and its sequence is invariable. orf2 is not always present and differs in the function of the system. It is sometimes named atd (accessory tropism determinant) or hrdC (helicase and RNase D C terminal). The order of these elements is changeable. The template repeat is transcribed and integrated at the place of the variable repeat by a reverse transcription process that also exchanges some adenines for random nucleotides. RT is the reverse transcriptase.
Insertion sequences.
An IS is a relatively small (0.7- to 2.5-kb) DNA segment. It contains one or two open reading frames (ORFs) encoding only proteins responsible for functions involved in its mobility (a transposase) and is bounded by short terminal IR sequences (Fig. 1A) (14–16). Insertion of an IS will always change the host genome, whereas excision of an IS can either restore the chromosome to its original state or create a mutation. Table 1 lists some examples of IS-mediated alterations in bacterial genomes.
TABLE 1.
Examples of prokaryotic genomic rearrangements induced by natural transposable elementsa
| Rearrangement | Element(s) (reference[s]) |
|||||
|---|---|---|---|---|---|---|
| IS | MITE | Transposon | Composite transposon | Conjugative or mobilizable transposon | Bacteriophage | |
| Change upon insertion of the element | ||||||
| Addition of genetic material | IS1186 (601) | BOX (44) | Tn6061 (602) | Tn2922 (603) | Tn916 (604) | D3112 (605) |
| Inactivation of genes | IS629, ISEc8 (29) | RUP, BOX (44) | Tn551 (606) | Tn4001(607) | Tn916 (608) | Mu (86) |
| Creation of gene fusion | RPE (53) | Tn916 (609) | ||||
| Activation of genes by addition of a −35 sequence | IS2 (18) | Tn4652 (610) | ||||
| Activation of genes due to the presence of an outward promoter | IS6110 (19) | Correia (52) | Gamma delta (611) | Tn10 (IS10) (612) | ||
| Activation of genes by leakage of promoter from genes inside the element | ISTosp1 (17) | Tn1000 (613) | ||||
| Change due to perturbation of gene regulation | IS1 (21, 23) | Tn315 (614) | ||||
| Change of DNA topology around insertion site that impacts gene expression | IS1 and IS5 (20, 22) | ERIC (615) | ||||
| Addition of a binding site that impacts gene expression | IS5 (24) | Correia (616) | Tn4652 (617) | |||
| Change of mRNA properties of genes adjacent to insertion site | ERIC (618) | |||||
| Change upon excision of the element | ||||||
| Nucleotide substitution | Tn916 (619) | |||||
| Nearly precise excision (DNA insertion) | IS629 (28) | Tn10 (620) | Tn916 (621) | |||
| Imprecise excision (DNA deletion) | IS629 (28, 29) | Tn7 (622) | Tn5, Tn10 (622) | Mu (622) | ||
| Change involving another DNA molecule | ||||||
| Adjacent deletion (in which the element is not deleted) | IS629 (28) | Tn3 (623) | Tn10 (624) | Mucts62 (625) | ||
| Large deletion | IS407A, ISBma2 (626) | Correia (627) | Tn5 (IS50) (628) | Tn5386-Tn916 (629) | Mu (630) | |
| Large duplication | IS200 (631) | Tn4651 (632) | Tn10 (633) | |||
| Inversion | IS905 (634) | Tn2660 (635) | Tn10 (622) | CampMu (636) | ||
| Large genomic rearrangement | IS407A, ISBma2 (626) | MITE (41) | DEH (637) | Mu (638) | ||
| Replicon fusion or cointegrate | IS21 (639) | Tn4430 (640) | Tn4400 (641) | NBU1 (642) | D108 (643) | |
The list is nonexhaustive.
Insertion of an IS in a chromosome changes the genome of the host organism, as it adds the transposase gene(s) and often duplicates a target sequence, creating a direct repeat. Additionally, insertion of an IS can modify the expression of some host genes. The disruption of a gene or its regulatory sequence can lead to gene inactivation. Depending on the gene that is inactivated, the direct or indirect cellular consequences of the IS insertion can vary from advantageous to deleterious. On the other hand, an IS inserting upstream of a gene can activate the expression of this gene in any of the several ways described below. Again, the consequences of this activation can be diverse, from beneficial to lethal. Transcription of a gene within an IS can carry on outside the IS and transcribe neighboring host genes (17). Alternatively, some ISs contain an outward-facing −35 hexamer promoter motif in or near their terminal inverted repeats; their integration into the genome at the correct distance from a −10 promoter sequence changes the regulation of the downstream gene(s) (18). An IS might also contain an entire outward-facing promoter in or near its inverted repeats (19). Upon insertion of the IS, this promoter can activate downstream host genes. Furthermore, insertion of an IS can change the topology of the DNA into which it is inserted and can sometimes introduce or disrupt a regulatory binding sequence, affecting the regulation of the downstream gene(s) (20–24). Interestingly, the transposition of some ISs can be regulated by certain natural conditions required to activate the transcription of otherwise silent operons (25). An IS can also induce phase variation by alternating insertion and precise excision at a specific locus within a gene (see “Excision/insertion of DNA elements,” below) (26). The distribution of ISs in the genome is not random, as there are more ISs where they are less disturbing, in the intergenic regions between convergently oriented genes (27). This distribution suggests that detrimental insertions outnumber beneficial insertions.
Incorrect excisions of ISs are mostly consequences of the action of some host proteins, mainly but not exclusively DNA replication or repair proteins, and result in the introduction of mutations into the host chromosome (see also “Genome Instability Due to Recombination at Repeated Sequences,” below). After a nearly precise excision, some IS DNA remains in the host chromosome, resulting in an insertion (28), whereas an imprecise excision removes some host DNA, resulting in a deletion (28, 29).
The interaction of an IS with another DNA molecule with which it shares identical sequences, either another copy of the same IS, a different transposable element, or some genomic DNA, can result in more important genomic rearrangements. Under these circumstances, there are two possible mechanisms leading to genome instability (30). The first mechanism is direct and involves the action of the transposase and the ends of different transposable elements (or similar sequences) in an alternative transposition process. The second mechanism is indirect and relies on host proteins, as it uses the host homologous or illegitimate recombination systems (see Instability Mediated by Homologous and Illegitimate Recombination, below). Overall, these two processes induce IS-dependent genome instability, such as adjacent deletions, in which DNA connected to one end of the element is deleted without affecting the element itself, large-scale deletions, duplications, insertions, and chromosomal rearrangements. Recombination of two elements displaying the same orientation would lead to a deletion, whereas recombination of two elements of opposite orientations would result in an inversion of the intervening sequence. Importantly, ISs can insert into plasmids or bacteriophages as well as into chromosomes. Recombination between two ISs on different DNA molecules or a failure to resolve structures during transposition can lead to replicon fusions or cointegrates. This includes the formation of Hfr strains if the recombination event is between ISs on a chromosome and a conjugative plasmid such as F (31). Such events enable the transfer of chromosomal DNA by conjugation (32, 33).
An IS is a small DNA molecule, but its insertion or excision can cause important genome instability in its host, especially when it involves recombination or transposition with other DNA sequences. ISs can be considered selfish parasites or symbiotic sequences helping their hosts to evolve (see “Horizontal Gene Transfer in Prokaryotes,” below).
Miniature inverted-repeat transposable elements.
MITEs are small, AT-rich DNA sequences (0.1 to 0.5 kb) containing terminal inverted repeats, often displaying a TA dinucleotide motif at their extremities and being surrounded by target-site duplications (Fig. 1B) (4, 34, 35). They often possess the recognition sequences necessary for their mobility but do not encode a transposase. MITEs are widespread in eukaryotic genomes, where they can achieve high transposition activity using transposases encoded by other autonomous elements (36). Mobilization of MITEs has also been shown in bacteria (37). The study of MITEs in prokaryotes began recently, and they have not yet been well defined. As a consequence, distinctive MITE-like sequences have been classed and named differently in various organisms. They are referred to as MITEs in several bacteria but also as Correia elements (CE/NEMIS/CREE/SRE) in Neisseria; RUP, BOX, and SPRITE in Streptococcus; RPE in Rickettsia; CIR in Caulobacter and Brucella; Nezha in cyanobacteria; ISM854-1 in Microcystis; and RU-1 (ERIC/IRU), RU-2 (YPAL), or RU-3 in enterobacteria (11, 35, 38–44; for a more complete list, see reference 4).
Examples of MITE-induced genome instability in prokaryotes are listed in Table 1. As for ISs, MITE insertion can add genetic material, including functional ORFs (45); inactivate a gene; or modulate the transcription of neighboring genes by introducing an outward-facing promoter or a regulatory binding site or by changing the DNA topology at the insertion site. Additionally, two MITEs can recombine, leading to the formation of large deletions or other chromosomal rearrangements (46, 47). Strikingly, due to their small size, two main types of MITE-specific genome instability can also occur. Frequently, a MITE encodes one or several ORFs, and its insertion into a host gene can result in an in-frame gene fusion and the formation of a new protein (48). Sometimes, an inserted ORF encodes a specific motif that will change the function or the localization of the protein. MITEs can also have an effect on the regulation or the stability of mRNAs generated by genes surrounding their insertion sites (35). For example, Correia elements can be cotranscribed with their adjacent genes and be targeted for cleavage by RNase III, changing the stability level of these transcripts and therefore gene expression levels (49, 50). The same element can also act as a transcriptional terminator (51) and maybe as a noncoding regulatory RNA (52).
MITEs have definite actions on the genome of their host, from slightly detrimental to maybe beneficial (48, 53). Further studies of MITEs in bacteria may reveal their origins and intrinsic cellular functions.
Repetitive extragenic palindromic sequences and bacterial interspersed mosaic elements.
REP sequences were first discovered to be distributed throughout the chromosomes of enteric bacteria (they have also been called PUs, for palindromic units) (34, 54, 55). REP-like sequences have now been widely found in other bacteria but seem to be absent from extrachromosomal elements. The E. coli chromosome contains nearly 600 REP sequences, which corresponds to 1% of its genome. They are highly repeated imperfect palindromes of 20 to 40 nucleotides that are generally in extragenic but transcribed genomic regions. About 25% of E. coli transcription units harbor REP sequences. They can be found as single occurrences but are more often organized in pairs or in clusters. A BIME is a pair of REP sequences in an inverse orientation separated by a short linker sequence containing other conserved sequence motifs (56, 57). The E. coli chromosome contains >250 BIMEs, mostly in GC-rich genomic regions.
REP sequences can influence the expression or the regulation of genes or operons. After transcription, some REP sequences can fold into stable RNA structures that protect upstream mRNAs from degradation by 3′-to-5′ exonucleases (58, 59). Therefore, REP sequences can control differential gene expression in an operon by modulating the stability of the different mRNA segments. Additionally, some BIMEs are involved in transcription attenuation using a Rho-dependent mechanism (57), and a subclass of REP sequences can act as transcription terminators (60). Strikingly, BIMEs have also been found to specifically interact with a number of proteins, which might indicate a role of these repetitive elements in DNA topology and/or in the organization or the structure of the bacterial nucleoid. BIMEs of one category are bound by the integration host factor (IHF); these structures have been called RIBs (reiterative ihf BIMEs) (61) or RIPs (repetitive IHF-binding palindromic elements) (62). Additionally, DNA gyrase binds and cleaves some BIMEs (56, 63–65). DNA polymerase I (Pol I) also binds certain BIMEs (56, 66). Finally, the nucleoid protein HU might interact with these repetitive elements (67). Notably, REP sequences have been shown to stimulate the innate immune system of mammalian cells (68).
The number and the location of BIMEs and REP sequences are variable as a function of the bacterial strain and species (69). A REP-associated transposase was found, suggesting that BIMEs might be nonautonomous mobilizable transposable elements (70). However, alternative mechanisms have been proposed recently to explain the apparent mobility of BIMEs (71).
BIMEs and REP sequences seem to have an important effect on genome instability, bacterial evolution, and speciation. They are hot spots for specific transpositions (72–75), and they have been found at the junctions of RecA-dependent and RecA-independent duplications (76, 77).
Transposons.
Transposons generally range in size from 2.5 to 60 kb and usually possess long terminal inverted repeats and one or several accessory genes that confer an advantageous phenotype to their bacterial host, such as antibiotic, heavy metal, or phage resistance; catabolic, vitamin, or antimicrobial compound synthesis pathways; or nitrogen fixation (Fig. 1C to E). Transposons comprise functional modules, defined as regions devoted to individual functions (Fig. 1C). Complex transposons have been classified according to their structures and properties. A composite or compound transposon is flanked on both sides by similar or identical ISs, at least one of which one encodes a functional transposase, permitting their transposition together with the sequence that separates them (Fig. 1D) (78). A conjugative transposon, also named an ICE, can transpose intracellularly or excise to transfer intercellularly by conjugation (Fig. 1E) (79–82). These elements have phage, plasmid, and transposon characteristics (e.g., ICEs can integrate and excise using an integrase enzyme) and are transmissible among bacteria. Mobilizable transposons or plasmids can be mobilized by conjugative elements but are not self-transmissible (83). Recently, a conjugative transposon from Bacillus subtilis was also shown to mobilize plasmids that did not have the usual characteristics of mobilizable plasmids (84).
Most transposon-induced genome instabilities are similar to genome instabilities that originate from ISs (Table 1). Some elements, such as the conjugative transposon Tn5397, have strong insertion site preferences (85). Upon insertion, a transposon can disrupt a gene or modify the regulation of neighboring genes. As a consequence, transposons became useful tools for mutagenesis. Transposons can also induce genomic rearrangements, such as deletions, duplications, or inversions, or the formation of cointegrates. However, an important change caused by natural transposons but not by ISs is the addition of accessory genetic material into the host chromosome, as described above.
Transposable bacteriophages.
Transposable bacteriophages are viruses that can transpose their DNA into a bacterial chromosome, plasmid, or prophage, often duplicating the sequence surrounding their insertion site during this process (Fig. 1F) (86–88). These temperate phages can stay in their host genomes as latent prophages (lysogenic cycle) or replicate actively (lytic cycle). They are mutator elements, as their integration into their host genome is nearly random (Mu phages). Therefore, transposable bacteriophages are useful tools to identify genes involved in different pathways by mutagenesis. Examples of the effect of bacteriophage transpositions on the bacterial genome are listed in Table 1. Insertion of this kind of element into a gene (or its regulatory sequence) might result in inactivation of the gene. Importantly, mutations created by these elements have a polar effect, so the downstream genes in the same operon will also be inactivated (89). Additionally, transposable bacteriophages can induce the formation of different genomic rearrangements: various sizes of deletions or inversions or the formation of cointegrates. Finally, these bacteriophages can stimulate the mobility of other bacteriophages or induce recombination between transposable elements (90, 91).
Genomic islands.
Genomic islands (GIs) or chromosomal islands are large DNA sequences specifically present in the genomes of certain bacterial strains but not in the genomes of their most closely related variants (92–103). They are generally integrated within a bacterial chromosome, but they can also be found on plasmids or in phages. Some ICEs, integrated plasmids, or prophages have been considered GIs. These islands usually encode a number of accessory genes offering a selective advantage to the cell, which enhances the bacterium's chances of survival or of colonization of a new niche. Introduction of a new GI can result in a total change of phenotype, behavior, or life-style of the receiving organism. Depending on the provided phenotypic advantages, a GI can be a pathogenicity island (such as Salmonella pathogenicity island 1 [SPI1] [104]), a fitness island (such as E. coli acid fitness island [AFI] [105]), a metabolic island (such as the Xanthomonas xanthan gum production island [106]), a resistance island (to antibiotics) (such as AbaR7 in Acinetobacter baumannii [107]), a symbiosis island (such as the Mesorhizobium loti strain R7A symbiosis island [108]), a saprophytic island (like the island encoding adhesins in some E. coli strains [94]), an ecological island (such as an island permitting phenol degradation in Pseudomonas putida [94, 109]), or a defense island (as in Shewanella sp. strain ANA-3 [110]). Strikingly, similar GIs may show distinct functions in different bacteria or under specific ecological conditions or life-styles. A bacterium can contain various GIs in its genome.
A GI is usually between 10 and 200 kb in length; smaller regions with similar characteristics have been called genomic islets. They often show evidence of horizontal gene transfer even though this capacity might have been lost. They are usually inserted into tRNA gene loci, which often act as integration sites for foreign DNA, mainly prophages. They are flanked by directly repeated sequences consisting of a few to more than a hundred nucleotides. These repeated sequences may have been generated during the chromosomal integration of the GI or of some mobile elements by recombination or by transposition. Additionally, the percentage of G+C content, the frequency of small repeats, and the codon usage of a GI are generally different from the rest of the chromosome, indicating that these sequences were imported by horizontal gene transfer. This hypothesis is consistent with the fact that the same GI can be found in distantly related bacterial species. Finally, most GIs encode functional or degenerated mobile elements (phage or plasmid genes, ISs, integrases, transposases, or restriction-modification or toxin-antitoxin [TA] gene complexes). These mobility genes can be involved in the formation, rearrangement, integration, deletion, and mobility of GIs. Some of these islands have mosaic-like structures, suggesting an evolution that required multiple acquisitions from various donors. Most GIs can excise by homologous recombination at the direct repeats or with the help of a GI-encoded integrase or IS elements. Sometimes a GI excises together with a part of or all of its surrounding tRNA, leading to the loss of this coding sequence (93). Interestingly, specific parts or a whole pathogenicity island can excise at a precise point of a pathological process to facilitate the next stage of an infection (as in Salmonella enterica serovar Enteritidis [111]). Following its excision, a GI can integrate into a different locus of the same chromosome or be transmitted to another cell by horizontal gene transfer (such as the 89K pathogenicity island in Streptococcus suis serotype 2 [112]). The bacterial background and its environment have a great impact on the transferability of GIs, which can increase under stress conditions (113) [see also “Mechanism of HGT. (ii) Natural limitations of HGT,” below].
GIs generally encode proteins playing novel roles in transport, DNA binding and modification, cell motility, cell defense, the cell surface, and pathogenicity (114). Regulators encoded on a GI can control the expression of genes anywhere on the host genome. Similarly, GI genes can be regulated by proteins encoded by the same GI, another GI, or the bacterial host genome, indicating that GIs are well adapted to their hosts. Therefore, GI genes can be regulated by some environmental signals, such as pH (115), osmolarity (116), temperature (117), cell density (118), or the concentration of specific elements (116, 119–121).
To summarize, with or without the help of plasmids or bacteriophages, mobile elements can move between organisms belonging to different species or genera. Additionally, they can incorporate into each other, which is an efficient method for accumulating resistance genes and improving their characteristics (122). Integration of an IS into a transposon may change the expression of genes in this transposon, and recombination at IS sites within or between transposons creates new elements. Importantly, insertions, fusions, and rearrangements between mobile elements seem to be at the origin of genomic islands, of the assembling and expression patterns of the arrays of genes carried by them, and sometimes of their transfer within or between organisms (15, 123). Therefore, transposable elements have an essential role in horizontal gene transfer and the spread of antibiotic resistance and pathogenicity determinants.
Inteins, Introns, and Retroelements
An intein is a mobile element encoding a peptide that splices out from a host protein after its translation. An intron is an intragenic element encoding an RNA that splices out after its transcription. There are two types of introns in bacteria: group I introns and group II introns. They are both transposable elements, and group II introns are also retroelements. A retroelement encodes a reverse transcriptase and functions through an RNA intermediate. There are three main retroelements in bacteria: group II introns, retrons, and diversity-generating retroelements (DGRs).
Self-splicing elements.
Inteins and introns encode self-splicing proteins and RNAs, respectively. Usually, self-splicing elements integrate into their new host at the exact inteinless or intronless DNA locus, using a process named homing (or retrohoming for group II introns). Alternatively, they can occasionally recombine, reverse splice, or retrotranspose into a new DNA locus. Full elements encode homing endonucleases, which are responsible for their mobility.
(i) Inteins.
Inteins are widespread in bacteria (124–136). They are peptides of 134 to 608 amino acids encoded in frame within host proteins. One host protein can contain several inteins. By posttranslational processing, an intein self-catalyzes its precise excision and the concomitant ligation of its flanking regions, resulting in the formation of a mature functional host protein. Inteins do not require cofactors or accessory proteins for protein splicing. They often encode a homing endonuclease domain that is essential only for their mobility. This domain is also encoded in frame with the intein and the host protein. Interestingly, a protein containing an intein can be encoded by two partial genes localized in different places in a genome (137, 138). Each gene encodes a part of the protein fused to a part of the intein so that the original gene is disrupted within the intein element. After translation, the intein fuses the two halves of the protein by trans-splicing, resulting in the formation of an active whole protein. Inteins were hypothesized to have a role in cell development or in the regulation of protein expression or activity. Since their discovery, inteins became very useful biotechnological tools (as self-cleavable affinity tags for protein purification or to ligate expressed protein by trans-splicing [131]).
Inteins generally interrupt the conserved regions of essential proteins. They can be found in proteins involved in DNA or RNA metabolism and biosynthesis (such as a ribonucleotide reductase in Anabaena [139]); cell division (such as an ATPase involved in chromatin remodeling in Deinococcus radiodurans [140]); transcription (such as the GyrA gyrase in mycobacteria [141]); and DNA replication (such as DnaX, the DNA polymerase subunit, in Synechocystis and the DnaB helicase in Rhodothermus marinus [142, 143]), repair, and recombination (such as RecA in mycobacteria [144]). There are several nonexclusive hypotheses for their localization. By choosing conserved sequences, inteins would increase their chances of mobility and of not being counterselected (as imprecise excisions of these elements will probably result in nonfunctional host proteins). On the other hand, expression of inteins at the same time as DNA repair and recombination proteins would help the cell to recover from DNA cleavage (unwanted cleavage or during the homing process).
Bacterial intein-like domains (BILs) also posttranslationally self-process their host proteins (145, 146). They are domains of 130 to 165 amino acids found in proteobacteria, actinobacteria, and the Bacillus-Clostridium group. Interestingly, the number of BILs per species is very variable, probably due to gene duplications, as BILs were not shown to be mobile elements. These domains can be present in nonconserved regions of hypervariable bacterial proteins such as secreted proteins (such as the FhaB-like protein in Pseudomonas syringae, FhaB being a secreted filamentous hemagglutinin [145]). BILs might have a role in generating protein diversity, cell development, and host microevolution.
(ii) Introns.
In bacteria, group I and group II introns are small mobile elements (0.2 to 1 kb and 0.7 to 3 kb, respectively) that are transcribed into self-splicing RNAs (147, 148). These introns are widespread within the bacterial kingdom but not very abundant.
(a) Group I introns.
A group I intron is transcribed into a structured self-splicing RNA with 10 helices capped by loops and joined by junctions (149). The site-specific homing endonuclease, which is used by the intron to invade another DNA molecule, is generally encoded within a terminal loop. Group I introns frequently lose this gene, limiting their mobility. They often insert into bacteriophages. In the bacterial chromosome, they are found in tRNAs, rRNAs, or essential genes. Importantly, in bacteriophage T4, a group I intron inserting into a new locus leads to the coconversion of the exons at the new insertion site (150). This action changes the sequences surrounding the new intron, with repercussions for the encoded protein or RNA. Furthermore, some group I introns cannot splice out, which may have consequences on cell growth (151).
(b) Group II introns.
Group II introns behave more as retroelements than as introns (152, 153). They are transcribed into highly structured RNAs with six distinct double-helical domains, which provide the catalytic activity for splicing. Additionally, they encode in their fourth domain a protein showing endonuclease, reverse transcriptase, and RNA maturase activities, providing mobility to the intron (154). Bacterial group II introns are often fragmented, meaning that part of the intron is missing or mixed with DNA from another origin (155). However, they mostly integrate into mobile elements, which increases their chances of proliferation. Their rare integrations into host genes inactivate a targeted gene only when the intron cannot splice out (155). Surprisingly, for bacteria, a group II intron can also undergo alternative splicing, which alters the sequence of the host protein (156). These introns might also be at the origin of gene conversion events (157). Finally, they can induce deletions, inversions, or other chromosomal rearrangements in their host genome, as is the case in Wolbachia bacterial endosymbionts (155, 157).
(iii) Homing endonucleases.
Homing endonucleases are considered to be the real mobile elements encoded within introns and inteins, but they can also be freestanding in intergenic regions (129, 133, 136, 158–166). A homing endonuclease would bring mobility to a splicing element in exchange for the capacity to target conserved genes without being detrimental to the host bacterium. A splicing element avoids being counterselected, as it does not disturb the function of its host protein. Furthermore, deletion of the element may be counterselected, as imprecise excision is likely to damage the host gene. Therefore, this association increases the chances of the homing endonuclease and the splicing element being maintained in a population and invading other bacteria by horizontal gene transfer. During the homing process, the endonuclease cleaves the target DNA. Gene conversion then occurs during the DNA repair process, when the splicing element is copied into the previously empty allele. Insertion of the splicing element disrupts the recognition site of the homing endonuclease, preventing new cleavage. Additionally, the splicing element can insert into a new target gene by illegitimate recombination.
Homing endonucleases are usually encoded by short genes (<1 kb). They recognize specific DNA sequences of 12 to 44 residues, allowing a few single-base-pair changes within this target sequence, which is often present only once by genome. This rare-cutting characteristic helps maximizing their mobility while minimizing nonspecific cleavage and makes them excellent biological tools. Some homing endonucleases need associated proteins to regulate their activity. Finally, certain homing endonucleases also encode a maturase activity (helping intron RNA splicing) and/or a reverse transcriptase activity (for the retrohoming of group II introns).
Retroelements.
(i) Retrons.
Retrons are rare retroelements of about 2 kb that are inserted into prophages and a wide variety of bacterial chromosomes (167–169). They are composed of at least three ORFs, encoding a reverse transcriptase and a peculiar DNA/RNA hybrid molecule that has been described as multicopy single-stranded DNA (msDNA) (Fig. 2). Usually, msDNA is a small, single-stranded cDNA molecule covalently bound to an RNA molecule, which folds together into a stable secondary structure. This molecule accumulates abundantly in its host, up to 1,000 molecules per cell.
So far, there has been no proof that retrons are mobile elements. However, truncated copies of msDNAs can be found inserted into some bacterial genomes (170, 171). Upon integration, retrons seem to replace various sizes of sequences of the host genomic DNA (172–174). Strikingly, overexpression of some msDNAs increases the number of frameshift and base substitution mutations in E. coli (175, 176). This increased mutagenic level results from the binding of most cellular mismatch repair proteins to the mismatches on msDNA molecules. Similarly, the frequency of recombination between donor and recipient DNA sequences during matings of Salmonella and E. coli cells increases when some msDNAs are overexpressed, as the interspecific recombination frequency is normally reduced by the action of mismatch repair proteins (177). The function of msDNA molecules is still unknown, but they could be involved in helping bacteria to increase their mutation rates when needed for survival.
(ii) Diversity-generating retroelements.
Diversity-generating retroelements (DGRs) rely on reverse transcription to create diversity in DNA sequences that encode proteins involved in ligand-receptor interactions, mainly extracellular, cell wall, or membrane proteins. The main studied example is in the BPP-1 bacteriophage of Bordetella species, but DGR sequence patterns have been found in a wide range of bacterial prophages, plasmids, and chromosomes (178–184). DGRs are usually composed of two repeats of about 150 bp and two ORFs (Fig. 3). The first repeat forms the 3′ end of a gene and is the variable repeat (VR), as its sequence can undergo nucleotide substitutions at variable hotspots. Generally, downstream of the gene containing the VR are the template repeat (TR), which has an invariant sequence, and the ORFs, one of them encoding a reverse transcriptase. Strikingly, mutations in the sequence of the variable repeat always exchange a dATP for a random deoxyribonucleotide. The mechanism creating this directed mutagenesis involves the reverse transcriptase and a unidirectional transfer of information from the TR to the VR. A DGR induces at a low frequency a very high variability in the sequence of the protein encoded by the targeted gene. The fact that a number of DGRs are found in surface proteins, such as the LdtA lipoprotein in Legionella, led to the suggestion that DGRs may play an important role in the interaction that a bacterium has with its environment (184).
More groups of bacterial retroelements have been identified only recently (185, 186). Future studies might reveal some unknown effects of these elements on genome instability.
Integrons
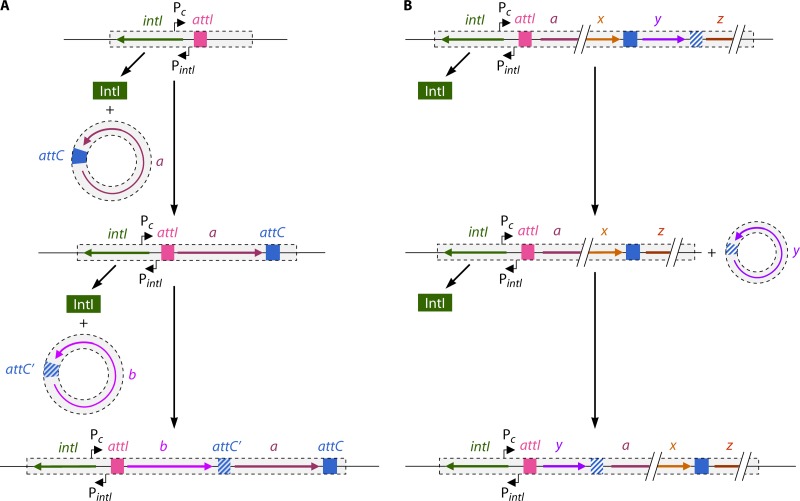
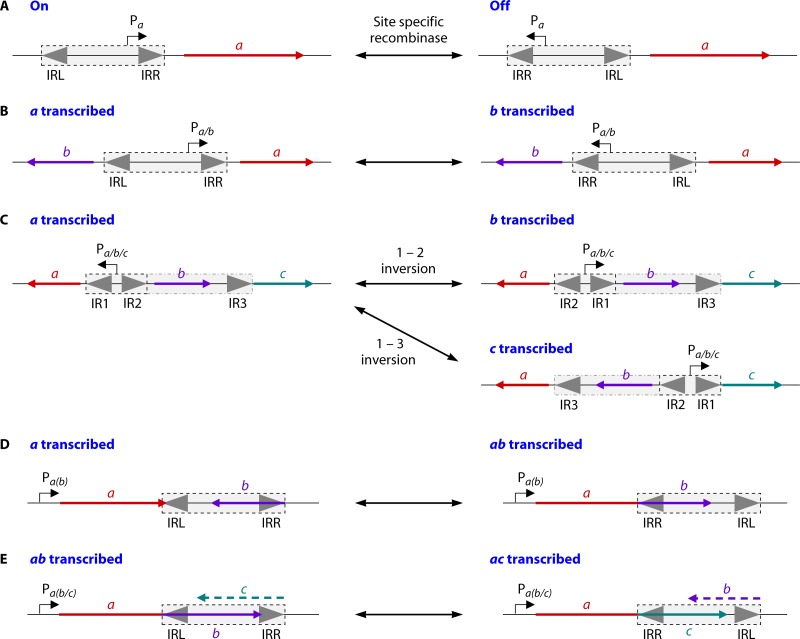
Integrons are relatively common non-self-transferable genetic elements that capture and rearrange single promoterless ORFs into an operon, allowing the appropriate expression of these genes (Fig. 4) (187–190). They are composed of a stable platform into which various gene cassettes that encode accessory functions are integrated. The stable part of the integron is generally composed of the intI gene and its associated promoter, a primary attI recombination site upstream of the promoter of intI, and a Pc promoter, located in the intI gene or the attI site (Fig. 4A). The intI gene encodes a site-specific tyrosine recombinase. The gene cassettes are free circular DNA molecules generally containing a single promoterless ORF and an attC recombination site (also called a 59-base element but with a size varying from 57 bp to 141 bp). The attC recombination site is a cassette-specific imperfect inverted repeat, variable in length and sequence, that can form secondary structures through self-pairing of the DNA strands (191). The IntI integrase recognizes the secondary structure of the bottom strand of the single-stranded attC site on the gene cassette and recombines it with the attI site in the integron (191–193), inserting the gene cassette just behind the Pc promoter in an orientation that usually results in the expression of the newly integrated gene. Integration of a single-stranded product may favor events following DNA transfer of a single strand via conjugation (i.e., in an intercellular event). Importantly, the next gene introduced into the integron will also be inserted at the attI site and will therefore be between the promoter and the previously integrated gene. Successive integrations result in the formation of an array of gene cassettes, which is the variable part of the integron. The further a gene is from the Pc promoter, the lower its chances are to be transcribed. IntI can also promote the excision of a gene cassette, permitting the mobility of a gene within the array in order to stimulate its expression (Fig. 4B) (194). Additional cassettes can also be acquired by horizontal gene transfer. A newly integrated cassette will be maintained in the integron if it confers an advantageous phenotype on the cell.
FIG 4.
Schematic organization of an active integron. An integron (represented as a rectangle) is constituted of a stable platform and a variable part. The stable platform is integrated into the host genome or a plasmid. It is composed of a site-specific recombinase gene (intI) and its promoter (PintI) as well as a primary recombination site, attI, upstream of the intI gene and the Pc promoter, located in the intI gene or the attI site. The variable part is formed by cassettes, each containing a gene and an attC recombination site. (A) Integration of gene cassettes into the stable platform of the integron. IntI mediates recombination between the attC site of the incoming cassette and the attI site of the integron so that the gene cassette integrates behind the Pc promoter, allowing gene expression. Successive integrations permit the formation of an array of gene cassettes, with the newly integrated cassette being nearest Pc. (B) Excision and reinsertion of a gene cassette. A gene in a cassette that is too far from Pc is not expressed anymore. IntI can mediate the excision of any gene cassette in the integron by recombination between the two attC sites surrounding the gene and its reinsertion in attI behind the Pc promoter. Therefore, the integron contains a reservoir of genes that can be rearranged and used by the cell under selective environments.
There are two main forms of integrons: mobile integrons (or resistance integrons) and superintegrons (or chromosomal integrons). Mobile integrons are carried by conjugative plasmids, transposons, or other mobile elements (46, 195, 196). In their array, gene cassettes have variable attC sites and encode a few proteins often involved in antibiotic resistance. Superintegrons are nonmobile and located in their host chromosome. Their very large numbers of cassettes have similar attC sites and express proteins involved in the cell's interactions with its environment or in undefined functions. These integrons can represent a significant fraction of the genome of their host (up to 3%). Importantly, the Pc promoter cannot control the expression of all genes present in a superintegron, and the expression of the IntI integrase is induced by cellular stress, especially DNA damage (via the SOS system [197]). These characteristics indicate that only the first few genes downstream of Pc are expressed and that the subsequent genes are silent, constituting a pool of information that the cell can rearrange, select, and use when necessary (Fig. 4B). However, a recent study demonstrated that some cassettes within the array can include a promoter that controls the transcription of several genes (198). To be able to maintain such large numbers of unused genes, superintegrons contain postsegregation killing systems (see “Postsegregation killing systems,” below).
Recombination generally occurs between an attI site and an attC site in a free gene cassette (199). However, two attC sites (or attI and attC) in an array can recombine to generate a cassette (200), and there are rare events of two attI sites in different integrons recombining, which leads to chromosomal rearrangements (201). A gene cassette can also occasionally integrate into another locus rather than within an integron, inducing the formation of a replicon fusion (202–205). Importantly, a cassette integrated into a nonspecific site cannot excise, becoming a permanent feature of its host's genome. The inserted gene will be transcribed only when placed downstream of a promoter. Finally, recombination, deletion, or duplication can occur between closely related or adjacent cassettes, leading to the formation of new gene cassettes (196).
Superintegrons add a large number of genes to the genomes of their hosts. They encode proteins that are essential for cell survival under specific conditions. Mobile integrons are easily transmitted between hosts and are at the origin of the rise of multidrug resistance in some bacteria. Finally, integrons are often associated with other mobile elements (190, 206).
Genetic Elements Controlling the Stability of Mobile Elements
Two different genetic elements can bring genome variability as well as control the stability of mobile elements (including bacteriophages and plasmids). Postsegregation killing systems can stabilize the mobile elements and integrons that they are associated with and attack other foreign DNA. Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) proteins (CRISPR-Cas systems) can protect their host cells against invasion by bacteriophages or plasmids.
Postsegregation killing systems.
Postsegregation killing systems are also called addiction modules because their loss leads to the death of their host bacterium. There are two essential components in a postsegregation killing system. The first element can attack the host organism or an invading agent, whereas the second element protects its host from attacks mediated by the first component. Upon integration into a new host, this system is tightly regulated so that protection occurs before any attack is possible, resulting in the survival of the new host. Following the loss of genes encoding a postsegregation killing system, attack elements override the protection elements, which is usually lethal for the previously hosting bacterium. Two kinds of postsegregation killing systems are widely present in prokaryotes: toxin-antitoxin (TA) systems and restriction-modification (RM) systems. Many postsegregation killing systems are mobile. Remarkably, some postsegregation killing systems have been perfectly coordinated with host development, cell biology, evolution, and speciation.
(i) Toxin-antitoxin systems.
TA systems are small mobile genetic modules found in bacterial chromosomes, viruses, and mobile elements (207–223). They are generally composed of two closely linked genes encoding a stable toxin and an unstable antitoxin. These genes are often autoregulated. Toxins are always small proteins (<130 amino acids), whereas antitoxins can be proteins or RNAs. Depending on the type of TA system, an antitoxin can either sequester its cognate toxin in a complex to stop its activity or inhibit its translation. Loss of a TA system by exclusion, segregation, recombination, or mutation can result in cell death or cell growth arrest, as the stable toxin outlives the unstable antitoxin and attacks the host cell. There are often multiple TA systems in a bacterial cell; some accumulate mutations which inactivate them. Toxin proteins can act differently on their host organism. They can impede cell division (224), stop DNA replication (225), cleave mRNAs (226), inhibit transcription (227) or translation (228), attack the cell membrane (229), stop cell wall biosynthesis (230), or decrease ATP synthesis (222).
Various roles have been proposed for TA systems; some are not exclusive, and others can be specific to certain TA systems. At first, these systems were found to participate in the maintenance of other mobile elements (plasmids, prophages, ICEs, integrons, or genomic islands), as their loss results in cell death, and they can exclude competitor elements (some toxins can attack invading elements [231] or act as abortive infection systems [232] or as antiaddictive modules, if the invading element bears a similar toxin [233], or the exclusion can be the result of plasmid-plasmid competition [234]). In the chromosome, they can stabilize large dispensable DNA regions that could otherwise be deleted (235). They have also been reported to be selfish elements that promote only their own survival or to be “junk” DNA from ancient mobile elements that will slowly be lost by the cell. On the other hand, TA systems also protect their host cell against attacks from phages and other mobile elements (233). Remarkably, some TA systems are completely integrated into their host regulatory network and now play a central role in prokaryotic cell biology. They have been shown to be involved in the development or the behavior of certain bacteria. For example, they can control cell death during fruiting body formation in Myxococcus xanthus or motility in E. coli (236). Some TA systems might also play roles in pathogenicity (219). Additionally, TA systems can have important functions in regulating the physiological states of bacterial populations and in stress responses (237). Under various stress stimuli (DNA damage, starvation, the presence of antibiotics or free radicals in the environment, high temperature, oxidative or osmotic shock, entrance into stationary phase, quorum sensing, or infecting phages), the toxins can be activated, often as a result of the degradation of antitoxins by stress-induced proteases or of the induction of toxin transcription. Depending on the nature of the stress and of the activated TA system(s), toxins can provoke altruistic programmed cell death to either release nutrients for other cells (226) or stop an infection (232), induce biofilm formation (221), arrest cell growth until improvement of the environmental conditions (238), or differentiate a subpopulation of cells into persisters (multidrug-resistant bacteria in a dormant state) (239). These stress responses can be determined by TA systems controlling DNA replication or the regulation of global or specific gene expression (240, 241). Finally, some toxins have a role in recycling damaged RNA to decrease cellular stress and release essential compounds for cell survival. Altogether, TA systems can have multiple roles and be perfectly integrated into the bacterial regulation system.
(ii) Restriction-modification systems.
Essentially, an RM system encodes a restriction endonuclease and its cognate modification activity, most often a methyltransferase (242–250). When several proteins are needed to perform these activities, their genes are usually located together on the bacterial chromosome, a virus, or a mobile element. Numerous chromosomal RM systems have been inactivated by insertions, deletions, or point mutations. The nuclease and the modification activities recognize and act upon a highly specific DNA sequence, different from one system to another. Modification of this sequence protects the genomic DNA from cleavage by the nuclease. A mutation in the modification activity or the loss of the whole system is often lethal for the host, as the bacterial DNA would then not be protected from the nuclease activity.
So far, several roles have been attributed to RM systems. First, these systems have been considered host defense mechanisms against invasions of phages, foreign DNA, and mobile elements. They attack invading elements that do not display the correct modifications, and no other competing genetic element can eliminate them without resulting in the death of the host cell (251, 252). According to the need of a bacterium, inactivation of a DNA restriction-modification system can enable an increase in the cell's capacity to incorporate foreign DNA into its genome, whereas activation of this system can protect the cell against a phage or DNA invasion. A temperate phage infection might be detrimental for an individual bacterium but useful at the population level, as lysogenization increases cellular genetic information, while lysis provides nutrients for the rest of the population. It has been hypothesized that DNA restriction enzymes from intracellular bacteria might also digest their host DNA to obtain precursor molecules for their own use (253). Second, these systems can stabilize other mobile elements in the cell population. RM genes are often linked to sequences resembling or being mobile genetic elements (within plasmids [254], bacteriophages [255], transposons [256], ICEs [257], integrons [242], or genomic islands [258] or near transposases [259], resolvases [259], invertases [260], integrases [261], topoisomerases [247], or phage-related sequences [262]). Third, they have been described as selfish elements that promote mainly their own survival. As well as using mobile elements as transporters, RM systems can move by themselves, probably as a result of their DNA cleavage activity. Following DNA restriction, an RM system can transpose into the location where its host repairs the break. Additionally, the death of their hosts releases DNA fragments encoding the RM systems into the environment, from where they can invade new competent cells. Fourth, they can increase bacterial diversity and phase and antigenic variation by inducing genomic rearrangement and homologous recombination during break repair or with incoming foreign DNA (restriction of the invading DNA might help separate beneficial and deleterious mutations) (243, 244). Fifth, they have been hypothesized to monitor the epigenetic DNA methylation level of their host in order to eliminate cells with unusual levels of methylation (248).
RM systems can be at the origin of various genome instabilities. DNA methylation may locally increase the DNA mutation level (263). Additionally, the action of an endonuclease can induce the SOS response system, which can increase mutagenesis in the host cell. Furthermore, repair after DNA restriction or during the insertion of an RM system can result in genomic rearrangement and phase or antigenic variation. An RM system can transpose into a new locus (264). It can simply insert into an operon-like gene cluster (like in the pur operon in Streptococcus suis [265]) or insert with a short or long and variable target duplication (such as an 8-bp sequence when type II RM genes insert into Xanthomonas oryzae pv. oryzae [247] or a 506-bp-long direct duplication after insertion of a type IIS system in Helicobacter pylori [266]). These long direct repeats can induce the formation of sequence amplification (like BamHI in Bacillus [267]) or of gene fusion, which can generate novel proteins (like the formation of an active type II M gene in H. pylori [247]). An RM system can also integrate by substitution (such as two type III RM homologues in H. pylori [266]). Insertions and substitutions may be associated with neighboring inversions (as in Pyrococcus [264]). Finally, a more complicated form of genomic rearrangement arising from the insertion of RM systems is the association of a substitution, an inversion, and a deletion (or insertion) next to each other (as in two RM systems in H. pylori [266]).
CRISPR-Cas systems.
CRISPR-Cas systems have been identified recently and studied intensively over the last decade (268–284). The main known role of these systems is to protect the cell by providing adaptive and hereditary immunity against previously encountered bacteriophages and plasmids. CRISPR-Cas systems have been found in the chromosomes of about half of the sequenced bacteria and nearly all archaea; they can also be found on plasmids, phages, and mobile elements. Around half of these genomes carry more than one CRISPR-Cas system (up to 18). Several systems present in the same organism can be similar or very different. A basic CRISPR-Cas system is composed of a leader sequence immediately followed by a CRISPR array and in the proximity of a group of cas genes (Fig. 5). Leader sequences and CRISPR arrays do not contain ORFs. A leader sequence is usually several hundred nucleotides in length with a large proportion of adenines and thymines. A CRISPR array is formed of a succession of short identical direct repeats (21 to 50 bp) separated by unique highly variable sequences of similar sizes, named spacers (20 to 84 bp). Importantly, spacer sequences can be identical to some bacteriophage, plasmid, or chromosomal sequences. These spacers encode the immunological memory of the system. The sequence of the leader and the sequence and number of repeats vary greatly between organisms or CRISPR-Cas systems (from two to several hundred repeats per array, but most loci have around 50 repeats). Similarly, the number of cas genes and the nature of the proteins which they encode are also dependent on the CRISPR-Cas system (generally 4 to 20 different genes). Usually, a specific CRISPR array is encountered with a cognate set of cas genes in differing genomic locations, gene orders, orientations, and groupings. Occasionally, some cas genes are at a different locus in the chromosome, but they would be present only when there is a CRISPR array in the genome. Cas is a large family of proteins carrying diverse but specific functional domains, such as integrase, endonuclease, exonuclease, RNase, helicase, polymerase, transcriptional regulator, or RNA- and DNA-binding domains.
FIG 5.
Schematic organization and mechanism of action of a basic CRISPR-Cas system (represented as an open rectangle). A CRISPR-Cas system is usually composed of cas genes (maroon arrows) and a leader sequence (blue rectangle) containing a promoter on the 5′ end of a CRISPR array, formed by short identical direct repeats (DR) (blue triangles) alternating with unique sequences (spacers) (colored octagons). (A) Adaptation is the first step of CRISPR-Cas immunity. A plasmid or bacteriophage DNA invades the bacterium. In rare cases, Cas proteins recognize this DNA as a threat (often thanks to a PAM sequence) (dashed open squares) and introduce a new repeat and spacer sequence into the CRISPR array between the leader sequence and the first repeat. This spacer corresponds to the protospacer in the foreign DNA (colored diamonds), a sequence near the PAM motif, if present. (B) The promoter in the leader sequence allows the transcription of the CRISPR array into a pre-crRNA (pre-CRISPR RNA). Cas proteins cleave the pre-crRNA into crRNAs, each containing a part of a repeat and a spacer. (C) Cas proteins and a specific crRNA target and inactivate the foreign DNA previously encountered, if present again in the cell. Some cellular proteins might help the Cas proteins in any of all these different stages.
There are three steps in the CRISPR-Cas immunity process: adaptation, crRNA (CRISPR RNA) expression, and interference (Fig. 5). Adaptation comprises the recognition and assimilation of a foreign DNA sequence by the CRISPR-Cas system. Cas proteins, with the potential help of the leader sequence and host proteins, can target and process the DNA of an invading plasmid or bacteriophage. They identify a specific sequence within this DNA, called the protospacer, and add it to the CRISPR array as a new oriented spacer. Most CRISPR-Cas systems recognize a very short sequence in the vicinity of the protospacer, named the protospacer-adjacent motif (PAM) (2 to 5 bp). Spacer integration generally occurs between the leader sequence and the first repeat of the CRISPR array and is accompanied by the duplication of a repeat. Occasionally, several protospacers from one foreign DNA sequence can be added into the CRISPR array, each with its own repeat sequence. This multiple acquisition increases the cell's level of resistance to a new invasion by this element (285). In a process called priming, a spacer already present in a CRISPR array before an infection with a virus containing the corresponding protospacer leads to a rapid acquisition of additional spacers recognizing this foreign element (286, 287). In the second step of this immunity pathway, the CRISPR array is generally transcribed from a promoter in the leader sequence, resulting in the formation of a nontranslated RNA, the pre-crRNA. Cas proteins and/or cellular ribonucleases process the pre-crRNA into fixed-sized crRNAs (35 to 46 bp), each containing a part of a repeat on its 5′ side, a part or all the spacer, and sometimes also a part of a repeat on the 3′ side. For this restriction step, certain CRISPR-Cas systems also use a trans-encoded transcript named trans-activating crRNA (tracrRNA). Finally, interference happens when the same foreign DNA sequence tries to invade anew the same cell or its offspring. The specific crRNA guides Cas proteins to the protospacer sequence on the invading DNA. The Cas-crRNA complex then inactivates this DNA by silencing or degradation. Notably, a specific group of Cas proteins, the Cmr proteins, seems to attack RNA and not DNA (288, 289). Cmr proteins are present in about 70% of archaea and 15% of bacteria. Importantly, to escape the cell defense mechanism provided by CRISPR-Cas systems, viruses constantly mutate their genomes by point mutation, deletion, or recombination (290, 291). Additionally, some bacterial prophages might encode their own defense system, for example, by using a protein that specifically binds and opens the DNA structure of the repeated sequences in the CRISPR array (268, 292) or by expressing an anti-CRISPR gene (293). Finally, CRISPR-Cas systems can also be inactivated by the action of mobile elements (280).
To be efficient, the CRISPR-Cas system must be kept under control. The size of a CRISPR array is restricted by occasional deletions of “old” repeat spacer units, which might be the result of homologous recombination between identical direct repeats (294). Furthermore, to avoid an autoimmune response, CRISPR-Cas systems need a way to distinguish foreign DNA from the DNA sequences of the CRIPSR arrays themselves. So far, two different strategies of DNA identification have been suggested. In some CRISPR-Cas systems, DNA cleavage of the chromosome is inhibited by the presence of the repeat sequence adjoining the spacer (295). Alternatively, CRISPR-Cas systems may recognize in the foreign DNA the presence of the PAM sequence at a set distance from the protospacer (281, 296, 297). As the genome does not carry PAM sequences, it is not regarded as a threat and therefore is not targeted. Remarkably, the expression of CRISPR-Cas systems can be regulated by some cellular transcription factors, such as the heat-stable nucleoid structuring protein (H-NS) and its antagonist LeuO in E. coli and Salmonella enterica (276, 298–300). Moreover, certain cellular factors and pathways can control the activation of some CRISPR-Cas systems. In Thermus thermophilus, phage infections induce the transcription of cas genes and CRISPR arrays by a sensing mechanism using the cell's cyclic AMP receptor proteins (301, 302). Stress, such as phage infection, the accumulation of misfolded proteins in the E. coli membrane, or the absence of ClpP in Streptococcus mutans, can also activate the expression of certain Cas proteins (303, 304).
CRISPR-Cas systems can be located in specialized regions of the genome encoding proteins involved in defense and stress response mechanisms (defense islands) (305). Even though the highly dynamic evolution pattern of cas genes would agree with a function of CRISPR-Cas systems in cell immunity (306), previous phylogenetic studies suggested that this might not be their main role and that these systems may have other cellular roles (307, 308). An important proportion of CRISPR array spacers correspond to bacterial chromosomal sequences, probably originating from immunity accidents (296). Eighteen percent of the organisms encoding a CRIPSR-Cas system display at least one self-targeting spacer. However, about half of these protospacers are located in elements that were probably introduced into the host genome by horizontal gene transfer (prophages, transposons, and plasmids). Other self-targeting spacers seem to be unstable in the array and can be deleted. Moreover, the presence of some self-targeting spacers can also result in mutations inactivating part of or the full CRISPR-Cas system or steer the evolution of the host genome. For example, a CRISPR spacer corresponding to the histidyl-tRNA synthetase (hisS) gene in Pelobacter carbinolicus might have induced the disappearance in this bacterium of genes encoding proteins with multiple closely spaced histidines (309). However, some self-targeting spacers might be used by CRISPR-Cas systems to regulate endogenous functions by controlling the expression of specific genes. In Pseudomonas aeruginosa cells containing a CRISPR-Cas system, the presence of a lysogenic bacteriophage results in the inhibition of biofilm formation and bacterial swarming, probably to avoid the propagation of the phage (310). In M. xanthus, the formation of fruiting bodies following starvation involves Cas proteins (311, 312). Moreover, in E. coli, one of the most conserved Cas proteins is a nuclease that physically interacts with DNA repair proteins (313). This protein and its CRISPR array have been proposed to have an important role in DNA repair and chromosomal segregation following DNA damage. Finally, repeats present in CRISPR arrays can be at the origin of large genomic rearrangements, which are evolutionarily important (314). Additional studies should determine whether all these activities of CRISPR-Cas systems are part of a defense mechanism or represent separate cellular roles.
Nowadays, CRISPR-Cas systems are becoming useful tools for a number of applications. Spoligotyping is based on differences between CRISPR-Cas systems to identify bacterial strains (315). This technique helps investigations of evolution and geographical and/or historical studies (316, 317) and permits the identification of microbial populations (318) or the analysis of pathogen outbreaks (319). New spacer repeat units are inserted in 5′ extremities of CRISPR arrays, which provide information on recent infections. On the other hand, 3′ extremities of CRISPR arrays correspond to older infections and are more conserved in evolution. As a consequence, studies of CRISPR arrays can reveal the sequence and identity of viruses that are new or difficult to access otherwise as well as information on the evolution of a population of viruses and/or bacteria in a studied environment and on the coevolutionary dynamics between viruses and their hosts. Additionally, industries using bacteria are domesticating CRISPR-Cas systems to naturally generate phage-resistant strains. This is particularly interesting for food industries, where there is a need to fight phage infections without genetically modifying the organism. The fact that CRISPR-Cas systems encoding Cmr proteins can target RNA has been used to design a system permitting the cleavage or silencing of a desired RNA target, creating a new way of impeding the expression of specific proteins in a cell (289). CRISPR-Cas systems can also become customized restriction enzymes for genome engineering (320–322).
The singularity of this system resides in the fact that cells acquire resistance to a specific foreign DNA that can be inherited by their offspring. Furthermore, CRISPR-Cas systems can be transmitted to other species by horizontal gene transfer, as they can also be present on plasmids or prophages (323, 324).
INSTABILITY MEDIATED BY HOMOLOGOUS AND ILLEGITIMATE RECOMBINATION
DNA replication, repair, and homologous recombination normally maintain genome stability. However, these processes can also induce genome instability and chromosomal rearrangement (325). Related and repeated sequences within the chromosome or specialized genetic elements play important roles in genome instability mediated by homologous or illegitimate recombination. Related sequences can be substrates for gene conversion. Recombination between inverted repeated sequences can lead to DNA sequence inversion, whereas recombination between directly repeated sequences can lead to duplication, amplification, or deletion. Finally, a deleted fragment can potentially reinsert at another locus in a genome, generating a translocation.
Mechanisms of Homologous and Illegitimate Recombination
Homologous recombination.
Homologous recombination is the exchange of genetic information between DNA molecules of identical or near-identical sequence (326–328). These homologous DNA sequences can be near or far apart on the chromosome or on different molecules. The minimal homology necessary for this process has been estimated to be between 20 and 100 bp (329). Imperfections in the homologous sequence dramatically decrease the recombination rate. Homologous recombination contributes both to the maintenance of genome stability and to genetic instability, as recombination can repair DNA damage and can reassort genetic information. Furthermore, it is an essential process for the integration of numerous horizontally transferred genes into their new host chromosome. Therefore, homologous recombination has a double role in the cell: it helps cell survival by maintaining genome integrity while promoting genome rearrangement that leads to diversity, evolution, and speciation.
In bacteria, homologous recombination has been studied most extensively in E. coli. Two principal pathways have been identified: the RecBCD-RecA pathway for double-strand break repair and the RecFOR-RecA pathway for single-strand gap repair. In both pathways, the RecA protein plays a central role. RecA binds to single-stranded DNA, forming a spiral filament that catalyzes a strand-exchange reaction with double-stranded DNA of identical or near-identical sequence. The binding of RecA stretches the single-stranded DNA molecule to 1.5 times the length of its equivalent double-stranded DNA. However, X-ray crystallography recently indicated that the stretching occurs between triplets of bases that retain the normal separation found in B-type DNA (330). It is presumably these triplets bound to the RecA filament that probe the structure of the double-stranded DNA molecule to find sequence identity. When it is found, strand exchange occurs within the RecA filament, generating a postsynaptic structure that remains stretched between normally separated triplets of base pairs. The process by which sequence identity is found (homology searching) remains mysterious. However, a recent single-molecule study has shown that the reaction is dramatically enhanced by a random-coil configuration (as opposed to a stretched-out configuration) of the targeted double-stranded DNA. This study has revealed that the RecA–single-stranded DNA filament makes multiple heterologous contacts with the random coil, leading to the discovery and stabilization of interactions that are homologous (331). In the RecBCD-RecA pathway, the RecBCD helicase-nuclease is responsible for generating single-stranded DNA and loading the RecA protein (327). In the RecFOR-RecA pathway, the RecF, RecO, and RecR proteins work together to facilitate the loading of the RecA protein onto single-stranded DNA via the displacement of single-strand-binding (SSB) protein (326).
Illegitimate recombination.
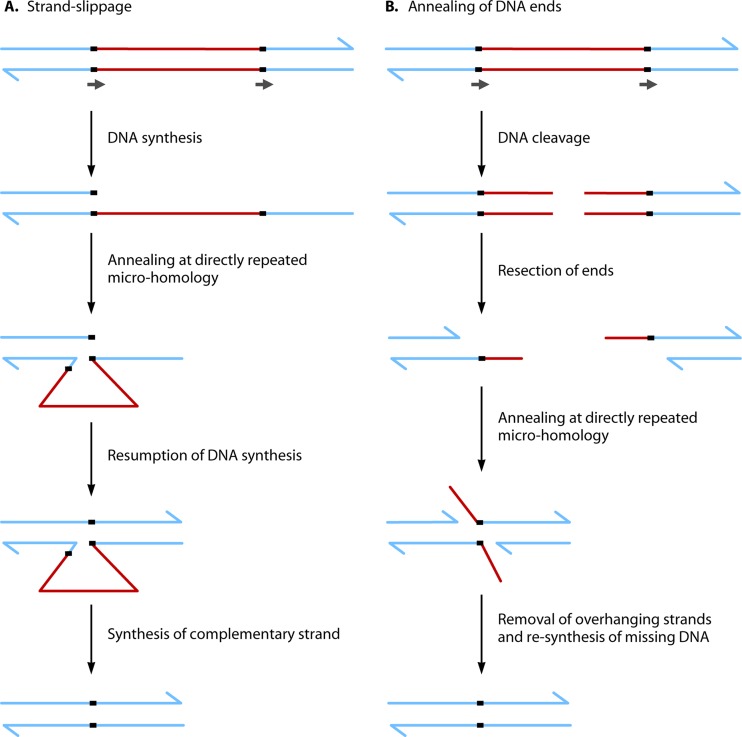
Illegitimate recombination refers to a collection of different reactions generally occurring at closely spaced DNA sequences that share little or no homology (332–335). This process is RecA independent and takes place when DNA strands anneal in aberrant configurations following a problem in DNA processing (Fig. 6). Stretches of a few base pairs of microhomology generally play a critical role in the efficiency of illegitimate recombination, as they promote DNA strand annealing at DNA ends formed either during DNA synthesis (strand slippage) or following DNA strand breakage (single-strand annealing). Illegitimate recombination events occurring by strand slippage or the annealing of DNA ends can be difficult or impossible to distinguish from the substrate and product structures. Illegitimate recombination occurring during DNA synthesis can be responsible for local sequence conversion, deletion, or duplication. It can arise during DNA replication (336, 337), transposition (91, 338), or gyrase- and topoisomerase I-mediated strand cleavage (339–341); as a consequence of UV or gamma irradiation (342–344); or following the transformation of cells with linear DNA sequences under circumstances where homologous recombination is not possible (345, 346). Single-strand annealing following a DNA break can occur after DNA degradation, leading to a local deletion. Mutations in the DNA Pol III or the mismatch repair system increase the rates of illegitimate recombination (347, 348). Some bacteria (e.g., B. subtilis, Mycobacterium tuberculosis, Mycobacterium smegmatis, and P. aeruginosa) encode a bona fide nonhomologous end-joining (NHEJ) system (349–353). This system consists of the Ku and LigD proteins, which act together to rejoin DNA ends at DNA sequences containing microhomologies. Bacterial NHEJ is thought to be particularly important in these bacteria to repair DNA, when a homologous template is not present to enable DNA double-strand break repair to occur by homologous recombination.
FIG 6.
Two classes of illegitimate recombination events. (A) Strand slippage. Strand annealing at regions of DNA microhomology (indicated by black arrows) can occur during DNA synthesis, resulting in deletions, duplications, and other rearrangements. This figure depicts the formation of a deletion by strand slippage. (B) Annealing of DNA ends. Strand annealing can also occur at DNA ends following resection and the formation of single-stranded regions. Microhomologies (indicated by black arrows) facilitate annealing. Importantly, the deletion events depicted here are identical whether produced by strand slippage or by annealing of DNA ends.
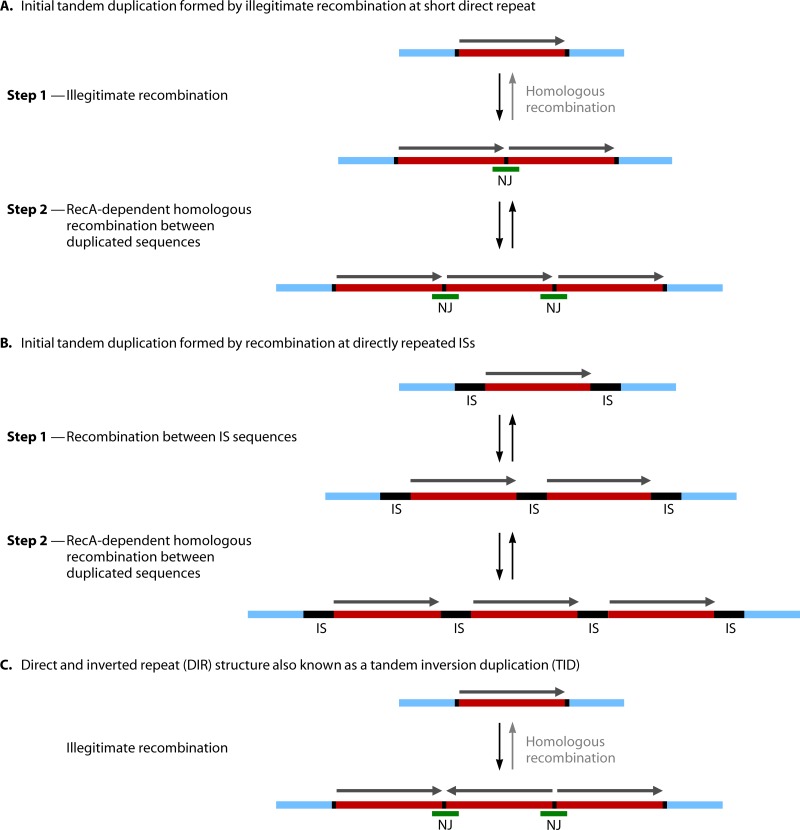
Gene Conversion
Gene conversion is a widespread mechanism of unidirectional transfer of genetic information that can occur as a consequence of homologous recombination when two related but different DNA sequences find themselves in the same cell (354, 355). The presence of these sequences can be the result of a gene transfer mechanism, or the cell can carry diverged copies of specific gene sequences. Gene conversion following gene transfer is responsible for the generation of combinations of alleles not originally present in the same cell, which accelerates genome divergence through evolution (see “HGT and evolution,” below). Intracellular gene conversions can mediate controlled variations at specific DNA sequences, called cassettes, resulting in phase or antigenic variation (356, 357) (see “Phase and Antigenic Variation in Bacteria,” below). The level of diversity introduced by gene conversion depends on the number of cassettes and their sequence variability. Gene conversion is sometimes accompanied by crossing-over, which can lead to a chromosomal inversion. Gene conversion seems to occur more frequently in pathogenic bacteria than in nonpathogenic bacteria (358). Gene conversion rates are influenced by the bacterial cell cycle, the presence of the mismatch repair system, and the level of iron in the environment (354).
Pilin antigenic variation in Neisseria species exemplifies how gene conversion can be used to facilitate the controlled replacement of one expressed gene sequence with DNA sequences stored at silent loci (357, 359). This exchange is mediated by a unidirectional gene conversion reaction, where information expressed from the pilE gene is replaced with that present in one of several silent pilS genes. This reaction retains the information at the pilS locus and is accomplished by a gap repair reaction mediated by a RecOR-RecA pathway (there is no RecF protein in Neisseria). A DNA sequence adjacent to the pilE gene is required for the reaction to proceed. This sequence, G3TG3T2G3TG3, on the lagging-strand template, has the potential to fold into a G-quadruplex structure. It is likely that the establishment of this quadruplex structure is required for the formation of single-stranded DNA at the pilE locus, which both initiates the reaction and marks pilE as the recipient of genetic information during this gene conversion (360).
Genome Instability Due to Recombination at Repeated Sequences
Tandem-repeat deletion or amplification.
Deletions and amplifications of almost any bacterial gene can occur at tandem-repeated sequences by homologous or illegitimate recombination, leading to changes in gene copy numbers. Tandem duplications of large chromosomal sections have been detected in many bacterial genomes, including those of B. subtilis, E. coli, Haemophilus influenzae, Klebsiella aerogenes, S. enterica, Salmonella Typhimurium, Streptococcus pneumoniae, and Streptomyces coelicolor. When they have been determined, tandem-duplication frequencies of approximately 10−3 to 10−4 per cell are common (361). Duplications can be further amplified by homologous recombination (Fig. 7) (362). The same sequence can be amplified up to 100 times. Unless selected for, these events are mostly undetected and revert to the unduplicated state without any noticeable consequence. However, they provide a fertile ground for selection to act upon in situations where gene dosage provides a growth advantage, for example, to increase bacterial resistance to a specific antibiotic (363). Gene amplifications bring solutions to selective problems by providing the population with more time and more cells necessary to facilitate further genetic adaptations. A substantial body of work on the nature of tandem-repeat duplications and their consequences was carried out in the 1970s. An excellent review of this early work was written by Anderson and Roth (361), where they described how these duplications can usually be tolerated because they preserve all the existing DNA sequences of the unduplicated part of the chromosome, while they may contain single novel junctions at their centers. These novel junctions can alter the nature of an expressed protein or the transcription level or control of a neighboring gene. Tandem-repeat instability varies as a function of the DNA sequence and the number of repeats (for example, CAG repeats are more instable than CTG repeats, and longer repeats are more instable [364]), their distance (365), their location in the genome (instability between repeats on the same molecules or on different molecules will result in different outcomes), and the cellular pathway leading to their instability (366). Additionally, the rate of tandem-repeat instability increases following DNA damage (367). Misalignment of repeated sequences might occur during DNA replication between the nascent strand and a second site on its template, on the nascent strand itself, or on the other sister chromosome. Tandem-repeat instability can result from illegitimate recombination, by strand-slippage or single-strand annealing, from homologous recombination between dispersed regions of homology such as IS elements, or by nonequal recombination between sister chromosomes or rolling-circle replication (Fig. 6 and 7) (for more details, see references 325, 333, 335, 362, 366, and 368). Importantly, illegitimate events create novel junction sequences, whereas the misaligned homologous recombination events do not (77).
FIG 7.
Two possible mechanisms for the formation of a tandem duplication and its subsequent amplification or reduction. (A) A tandem duplication can be formed by a strand slippage mechanism where a DNA sequence is copied and microhomology is then used to reinvade and copy the same DNA sequence again. This process generates a novel junction sequence (NJ) between the two repeated copies of DNA sequence. The duplicated sequence can then be amplified or reduced by RecA-mediated homologous recombination. (B) A tandem duplication can occur by homologous recombination between repetitive sequences such as insertion sequences (ISs). This reaction does not generate novel junction sequences. Again, the duplicated sequence can then be amplified or reduced by RecA-mediated homologous recombination. (C) DIR/TID structures can be formed by strand slippage during DNA replication or DNA repair. These structures consist of two overlapping DNA palindromes and may not be stable enough to persist for long. Symmetric deletions that reduce the symmetry of the palindrome centers have been documented, which may occur via subsequent rounds of strand slippage.
(i) Tandem-repeat variation by illegitimate recombination: microsatellite instability and contingency loci.
Microsatellites are repetitive sequences composed of small repeated units (usually 1 to 5 bp in length). These sequences have been shown to be unstable in bacteria, where their expansion and contraction probably result from strand slippage (364, 366, 369, 370). The mutation frequency in microsatellites is around 10−4. The rates of microsatellite instability can be modulated by different cellular processes using DNA synthesis, such as DNA replication, recombination, and repair. Furthermore, it has been shown recently that mismatch repair at these sequences can stimulate strand slippage at a longer directly repeated sequence located several kilobases away (367). Microsatellites occur naturally in numerous bacteria, and their expansions and contractions can regulate specific gene expression or alter coding sequences, resulting in phase or antigenic variation (see “Phase and Antigenic Variation in Bacteria,” below). This is particularly important for the control of expression at contingency loci, where gene expression can be reversibly activated and inactivated in a way that is beneficial for a pathogenic bacterium as it attempts to evade the defense strategies of its host (371). Two well-studied examples of contingency loci are antigenic variations at the fimbrial genes hifA and hifB of H. influenzae and at the opaE adhesin-invasin opacity locus of Neisseria gonorrhoeae (371, 372). In the case of the fimbriae of H. influenzae, a dinucleotide repeat sequence, (TA)n, lies between the −10 and −35 recognition sequences of overlapping divergent promoters of the hifA and hifB genes. Expansions and contractions of the TA repeat number alternate optimal spacing for the promoters of hifA and hifB, controlling fimbrial phase variation (373). In the case of the opacity genes of N. gonorrhoeae, the opaE genes carry multiple copies of the pentameric sequence CTCTT in the leader region of their ORF. Variations in the number of CTCTT repeats determine whether or not a particular copy of the gene is translated. In this way, the bacterium ensures a changing pattern of expression of different opacity variants (374).
(ii) Tandem-repeat variation by homologous recombination.
It is likely that the majority of tandem duplications are initiated by illegitimate events, but the phenomenon of resistance transfer factor transitioning in E. coli is a good example of an event initiated by homologous recombination at directly repeated sequences (e.g., ISs) (368, 375). The r-determinant part of a plasmid encoding antibiotic resistance can be amplified under selection for increased drug resistance and returned to a single copy when the selection is relaxed. There is substantial evidence that in all situations of large tandem duplication, amplification, or reduction, reactions are mediated by RecA-catalyzed homologous recombination (Fig. 7) (376). Over the past decade or more, there has been an active debate regarding whether the amplification reaction is stimulated when cells are held in the stationary phase for prolonged periods of time (377–379).
Hairpin structure-stimulated strand slippage.
DNA palindromes are sequences that are identical when read forwards and backwards. They promote illegitimate recombination by strand slippage by forming hairpin structures in single-stranded DNA that bring together sequences sharing microhomology. Deletions of palindromes by strand slippage preferentially occur on the lagging strand of the replication fork, consistent with the formation of DNA hairpins in the single-stranded regions generated between the Okazaki fragments (380, 381). Importantly, SbcCD is a nuclease that can cleave these hairpins and channel the DNA down the repair pathway of homologous recombination that is faithful and accurate (382, 383). However, it appears that if the palindrome is flanked by directly repeated sequences, its cleavage by SbcCD can also stimulate genomic instability mediated by a strand-annealing pathway, resulting mainly in deletions (384). A similar stimulation of instability through the action of SbcCD has been observed in E. coli in the presence of CAG/CTG trinucleotide repeats (364).
Palindromes and other closely spaced inverted-repeat sequences (quasipalindromes) can also stimulate the formation of a particular tandem duplication that consists of both direct and inverted repeats (DIRs) (385), also described as a tandem inversion duplication (TID) (Fig. 7C) (77). Models for how these structures can be generated have been proposed and involve various strand slippage reactions during DNA replication and DNA repair (77, 385, 386). The structure formed consists of two overlapping DNA palindromes and is itself likely to be prone to hairpin structure-stimulated strand slippage that reduces the symmetry of the palindrome centers. This mechanism of symmetry reduction via strand slippage within palindromes has been documented previously (381).
Control of genomic instability by DNA repair.
Homologous recombination contributes to genome stability, as unrepaired DNA double-strand breaks are a potential source of aberrant reactions that can lead to the formation of new DNA junctions and chromosomal rearrangements by illegitimate events. One such reaction is the formation of inverted chromosome dimers at the site of a palindromic sequence in E. coli recA sbcDC mutants (387). In the presence of SbcCD and RecA, homologous recombination repairs breaks in a way that avoids the formation of these inverted chromosome dimers. Moreover, DNA palindromes are hot spots for deletion formation by illegitimate recombination (see “Hairpin structure-stimulated strand slippage,” above). However, SbcCD and homologous recombination may limit the frequency of these deletions (388). Supporting this hypothesis are the facts that RecBCD and SbcCD decrease the level of homology-facilitated illegitimate recombination in Acinetobacter baylyi (389) and that rates of illegitimate inversions are elevated in E. coli sbcC mutants (390).
Recombination has a dual cellular function. It protects the genome and maintains genome stability as well as increases genome instability, leading to deletion, duplication, amplification, inversion, and translocation. These instabilities develop diversity through phase and antigenic variation and induce speciation and evolution. The role of recombination in genome instability is highly dependent on related or repeated sequences, which are various and numerous in a bacterial genome.
Site-Specific Recombination
Site-specific recombination generally uses a defined recombinase to recognize and recombine rare specific sites carried at the extremities of a DNA sequence element. This type of recombination requires a precise and oriented proximity of the recombination sites and is conservative (no loss or gain of DNA). The relative orientation of the recombination sites within a genome determines the outcome of this process. Recombination of a molecule containing inverted repeated sites results in an inversion of the internal DNA sequence element. On the other hand, recombination within one DNA molecule carrying directly repeated sites results in the formation of two separate molecules, whereas recombination of two DNA molecules carrying similar sites leads to the fusion of these molecules. There are three main site-specific recombination systems in bacteria: site-specific inversion systems, excision or insertion of DNA elements, and developmentally regulated gene rearrangements.
Site-specific inversion systems.
Site-specific inversion systems are widely spread in bacterial chromosomes, plasmids, and phages (391–395). Their sizes differ from a little more than a hundred base pairs to 35 kb. They play important roles in the synthesis and regulation of cell surface proteins (pili, flagella, surface layer proteins, and variable surface antigens) as well as type I restriction-modification systems. They bring a selective advantage to their hosts by increasing the adaptability of the organism, which might permit cell survival in differing environments, changes in pathogenicity levels, escape from an immune system, or protection against viruses. At a frequency of generally 10−3 to 10−5 per cell per generation, an invertase (or recombinase) recognizes and inverts sequences bracketed by two terminal inverted repeats that delimit the element (Fig. 8). The invertase gene can be located within the inverted fragment, in its vicinity, or somewhere else in the chromosome. The genotypic and phenotypic consequences of a site-specific inversion depend on the position and characteristics of the element (Fig. 8). In the following examples, the term “gene” is used to represent a gene or several genes in the same operon. Some site-specific inversion elements can, in one orientation, add a transcriptional terminator between a gene and its promoter, hindering the transcription of this gene and resulting in phase variation (396). Additionally, numerous invertible elements encompass an outward-facing promoter. One of these elements situated upstream of a gene can act as an on/off transcription switch for this region (Fig. 8A) (397). When located between two genes, this element can allow the alternate transcription of one of the two genes selectively (Fig. 8B) (398), inducing antigenic variation by the formation of different encoded proteins. A cluster of invertible segments can also be found, permitting the relative mobility of a promoter sequence upstream of various genes (Fig. 8C) (399). Site-specific inversion elements can also be located within a coding region, where their inversion results in the substitution of part of the gene (generally the 3′ end) (Fig. 8D and E) (400). Here again, a cluster of invertible segments recombining individually or in groups can lead to the formation of numerous variable proteins (401). Importantly, site-specific inversion elements can also indirectly change the cell phenotype by controlling the transcription of regulatory proteins (402). Several invertases can act on the same site-specific inversion element (403). Some invertases can promote only one direction of the reaction (for example, to promote the switch from on to off but not off to on) (404), can control the inversion levels of various site-specific inversion elements (405), or can be transcriptionally or posttranscriptionally regulated (406, 407). Strikingly, inversion levels can be modulated by host regulatory proteins or external factors (408). In summary, site-specific inversion elements help bacterial survival by permitting the presence of different individual cell phenotypes within one clonal population (see “Phase and Antigenic Variation in Bacteria,” below).
FIG 8.
Schematic organization of various examples of site-specific inversion systems. Inversion of the element results in the activation or inactivation of the transcription of the neighboring gene (A); the expression of either the a or the b gene (B); the expression of the a, b, or c gene selectively (C); the expression of a short or a longer protein (D); or the transcription of a gene encoding a protein with a different carboxyl-terminal domain (E).
Excision/insertion of DNA elements.
Some transposable elements, bacteriophages, and plasmids use site-specific recombination to change location or state (409). In some cases, excision and specific reintegration of an element can control phase variation in its host. For example, in Pseudomonas atlantica, the presence of an unstable IS492 element in its chromosome inactivates the synthesis of extracellular polysaccharides, whereas its precise excision as an independent circular molecule allows the production of these polysaccharides (26, 410–412). Therefore, the frequency of excision and insertion of IS492 directly regulates the production of these molecules, which are important in biofilm formation. Similarly, the formation of a specific lipopolysaccharide in Legionella pneumophila depends on the presence of a 30-kb unstable genetic element in its chromosome (413). This element probably originates from a phage and can excise as a replicating plasmid, consequently stopping its host's production of lipopolysaccharide.
Developmentally regulated gene rearrangements.
In bacteria, several developmentally regulated gene rearrangements result from site-specific recombination events (414–418). These rearrangements can occur in the mother cell during the sporulation process in B. subtilis and Clostridium difficile or during the formation of heterocysts, which are cells specialized for nitrogen fixation in filamentous cyanobacteria. In each case, a DNA element (with a size of between 4 kb and 55 kb) interrupts a coding sequence that is essential for cell specialization. Environmental signals and cell regulation induce the activity of a site-specific recombinase, which catalyzes a gene rearrangement by excising the element to form a nonreplicating circular DNA molecule and ligating the ends of the previously interrupted chromosomal gene. This excision is nonreversible, but these specialized cells (mother cells or heterocysts) cannot divide, so the loss of DNA does not affect another generation. Interestingly, these inserted elements seem to have a phage origin (416, 419). Their mechanism of action is reminiscent of the response of a lysogenic lambda phage when its host is under threat, which is to enter a lytic cycle by excising its DNA from the host chromosome (414). Remarkably, in B. subtilis, excision of the inserted element simply removes a block to the formation of molecules essential for sporulation, whereas in C. difficile, the excision of this element is an indispensable step in the sporulation process, as a strain in which this element is already deleted cannot sporulate (416).
Site-specific recombination is a simple but efficient form of genome rearrangement. Because it can lead to phase and antigenic variation, the consequences of this process are very diverse and can be essential for cell survival (see “Phase and Antigenic Variation in Bacteria,” below).
CONSEQUENCES OF GENOME INSTABILITY IN BACTERIA
Phase and Antigenic Variation in Bacteria
Phase and antigenic variation involves several programmed genetic or epigenetic processes found in numerous bacteria living in various environments (see references 371, 391–393, 395, and 420–426 and references therein). It induces specific, heritable, and reversible changes in the cell phenotype at rates higher than those of random mutation (10−1 to 10−5 events per cell per generation, depending on the system). Phase variation can modulate the level of expression of a gene or an operon (often in an on/off manner but sometimes gradually), whereas antigenic variation results in the production of a number of alternative proteins. Phase variation of several genes can create antigenic variation. Phase and antigenic variation can lead to the appearance of one or several different subpopulations. When required, the ability to combine the variation of several genes or exchange different parts of a gene leads to the possibility of formation of a very large number of phenotypes.
Mechanisms of phase and antigenic variation.
Various genetic processes can be at the origin of phase and antigenic variation (395, 424, 425). A first class of mechanisms is dependent on the cellular DNA replication, recombination, and repair systems (354, 362, 366). For example, at contingency loci, the number of short repeated sequences within a promoter or an ORF can vary following strand slippage during DNA replication or repair, leading to a transcriptional or translational switch in the expression of this gene or operon (335, 371) [see “Tandem-repeat variation. (i) Tandem-repeat variation by illegitimate recombination: microsatellite instability and contingency loci,” above]. Additionally, gene conversion or allele replacement permits the expression of one interchangeable gene (or part of a gene) out of a pool of silent genes, resulting mainly in antigenic variation (354) (see “Gene Conversion,” above). Multiple MITEs encoding outward-facing promoters of different strengths can recombine within a genome and induce phase variation (52). A gene can be duplicated or amplified by illegitimate and/or homologous recombination, which changes the level of expression of the gene product (362, 376) (see “Tandem-repeat deletion and amplification,” above). When not selected for, the duplication can be deleted by recombination at repeated sequences such as ISs. On the other hand, site-specific inversion systems control phase and antigenic variation independently of the cellular DNA replication, recombination, and repair pathways (see “Site-specific inversion systems,” above). Similarly, ISs, prophages, and other elements, such as a plasmid containing prophage genes, induce phase variation by excision and specific reintegration (25, 410, 413, 427) (see “Excision/insertion of DNA elements,” above). Finally, DGRs use their encoded reverse transcriptase to create antigenic variation [see “Retroelements. (ii) Diversity-generating retroelements,” above]. Notably, bacterial species generally have their preferred variation mechanisms.
Roles of phase and antigenic variation.
Most phase- and antigenic-variable proteins have important roles in mediating interactions between bacteria and their environments (393, 395). They control the formation of the capsule (428), the pili (429), flagella (430), adhesins (431), antifungal metabolites (432), iron acquisition factors (433), and other surface-exposed molecules (432, 434), sometimes affecting motility (432) or colony morphology (434) and opacity (374). Nevertheless, these proteins can also be involved in general cellular pathways, such as DNA restriction-modification (435), gene regulation (436), or metabolism (437). Phase and antigenic variation is thought to be essential for commensal, symbiotic, and pathogenic bacteria, as these processes can play an active role during the invasion (438) and infection (439) of an organism and help these bacteria to face the challenges raised by their hosts (440). However, this variation is also happening in bacteria that are not host associated. Alteration of the cell surface structure can change the capacity of adhesion of a bacterium, permitting the colonization of different environments (different organisms, tissues, or habitats, in or outside a host) and biofilm formation or detachment. Importantly, the level of virulence of some pathogenic bacteria is determined by phase or antigenic variation (441). Moreover, thanks to this variation, bacteria residing in an organism can evade host innate and acquired immune mechanisms (442) and avoid being targeted by cross-immunity (443). These escapes increase the duration of infection and, hence, the chances of spreading the bacteria to new hosts, as well as facilitating chronic infections and allowing several successive colonization events of the same host by similar bacterial strains. As a consequence, these bacteria conserve a larger population of hosts susceptible to infection and increase their chances of exchanging genetic information with other strains sharing the same host. In summary, regulation of cell metabolism and gene expression by phase and antigenic variation (directly or via the control of DNA methylation) helps the bacterium to save energy and to respond to stress signals and can govern some of the cell properties (virulence, biofilm formation, colonization, and sensitivity to bacteriophage invasions, etc.).
Regulation of phase and antigenic variation.
Phase and antigenic variation is key to a survival strategy based on heterogeneity. It generates diverse subpopulations that can be used either to survive potentially changing and stressful conditions or to give the opportunity for a few bacteria to colonize a new environment. In specific surroundings, several of these subpopulations might be needed to increase the fitness of a mixed but stable population. Importantly, even if a specific variation confers a growth disadvantage under the existing conditions, rearrangements constantly create specific phenotypes so that, when needed, cells with the right characteristics are always present in the population. Phase and antigenic variation is mediated by random events, as no prediction can be made regarding which gene in which bacterium will vary and when it will happen. However, the resulting phenotype is not random, since phase and antigenic variation is predictably encoded within the bacterial genome. The numbers and the roles of phase or antigenic variation genes in a bacterial genome are very different depending on the organism. Moreover, the consequences of variation depend on the environment and the nature of the proteins controlled by this system. The expression of phase-variable genes and the mechanism of phase variation can be controlled in a complex manner by cellular regulatory proteins (444). Due to their mechanisms, some variations can happen only at certain stages of the cell cycle (growing dividing cells or nondividing cells [391, 392, 426]). Furthermore, depending on the nature of the system, the level of variation can be controlled by the regulation of the recombination mechanism, recombinase expression, repair systems, or accessory proteins.
Some environmental or intracellular factors can influence the timing and frequencies of this variation, therefore controlling the level of the appearance of subpopulations when required for survival (393, 420). These factors are different as a function of the variation mechanism and can totally repress phase variation. Environmental or intracellular stress conditions that result in DNA supercoiling can modify phase variation switching frequency (445). Some systems are regulated by information about the cell's location, outside or within a host or a tissue, that can be given by the temperature (446, 447) or the composition of the environment (pH [448], carbon sources [449], oxygen [450] and iron [451] levels, amino acid concentrations [446], or the presence of specific elements [452]). Certain mechanisms answer to eukaryotic host-specific signal molecules, such as the presence of sialic acid, which is released by the host as a defense mechanism (453). Therefore, bacteria have evolved a survival system by which, following invasion, they can use the responses of their hosts to modulate their genome accordingly.
Phase and antigenic variation is essential for bacterial survival when there is no time or no suitable environmental signal to use classical regulatory systems. Accordingly, bacteria with a small genome seem to encode fewer two-component systems and more variation systems (371).
Horizontal Gene Transfer in Prokaryotes
Horizontal gene transfer (HGT) is a process that brings nonparental genetic information into a cell. At present, HGT events are rare and affect only a limited portion of a genome at a time, but they can have major consequences (454). HGT has been at the origin of animated scientific and philosophical debates for several decades (455–484).
Benefits of HGT.
Overall, HGT might be an advantageous process, as no cell has yet excluded it by changing its genetic code (460). Importantly, most HGT events probably have a neutral or deleterious effect on their new host and are rapidly lost; only an advantageous HGT event can be fixed in a population (485). HGT can quickly bring together systems that have already evolved and are ready to work under various conditions. It is a risky strategy, but the evolution of new beneficial genes is long and rare, so sharing could be better than remaking. In addition, HGT can change the level of expression of genes (generating higher, lower, constitutive, or different regulation). It can activate the transcription of cryptic genes, sometimes as part of a regulation process (25). Cryptic genes might be a genetic reservoir of information ready to be activated by mutations (486). HGT permits an individual bacterium to maintain a compact genome, whereas a huge number of accessory genes is available at the population level. Baumdicker and collaborators have proposed “the infinitely many genes model,” which is based on the possibility that individual genomes have access to an unbounded reservoir of novel genes (487). Furthermore, HGT can create chromosomal rearrangement, plasmid integration, deletion, insertion, novel gene fusion (bringing novel function), or duplication that can open new possibilities for future evolution. Through these events, the main role of HGT is in the initial acquisition of pathways; its role in pathway variation is small (488).
The beneficial effects of HGT are greater when a population grows in a stressful environment. HGT can facilitate niche adaptation and is important for bacterial mutagenesis and the maintenance of genetic heterogeneity. It has an essential role in the evolution and speciation of prokaryotes. Thanks to this process, cells can gain genetic information and increase their genome plasticity by the introduction of mobile elements. Remarkably, some genetic variations brought about by HGT might not be totally random but can happen with statistical reproducibility (489).
Mechanism of HGT.
Genes introduced by HGT can be new to the host bacterium (or come back after being lost) or can be paralogues of existing genes or substitutes for them. These genes could confer a novel pathway essential for cell survival or colonization of a new niche or could encode a protein more efficient than the one originally produced by the cell.
(i) Agents mediating HGT.
In bacteria, HGT can be mediated by various mechanisms: by transformation, conjugation, or transduction or by using gene transfer agents (GTAs), nanotubes, or membrane vesicles (MVs). Transformation is a process by which a cell takes up DNA from its environment. It occurs when the recipient bacterium is competent, a particular physiological state natural to some bacteria. Conjugation permits the direct transfer of DNA between two cells bridged by a pilus. It requires the presence of a conjugative plasmid or a chromosomally integrated conjugative element (ICE) in the donor cell. Transduction uses bacteriophages to transport DNA from one cell to another. GTAs are natural vectors that convey genomic DNA in a transduction-like manner (481, 490–493). They are host-encoded virus-like elements that cannot induce cell lysis but package and transport random fragments of the host chromosome. So far, GTAs have been described in proteobacteria and spirochetes. Nanotubes are tubular protrusions that join neighboring cells grown on solid surfaces (494). They have been suggested to permit the exchange of cellular molecules, including nonconjugative plasmids, within and between species. Finally, extracellular outer MVs are naturally produced by numerous Gram-negative bacteria (495–497). These spherical vesicles can transport proteins and/or DNA (from the host chromosome, a plasmid, or a phage) to a new host. Importantly, in a number of these processes, homologous recombination is essential for the integration of the horizontally transferred genes into their new host chromosome.
(ii) Natural limitations of HGT.
The existence of many different kinds of transfer mechanism ensures that no bacterium is completely immune to HGT. Nevertheless, the level of HGT depends on the organization of the recipient cell and on its environment. Agents mediating HGT have restricted ranges. Not all cells are competent, and phages and plasmids have specific hosts. A bacteriophage might also destroy its host during HGT. Additionally, HGT efficiencies depend on the level of DNA stability provided by the transport carrier and on the physical distance between the donor and the recipient cell. Most transfers occur between cells residing in the same habitats. Furthermore, the majority of HGT events will be quickly lost, as the new DNA has to be incorporated into the total genome of the recipient cell and be expressed into a useful product. The genome integration process can be carried out by homologous recombination, illegitimate incorporation at a double-strand break, or specialized genetic elements, such as mobile elements, MITEs, plasmids, phages, integrons, or genomic islands. Some elements integrate randomly, whereas others have specific targets. The frequency of recent HGT correlates linearly with the GC content and the genome and proteome sequence similarities between the transfer donor and the recipient cells (498). The cell's genome size, carbon utilization, and oxygen tolerance are also important factors (499). Homologous sequences are necessary to integrate DNA into a new host genome by homologous recombination. However, if available, the nonhomologous end-joining pathway seems to be able to help a bacterium to overcome this sequence similarity barrier (498). Once integrated into the genome, the regulation, transcription, and translation apparatus of the new host might not recognize horizontally transferred genes from a very different organism. For example, the new DNA might contain suboptimal codon frequencies that do not fit the tRNA pool of the recipient cell. Finally, the new genetic information should pass the test of natural selection. A large number of bacterial pseudogenes are horizontally transferred genes that were not useful at the time of their acquisition (500). To be fixed in a large population, the transferred DNA might need to bring more advantages than problems. In most cases, it cannot be toxic or disrupt a gene encoding an important cellular function when integrating. Often, it must be expressed at a functional level without decreasing the fitness of the cell or hindering the function of other cellular components. The encoded information usually needs to be new and useful as it is or to be an improvement on the information previously held by the cell. Strikingly, some eukaryotic genes can be found in parasitic or symbiotic bacteria (501), and some hyperthermophilic bacteria have acquired various genes from archaea (502), demonstrating that HGT can cross major phylogenetic barriers. Once successfully in the genome, the transferred sequence slowly acquires the characteristics of its new host genome, which increases its stability and might change its expression level or function.
(iii) Cellular mechanisms to fight HGT.
Most cells actively fight HGT by acting against the invasion of agents mediating this process, such as viruses, mobile elements, or conjugative plasmids. These various defense mechanisms include differential recognition by DNA uptake systems, CRISPR-Cas systems, restriction-modification systems, toxin-antitoxin systems, endogenous nucleases, and mismatch repair systems. Paradoxically, most of these systems would have been introduced into the cell genome by HGT. Additionally, some cells can specifically silence certain foreign genes by the binding of histone-like nucleotide structuring (H-NS) proteins (503).
(iv) Mechanisms of propagation of HGT elements.
An HGT element can use another HGT element to be transferred. For example, an IS element can move into a conjugative plasmid to be transported into another host. Some HGT elements avoid being lost by using postsegregation or postdisturbance killing mechanisms (see “Postsegregation killing systems,” above). Moreover, the use of certain antibiotics induces the SOS system, which activates the transfer of bacteriophages and ICEs, resulting in the spread of antibiotic resistance genes by HGT (504, 505). Some HGT induces biofilm formation, which improves the capacity of transferring DNA (506). Finally, in order to limit competition, specific HGT elements, such as phages, restriction-modification systems (243), toxin-antitoxin systems, CRISPR-Cas systems (507), or ISs (29), can fight other invading genetic elements.
HGT in contemporary organisms.
(i) Methods used to detect HGT.
Phylogenetic and/or genomic analyses can be used to determine whether a gene has been subject to HGT. With these types of analyses, it might be difficult to determine whether a gene was introduced into a bacterium by HGT or lost from its closely related organisms, so a combination of analyses might be more reliable (470). Phylogenetic analyses are based on anomalies in gene tree comparisons, indicating that a gene has a different origin than the rest of the genome. Genomic analyses rely on specific characteristics of the studied genome (frequencies of nucleotides, codons, and amino acids and gene distribution patterns). Additionally, regions adjacent to genes susceptible to have been horizontally transferred can be analyzed for potential relics of sequences that helped their integration. Importantly, genomic analyses can indicate only recent HGT events, as transferred sequences will progressively acquire their host characteristics by directional mutation pressure.
(ii) Levels of HGT in contemporary organisms.
Several studies have concluded that, depending on the bacterial strain, up to 20% of genes in a prokaryotic genome were recently introduced by HGT (459, 508, 509). According to Dutta and Pan, these numbers are underestimates, whereas they are overestimates for Kurland and collaborators and for Gao and Gupta (464, 483, 510, 511). In addition, a study of 181 sequenced prokaryotic genomes indicated that at least 81% of the genes in each of these genomes had been involved in HGT at one point in their history (512).
(iii) Genes susceptible to HGT.
The type and number of genes acquired by HGT are limited by the environment and the selective pressures exerted on the cells. All functional categories of genes are susceptible to HGT (513). However, some categories of genes are inherited more often than others, and some genes might be toxic to their new host and so cannot be transferred to them. Moreover, high gene expression levels can result in a fitness cost that limits HGT (514). Lerat and collaborators indicated that single-copy orthologous genes would be resistant to HGT (515), but this affirmation was contested by Bapteste and collaborators (516). Basically, genes that encode proteins that interact with other cellular molecules are less transferable, unless all the proteins of the pathway are encoded in a transferable operon (517, 518). Therefore, a protein with high interactivity can be displaced only by a similar protein from a closely related organism, whereas a modular element can be transferred from a phylogenetically more distant donor. As a consequence, HGT is more frequent for enzymes involved in peripheral cellular mechanisms than for enzymes involved in reactions central to cell survival. Most genes encoding molecules involved in replication, transcription, translation, and housekeeping are rarely acquired by horizontal transfer (519, 520). Conversely, metabolic and regulatory networks are shaped by HGT that can provide entire genetic pathways (521). However, the size of a bacterial genome is confined, so a cell that has acquired genetic material by HGT should lose an equivalent portion of its genome in the same HGT event or by decay and gene loss. Therefore, to be fixed in a population, a transferred gene must bring an advantage to its new host, which rarely occurs. Examples of new characteristics that can be introduced by HGT include metabolic properties, detoxification of heavy metals, fermentation of exotic carbon sources, defense mechanisms, antibiotic resistance, pathogenic functions, virulence attributes, quorum sensing, aerobic respiration, photosynthesis, thermophily, and halophily.
(iv) Relationships between HGT and an organism's life-style.
The life-style of an organism can determine the amount and origins of HGT that it will receive. The rates of HGT are higher when a bacterium is in a biofilm community than when it is in a planktonic state (506). Moreover, some pairs of bacterial species were identified to be linked by a highway of gene sharing, meaning that numerous different genes were horizontally transferred between these cells (522). Cyanobacteria living in extreme environments contain more mobile elements, which increase their genome plasticity and their chances of survival (11).
HGT is a major determinant of the integrity and size of some prokaryotic genomes. A number of common particularities characterizes genomes of obligate intracellular pathogens and symbionts as well as some extracellular symbionts that are physically isolated from the rest of the bacterial community (157, 459, 509, 523–542). These genomes are much smaller than the genomes of free-living bacteria (from 4 to almost 30 times smaller than the E. coli genome), have a strong A+T bias, and have very few mobile elements, regulatory systems, and pseudogenes. Additionally, these prokaryotes have spontaneous mutation rates at least 10 times higher than those of free-living bacteria (543). Some obligate intracellular bacteria also have a high copy number of their genome (544). Almost any functional category of gene can be lost in an obligate intracellular bacterium, but there is a common pattern. These bacteria usually preserve genes involved in essential processes, such as transcription, translation, and DNA replication, as well as chaperones and genes devoted to interactions with their hosts. Conversely, genes that are often lost encode proteins involved in cell envelope biogenesis, regulatory systems, metabolism (except for proteins needed for survival), and DNA repair and recombination. Repeated DNA, mobile elements, redundant pathways, and duplicated genes are almost always lost (545). Interestingly, the fact that comparable genome characteristics were visible in the majority of obligate intracellular symbionts and pathogens indicates that similar evolutionary forces led these bacterial genomes in this direction. Studies of the genomes of numerous bacteria that spent different lengths of time as obligate intracellular microorganisms indicated the dynamics of genome modulations leading to the speciation of these bacteria. Rapid evolutionary changes often occur immediately after host restriction. However, the reduction of the genome size and the proportion of A+T content in the genome increase with the time of association between a bacterium and its host. The genome of a bacterium that recently became an obligate intracellular organism contains a large quantity of mobile elements and pseudogenes. These mobile elements, along with repeated sequences, inactivate genes and induce inversions, deletions, and numerous chromosomal rearrangements (homologous recombination dependent or independent). As the population is small, isolated, and in a stable and rich environment, the pressure exerted by selection is relaxed, so transpositions and mutations are not counterselected and become fixed in the population, resulting in the proliferation of mobile elements and pseudogenes. These mutations might be beneficial (eliminating proteins that could be recognized by the host immune system), neutral (pathways that are redundant or not needed anymore), or even slightly deleterious. Once inactivated, these genes are deleted, as there is a bias toward deletion in bacteria (546). The isolation of these bacteria in a host cell dramatically reduces their opportunities to gain new genetic material by HGT. Therefore, gene losses are almost irreversible, and the genome shrinks. Once some DNA repair genes are inactivated, the mutation rate increases even more, as does the rate of transitions of GC into AT, resulting in an A+T-rich genome. Moreover, mutations in recombination genes decrease further the chances of genetic exchange. After this first phase of intense genome reduction, mobile elements themselves undergo inactivation and loss or will become lethal. Subsequently, there is a gradual gene loss following gene inactivation by mutation. During this process, genes are mainly lost, but not shortened, and the size of intergenic regions principally decreases only in the very small genomes of bacteria undergoing an ancient association with their host. In summary, there are two steps that lead to the reduction of the size of these genomes. First, mobile elements are responsible for a large part of the deletions and chromosomal rearrangements, resulting in the shrinking of the genome. Second, the isolation of these organisms results in the absence of HGT, so the lost DNA cannot be replaced.
Strikingly, despite being small, the genomes of some pathogenic and parasitic obligate intracellular bacteria have much more repetitive DNA and mobile elements than others (530, 547). These bacteria can switch hosts or infect host cells that contain other intracellular microorganisms. Within their hosts, coinfecting bacteria can exchange genetic material by HGT (548). Therefore, these obligate intracellular microorganisms can escape some of the genetic confinement of their ecological niche. Similarly, genomes of obligate intra-amoebal microorganisms do not seem to shrink as much as the genomes of other obligate intracellular bacteria (549, 550). Here, amoebae act as reservoirs containing large communities of microorganisms sharing an ecological niche. They constitute a place where these bacteria can exchange DNA or acquire DNA from organisms that have been degraded by the amoebae (551).
(v) Selfishness of mobile elements.
Mobile elements have been described as selfish entities or genetic parasites because they usually survive by invading a host genome without providing a beneficial phenotype to that host (243, 511, 552–554). This ability results from the fact that mobile elements can create copies of themselves, to spread quicker than the host genes and promote their own survival within and between cells by HGT. Additionally, mobile elements often encode postsegregation killing systems, which are lethal for host cells that eliminate them. To decrease their chances of inactivation by mutation, some spliceable mobile elements, such as introns and inteins, insert into or near nucleotide sequences encoding residues that are functionally critical for host survival. However, some mobile elements may provide short- or long-term advantages to their host cell. To increase the efficiency of their invasions, some mobile elements carry genes encoding phenotypic benefits for their host. Postsegregation killing systems can fight infections by new mobile elements, plasmids, or phages and can act as part of cell biology pathways or altruistic cell death strategies (for further examples, see “Postsegregation killing systems,” above). Finally, as mutators, mobile elements can induce variation for future evolution. Therefore, mobile elements should not be considered entirely selfish, but the relationship between a mobile element and its host might be understood as varying from extreme parasitism to mutualism (555).
HGT and evolution.
The role of HGT in evolution was first ignored and then considered to be a minor player as a consequence of rare events. However, in the era of genomics, HGT is now recognized as a major force in evolution, alongside genomic mutation, gene loss, gene duplication, and recombinational events (see references 457, 467, 509, and 556–564 as well as references cited at the beginning of “Horizontal Gene Transfer in Prokaryotes,” above). HGT is now thought to have been essential for the origin and development of life on the planet and to still be very important for the transfer of optimized pathways of genetic information, resulting in what has been called “evolutionary genetic quantum leaps,” and for increasing genome plasticity, leading to adaptation, genomic diversification, and speciation. Additionally, HGT and especially mobile elements provide important mechanisms of evolution of new genes (by bringing small insertions or deletions, by formation and activation of pseudogenes or new hybrid genes, by induction of genome duplications forming paralogous genes, or by genome remodeling following chromosomal rearrangements).
(i) HGT and the beginning of life on Earth.
HGT might have had an essential role in the development and evolution of very early life forms on the planet. The theory of the universal common ancestor presented by Woese describes the first living organisms as single communal evolutionary units in which HGT and mutation rates were very high, resulting in high evolution rates (565). These primitive organisms would have consisted of a pool of constantly exchanged genes. Evolution was then a communal process; there was no individual lineage (460). The genetic code would have been an innovation of this time (566). All modern organisms would have descended from this universal ancestral community of genes. Such a theory is hard to confirm or falsify and has been contested by Poole, who thinks that extreme rates of HGT without barriers may be disruptive to evolutionary transitions (478).
According to the selfish-operon hypothesis presented by Lawrence and Roth, HGT would also be accountable for the formation of operons, as genes necessary to carry a single pathway increase their chances of cotransfer when clustered in an operon (567, 568). Alternatively, the formation of operons might be the result of biophysical constraints (569) or could be driven by random gene deletions (for conserved genes) (570), or HGT might simply promote the prevalence of preexisting operons formed when gene regulation is complex (571). Arguing against a role of HGT in operon formation, analyses indicate that essential genes are particularly abundant in E. coli operons, while HGT events exchange mostly nonessential genes, and that horizontally transferred genes would have the least chances to be members of an operon (572, 573).
(ii) HGT and the “tree of life.”
A traditional eukaryotic “tree of life” is grounded on vertical inheritance (genes passed directly from parents to offspring). However, prokaryotic genomes have been extensively manipulated by HGT, so their histories are different from the histories of all their genes. In other words, it is problematic to determinate the lineage of an organism when most of its genes have different origins. Therefore, a number of scientists think that origins and relationships between prokaryote species cannot be represented as a tree and have proposed various alternative ways to describe the evolution of bacteria and life, usually as a forest, a network, or a ring (473, 516, 574–581). Conversely, numerous scientists think that the universal tree might still be an appropriate concept and have succeeded in constructing a tree of life despite or even based on HGT (468, 483, 511, 518, 561, 582–591). Remarkably, as well as being used to construct phylogenetic trees, 16S rRNAs were used by microbial ecologists to predict the ecological functions of a microbe. This methodology is now thought to be fruitless, as the presence of HGT unlinks the function and the genotype of bacteria (480, 592).
(iii) HGT and bacterial species.
Exchanges of genetic material due to HGT promote microbial diversification and speciation, for example, by changing the ecological niche of a microorganism. HGT also contributes to controversies concerning whether bacteria can be divided into species and what would then be the boundaries of these species (463, 593–597). Traditionally, species are ecologically distinct organisms that went through an irreversible divergence and for which diversity is limited by barriers to outcrossing. For Sonea and Mathieu, the lack of reproductive isolation in prokaryotes results in the absence of bacterial species (462). Some new microorganism-specific classification systems have even been proposed (475, 598). However, other scientists argue that the complexity added by HGT does not impede the classification of bacteria within species, as the vast majority of a bacterial genome is still vertically inherited (457, 483).
(iv) HGT, CRISPR-Cas, Lamarckism, and Darwinism.
The CRISPR-Cas system and the process of HGT integrate environmental information into a bacterial genome, permitting inherited adaptation to an environmental stimulus. For this reason, these two mechanisms have been proposed to be Lamarckian and quasi-Lamarckian, respectively (474, 484, 599, 600). Stress-induced mutagenesis, including that occurring following the activation of mobile elements, has also been suggested to be a quasi-Lamarckian process. Basically, the theory of Lamarckism states that evolution is driven primarily by nonrandomly acquired inheritable changes affected directly by the use of systems. On the other hand, the theory of Darwinism considers that random mutations provide evolutionary materials that can be lost or fixed by natural selection, leading to the evolution of organisms that are best adapted to their environment. According to Koonin and Wolf, evolution would use a continuum of processes within a range starting from total randomness up to systems perfectly targeted to specific responses to the cell environment (484, 599). The CRISPR-Cas system, HGT, and stress-induced mutagenesis can still be considered in accordance with Darwin's original ideas of evolution by variation and selection (484, 591). Furthermore, these processes seem to be Lamarckian only at the organismal level, as HGT genes evolve according to Darwinian processes; variations within genes are random and will be purged by natural selection, regardless of the origin of these genes (478).
In summary, HGT is the bacterial way of sharing genetic information. It is a powerful and complex process that leads to variation, evolution, and speciation. It might also be at the origin of the diversity of life on Earth.
CONCLUSION
Bacterial genome integrity is constantly threatened by external agents, such as mobile elements or phages, as well as by the operation of their own DNA replication and repair systems at related or repeated sequences. However, the large number of bacteria transforms genome instability into a driving force for bacterial survival, diversification, adaptation, speciation, and evolution. A growing bacterial population develops a balance between genome maintenance and instability that depends on the type of bacterium, the cell cycle, and the environment. Furthermore, bacteria utilize genome instability to increase their gene diversity and control gene expression and the response to various stresses. Further studies on these genome instability processes will lead to a better understanding of their role and action. Over the last 15 years, the development of genomic technology has allowed the discovery of several new genome instability processes, and it would not be surprising if there were more to come.
ACKNOWLEDGMENTS
We thank Meriem El Karoui and Garry Blakely for critical comments on the manuscript. Bacterial genome instability is a broad subject that has been studied for numerous years, leading to the publication of countless excellent articles and reviews. Unfortunately, we could not cite them all, and we apologize to the deserving colleagues who were not mentioned here.
We are grateful for funding from the Medical Research Council.
REFERENCES
- 1.Whitman WB, Coleman DC, Wiebe WJ. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U. S. A. 95:6578–6583. 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krawiec S, Riley M. 1990. Organization of the bacterial chromosome. Microbiol. Rev. 54:502–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saier MH., Jr 2008. The bacterial chromosome. Crit. Rev. Biochem. Mol. Biol. 43:89–134. 10.1080/10409230801921262 [DOI] [PubMed] [Google Scholar]
- 4.Delihas N. 2011. Impact of small repeat sequences on bacterial genome evolution. Genome Biol. Evol. 3:959–973. 10.1093/gbe/evr077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rebollo JE, Francois V, Louarn JM. 1988. Detection and possible role of two large nondivisible zones on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. U. S. A. 85:9391–9395. 10.1073/pnas.85.24.9391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esnault E, Valens M, Espéli O, Boccard F. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet. 3:e226. 10.1371/journal.pgen.0030226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shapiro JA. 2009. Letting Escherichia coli teach me about genome engineering. Genetics 183:1205–1214. 10.1534/genetics.109.110007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leavis HL, Willems RJ, van Wamel WJ, Schuren FH, Caspers MP, Bonten MJ. 2007. Insertion sequence-driven diversification creates a globally dispersed emerging multiresistant subspecies of E. faecium. PLoS Pathog. 3:e7. 10.1371/journal.ppat.0030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bickhart DM, Gogarten JP, Lapierre P, Tisa LS, Normand P, Benson DR. 2009. Insertion sequence content reflects genome plasticity in strains of the root nodule actinobacterium Frankia. BMC Genomics 10:468. 10.1186/1471-2164-10-468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qiu N, He J, Wang Y, Cheng G, Li M, Sun M, Yu Z. 2010. Prevalence and diversity of insertion sequences in the genome of Bacillus thuringiensis YBT-1520 and comparison with other Bacillus cereus group members. FEMS Microbiol. Lett. 310:9–16. 10.1111/j.1574-6968.2010.02033.x [DOI] [PubMed] [Google Scholar]
- 11.Lin S, Haas S, Zemojtel T, Xiao P, Vingron M, Li R. 2011. Genome-wide comparison of cyanobacterial transposable elements, potential genetic diversity indicators. Gene 473:139–149. 10.1016/j.gene.2010.11.011 [DOI] [PubMed] [Google Scholar]
- 12.Craig NL. 1991. Tn7: a target site-specific transposon. Mol. Microbiol. 5:2569–2573. 10.1111/j.1365-2958.1991.tb01964.x [DOI] [PubMed] [Google Scholar]
- 13.Lodge JK, Weston-Hafer K, Berg DE. 1988. Transposon Tn5 target specificity: preference for insertion at G/C pairs. Genetics 120:645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahillon J, Léonard C, Chandler M. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. 150:675–687. 10.1016/S0923-2508(99)00124-2 [DOI] [PubMed] [Google Scholar]
- 16.Siguier P, Filée J, Chandler M. 2006. Insertion sequences in prokaryotic genomes. Curr. Opin. Microbiol. 9:526–531. 10.1016/j.mib.2006.08.005 [DOI] [PubMed] [Google Scholar]
- 17.Luque I, újar A, Jia L, Zabulon G, de Marsac NT, Flores E, Houmard J. 2006. Regulated expression of glutamyl-tRNA synthetase is directed by a mobile genetic element in the cyanobacterium Tolypothrix sp. PCC 7601. Mol. Microbiol. 60:1276–1288. 10.1111/j.1365-2958.2006.05170.x [DOI] [PubMed] [Google Scholar]
- 18.Bowen SW, Hassan HM. 1993. Characterization of cis-acting regulatory mutations causing anaerobic expression of the sodA gene in Escherichia coli. Arch. Biochem. Biophys. 302:372–379. 10.1006/abbi.1993.1226 [DOI] [PubMed] [Google Scholar]
- 19.Safi H, Barnes PF, Lakey DL, Shams H, Samten B, Vankayalapati R, Howard ST. 2004. IS6110 functions as a mobile, monocyte-activated promoter in Mycobacterium tuberculosis. Mol. Microbiol. 52:999–1012. 10.1111/j.1365-2958.2004.04037.x [DOI] [PubMed] [Google Scholar]
- 20.Reynolds AE, Mahadevan S, LeGrice SF, Wright A. 1986. Enhancement of bacterial gene expression by insertion elements or by mutation in a CAP-cAMP binding site. J. Mol. Biol. 191:85–95. 10.1016/0022-2836(86)90424-9 [DOI] [PubMed] [Google Scholar]
- 21.Ozawa Y, Mizushima S, Mizuno T. 1990. Osmoregulatory expression of the ompC gene in Escherichia coli K-12; IS1 insertion in the upstream regulatory region results in constitutive activation of the promoter. FEMS Microbiol. Lett. 56:295–299 [DOI] [PubMed] [Google Scholar]
- 22.Singh J, Mukerji M, Mahadevan S. 1995. Transcriptional activation of the Escherichia coli bgl operon: negative regulation by DNA structural elements near the promoter. Mol. Microbiol. 17:1085–1092. 10.1111/j.1365-2958.1995.mmi_17061085.x [DOI] [PubMed] [Google Scholar]
- 23.Fernández A, Gil E, Cartelle M, Pérez A, Beceiro A, Mallo S, Tomás MM, Pérez-Llarena FJ, Villanueva R, Bou G. 2007. Interspecies spread of CTX-M-32 extended-spectrum beta-lactamase and the role of the insertion sequence IS1 in down-regulating bla CTX-M gene expression. J. Antimicrob. Chemother. 59:841–847. 10.1093/jac/dkm030 [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Saier MH., Jr 2009. A novel mechanism of transposon-mediated gene activation. PLoS Genet. 5:e1000689. 10.1371/journal.pgen.1000689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall BG. 1999. Transposable elements as activators of cryptic genes in E. coli. Genetica 107:181–187. 10.1023/A:1003936706129 [DOI] [PubMed] [Google Scholar]
- 26.Perkins-Balding D, Duval-Valentin G, Glasgow AC. 1999. Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J. Bacteriol. 181:4937–4948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plague GR. 2010. Intergenic transposable elements are not randomly distributed in bacteria. Genome Biol. Evol. 2:584–590. 10.1093/gbe/evq040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kusumoto M, Ooka T, Nishiya Y, Ogura Y, Saito T, Sekine Y, Iwata T, Akiba M, Hayashi T. 2011. Insertion sequence-excision enhancer removes transposable elements from bacterial genomes and induces various genomic deletions. Nat. Commun. 2:152. 10.1038/ncomms1152 [DOI] [PubMed] [Google Scholar]
- 29.Ooka T, Ogura Y, Asadulghani M, Ohnishi M, Nakayama K, Terajima J, Watanabe H, Hayashi T. 2009. Inference of the impact of insertion sequence (IS) elements on bacterial genome diversification through analysis of small-size structural polymorphisms in Escherichia coli O157 genomes. Genome Res. 19:1809–1816. 10.1101/gr.089615.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gray YH. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16:461–468. 10.1016/S0168-9525(00)02104-1 [DOI] [PubMed] [Google Scholar]
- 31.Virolle MJ, Gélugne JP, Béjar S, Bouché JP. 1983. Origin of Escherichia coli K-12 Hfr B7. J. Bacteriol. 153:610–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smillie C, Garcillán-Barcia MP, Francia MV, Rocha EP, de la Cruz F. 2010. Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74:434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lesic B, Zouine M, Ducos-Galand M, Huon C, Rosso ML, Prévost MC, Mazel D, Carniel E. 2012. A natural system of chromosome transfer in Yersinia pseudotuberculosis. PLoS Genet. 8:e1002529. 10.1371/journal.pgen.1002529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachellier S, Clément JM, Hofnung M. 1999. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol. 150:627–639. 10.1016/S0923-2508(99)00128-X [DOI] [PubMed] [Google Scholar]
- 35.Delihas N. 2008. Small mobile sequences in bacteria display diverse structure/function motifs. Mol. Microbiol. 67:475–481. 10.1111/j.1365-2958.2007.06068.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR. 2009. Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science 325:1391–1394. 10.1126/science.1175688 [DOI] [PubMed] [Google Scholar]
- 37.Bardaji L, Anorga M, Jackson RW, Martinez-Bilbao A, Yanguas-Casas N, Murillo J. 2011. Miniature transposable sequences are frequently mobilized in the bacterial plant pathogen Pseudomonas syringae pv. phaseolicola. PLoS One 6:e25773. 10.1371/journal.pone.0025773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mennecier S, Servant P, Coste G, Bailone A, Sommer S. 2006. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 59:317–325. 10.1111/j.1365-2958.2005.04936.x [DOI] [PubMed] [Google Scholar]
- 39.Xu M, Kong R, Xu X. 2006. A miniature insertion element transposable in Microcystis sp. FACHB 854. Prog. Nat. Sci. 16:486–491. 10.1080/10020070612330024 [DOI] [Google Scholar]
- 40.Chen Y, Zhou F, Li G, Xu Y. 2008. A recently active miniature inverted-repeat transposable element, Chunjie, inserted into an operon without disturbing the operon structure in Geobacter uraniireducens Rf4. Genetics 179:2291–2297. 10.1534/genetics.108.089995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naito M, Hirakawa H, Yamashita A, Ohara N, Shoji M, Yukitake H, Nakayama K, Toh H, Yoshimura F, Kuhara S, Hattori M, Hayashi T. 2008. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis. DNA Res. 15:215–225. 10.1093/dnares/dsn013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Romine MF, Carlson TS, Norbeck AD, McCue LA, Lipton MS. 2008. Identification of mobile elements and pseudogenes in the Shewanella oneidensis MR-1 genome. Appl. Environ. Microbiol. 74:3257–3265. 10.1128/AEM.02720-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou F, Tran T, Xu Y. 2008. Nezha, a novel active miniature inverted-repeat transposable element in cyanobacteria. Biochem. Biophys. Res. Commun. 365:790–794. 10.1016/j.bbrc.2007.11.038 [DOI] [PubMed] [Google Scholar]
- 44.Croucher NJ, Vernikos GS, Parkhill J, Bentley SD. 2011. Identification, variation and transcription of pneumococcal repeat sequences. BMC Genomics 12:120. 10.1186/1471-2164-12-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Palmenaer D, Vermeiren C, Mahillon J. 2004. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol. Microbiol. 53:457–467. 10.1111/j.1365-2958.2004.04146.x [DOI] [PubMed] [Google Scholar]
- 46.Gillings MR, Labbate M, Sajjad A, Giguère NJ, Holley MP, Stokes HW. 2009. Mobilization of a Tn402-like class 1 integron with a novel cassette array via flanking miniature inverted-repeat transposable element-like structures. Appl. Environ. Microbiol. 75:6002–6004. 10.1128/AEM.01033-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poirel L, Carrër A, Pitout JD, Nordmann P. 2009. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 53:2492–2498. 10.1128/AAC.00033-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delihas N. 2007. Enterobacterial small mobile sequences carry open reading frames and are found intragenically—evolutionary implications for formation of new peptides. Gene Regul. Syst. Biol. 1:191–205 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2759123/ [PMC free article] [PubMed] [Google Scholar]
- 49.Mazzone M, De Gregorio E, Lavitola A, Pagliarulo C, Alifano P, Di Nocera PP. 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 278:211–222. 10.1016/S0378-1119(01)00725-9 [DOI] [PubMed] [Google Scholar]
- 50.De Gregorio E, Abrescia C, Carlomagno MS, Di Nocera PP. 2003. Ribonuclease III-mediated processing of specific Neisseria meningitidis mRNAs. Biochem. J. 374:799–805. 10.1042/BJ20030533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Francis F, Ramirez-Arcos S, Salimnia H, Victor C, Dillon JR. 2000. Organization and transcription of the division cell wall (dcw) cluster in Neisseria gonorrhoeae. Gene 251:141–151. 10.1016/S0378-1119(00)00200-6 [DOI] [PubMed] [Google Scholar]
- 52.Siddique A, Buisine N, Chalmers R. 2011. The transposon-like Correia elements encode numerous strong promoters and provide a potential new mechanism for phase variation in the meningococcus. PLoS Genet. 7:e1001277. 10.1371/journal.pgen.1001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abergel C, Blanc G, Monchois V, Renesto P, Sigoillot C, Ogata H, Raoult D, Claverie JM. 2006. Impact of the excision of an ancient repeat insertion on Rickettsia conorii guanylate kinase activity. Mol. Biol. Evol. 23:2112–2122. 10.1093/molbev/msl082 [DOI] [PubMed] [Google Scholar]
- 54.Stern MJ, Ames GF, Smith NH, Robinson EC, Higgins CF. 1984. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell 37:1015–1026. 10.1016/0092-8674(84)90436-7 [DOI] [PubMed] [Google Scholar]
- 55.Lupski JR, Weinstock GM. 1992. Short, interspersed repetitive DNA sequences in prokaryotic genomes. J. Bacteriol. 174:4525–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gilson E, Saurin W, Perrin D, Bachellier S, Hofnung M. 1991. Palindromic units are part of a new bacterial interspersed mosaic element (BIME). Nucleic Acids Res. 19:1375–1383. 10.1093/nar/19.7.1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Espéli O, Moulin L, Boccard F. 2001. Transcription attenuation associated with bacterial repetitive extragenic BIME elements. J. Mol. Biol. 314:375–386. 10.1006/jmbi.2001.5150 [DOI] [PubMed] [Google Scholar]
- 58.Newbury SF, Smith NH, Robinson EC, Hiles ID, Higgins CF. 1987. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell 48:297–310. 10.1016/0092-8674(87)90433-8 [DOI] [PubMed] [Google Scholar]
- 59.Stern MJ, Prossnitz E, Ames GF. 1988. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol. Microbiol. 2:141–152. 10.1111/j.1365-2958.1988.tb00015.x [DOI] [PubMed] [Google Scholar]
- 60.Gilson E, Rousset JP, Clément JM, Hofnung M. 1986. A subfamily of E. coli palindromic units implicated in transcription termination? Ann. Inst. Pasteur Microbiol. 137B:259–270 [DOI] [PubMed] [Google Scholar]
- 61.Boccard F, Prentki P. 1993. Specific interaction of IHF with RIBs, a class of bacterial repetitive DNA elements located at the 3′ end of transcription units. EMBO J. 12:5019–5027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oppenheim AB, Rudd KE, Mendelson I, Teff D. 1993. Integration host factor binds to a unique class of complex repetitive extragenic DNA sequences in Escherichia coli. Mol. Microbiol. 10:113–122. 10.1111/j.1365-2958.1993.tb00908.x [DOI] [PubMed] [Google Scholar]
- 63.Yang Y, Ames GF. 1988. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc. Natl. Acad. Sci. U. S. A. 85:8850–8854. 10.1073/pnas.85.23.8850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bachellier S, Saurin W, Perrin D, Hofnung M, Gilson E. 1994. Structural and functional diversity among bacterial interspersed mosaic elements (BIMEs). Mol. Microbiol. 12:61–70. 10.1111/j.1365-2958.1994.tb00995.x [DOI] [PubMed] [Google Scholar]
- 65.Espéli O, Boccard F. 1997. In vivo cleavage of Escherichia coli BIME-2 repeats by DNA gyrase: genetic characterization of the target and identification of the cut site. Mol. Microbiol. 26:767–777. 10.1046/j.1365-2958.1997.6121983.x [DOI] [PubMed] [Google Scholar]
- 66.Gilson E, Perrin D, Hofnung M. 1990. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 18:3941–3952. 10.1093/nar/18.13.3941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macvanin M, Edgar R, Cui F, Trostel A, Zhurkin V, Adhya S. 2012. Noncoding RNAs binding to the nucleoid protein HU in Escherichia coli. J. Bacteriol. 194:6046–6055. 10.1128/JB.00961-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Magnusson M, Tobes R, Sancho J, Pareja E. 2007. Natural DNA repetitive extragenic sequences from gram-negative pathogens strongly stimulate TLR9. J. Immunol. 179:31–35 http://www.jimmunol.org/content/179/1/31.long [DOI] [PubMed] [Google Scholar]
- 69.Bachellier S, Clément JM, Hofnung M, Gilson E. 1997. Bacterial interspersed mosaic elements (BIMEs) are a major source of sequence polymorphism in Escherichia coli intergenic regions including specific associations with a new insertion sequence. Genetics 145:551–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nunvar J, Huckova T, Licha I. 2010. Identification and characterization of repetitive extragenic palindromes (REP)-associated tyrosine transposases: implications for REP evolution and dynamics in bacterial genomes. BMC Genomics 11:44. 10.1186/1471-2164-11-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ton-Hoang B, Siguier P, Quentin Y, Onillon S, Marty B, Fichant G, Chandler M. 2012. Structuring the bacterial genome: Y1-transposases associated with REP-BIME sequences. Nucleic Acids Res. 40:3596–3609. 10.1093/nar/gkr1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Clément JM, Wilde C, Bachellier S, Lambert P, Hofnung M. 1999. IS1397 is active for transposition into the chromosome of Escherichia coli K-12 and inserts specifically into palindromic units of bacterial interspersed mosaic elements. J. Bacteriol. 181:6929–6936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilde C, Bachellier S, Hofnung M, Clément JM. 2001. Transposition of IS1397 in the family Enterobacteriaceae and first characterization of ISKpnI, a new insertion sequence associated with Klebsiella pneumoniae palindromic units. J. Bacteriol. 183:4395–4404. 10.1128/JB.183.15.4395-4404.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilde C, Escartin F, Kokeguchi S, Latour-Lambert P, Lectard A, Clément JM. 2003. Transposases are responsible for the target specificity of IS1397 and ISKpnI for two different types of palindromic units (PUs). Nucleic Acids Res. 31:4345–4353. 10.1093/nar/gkg494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tobes R, Pareja E. 2006. Bacterial repetitive extragenic palindromic sequences are DNA targets for insertion sequence elements. BMC Genomics 7:62. 10.1186/1471-2164-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shyamala V, Schneider E, Ames GF. 1990. Tandem chromosomal duplications: role of REP sequences in the recombination event at the join-point. EMBO J. 9:939–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reams AB, Kofoid E, Kugelberg E, Roth JR. 2012. Multiple pathways of duplication formation with and without recombination (RecA) in Salmonella enterica. Genetics 192:397–415. 10.1534/genetics.112.142570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reznikoff WS. 2008. Transposon Tn5. Annu. Rev. Genet. 42:269–286. 10.1146/annurev.genet.42.110807.091656 [DOI] [PubMed] [Google Scholar]
- 79.Salyers AA, Shoemaker NB, Stevens AM, Li LY. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scott JR, Churchward GG. 1995. Conjugative transposition. Annu. Rev. Microbiol. 49:367–397. 10.1146/annurev.mi.49.100195.002055 [DOI] [PubMed] [Google Scholar]
- 81.Pembroke JT, MacMahon C, McGrath B. 2002. The role of conjugative transposons in the Enterobacteriaceae. Cell. Mol. Life Sci. 59:2055–2064. 10.1007/s000180200005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- 83.Adams V, Lyras D, Farrow KA, Rood JI. 2002. The clostridial mobilisable transposons. Cell. Mol. Life Sci. 59:2033–2043. 10.1007/s000180200003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee CA, Thomas J, Grossman AD. 2012. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J. Bacteriol. 194:3165–3172. 10.1128/JB.00301-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang H, Smith MC, Mullany P. 2006. The conjugative transposon Tn5397 has a strong preference for integration into its Clostridium difficile target site. J. Bacteriol. 188:4871–4878. 10.1128/JB.00210-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taylor AL. 1963. Bacteriophage-induced mutation in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 50:1043–1051. 10.1073/pnas.50.6.1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bukhari AI. 1976. Bacteriophage Mu as a transposition element. Annu. Rev. Genet. 10:389–412. 10.1146/annurev.ge.10.120176.002133 [DOI] [PubMed] [Google Scholar]
- 88.Kleckner N. 1981. Transposable elements in prokaryotes. Annu. Rev. Genet. 15:341–404. 10.1146/annurev.ge.15.120181.002013 [DOI] [PubMed] [Google Scholar]
- 89.Daniell E, Abelson J. 1973. Lac messenger RNA in lacZ gene mutants of Escherichia coli caused by insertion of bacteriophage Mu. J. Mol. Biol. 76:319–322. 10.1016/0022-2836(73)90395-1 [DOI] [PubMed] [Google Scholar]
- 90.Faelen M, Toussaint A, De Lafonteyne J. 1975. Model for the enhancement of lambda-gal integration into partially induced Mu-1 lysogens. J. Bacteriol. 121:873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cameron RK, Ulycznyj PI, DuBow MS. 1995. Mu transposase-stimulated illegitimate recombination of Tn3kan- and IS101-containing plasmids. Res. Microbiol. 146:601–616. 10.1016/0923-2508(96)81059-X [DOI] [PubMed] [Google Scholar]
- 92.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089–1097. 10.1046/j.1365-2958.1997.3101672.x [DOI] [PubMed] [Google Scholar]
- 93.Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641–679. 10.1146/annurev.micro.54.1.641 [DOI] [PubMed] [Google Scholar]
- 94.Hacker J, Carniel E. 2001. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2:376–381. 10.1093/embo-reports/kve097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hentschel U, Hacker J. 2001. Pathogenicity islands: the tip of the iceberg. Microbes Infect. 3:545–548. 10.1016/S1286-4579(01)01410-1 [DOI] [PubMed] [Google Scholar]
- 96.Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414–424. 10.1038/nrmicro884 [DOI] [PubMed] [Google Scholar]
- 97.Schmidt H, Hensel M. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14–56. 10.1128/CMR.17.1.14-56.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gal-Mor O, Finlay BB. 2006. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell. Microbiol. 8:1707–1719. 10.1111/j.1462-5822.2006.00794.x [DOI] [PubMed] [Google Scholar]
- 99.Manson JM, Gilmore MS. 2006. Pathogenicity island integrase cross-talk: a potential new tool for virulence modulation. Mol. Microbiol. 61:555–559. 10.1111/j.1365-2958.2006.05262.x [DOI] [PubMed] [Google Scholar]
- 100.Juhas M, van der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33:376–393. 10.1111/j.1574-6976.2008.00136.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Novick RP, Christie GE, Penadés JR. 2010. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 8:541–551. 10.1038/nrmicro2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dutta C, Paul S. 2012. Microbial lifestyle and genome signatures. Curr. Genomics 13:153–162. 10.2174/138920212799860698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Daccord A, Ceccarelli D, Rodrigue S, Burrus V. 2013. Comparative analysis of mobilizable genomic islands. J. Bacteriol. 195:606–614. 10.1128/JB.01985-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Que F, Wu S, Huang R. 2013. Salmonella pathogenicity island 1 (SPI-1) at work. Curr. Microbiol. 66:582–587. 10.1007/s00284-013-0307-8 [DOI] [PubMed] [Google Scholar]
- 105.Mates AK, Sayed AK, Foster JW. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J. Bacteriol. 189:2759–2768. 10.1128/JB.01490-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lima WC, Paquola AC, Varani AM, Van Sluys MA, Menck CF. 2008. Laterally transferred genomic islands in Xanthomonadales related to pathogenicity and primary metabolism. FEMS Microbiol. Lett. 281:87–97. 10.1111/j.1574-6968.2008.01083.x [DOI] [PubMed] [Google Scholar]
- 107.Sung JY, Koo SH, Cho HH, Kwon KC. 2012. AbaR7, a genomic resistance island found in multidrug-resistant Acinetobacter baumannii isolates in Daejeon, Korea. Ann. Lab. Med. 32:324–330. 10.3343/alm.2012.32.5.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, Elliot RM, Fleetwood DJ, McCallum NG, Rossbach U, Stuart GS, Weaver JE, Webby RJ, De Bruijn FJ, Ronson CW. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086–3095. 10.1128/JB.184.11.3086-3095.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ravatn R, Studer S, Springael D, Zehnder AJ, van der Meer JR. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360–4369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Makarova KS, Wolf YI, Snir S, Koonin EV. 2011. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J. Bacteriol. 193:6039–6056. 10.1128/JB.05535-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Quiroz TS, Nieto PA, Tobar HE, Salazar-Echegarai FJ, Lizana RJ, Quezada CP, Santiviago CA, Araya DV, Riedel CA, Kalergis AM, Bueno SM. 2011. Excision of an unstable pathogenicity island in Salmonella enterica serovar Enteritidis is induced during infection of phagocytic cells. PLoS One 6:e26031. 10.1371/journal.pone.0026031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li M, Shen X, Yan J, Han H, Zheng B, Liu D, Cheng H, Zhao Y, Rao X, Wang C, Tang J, Hu F, Gao GF. 2011. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol. Microbiol. 79:1670–1683. 10.1111/j.1365-2958.2011.07553.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. 2004. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J. Bacteriol. 186:3086–3096. 10.1128/JB.186.10.3086-3096.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Merkl R. 2006. A comparative categorization of protein function encoded in bacterial or archeal genomic islands. J. Mol. Evol. 62:1–14. 10.1007/s00239-004-0311-5 [DOI] [PubMed] [Google Scholar]
- 115.Scott DR, Marcus EA, Wen Y, Oh J, Sachs G. 2007. Gene expression in vivo shows that Helicobacter pylori colonizes an acidic niche on the gastric surface. Proc. Natl. Acad. Sci. U. S. A. 104:7235–7240. 10.1073/pnas.0702300104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Njoroge JW, Gruber C, Sperandio V. 2013. The interacting Cra and KdpE regulators are involved in the expression of multiple virulence factors in enterohemorrhagic Escherichia coli. J. Bacteriol. 195:2499–2508. 10.1128/JB.02252-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wu DQ, Ye J, Ou HY, Wei X, Huang X, He YW, Xu Y. 2011. Genomic analysis and temperature-dependent transcriptome profiles of the rhizosphere originating strain Pseudomonas aeruginosa M18. BMC Genomics 12:438. 10.1186/1471-2164-12-438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choi J, Shin D, Ryu S. 2007. Implication of quorum sensing in Salmonella enterica serovar Typhimurium virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect. Immun. 75:4885–4890. 10.1128/IAI.01942-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Prouty AM, Gunn JS. 2000. Salmonella enterica serovar Typhimurium invasion is repressed in the presence of bile. Infect. Immun. 68:6763–6769. 10.1128/IAI.68.12.6763-6769.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ellermeier JR, Slauch JM. 2008. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J. Bacteriol. 190:476–486. 10.1128/JB.00926-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Maddox SM, Coburn PS, Shankar N, Conway T. 2012. Transcriptional regulator PerA influences biofilm-associated, platelet binding, and metabolic gene expression in Enterococcus faecalis. PLoS One 7:e34398. 10.1371/journal.pone.0034398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rice LB. 2002. Association of different mobile elements to generate novel integrative elements. Cell. Mol. Life Sci. 59:2023–2032. 10.1007/s000180200002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Parks AR, Peters JE. 2007. Transposon Tn7 is widespread in diverse bacteria and forms genomic islands. J. Bacteriol. 189:2170–2173. 10.1128/JB.01536-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Davis EO, Sedgwick SG, Colston MJ. 1991. Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J. Bacteriol. 173:5653–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davis EO, Jenner PJ, Brooks PC, Colston MJ, Sedgwick SG. 1992. Protein splicing in the maturation of M. tuberculosis recA protein: a mechanism for tolerating a novel class of intervening sequence. Cell 71:201–210. 10.1016/0092-8674(92)90349-H [DOI] [PubMed] [Google Scholar]
- 126.Cooper AA, Stevens TH. 1993. Protein splicing: excision of intervening sequences at the protein level. Bioessays 15:667–674. 10.1002/bies.950151006 [DOI] [PubMed] [Google Scholar]
- 127.Colston MJ, Davis EO. 1994. The ins and outs of protein splicing elements. Mol. Microbiol. 12:359–363. 10.1111/j.1365-2958.1994.tb01025.x [DOI] [PubMed] [Google Scholar]
- 128.Perler FB, Davis EO, Dean GE, Gimble FS, Jack WE, Neff N, Noren CJ, Thorner J, Belfort M. 1994. Protein splicing elements: inteins and exteins—a definition of terms and recommended nomenclature. Nucleic Acids Res. 22:1125–1127. 10.1093/nar/22.7.1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Belfort M, Reaban ME, Coetzee T, Dalgaard JZ. 1995. Prokaryotic introns and inteins: a panoply of form and function. J. Bacteriol. 177:3897–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu XQ. 2000. Protein-splicing intein: genetic mobility, origin, and evolution. Annu. Rev. Genet. 34:61–76. 10.1146/annurev.genet.34.1.61 [DOI] [PubMed] [Google Scholar]
- 131.Paulus H. 2000. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 69:447–496. 10.1146/annurev.biochem.69.1.447 [DOI] [PubMed] [Google Scholar]
- 132.Pietrokovski S. 2001. Intein spread and extinction in evolution. Trends Genet. 17:465–472. 10.1016/S0168-9525(01)02365-4 [DOI] [PubMed] [Google Scholar]
- 133.Gogarten JP, Senejani AG, Zhaxybayeva O, Olendzenski L, Hilario E. 2002. Inteins: structure, function, and evolution. Annu. Rev. Microbiol. 56:263–287. 10.1146/annurev.micro.56.012302.160741 [DOI] [PubMed] [Google Scholar]
- 134.Paulus H. 2003. Inteins as targets for potential antimycobacterial drugs. Front. Biosci. 8:s1157–s1165. 10.2741/1195 [DOI] [PubMed] [Google Scholar]
- 135.Raghavan R, Minnick MF. 2009. Group I introns and inteins: disparate origins but convergent parasitic strategies. J. Bacteriol. 191:6193–6202. 10.1128/JB.00675-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Barzel A, Naor A, Privman E, Kupiec M, Gophna U. 2011. Homing endonucleases residing within inteins: evolutionary puzzles awaiting genetic solutions. Biochem. Soc. Trans. 39:169–173. 10.1042/BST0390169 [DOI] [PubMed] [Google Scholar]
- 137.Wu H, Hu Z, Liu XQ. 1998. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U. S. A. 95:9226–9231. 10.1073/pnas.95.16.9226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Iwai H, Züger S, Jin J, Tam PH. 2006. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 580:1853–1858. 10.1016/j.febslet.2006.02.045 [DOI] [PubMed] [Google Scholar]
- 139.Gleason FK, Olszewski NE. 2002. Isolation of the gene for the B12-dependent ribonucleotide reductase from Anabaena sp. strain PCC 7120 and expression in Escherichia coli. J. Bacteriol. 184:6544–6550. 10.1128/JB.184.23.6544-6550.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44–79. 10.1128/MMBR.65.1.44-79.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fsihi H, Vincent V, Cole ST. 1996. Homing events in the gyrA gene of some mycobacteria. Proc. Natl. Acad. Sci. U. S. A. 93:3410–3415. 10.1073/pnas.93.8.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Liu XQ, Hu Z. 1997. Identification and characterization of a cyanobacterial DnaX intein. FEBS Lett. 408:311–314. 10.1016/S0014-5793(97)00393-1 [DOI] [PubMed] [Google Scholar]
- 143.Liu XQ, Hu Z. 1997. A DnaB intein in Rhodothermus marinus: indication of recent intein homing across remotely related organisms. Proc. Natl. Acad. Sci. U. S. A. 94:7851–7856. 10.1073/pnas.94.15.7851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Frischkorn K, Sander P, Scholz M, Teschner K, Prammananan T, Böttger EC. 1998. Investigation of mycobacterial recA function: protein introns in the RecA of pathogenic mycobacteria do not affect competency for homologous recombination. Mol. Microbiol. 29:1203–1214. 10.1046/j.1365-2958.1998.01003.x [DOI] [PubMed] [Google Scholar]
- 145.Amitai G, Belenkiy O, Dassa B, Shainskaya A, Pietrokovski S. 2003. Distribution and function of new bacterial intein-like protein domains. Mol. Microbiol. 47:61–73. 10.1046/j.1365-2958.2003.03283.x [DOI] [PubMed] [Google Scholar]
- 146.Dori-Bachash M, Dassa B, Peleg O, Pineiro SA, Jurkevitch E, Pietrokovski S. 2009. Bacterial intein-like domains of predatory bacteria: a new domain type characterized in Bdellovibrio bacteriovorus. Funct. Integr. Genomics 9:153–166. 10.1007/s10142-008-0106-7 [DOI] [PubMed] [Google Scholar]
- 147.Tourasse NJ, Kolstø AB. 2008. Survey of group I and group II introns in 29 sequenced genomes of the Bacillus cereus group: insights into their spread and evolution. Nucleic Acids Res. 36:4529–4548. 10.1093/nar/gkn372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Edgell DR, Chalamcharla VR, Belfort M. 2011. Learning to live together: mutualism between self-splicing introns and their hosts. BMC Biol. 9:22. 10.1186/1741-7007-9-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Haugen P, Simon DM, Bhattacharya D. 2005. The natural history of group I introns. Trends Genet. 21:111–119. 10.1016/j.tig.2004.12.007 [DOI] [PubMed] [Google Scholar]
- 150.Mueller JE, Smith D, Belfort M. 1996. Exon coconversion biases accompanying intron homing: battle of the nucleases. Genes Dev. 10:2158–2166. 10.1101/gad.10.17.2158 [DOI] [PubMed] [Google Scholar]
- 151.Everett KD, Kahane S, Bush RM, Friedman MG. 1999. An unspliced group I intron in 23S rRNA links Chlamydiales, chloroplasts, and mitochondria. J. Bacteriol. 181:4734–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Robart AR, Zimmerly S. 2005. Group II intron retroelements: function and diversity. Cytogenet. Genome Res. 110:589–597. 10.1159/000084992 [DOI] [PubMed] [Google Scholar]
- 153.Toro N, Jiménez-Zurdo JI, García-Rodríguez FM. 2007. Bacterial group II introns: not just splicing. FEMS Microbiol. Rev. 31:342–358. 10.1111/j.1574-6976.2007.00068.x [DOI] [PubMed] [Google Scholar]
- 154.Ferat JL, Michel F. 1993. Group II self-splicing introns in bacteria. Nature 364:358–361. 10.1038/364358a0 [DOI] [PubMed] [Google Scholar]
- 155.Dai L, Zimmerly S. 2002. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 30:1091–1102. 10.1093/nar/30.5.1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Robart AR, Montgomery NK, Smith KL, Zimmerly S. 2004. Principles of 3′ splice site selection and alternative splicing for an unusual group II intron from Bacillus anthracis. RNA 10:854–862. 10.1261/rna.5246804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Leclercq S, Giraud I, Cordaux R. 2011. Remarkable abundance and evolution of mobile group II introns in Wolbachia bacterial endosymbionts. Mol. Biol. Evol. 28:685–697. 10.1093/molbev/msq238 [DOI] [PubMed] [Google Scholar]
- 158.Belfort M. 1991. Self-splicing introns in prokaryotes: migrant fossils? Cell 64:9–11. 10.1016/0092-8674(91)90201-9 [DOI] [PubMed] [Google Scholar]
- 159.Belfort M, Roberts RJ. 1997. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 25:3379–3388. 10.1093/nar/25.17.3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jurica MS, Stoddard BL. 1999. Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci. 55:1304–1326. 10.1007/s000180050372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gimble FS. 2000. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett. 185:99–107. 10.1111/j.1574-6968.2000.tb09046.x [DOI] [PubMed] [Google Scholar]
- 162.Stoddard BL. 2005. Homing endonuclease structure and function. Q. Rev. Biophys. 38:49–95. 10.1017/S0033583505004063 [DOI] [PubMed] [Google Scholar]
- 163.Gogarten JP, Hilario E. 2006. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol. Biol. 6:94. 10.1186/1471-2148-6-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Barzel A, Obolski U, Gogarten JP, Kupiec M, Hadany L. 2011. Home and away—the evolutionary dynamics of homing endonucleases. BMC Evol. Biol. 11:324. 10.1186/1471-2148-11-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Hafez M, Hausner G. 2012. Homing endonucleases: DNA scissors on a mission. Genome 55:553–569. 10.1139/g2012-049 [DOI] [PubMed] [Google Scholar]
- 166.Taylor GK, Stoddard BL. 2012. Structural, functional and evolutionary relationships between homing endonucleases and proteins from their host organisms. Nucleic Acids Res. 40:5189–5200. 10.1093/nar/gks226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lampson BC, Inouye M, Inouye S. 2005. Retrons, msDNA, and the bacterial genome. Cytogenet. Genome Res. 110:491–499. 10.1159/000084982 [DOI] [PubMed] [Google Scholar]
- 168.Inouye S, Inouye M. 1995. Structure, function, and evolution of bacterial reverse transcriptase. Virus Genes 11:81–94. 10.1007/BF01728650 [DOI] [PubMed] [Google Scholar]
- 169.Travisano M, Inouye M. 1995. Retrons: retroelements of no known function. Trends Microbiol. 3:209–212. 10.1016/S0966-842X(00)88924-6 [DOI] [PubMed] [Google Scholar]
- 170.Lampson BC, Rice SA. 1997. Repetitive sequences found in the chromosome of the myxobacterium Nannocystis exedens are similar to msDNA: a possible retrotransposition event in bacteria. Mol. Microbiol. 23:813–823. 10.1046/j.1365-2958.1997.2671627.x [DOI] [PubMed] [Google Scholar]
- 171.Lampson BC, Xu C, Rice SA, Inouye S. 2002. A partial copy of msDNA from a new retron element is likely a retrotransposed DNA found in the myxobacterium Nannocystis exedens. Gene 299:251–261. 10.1016/S0378-1119(02)00977-0 [DOI] [PubMed] [Google Scholar]
- 172.Herzer PJ, Inouye S, Inouye M. 1992. Retron-Ec107 is inserted into the Escherichia coli genome by replacing a palindromic 34bp intergenic sequence. Mol. Microbiol. 6:345–354. 10.1111/j.1365-2958.1992.tb01477.x [DOI] [PubMed] [Google Scholar]
- 173.Lim D. 1995. Analysis of a retron EC86 and EC67 insertion site in Escherichia coli. Plasmid 34:58–61. 10.1006/plas.1995.1033 [DOI] [PubMed] [Google Scholar]
- 174.Dodd IB, Egan JB. 1996. The Escherichia coli retrons Ec67 and Ec86 replace DNA between the cos site and a transcription terminator of a 186-related prophage. Virology 219:115–124. 10.1006/viro.1996.0228 [DOI] [PubMed] [Google Scholar]
- 175.Maas WK, Wang C, Lima T, Zubay G, Lim D. 1994. Multicopy single-stranded DNAs with mismatched base pairs are mutagenic in Escherichia coli. Mol. Microbiol. 14:437–441. 10.1111/j.1365-2958.1994.tb02178.x [DOI] [PubMed] [Google Scholar]
- 176.Mao JR, Inouye S, Inouye M. 1996. Enhancement of frame-shift mutation by the overproduction of msDNA in Escherichia coli. FEMS Microbiol. Lett. 144:109–115. 10.1111/j.1574-6968.1996.tb08516.x [DOI] [PubMed] [Google Scholar]
- 177.Maas WK, Wang C, Lima T, Hach A, Lim D. 1996. Multicopy single-stranded DNA of Escherichia coli enhances mutation and recombination frequencies by titrating MutS protein. Mol. Microbiol. 19:505–509. 10.1046/j.1365-2958.1996.392921.x [DOI] [PubMed] [Google Scholar]
- 178.Doulatov S, Hodes A, Dai L, Mandhana N, Liu M, Deora R, Simons RW, Zimmerly S, Miller JF. 2004. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature 431:476–481. 10.1038/nature02833 [DOI] [PubMed] [Google Scholar]
- 179.Medhekar B, Miller JF. 2007. Diversity-generating retroelements. Curr. Opin. Microbiol. 10:388–395. 10.1016/j.mib.2007.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guo H, Tse LV, Barbalat R, Sivaamnuaiphorn S, Xu M, Doulatov S, Miller JF. 2008. Diversity-generating retroelement homing regenerates target sequences for repeated rounds of codon rewriting and protein diversification. Mol. Cell 31:813–823. 10.1016/j.molcel.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Guo H, Tse LV, Nieh AW, Czornyj E, Williams S, Oukil S, Liu VB, Miller JF. 2011. Target site recognition by a diversity-generating retroelement. PLoS Genet. 7:e1002414. 10.1371/journal.pgen.1002414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Schillinger T, Lisfi M, Chi J, Cullum J, Zingler N. 2012. Analysis of a comprehensive dataset of diversity generating retroelements generated by the program DiGReF. BMC Genomics 13:430. 10.1186/1471-2164-13-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Schillinger T, Zingler N. 2012. The low incidence of diversity-generating retroelements in sequenced genomes. Mob. Genet. Elements 2:287–291. 10.4161/mge.23244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Arambula D, Wong W, Medhekar BA, Guo H, Gingery M, Czornyj E, Liu M, Dey S, Ghosh P, Miller JF. 2013. Surface display of a massively variable lipoprotein by a Legionella diversity-generating retroelement. Proc. Natl. Acad. Sci. U. S. A. 110:8212–8217. 10.1073/pnas.1301366110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kojima KK, Kanehisa M. 2008. Systematic survey for novel types of prokaryotic retroelements based on gene neighborhood and protein architecture. Mol. Biol. Evol. 25:1395–1404. 10.1093/molbev/msn081 [DOI] [PubMed] [Google Scholar]
- 186.Simon DM, Zimmerly S. 2008. A diversity of uncharacterized reverse transcriptases in bacteria. Nucleic Acids Res. 36:7219–7229. 10.1093/nar/gkn867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mazel D. 2006. Integrons: agents of bacterial evolution. Nat. Rev. Microbiol. 4:608–620. 10.1038/nrmicro1462 [DOI] [PubMed] [Google Scholar]
- 188.Boucher Y, Labbate M, Koenig JE, Stokes HW. 2007. Integrons: mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 15:301–309. 10.1016/j.tim.2007.05.004 [DOI] [PubMed] [Google Scholar]
- 189.Cambray G, Guerout AM, Mazel D. 2010. Integrons. Annu. Rev. Genet. 44:141–166. 10.1146/annurev-genet-102209-163504 [DOI] [PubMed] [Google Scholar]
- 190.Domingues S, da Silva GJ, Nielsen KM. 2012. Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mob. Genet. Elements 2:211–223. 10.4161/mge.22967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Bouvier M, Demarre G, Mazel D. 2005. Integron cassette insertion: a recombination process involving a folded single strand substrate. EMBO J. 24:4356–4367. 10.1038/sj.emboj.7600898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Collis CM, Kim MJ, Stokes HW, Hall RM. 2002. Integron-encoded IntI integrases preferentially recognize the adjacent cognate attI site in recombination with a 59-be site. Mol. Microbiol. 46:1415–1427. 10.1046/j.1365-2958.2002.03260.x [DOI] [PubMed] [Google Scholar]
- 193.Loot C, Ducos-Galand M, Escudero JA, Bouvier M, Mazel D. 2012. Replicative resolution of integron cassette insertion. Nucleic Acids Res. 40:8361–8370. 10.1093/nar/gks620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Collis CM, Hall RM. 1992. Gene cassettes from the insert region of integrons are excised as covalently closed circles. Mol. Microbiol. 6:2875–2885. 10.1111/j.1365-2958.1992.tb01467.x [DOI] [PubMed] [Google Scholar]
- 195.Liebert CA, Hall RM, Summers AO. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63:507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Partridge SR, Tsafnat G, Coiera E, Iredell JR. 2009. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 33:757–784. 10.1111/j.1574-6976.2009.00175.x [DOI] [PubMed] [Google Scholar]
- 197.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, Gonzalez-Zorn B, Barbé J, Ploy MC, Mazel D. 2009. The SOS response controls integron recombination. Science 324:1034. 10.1126/science.1172914 [DOI] [PubMed] [Google Scholar]
- 198.Michael CA, Labbate M. 2010. Gene cassette transcription in a large integron-associated array. BMC Genet. 11:82. 10.1186/1471-2156-11-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Collis CM, Grammaticopoulos G, Briton J, Stokes HW, Hall RM. 1993. Site-specific insertion of gene cassettes into integrons. Mol. Microbiol. 9:41–52. 10.1111/j.1365-2958.1993.tb01667.x [DOI] [PubMed] [Google Scholar]
- 200.Collis CM, Hall RM. 1992. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J. Bacteriol. 174:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Collis CM, Recchia GD, Kim MJ, Stokes HW, Hall RM. 2001. Efficiency of recombination reactions catalyzed by class 1 integron integrase IntI1. J. Bacteriol. 183:2535–2542. 10.1128/JB.183.8.2535-2542.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Francia MV, de la Cruz F, Garcia Lobo JM. 1993. Secondary-sites for integration mediated by the Tn21 integrase. Mol. Microbiol. 10:823–828. 10.1111/j.1365-2958.1993.tb00952.x [DOI] [PubMed] [Google Scholar]
- 203.Recchia GD, Stokes HW, Hall RM. 1994. Characterisation of specific and secondary recombination sites recognised by the integron DNA integrase. Nucleic Acids Res. 22:2071–2078. 10.1093/nar/22.11.2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Recchia GD, Hall RM. 1995. Plasmid evolution by acquisition of mobile gene cassettes: plasmid pIE723 contains the aadB gene cassette precisely inserted at a secondary site in the IncQ plasmid RSF1010. Mol. Microbiol. 15:179–187. 10.1111/j.1365-2958.1995.tb02232.x [DOI] [PubMed] [Google Scholar]
- 205.Francia MV, Garcia Lobo JM. 1996. Gene integration in the Escherichia coli chromosome mediated by Tn21 integrase (Int21). J. Bacteriol. 178:894–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Doublet B, Praud K, Weill FX, Cloeckaert A. 2009. Association of IS26-composite transposons and complex In4-type integrons generates novel multidrug resistance loci in Salmonella genomic island 1. J. Antimicrob. Chemother. 63:282–289. 10.1093/jac/dkn500 [DOI] [PubMed] [Google Scholar]
- 207.Engelberg-Kulka H, Glaser G. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53:43–70. 10.1146/annurev.micro.53.1.43 [DOI] [PubMed] [Google Scholar]
- 208.Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499. 10.1126/science.1088157 [DOI] [PubMed] [Google Scholar]
- 209.Gerdes K, Christensen SK, Løbner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382. 10.1038/nrmicro1147 [DOI] [PubMed] [Google Scholar]
- 210.Pandey DP, Gerdes K. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966–976. 10.1093/nar/gki201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Condon C. 2006. Shutdown decay of mRNA. Mol. Microbiol. 61:573–583. 10.1111/j.1365-2958.2006.05270.x [DOI] [PubMed] [Google Scholar]
- 212.Gerdes K, Wagner EG. 2007. RNA antitoxins. Curr. Opin. Microbiol. 10:117–124. 10.1016/j.mib.2007.03.003 [DOI] [PubMed] [Google Scholar]
- 213.Magnuson RD. 2007. Hypothetical functions of toxin-antitoxin systems. J. Bacteriol. 189:6089–6092. 10.1128/JB.00958-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fozo EM, Hemm MR, Storz G. 2008. Small toxic proteins and the antisense RNAs that repress them. Microbiol. Mol. Biol. Rev. 72:579–589. 10.1128/MMBR.00025-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Makarova KS, Wolf YI, Koonin EV. 2009. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol. Direct 4:19. 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Van Melderen L, Saavedra De Bast M. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. 10.1371/journal.pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13:781–785. 10.1016/j.mib.2010.10.006 [DOI] [PubMed] [Google Scholar]
- 218.Blower TR, Salmond GP, Luisi BF. 2011. Balancing at survival's edge: the structure and adaptive benefits of prokaryotic toxin-antitoxin partners. Curr. Opin. Struct. Biol. 21:109–118. 10.1016/j.sbi.2010.10.009 [DOI] [PubMed] [Google Scholar]
- 219.Bukowski M, Rojowska A, Wladyka B. 2011. Prokaryotic toxin-antitoxin systems—the role in bacterial physiology and application in molecular biology. Acta Biochim. Pol. 58:1–9 http://www.actabp.pl/pdf/1_2011/1.pdf [PubMed] [Google Scholar]
- 220.Guglielmini J, Van Melderen L. 2011. Bacterial toxin-antitoxin systems: translation inhibitors everywhere. Mob. Genet. Elements 1:283–290. 10.4161/mge.18477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77:5577–5583. 10.1128/AEM.05068-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 45:61–79. 10.1146/annurev-genet-110410-132412 [DOI] [PubMed] [Google Scholar]
- 223.Schuster CF, Bertram R. 2013. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol. Lett. 340:73–85. 10.1111/1574-6968.12074 [DOI] [PubMed] [Google Scholar]
- 224.Tan Q, Awano N, Inouye M. 2011. YeeV is an Escherichia coli toxin that inhibits cell division by targeting the cytoskeleton proteins, FtsZ and MreB. Mol. Microbiol. 79:109–118. 10.1111/j.1365-2958.2010.07433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Guérout AM, Iqbal N, Mine N, Ducos-Galand M, Van Melderen L, Mazel D. 2013. Characterization of the phd-doc and ccd toxin-antitoxin cassettes from Vibrio superintegrons. J. Bacteriol. 195:2270–2283. 10.1128/JB.01389-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ramisetty BC, Natarajan B, Santhosh RS. mazEF-mediated programmed cell death in bacteria: “What is this?”. Crit. Rev. Microbiol. 2013 Jun 25; doi: 10.3109/1040841X.2013.804030. [DOI] [PubMed] [Google Scholar]
- 227.Robson J, McKenzie JL, Cursons R, Cook GM, Arcus VL. 2009. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 390:353–367. 10.1016/j.jmb.2009.05.006 [DOI] [PubMed] [Google Scholar]
- 228.Liu M, Zhang Y, Inouye M, Woychik NA. 2008. Bacterial addiction module toxin Doc inhibits translation elongation through its association with the 30S ribosomal subunit. Proc. Natl. Acad. Sci. U. S. A. 105:5885–5890. 10.1073/pnas.0711949105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Unoson C, Wagner EG. 2008. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 70:258–270. 10.1111/j.1365-2958.2008.06416.x [DOI] [PubMed] [Google Scholar]
- 230.Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. 2011. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 9:e1001033. 10.1371/journal.pbio.1001033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Blower TR, Fineran PC, Johnson MJ, Toth IK, Humphreys DP, Salmond GP. 2009. Mutagenesis and functional characterization of the RNA and protein components of the toxIN abortive infection and toxin-antitoxin locus of Erwinia. J. Bacteriol. 191:6029–6039. 10.1128/JB.00720-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106:894–899. 10.1073/pnas.0808832106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Saavedra De Bast M, Mine N, Van Melderen L. 2008. Chromosomal toxin-antitoxin systems may act as antiaddiction modules. J. Bacteriol. 190:4603–4609. 10.1128/JB.00357-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cooper TF, Heinemann JA. 2000. Postsegregational killing does not increase plasmid stability but acts to mediate the exclusion of competing plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:12643–12648. 10.1073/pnas.220077897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Christensen-Dalsgaard M, Gerdes K. 2006. Two higBA loci in the Vibrio cholerae superintegron encode mRNA cleaving enzymes and can stabilize plasmids. Mol. Microbiol. 62:397–411. 10.1111/j.1365-2958.2006.05385.x [DOI] [PubMed] [Google Scholar]
- 236.González Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. 2006. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022). J. Bacteriol. 188:305–316. 10.1128/JB.188.1.305-316.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Christensen SK, Pedersen K, Hansen FG, Gerdes K. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809–819. 10.1016/S0022-2836(03)00922-7 [DOI] [PubMed] [Google Scholar]
- 238.Yamaguchi Y, Inouye M. 2011. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 9:779–790. 10.1038/nrmicro2651 [DOI] [PubMed] [Google Scholar]
- 239.Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc. Natl. Acad. Sci. U. S. A. 107:12541–12546. 10.1073/pnas.1004333107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yamaguchi Y, Inouye M. 2009. mRNA interferases, sequence-specific endoribonucleases from the toxin-antitoxin systems. Prog. Mol. Biol. Transl. Sci. 85:467–500. 10.1016/S0079-6603(08)00812-X [DOI] [PubMed] [Google Scholar]
- 241.Kim Y, Wang X, Zhang XS, Grigoriu S, Page R, Peti W, Wood TK. 2010. Escherichia coli toxin/antitoxin pair MqsR/MqsA regulate toxin CspD. Environ. Microbiol. 12:1105–1121. 10.1111/j.1462-2920.2009.02147.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. 1999. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 9:649–656. 10.1016/S0959-437X(99)00026-X [DOI] [PubMed] [Google Scholar]
- 243.Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742–3756. 10.1093/nar/29.18.3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Blumenthal RM, Cheng X. 2002. Restriction-modification systems, p 177–225 In Streips UN, Yasbin RE. (ed), Modern microbial genetics, 2nd ed. Wiley-Liss, Inc., New York, NY [Google Scholar]
- 245.Kobayashi I. 2002. Life cycle of restriction-modification gene complexes, powers in genome evolution. Int. Congr. Ser. 1246:191–200. 10.1016/S0531-5131(02)01142-1 [DOI] [Google Scholar]
- 246.Tock MR, Dryden DT. 2005. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8:466–472. 10.1016/j.mib.2005.06.003 [DOI] [PubMed] [Google Scholar]
- 247.Furuta Y, Abe K, Kobayashi I. 2010. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 38:2428–2443. 10.1093/nar/gkp1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ishikawa K, Fukuda E, Kobayashi I. 2010. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 17:325–342. 10.1093/dnares/dsq027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Furuta Y, Kobayashi I. 2013. Restriction-modification systems as mobile epigenetic elements, p 85–103 In Roberts AP, Mullany P. (ed), Bacterial integrative mobile genetic elements. Landes Bioscience, Austin, TX [Google Scholar]
- 250.Vasu K, Nagaraja V. 2013. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 77:53–72. 10.1128/MMBR.00044-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Naito Y, Naito T, Kobayashi I. 1998. Selfish restriction modification genes: resistance of a resident R/M plasmid to displacement by an incompatible plasmid mediated by host killing. Biol. Chem. 379:429–436. 10.1515/bchm.1998.379.4-5.429 [DOI] [PubMed] [Google Scholar]
- 252.Nakayama Y, Kobayashi I. 1998. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. U. S. A. 95:6442–6447. 10.1073/pnas.95.11.6442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Gumulak-Smith J, Teachman A, Tu AH, Simecka JW, Lindsey JR, Dybvig K. 2001. Variations in the surface proteins and restriction enzyme systems of Mycoplasma pulmonis in the respiratory tract of infected rats. Mol. Microbiol. 40:1037–1044. 10.1046/j.1365-2958.2001.02464.x [DOI] [PubMed] [Google Scholar]
- 254.Glatman LI, Moroz AF, Yablokova MB, Rebentish BA, Kcholmina GV. 1980. A novel plasmid-mediated DNA restriction-modification system in E. coli. Plasmid 4:350–351. 10.1016/0147-619X(80)90072-4 [DOI] [PubMed] [Google Scholar]
- 255.Iida S, Meyer J, Bachï B, Stålhammar-Carlemalm M, Schrickel S, Bickle TA, Arber W. 1983. DNA restriction-modification genes of phage P1 and plasmid p15B. Structure and in vitro transcription. J. Mol. Biol. 165:1–18 [DOI] [PubMed] [Google Scholar]
- 256.Rochepeau P, Selinger LB, Hynes MF. 1997. Transposon-like structure of a new plasmid-encoded restriction-modification system in Rhizobium leguminosarum VF39SM. Mol. Gen. Genet. 256:387–396. 10.1007/s004380050582 [DOI] [PubMed] [Google Scholar]
- 257.Balado M, Lemos ML, Osorio CR. 2013. Integrating conjugative elements of the SXT/R391 family from fish-isolated Vibrios encode restriction-modification systems that confer resistance to bacteriophages. FEMS Microbiol. Ecol. 83:457–467. 10.1111/1574-6941.12007 [DOI] [PubMed] [Google Scholar]
- 258.Jermyn WS, Boyd EF. 2005. Molecular evolution of Vibrio pathogenicity island-2 (VPI-2): mosaic structure among Vibrio cholerae and Vibrio mimicus natural isolates. Microbiology 151:311–322. 10.1099/mic.0.27621-0 [DOI] [PubMed] [Google Scholar]
- 259.Brassard S, Paquet H, Roy PH. 1995. A transposon-like sequence adjacent to the AccI restriction-modification operon. Gene 157:69–72. 10.1016/0378-1119(94)00734-A [DOI] [PubMed] [Google Scholar]
- 260.Vaišvila R, Vilkaitis G, Janulaitis A. 1995. Identification of a gene encoding a DNA invertase-like enzyme adjacent to the PaeR7I restriction-modification system. Gene 157:81–84. 10.1016/0378-1119(94)00793-R [DOI] [PubMed] [Google Scholar]
- 261.Lee KF, Shaw PC, Picone SJ, Wilson GG, Lunnen KD. 1998. Sequence comparison of the EcoHK31I and EaeI restriction-modification systems suggests an intergenic transfer of genetic material. Biol. Chem. 379:437–441. 10.1515/bchm.1998.379.4-5.437 [DOI] [PubMed] [Google Scholar]
- 262.Chaturvedi D, Chakravorty M. 2003. Restriction-modification system in bacteriophage MB78. Biochem. Biophys. Res. Commun. 303:884–890. 10.1016/S0006-291X(03)00421-2 [DOI] [PubMed] [Google Scholar]
- 263.Lieb M. 1991. Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics 128:23–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Chinen A, Uchiyama I, Kobayashi I. 2000. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene 259:109–121. 10.1016/S0378-1119(00)00459-5 [DOI] [PubMed] [Google Scholar]
- 265.Sekizaki T, Otani Y, Osaki M, Takamatsu D, Shimoji Y. 2001. Evidence for horizontal transfer of SsuDAT1I restriction-modification genes to the Streptococcus suis genome. J. Bacteriol. 183:500–511. 10.1128/JB.183.2.500-511.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Nobusato A, Uchiyama I, Ohashi S, Kobayashi I. 2000. Insertion with long target duplication: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene 259:99–108. 10.1016/S0378-1119(00)00456-X [DOI] [PubMed] [Google Scholar]
- 267.Sadykov M, Asami Y, Niki H, Handa N, Itaya M, Tanokura M, Kobayashi I. 2003. Multiplication of a restriction-modification gene complex. Mol. Microbiol. 48:417–427. 10.1046/j.1365-2958.2003.03464.x [DOI] [PubMed] [Google Scholar]
- 268.Sorek R, Kunin V, Hugenholtz P. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6:181–186. 10.1038/nrmicro1793 [DOI] [PubMed] [Google Scholar]
- 269.van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. 2009. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem. Sci. 34:401–407. 10.1016/j.tibs.2009.05.002 [DOI] [PubMed] [Google Scholar]
- 270.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. 10.1016/j.cell.2009.01.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Deveau H, Garneau JE, Moineau S. 2010. CRISPR/Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64:475–493. 10.1146/annurev.micro.112408.134123 [DOI] [PubMed] [Google Scholar]
- 272.Horvath P, Barrangou R. 2010. CRISPR/Cas, the immune system of bacteria and archaea. Science 327:167–170. 10.1126/science.1179555 [DOI] [PubMed] [Google Scholar]
- 273.Karginov FV, Hannon GJ. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37:7–19. 10.1016/j.molcel.2009.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190. 10.1038/nrg2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Vale PF, Little TJ. 2010. CRISPR-mediated phage resistance and the ghost of coevolution past. Proc. Biol. Sci. 277:2097–2103. 10.1098/rspb.2010.0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, Mastop M, Wagner EG, Schnetz K, Van Der Oost J, Wagner R, Brouns SJ. 2010. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77:1380–1393. 10.1111/j.1365-2958.2010.07315.x [DOI] [PubMed] [Google Scholar]
- 277.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. 2011. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 392:277–289. 10.1515/BC.2011.042 [DOI] [PubMed] [Google Scholar]
- 278.Bhaya D, Davison M, Barrangou R. 2011. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45:273–297. 10.1146/annurev-genet-110410-132430 [DOI] [PubMed] [Google Scholar]
- 279.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Shah SA, Garrett RA. 2011. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res. Microbiol. 162:27–38. 10.1016/j.resmic.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 281.Terns MP, Terns RM. 2011. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 14:321–327. 10.1016/j.mib.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Fineran PC, Charpentier E. 2012. Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology 434:202–209. 10.1016/j.virol.2012.10.003 [DOI] [PubMed] [Google Scholar]
- 283.Richter C, Chang JT, Fineran PC. 2012. Function and regulation of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR associated (Cas) systems. Viruses 4:2291–2311. 10.3390/v4102291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Barrangou R. 2013. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 4:267–278. 10.1002/wrna.1159 [DOI] [PubMed] [Google Scholar]
- 285.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 286.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. 2012. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat. Commun. 3:945. 10.1038/ncomms1937 [DOI] [PubMed] [Google Scholar]
- 287.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. 2012. CRISPR interference directs strand specific spacer acquisition. PLoS One 7:e35888. 10.1371/journal.pone.0035888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. 2009. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 139:945–956. 10.1016/j.cell.2009.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, III, Graveley BR, Terns RM, Terns MP. 2012. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell 45:292–302. 10.1016/j.molcel.2011.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Andersson AF, Banfield JF. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047–1050. 10.1126/science.1157358 [DOI] [PubMed] [Google Scholar]
- 291.Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190:1390–1400. 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Peng X, Brügger K, Shen B, Chen L, She Q, Garrett RA. 2003. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J. Bacteriol. 185:2410–2417. 10.1128/JB.185.8.2410-2417.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493:429–432. 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Horvath P, Romero DA, Coûté-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190:1401–1412. 10.1128/JB.01415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571. 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26:335–340. 10.1016/j.tig.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sashital DG, Wiedenheft B, Doudna JA. 2012. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell 46:606–615. 10.1016/j.molcel.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 77:1367–1379. 10.1111/j.1365-2958.2010.07265.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Pul U, Wurm R, Arslan Z, Geissen R, Hofmann N, Wagner R. 2010. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol. Microbiol. 75:1495–1512. 10.1111/j.1365-2958.2010.07073.x [DOI] [PubMed] [Google Scholar]
- 300.Medina-Aparicio L, Rebollar-Flores JE, Gallego-Hernández AL, Vázquez A, Olvera L, Gutiérrez-Ríos RM, Calva E, Hernández-Lucas I. 2011. The CRISPR/Cas immune system is an operon regulated by LeuO, H-NS, and leucine-responsive regulatory protein in Salmonella enterica serovar Typhi. J. Bacteriol. 193:2396–2407. 10.1128/JB.01480-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Shinkai A, Kira S, Nakagawa N, Kashihara A, Kuramitsu S, Yokoyama S. 2007. Transcription activation mediated by a cyclic AMP receptor protein from Thermus thermophilus HB8. J. Bacteriol. 189:3891–3901. 10.1128/JB.01739-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Agari Y, Sakamoto K, Tamakoshi M, Oshima T, Kuramitsu S, Shinkai A. 2010. Transcription profile of Thermus thermophilus CRISPR systems after phage infection. J. Mol. Biol. 395:270–281. 10.1016/j.jmb.2009.10.057 [DOI] [PubMed] [Google Scholar]
- 303.Chattoraj P, Banerjee A, Biswas S, Biswas I. 2010. ClpP of Streptococcus mutans differentially regulates expression of genomic islands, mutacin production, and antibiotic tolerance. J. Bacteriol. 192:1312–1323. 10.1128/JB.01350-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, DeLisa MP. 2011. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 79:584–599. 10.1111/j.1365-2958.2010.07482.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Koonin EV, Makarova KS. 2009. CRISPR-Cas: an adaptive immunity system in prokaryotes. F1000 Biol. Rep. 1:95 http://f1000.com/prime/reports/b/1/95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Takeuchi N, Wolf YI, Makarova KS, Koonin EV. 2012. Nature and intensity of selection pressure on CRISPR-associated genes. J. Bacteriol. 194:1216–1225. 10.1128/JB.06521-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Touchon M, Rocha EP. 2010. The small, slow and specialized CRISPR and anti-CRISPR of Escherichia and Salmonella. PLoS One 5:e11126. 10.1371/journal.pone.0011126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Touchon M, Charpentier S, Clermont O, Rocha EP, Denamur E, Branger C. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J. Bacteriol. 193:2460–2467. 10.1128/JB.01307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Aklujkar M, Lovley DR. 2010. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol. Biol. 10:230. 10.1186/1471-2148-10-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210–219. 10.1128/JB.00797-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J. Bacteriol. 189:3738–3750. 10.1128/JB.00187-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Babu M, Beloglazova N, Flick R, Graham C, Skarina T, Nocek B, Gagarinova A, Pogoutse O, Brown G, Binkowski A, Phanse S, Joachimiak A, Koonin EV, Savchenko A, Emili A, Greenblatt J, Edwards AM, Yakunin AF. 2011. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol. Microbiol. 79:484–502. 10.1111/j.1365-2958.2010.07465.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.DeBoy RT, Mongodin EF, Emerson JB, Nelson KE. 2006. Chromosome evolution in the Thermotogales: large-scale inversions and strain diversification of CRISPR sequences. J. Bacteriol. 188:2364–2374. 10.1128/JB.188.7.2364-2374.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Mokrousov I, Limeschenko E, Vyazovaya A, Narvskaya O. 2007. Corynebacterium diphtheriae spoligotyping based on combined use of two CRISPR loci. Biotechnol. J. 2:901–906. 10.1002/biot.200700035 [DOI] [PubMed] [Google Scholar]
- 316.Vergnaud G, Li Y, Gorgé O, Cui Y, Song Y, Zhou D, Grissa I, Dentovskaya SV, Platonov ME, Rakin A, Balakhonov SV, Neubauer H, Pourcel C, Anisimov AP, Yang R. 2007. Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA. Adv. Exp. Med. Biol. 603:327–338. 10.1007/978-0-387-72124-8_30 [DOI] [PubMed] [Google Scholar]
- 317.Cui Y, Li Y, Gorgé O, Platonov ME, Yan Y, Guo Z, Pourcel C, Dentovskaya SV, Balakhonov SV, Wang X, Song Y, Anisimov AP, Vergnaud G, Yang R. 2008. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One 3:e2652. 10.1371/journal.pone.0002652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Ginevra C, Jacotin N, Diancourt L, Guigon G, Arquilliere R, Meugnier H, Descours G, Vandenesch F, Etienne J, Lina G, Caro V, Jarraud S. 2012. Legionella pneumophila sequence type 1/Paris pulsotype subtyping by spoligotyping. J. Clin. Microbiol. 50:696–701. 10.1128/JCM.06180-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Sheen P, Couvin D, Grandjean L, Zimic M, Dominguez M, Luna G, Gilman RH, Rastogi N, Moore DA. 2013. Genetic diversity of Mycobacterium tuberculosis in Peru and exploration of phylogenetic associations with drug resistance. PLoS One 8:e65873. 10.1371/journal.pone.0065873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31:233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Godde JS, Bickerton A. 2006. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 62:718–729. 10.1007/s00239-005-0223-z [DOI] [PubMed] [Google Scholar]
- 324.Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int. J. Food Microbiol. 131:62–70. 10.1016/j.ijfoodmicro.2008.05.030 [DOI] [PubMed] [Google Scholar]
- 325.Roth JR, Benson N, Galitski T, Haack K, Lawrence JG, Miesel L. 1996. Rearrangements of the bacterial chromosome: formation and applications, p 2256–2276 In Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 2 ASM Press, Washington, DC [Google Scholar]
- 326.Michel B, Boubakri H, Baharoglu Z, LeMasson M, Lestini R. 2007. Recombination proteins and rescue of arrested replication forks. DNA Repair (Amst.) 6:967–980. 10.1016/j.dnarep.2007.02.016 [DOI] [PubMed] [Google Scholar]
- 327.Dillingham MS, Kowalczykowski SC. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72:642–671. 10.1128/MMBR.00020-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Michel B, Leach D. 11 September 2012. Chapter 7.2.7. Homologous recombination—enzymes and pathways. In Curtiss R, III, Kaper JB, Squires CL, Karp PD, Neidhardt FC, Slauch JM. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. 10.1128/ecosalplus.7.2.7 [DOI] [Google Scholar]
- 329.Radding C. 1988. Homologous pairing and strand exchange promoted by Escherichia coli RecA protein, p 193–229 In Kucherlapati R, Smith GR. (ed), Genetic recombination. American Society for Microbiology, Washington, DC [Google Scholar]
- 330.Chen Z, Yang H, Pavletich NP. 2008. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453:489–494. 10.1038/nature06971 [DOI] [PubMed] [Google Scholar]
- 331.Forget AL, Kowalczykowski SC. 2012. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 482:423–427. 10.1038/nature10782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ehrlich SD, Bierne H, d'Alencon E, Vilette D, Petranovic M, Noirot P, Michel B. 1993. Mechanisms of illegitimate recombination. Gene 135:161–166. 10.1016/0378-1119(93)90061-7 [DOI] [PubMed] [Google Scholar]
- 333.Rocha EP. 2003. An appraisal of the potential for illegitimate recombination in bacterial genomes and its consequences: from duplications to genome reduction. Genome Res. 13:1123–1132. 10.1101/gr.966203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ikeda H, Shiraishi K, Ogata Y. 2004. Illegitimate recombination mediated by double-strand break and end-joining in Escherichia coli. Adv. Biophys. 38:3–20. 10.1016/S0065-227X(04)80031-5 [DOI] [PubMed] [Google Scholar]
- 335.Lovett ST. 2004. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 52:1243–1253. 10.1111/j.1365-2958.2004.04076.x [DOI] [PubMed] [Google Scholar]
- 336.Michel B, Ehrlich SD. 1986. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 5:3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Michel B, Ehrlich SD. 1986. Illegitimate recombination at the replication origin of bacteriophage M13. Proc. Natl. Acad. Sci. U. S. A. 83:3386–3390. 10.1073/pnas.83.10.3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Shapiro JA, Leach D. 1990. Action of a transposable element in coding sequence fusions. Genetics 126:293–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ikeda H, Moriya K, Matsumoto T. 1981. In vitro study of illegitimate recombination: involvement of DNA gyrase. Cold Spring Harb. Symp. Quant. Biol. 45(Part 1):399–408 [DOI] [PubMed] [Google Scholar]
- 340.Shuman S. 1989. Vaccinia DNA topoisomerase I promotes illegitimate recombination in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:3489–3493. 10.1073/pnas.86.10.3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Ashizawa Y, Yokochi T, Ogata Y, Shobuike Y, Kato J, Ikeda H. 1999. Mechanism of DNA gyrase-mediated illegitimate recombination: characterization of Escherichia coli gyrA mutations that confer hyper-recombination phenotype. J. Mol. Biol. 289:447–458. 10.1006/jmbi.1999.2758 [DOI] [PubMed] [Google Scholar]
- 342.Yi TM, Stearns D, Demple B. 1988. Illegitimate recombination in an Escherichia coli plasmid: modulation by DNA damage and a new bacterial gene. J. Bacteriol. 170:2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Hanada K, Yamashita T, Shobuike Y, Ikeda H. 2001. Role of DnaB helicase in UV-induced illegitimate recombination in Escherichia coli. J. Bacteriol. 183:4964–4969. 10.1128/JB.183.17.4964-4969.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Shanado Y, Hanada K, Ikeda H. 2001. Suppression of gamma ray-induced illegitimate recombination in Escherichia coli by the DNA-binding protein H-NS. Mol. Genet. Genomics 265:242–248. 10.1007/s004380000399 [DOI] [PubMed] [Google Scholar]
- 345.Conley EC, Saunders VA, Jackson V, Saunders JR. 1986. Mechanism of intramolecular recyclization and deletion formation following transformation of Escherichia coli with linearized plasmid DNA. Nucleic Acids Res. 14:8919–8932. 10.1093/nar/14.22.8919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Chayot R, Montagne B, Mazel D, Ricchetti M. 2010. An end-joining repair mechanism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 107:2141–2146. 10.1073/pnas.0906355107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Heale SM, Petes TD. 1995. The stabilization of repetitive tracts of DNA by variant repeats requires a functional DNA mismatch repair system. Cell 83:539–545. 10.1016/0092-8674(95)90093-4 [DOI] [PubMed] [Google Scholar]
- 348.Saveson CJ, Lovett ST. 1997. Enhanced deletion formation by aberrant DNA replication in Escherichia coli. Genetics 146:457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d'Adda di Fagagna F, Devine KM, Bowater RP, Jeggo PA, Jackson SP, Doherty AJ. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686–1689. 10.1126/science.1074584 [DOI] [PubMed] [Google Scholar]
- 350.Pitcher RS, Brissett NC, Doherty AJ. 2007. Nonhomologous end-joining in bacteria: a microbial perspective. Annu. Rev. Microbiol. 61:259–282. 10.1146/annurev.micro.61.080706.093354 [DOI] [PubMed] [Google Scholar]
- 351.Shuman S, Glickman MS. 2007. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 5:852–861. 10.1038/nrmicro1768 [DOI] [PubMed] [Google Scholar]
- 352.Ramsden DA. 2011. Polymerases in nonhomologous end joining: building a bridge over broken chromosomes. Antioxid. Redox Signal. 14:2509–2519. 10.1089/ars.2010.3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.de Vega M. 2013. The minimal Bacillus subtilis nonhomologous end joining repair machinery. PLoS One 8:e64232. 10.1371/journal.pone.0064232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Santoyo G, Romero D. 2005. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol. Rev. 29:169–183. 10.1016/j.fmrre.2004.10.004 [DOI] [PubMed] [Google Scholar]
- 355.Palmer GH, Bankhead T, Lukehart SA. 2009. ‘Nothing is permanent but change'—antigenic variation in persistent bacterial pathogens. Cell. Microbiol. 11:1697–1705. 10.1111/j.1462-5822.2009.01366.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Davidsen T, Tønjum T. 2006. Meningococcal genome dynamics. Nat. Rev. Microbiol. 4:11–22. 10.1038/nrmicro1324 [DOI] [PubMed] [Google Scholar]
- 357.Cahoon LA, Seifert HS. 2011. Focusing homologous recombination: pilin antigenic variation in the pathogenic Neisseria. Mol. Microbiol. 81:1136–1143. 10.1111/j.1365-2958.2011.07773.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Morris RT, Drouin G. 2007. Ectopic gene conversions in bacterial genomes. Genome 50:975–984. 10.1139/G07-076 [DOI] [PubMed] [Google Scholar]
- 359.Mehr IJ, Seifert HS. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697–710. 10.1046/j.1365-2958.1998.01089.x [DOI] [PubMed] [Google Scholar]
- 360.Cahoon LA, Seifert HS. 2009. An alternative DNA structure is necessary for pilin antigenic variation in Neisseria gonorrhoeae. Science 325:764–767. 10.1126/science.1175653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Anderson RP, Roth JR. 1977. Tandem genetic duplications in phage and bacteria. Annu. Rev. Microbiol. 31:473–505. 10.1146/annurev.mi.31.100177.002353 [DOI] [PubMed] [Google Scholar]
- 362.Andersson DI, Hughes D. 2009. Gene amplification and adaptive evolution in bacteria. Annu. Rev. Genet. 43:167–195. 10.1146/annurev-genet-102108-134805 [DOI] [PubMed] [Google Scholar]
- 363.Ives CL, Bott KF. 1990. Characterization of chromosomal DNA amplifications with associated tetracycline resistance in Bacillus subtilis. J. Bacteriol. 172:4936–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Zahra R, Blackwood JK, Sales J, Leach DR. 2007. Proofreading and secondary structure processing determine the orientation dependence of CAG × CTG trinucleotide repeat instability in Escherichia coli. Genetics 176:27–41. 10.1534/genetics.106.069724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Bi X, Liu LF. 1996. A replicational model for DNA recombination between direct repeats. J. Mol. Biol. 256:849–858. 10.1006/jmbi.1996.0131 [DOI] [PubMed] [Google Scholar]
- 366.Bichara M, Wagner J, Lambert IB. 2006. Mechanisms of tandem repeat instability in bacteria. Mutat. Res. 598:144–163. 10.1016/j.mrfmmm.2006.01.020 [DOI] [PubMed] [Google Scholar]
- 367.Blackwood JK, Okely EA, Zahra R, Eykelenboom JK, Leach DR. 2010. DNA tandem repeat instability in the Escherichia coli chromosome is stimulated by mismatch repair at an adjacent CAG.CTG trinucleotide repeat. Proc. Natl. Acad. Sci. U. S. A. 107:22582–22586. 10.1073/pnas.1012906108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Peterson BC, Rownd RH. 1985. Recombination sites in plasmid drug resistance gene amplification. J. Bacteriol. 164:1359–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Kang S, Jaworski A, Ohshima K, Wells RD. 1995. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat. Genet. 10:213–218. 10.1038/ng0695-213 [DOI] [PubMed] [Google Scholar]
- 370.Iyer RR, Pluciennik A, Rosche WA, Sinden RR, Wells RD. 2000. DNA polymerase III proofreading mutants enhance the expansion and deletion of triplet repeat sequences in Escherichia coli. J. Biol. Chem. 275:2174–2184. 10.1074/jbc.275.3.2174 [DOI] [PubMed] [Google Scholar]
- 371.Moxon R, Bayliss C, Hood D. 2006. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 40:307–333. 10.1146/annurev.genet.40.110405.090442 [DOI] [PubMed] [Google Scholar]
- 372.Bayliss CD, Field D, Moxon ER. 2001. The simple sequence contingency loci of Haemophilus influenzae and Neisseria meningitidis. J. Clin. Invest. 107:657–662. 10.1172/JCI12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.van Ham SM, van Alphen L, Mooi FR, van Putten JP. 1993. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell 73:1187–1196. 10.1016/0092-8674(93)90647-9 [DOI] [PubMed] [Google Scholar]
- 374.Stern A, Brown M, Nickel P, Meyer TF. 1986. Opacity genes in Neisseria gonorrhoeae: control of phase and antigenic variation. Cell 47:61–71. 10.1016/0092-8674(86)90366-1 [DOI] [PubMed] [Google Scholar]
- 375.Huffman GA, Rownd RH. 1984. Transition of deletion mutants of the composite resistance plasmid NR1 in Escherichia coli and Salmonella typhimurium. J. Bacteriol. 159:488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Tlsty TD, Albertini AM, Miller JH. 1984. Gene amplification in the lac region of E. coli. Cell 37:217–224. 10.1016/0092-8674(84)90317-9 [DOI] [PubMed] [Google Scholar]
- 377.Foster PL. 1999. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu. Rev. Genet. 33:57–88. 10.1146/annurev.genet.33.1.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Rosenberg SM, Hastings PJ. 2004. Adaptive point mutation and adaptive amplification pathways in the Escherichia coli Lac system: stress responses producing genetic change. J. Bacteriol. 186:4838–4843. 10.1128/JB.186.15.4838-4843.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Quinones-Soto S, Roth JR. 2011. Effect of growth under selection on appearance of chromosomal mutations in Salmonella enterica. Genetics 189:37–53. 10.1534/genetics.111.130187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Trinh TQ, Sinden RR. 1991. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 352:544–547. 10.1038/352544a0 [DOI] [PubMed] [Google Scholar]
- 381.Pinder DJ, Blake CE, Lindsey JC, Leach DR. 1998. Replication strand preference for deletions associated with DNA palindromes. Mol. Microbiol. 28:719–727 [DOI] [PubMed] [Google Scholar]
- 382.Leach DR, Okely EA, Pinder DJ. 1997. Repair by recombination of DNA containing a palindromic sequence. Mol. Microbiol. 26:597–606. 10.1046/j.1365-2958.1997.6071957.x [DOI] [PubMed] [Google Scholar]
- 383.Eykelenboom JK, Blackwood JK, Okely E, Leach DR. 2008. SbcCD causes a double-strand break at a DNA palindrome in the Escherichia coli chromosome. Mol. Cell 29:644–651. 10.1016/j.molcel.2007.12.020 [DOI] [PubMed] [Google Scholar]
- 384.Bzymek M, Lovett ST. 2001. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics 158:527–540 http://www.genetics.org/content/158/2/527.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Pinder DJ, Blake CE, Leach DR. 1997. DIR: a novel DNA rearrangement associated with inverted repeats. Nucleic Acids Res. 25:523–529. 10.1093/nar/25.3.523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Brewer BJ, Payen C, Raghuraman MK, Dunham MJ. 2011. Origin-dependent inverted-repeat amplification: a replication-based model for generating palindromic amplicons. PLoS Genet. 7:e1002016. 10.1371/journal.pgen.1002016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Darmon E, Eykelenboom JK, Lincker F, Jones LH, White M, Okely E, Blackwood JK, Leach DR. 2010. E. coli SbcCD and RecA control chromosomal rearrangement induced by an interrupted palindrome. Mol. Cell 39:59–70. 10.1016/j.molcel.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Leach DR. 1994. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16:893–900. 10.1002/bies.950161207 [DOI] [PubMed] [Google Scholar]
- 389.Harms K, Wackernagel W. 2008. The RecBCD and SbcCD DNases suppress homology-facilitated illegitimate recombination during natural transformation of Acinetobacter baylyi. Microbiology 154:2437–2445. 10.1099/mic.0.2008/018382-0 [DOI] [PubMed] [Google Scholar]
- 390.Slupska MM, Chiang JH, Luther WM, Stewart JL, Amii L, Conrad A, Miller JH. 2000. Genes involved in the determination of the rate of inversions at short inverted repeats. Genes Cells 5:425–437. 10.1046/j.1365-2443.2000.00341.x [DOI] [PubMed] [Google Scholar]
- 391.Dybvig K. 1993. DNA rearrangements and phenotypic switching in prokaryotes. Mol. Microbiol. 10:465–471. 10.1111/j.1365-2958.1993.tb00919.x [DOI] [PubMed] [Google Scholar]
- 392.Henderson IR, Owen P, Nataro JP. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919–932. 10.1046/j.1365-2958.1999.01555.x [DOI] [PubMed] [Google Scholar]
- 393.van der Woude MW, Bäumler AJ. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17:581–611. 10.1128/CMR.17.3.581-611.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Grindley ND, Whiteson KL, Rice PA. 2006. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 75:567–605. 10.1146/annurev.biochem.73.011303.073908 [DOI] [PubMed] [Google Scholar]
- 395.Wisniewski-Dyé F, Vial L. 2008. Phase and antigenic variation mediated by genome modifications. Antonie Van Leeuwenhoek 94:493–515. 10.1007/s10482-008-9267-6 [DOI] [PubMed] [Google Scholar]
- 396.Emerson JE, Reynolds CB, Fagan RP, Shaw HA, Goulding D, Fairweather NF. 2009. A novel genetic switch controls phase variable expression of CwpV, a Clostridium difficile cell wall protein. Mol. Microbiol. 74:541–556. 10.1111/j.1365-2958.2009.06812.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Abraham JM, Freitag CS, Clements JR, Eisenstein BI. 1985. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 82:5724–5727. 10.1073/pnas.82.17.5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. U. S. A. 101:14919–14924. 10.1073/pnas.0404172101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Dworkin J, Blaser MJ. 1997. Nested DNA inversion as a paradigm of programmed gene rearrangement. Proc. Natl. Acad. Sci. U. S. A. 94:985–990. 10.1073/pnas.94.3.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Zhang XL, Morris C, Hackett J. 1997. Molecular cloning, nucleotide sequence, and function of a site-specific recombinase encoded in the major ‘pathogenicity island' of Salmonella typhi. Gene 202:139–146. 10.1016/S0378-1119(97)00466-6 [DOI] [PubMed] [Google Scholar]
- 401.Cerdeño-Tárraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463–1465. 10.1126/science.1107008 [DOI] [PubMed] [Google Scholar]
- 402.Yamamoto S, Kutsukake K. 2006. FljA-mediated posttranscriptional control of phase 1 flagellin expression in flagellar phase variation of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:958–967. 10.1128/JB.188.3.958-967.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.McClain MS, Blomfield IC, Eisenstein BI. 1991. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J. Bacteriol. 173:5308–5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Kulasekara HD, Blomfield IC. 1999. The molecular basis for the specificity of fimE in the phase variation of type 1 fimbriae of Escherichia coli K-12. Mol. Microbiol. 31:1171–1181. 10.1046/j.1365-2958.1999.01257.x [DOI] [PubMed] [Google Scholar]
- 405.Nakayama-Imaohji H, Hirakawa H, Ichimura M, Wakimoto S, Kuhara S, Hayashi T, Kuwahara T. 2009. Identification of the site-specific DNA invertase responsible for the phase variation of SusC/SusD family outer membrane proteins in Bacteroides fragilis. J. Bacteriol. 191:6003–6011. 10.1128/JB.00687-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Olsen PB, Klemm P. 1994. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett. 116:95–100. 10.1111/j.1574-6968.1994.tb06681.x [DOI] [PubMed] [Google Scholar]
- 407.Joyce SA, Dorman CJ. 2002. A Rho-dependent phase-variable transcription terminator controls expression of the FimE recombinase in Escherichia coli. Mol. Microbiol. 45:1107–1117. 10.1046/j.1365-2958.2002.03081.x [DOI] [PubMed] [Google Scholar]
- 408.Blomfield IC, Calie PJ, Eberhardt KJ, McClain MS, Eisenstein BI. 1993. Lrp stimulates phase variation of type 1 fimbriation in Escherichia coli K-12. J. Bacteriol. 175:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Campbell AM. 1992. Chromosomal insertion sites for phages and plasmids. J. Bacteriol. 174:7495–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Bartlett DH, Silverman M. 1989. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J. Bacteriol. 171:1763–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Higgins BP, Carpenter CD, Karls AC. 2007. Chromosomal context directs high-frequency precise excision of IS492 in Pseudoalteromonas atlantica. Proc. Natl. Acad. Sci. U. S. A. 104:1901–1906. 10.1073/pnas.0608633104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Higgins BP, Popkowski AC, Caruana PR, Karls AC. 2009. Site-specific insertion of IS492 in Pseudoalteromonas atlantica. J. Bacteriol. 191:6408–6414. 10.1128/JB.00771-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Lüneberg E, Mayer B, Daryab N, Kooistra O, Zähringer U, Rohde M, Swanson J, Frosch M. 2001. Chromosomal insertion and excision of a 30 kb unstable genetic element is responsible for phase variation of lipopolysaccharide and other virulence determinants in Legionella pneumophila. Mol. Microbiol. 39:1259–1271. 10.1111/j.1365-2958.2001.02314.x [DOI] [PubMed] [Google Scholar]
- 414.Haselkorn R. 1992. Developmentally regulated gene rearrangements in prokaryotes. Annu. Rev. Genet. 26:113–130. 10.1146/annurev.ge.26.120192.000553 [DOI] [PubMed] [Google Scholar]
- 415.Carrasco CD, Buettner JA, Golden JW. 1995. Programmed DNA rearrangement of a cyanobacterial hupL gene in heterocysts. Proc. Natl. Acad. Sci. U. S. A. 92:791–795. 10.1073/pnas.92.3.791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Haraldsen JD, Sonenshein AL. 2003. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol. Microbiol. 48:811–821. 10.1046/j.1365-2958.2003.03471.x [DOI] [PubMed] [Google Scholar]
- 417.Carrasco CD, Holliday SD, Hansel A, Lindblad P, Golden JW. 2005. Heterocyst-specific excision of the Anabaena sp. strain PCC 7120 hupL element requires xisC. J. Bacteriol. 187:6031–6038. 10.1128/JB.187.17.6031-6038.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Henson BJ, Watson LE, Barnum SR. 2005. Characterization of a 4 kb variant of the nifD element in Anabaena sp. strain ATCC 33047. Curr. Microbiol. 50:129–132. 10.1007/s00284-004-4338-z [DOI] [PubMed] [Google Scholar]
- 419.Krogh S, O'Reilly M, Nolan N, Devine KM. 1996. The phage-like element PBSX and part of the skin element, which are resident at different locations on the Bacillus subtilis chromosome, are highly homologous. Microbiology 142(Part 8):2031–2040 [DOI] [PubMed] [Google Scholar]
- 420.Hallet B. 2001. Playing Dr Jekyll and Mr Hyde: combined mechanisms of phase variation in bacteria. Curr. Opin. Microbiol. 4:570–581. 10.1016/S1369-5274(00)00253-8 [DOI] [PubMed] [Google Scholar]
- 421.Frank SA, Barbour AG. 2006. Within-host dynamics of antigenic variation. Infect. Genet. Evol. 6:141–146. 10.1016/j.meegid.2004.10.005 [DOI] [PubMed] [Google Scholar]
- 422.van der Woude MW. 2006. Re-examining the role and random nature of phase variation. FEMS Microbiol. Lett. 254:190–197. 10.1111/j.1574-6968.2005.00038.x [DOI] [PubMed] [Google Scholar]
- 423.Davidson CJ, Surette MG. 2008. Individuality in bacteria. Annu. Rev. Genet. 42:253–268. 10.1146/annurev.genet.42.110807.091601 [DOI] [PubMed] [Google Scholar]
- 424.Bayliss CD. 2009. Determinants of phase variation rate and the fitness implications of differing rates for bacterial pathogens and commensals. FEMS Microbiol. Rev. 33:504–520. 10.1111/j.1574-6976.2009.00162.x [DOI] [PubMed] [Google Scholar]
- 425.Deitsch KW, Lukehart SA, Stringer JR. 2009. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat. Rev. Microbiol. 7:493–503. 10.1038/nrmicro2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.van der Woude MW. 2011. Phase variation: how to create and coordinate population diversity. Curr. Opin. Microbiol. 14:205–211. 10.1016/j.mib.2011.01.002 [DOI] [PubMed] [Google Scholar]
- 427.Scott J, Thompson-Mayberry P, Lahmamsi S, King CJ, McShan WM. 2008. Phage-associated mutator phenotype in group A Streptococcus. J. Bacteriol. 190:6290–6301. 10.1128/JB.01569-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Bacon DJ, Szymanski CM, Burr DH, Silver RP, Alm RA, Guerry P. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769–777. 10.1046/j.1365-2958.2001.02431.x [DOI] [PubMed] [Google Scholar]
- 429.Blyn LB, Braaten BA, White-Ziegler CA, Rolfson DH, Low DA. 1989. Phase-variation of pyelonephritis-associated pili in Escherichia coli: evidence for transcriptional regulation. EMBO J. 8:613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Liu B, Hu B, Zhou Z, Guo D, Guo X, Ding P, Feng L, Wang L. 2012. A novel non-homologous recombination-mediated mechanism for Escherichia coli unilateral flagellar phase variation. Nucleic Acids Res. 40:4530–4538. 10.1093/nar/gks040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Goodwin AC, Weinberger DM, Ford CB, Nelson JC, Snider JD, Hall JD, Paules CI, Peek RM, Jr, Forsyth MH. 2008. Expression of the Helicobacter pylori adhesin SabA is controlled via phase variation and the ArsRS signal transduction system. Microbiology 154:2231–2240. 10.1099/mic.0.2007/016055-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.van den Broek D, Chin-A-Woeng TF, Eijkemans K, Mulders IH, Bloemberg GV, Lugtenberg BJ. 2003. Biocontrol traits of Pseudomonas spp. are regulated by phase variation. Mol. Plant Microbe Interact. 16:1003–1012. 10.1094/MPMI.2003.16.11.1003 [DOI] [PubMed] [Google Scholar]
- 433.Lewis LA, Gipson M, Hartman K, Ownbey T, Vaughn J, Dyer DW. 1999. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32:977–989. 10.1046/j.1365-2958.1999.01409.x [DOI] [PubMed] [Google Scholar]
- 434.Rosengarten R, Wise KS. 1990. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science 247:315–318. 10.1126/science.1688663 [DOI] [PubMed] [Google Scholar]
- 435.Hoskisson PA, Smith MC. 2007. Hypervariation and phase variation in the bacteriophage ‘resistome'. Curr. Opin. Microbiol. 10:396–400. 10.1016/j.mib.2007.04.003 [DOI] [PubMed] [Google Scholar]
- 436.Hendrixson DR. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646–1659. 10.1111/j.1365-2958.2006.05336.x [DOI] [PubMed] [Google Scholar]
- 437.Iqbal S, Parker G, Davidson H, Moslehi-Rahmani E, Robson RL. 2004. Reversible phase variation in the phnE gene, which is required for phosphonate metabolism in Escherichia coli K-12. J. Bacteriol. 186:6118–6123. 10.1128/JB.186.18.6118-6123.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Faulstich M, Böttcher JP, Meyer TF, Fraunholz M, Rudel T. 2013. Pilus phase variation switches gonococcal adherence to invasion by caveolin-1-dependent host cell signaling. PLoS Pathog. 9:e1003373. 10.1371/journal.ppat.1003373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Lindbäck T, Secic I, Rørvik LM. 2011. A contingency locus in prfA in a Listeria monocytogenes subgroup allows reactivation of the PrfA virulence regulator during infection in mice. Appl. Environ. Microbiol. 77:3478–3483. 10.1128/AEM.02708-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Clark SE, Eichelberger KR, Weiser JN. 2013. Evasion of killing by human antibody and complement through multiple variations in the surface oligosaccharide of Haemophilus influenzae. Mol. Microbiol. 88:603–618. 10.1111/mmi.12214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Gunther NW, IV, Snyder JA, Lockatell V, Blomfield I, Johnson DE, Mobley HL. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 70:3344–3354. 10.1128/IAI.70.7.3344-3354.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Clark SE, Snow J, Li J, Zola TA, Weiser JN. 2012. Phosphorylcholine allows for evasion of bactericidal antibody by Haemophilus influenzae. PLoS Pathog. 8:e1002521. 10.1371/journal.ppat.1002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Norris TL, Bäumler AJ. 1999. Phase variation of the lpf operon is a mechanism to evade cross-immunity between Salmonella serotypes. Proc. Natl. Acad. Sci. U. S. A. 96:13393–13398. 10.1073/pnas.96.23.13393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Schwan WR. 2011. Regulation of fim genes in uropathogenic Escherichia coli. World J. Clin. Infect. Dis. 1:17–25. 10.5495/wjcid.v1.i1.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Dove SL, Dorman CJ. 1994. The site-specific recombination system regulating expression of the type 1 fimbrial subunit gene of Escherichia coli is sensitive to changes in DNA supercoiling. Mol. Microbiol. 14:975–988. 10.1111/j.1365-2958.1994.tb01332.x [DOI] [PubMed] [Google Scholar]
- 446.Gally DL, Bogan JA, Eisenstein BI, Blomfield IC. 1993. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J. Bacteriol. 175:6186–6193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Ratiner YA. 1999. Temperature-dependent flagellar antigen phase variation in Escherichia coli. Res. Microbiol. 150:457–463. 10.1016/S0923-2508(99)00111-4 [DOI] [PubMed] [Google Scholar]
- 448.Moran AP, Knirel YA, Senchenkova SN, Widmalm G, Hynes SO, Jansson PE. 2002. Phenotypic variation in molecular mimicry between Helicobacter pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. Acid-induced phase variation in Lewis(x) and Lewis(y) expression by H. pylori lipopolysaccharides. J. Biol. Chem. 277:5785–5795. 10.1074/jbc.M108574200 [DOI] [PubMed] [Google Scholar]
- 449.Stinson RS, Talburt DE. 1978. Antigenic variation and increase in pathogenicity in Pseudomonas aeruginosa as a result of growth on glucose or N-hexadecane as sole carbon source. Can. J. Microbiol. 24:675–679. 10.1139/m78-113 [DOI] [PubMed] [Google Scholar]
- 450.Lane MC, Li X, Pearson MM, Simms AN, Mobley HL. 2009. Oxygen-limiting conditions enrich for fimbriate cells of uropathogenic Proteus mirabilis and Escherichia coli. J. Bacteriol. 191:1382–1392. 10.1128/JB.01550-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Serkin CD, Seifert HS. 2000. Iron availability regulates DNA recombination in Neisseria gonorrhoeae. Mol. Microbiol. 37:1075–1086. 10.1046/j.1365-2958.2000.02058.x [DOI] [PubMed] [Google Scholar]
- 452.Garrison-Schilling KL, Grau BL, McCarter KS, Olivier BJ, Comeaux NE, Pettis GS. 2011. Calcium promotes exopolysaccharide phase variation and biofilm formation of the resulting phase variants in the human pathogen Vibrio vulnificus. Environ. Microbiol. 13:643–654. 10.1111/j.1462-2920.2010.02369.x [DOI] [PubMed] [Google Scholar]
- 453.Sohanpal BK, El-Labany S, Lahooti M, Plumbridge JA, Blomfield IC. 2004. Integrated regulatory responses of fimB to N-acetylneuraminic (sialic) acid and GlcNAc in Escherichia coli K-12. Proc. Natl. Acad. Sci. U. S. A. 101:16322–16327. 10.1073/pnas.0405821101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Juhas M. 18 July 2013. Horizontal gene transfer in human pathogens. Crit. Rev. Microbiol. 10.3109/1040841X.2013.804031 [DOI] [PubMed] [Google Scholar]
- 455.Watanabe T. 1963. Infective heredity of multiple drug resistance in bacteria. Bacteriol. Rev. 27:87–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Doolittle WF. 1999. Lateral genomics. Trends Cell Biol. 9:M5–M8. 10.1016/S0962-8924(99)01664-5 [DOI] [PubMed] [Google Scholar]
- 457.Kurland CG. 2000. Something for everyone. Horizontal gene transfer in evolution. EMBO Rep. 1:92–95. 10.1093/embo-reports/kvd042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Levin BR, Bergstrom CT. 2000. Bacteria are different: observations, interpretations, speculations, and opinions about the mechanisms of adaptive evolution in prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 97:6981–6985. 10.1073/pnas.97.13.6981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. 10.1038/35012500 [DOI] [PubMed] [Google Scholar]
- 460.Woese CR. 2000. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. U. S. A. 97:8392–8396. 10.1073/pnas.97.15.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55:709–742. 10.1146/annurev.micro.55.1.709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Sonea S, Mathieu LG. 2001. Evolution of the genomic systems of prokaryotes and its momentous consequences. Int. Microbiol. 4:67–71. 10.1007/s101230100015 [DOI] [PubMed] [Google Scholar]
- 463.Cohan FM. 2002. Sexual isolation and speciation in bacteria. Genetica 116:359–370. 10.1023/A:1021232409545 [DOI] [PubMed] [Google Scholar]
- 464.Dutta C, Pan A. 2002. Horizontal gene transfer and bacterial diversity. J. Biosci. 27:27–33. 10.1007/BF02703681 [DOI] [PubMed] [Google Scholar]
- 465.Gogarten JP, Doolittle WF, Lawrence JG. 2002. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19:2226–2238. 10.1093/oxfordjournals.molbev.a004046 [DOI] [PubMed] [Google Scholar]
- 466.Woese CR. 2002. On the evolution of cells. Proc. Natl. Acad. Sci. U. S. A. 99:8742–8747. 10.1073/pnas.132266999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Boucher Y, Douady CJ, Papke RT, Walsh DA, Boudreau ME, Nesbo CL, Case RJ, Doolittle WF. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283–328. 10.1146/annurev.genet.37.050503.084247 [DOI] [PubMed] [Google Scholar]
- 468.Brown JR. 2003. Ancient horizontal gene transfer. Nat. Rev. Genet. 4:121–132. 10.1038/nrg1000 [DOI] [PubMed] [Google Scholar]
- 469.Lawrence JG, Hendrickson H. 2003. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 50:739–749. 10.1046/j.1365-2958.2003.03778.x [DOI] [PubMed] [Google Scholar]
- 470.Gogarten JP, Townsend JP. 2005. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3:679–687. 10.1038/nrmicro1204 [DOI] [PubMed] [Google Scholar]
- 471.Pal C, Papp B, Lercher MJ. 2005. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet. 37:1372–1375. 10.1038/ng1686 [DOI] [PubMed] [Google Scholar]
- 472.Susko E, Leigh J, Doolittle WF, Bapteste E. 2006. Visualizing and assessing phylogenetic congruence of core gene sets: a case study of the gamma-proteobacteria. Mol. Biol. Evol. 23:1019–1030. 10.1093/molbev/msj113 [DOI] [PubMed] [Google Scholar]
- 473.Doolittle WF, Bapteste E. 2007. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl. Acad. Sci. U. S. A. 104:2043–2049. 10.1073/pnas.0610699104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Goldenfeld N, Woese C. 2007. Biology's next revolution. Nature 445:369. 10.1038/445369a [DOI] [PubMed] [Google Scholar]
- 475.Bapteste E, Boucher Y. 2008. Lateral gene transfer challenges principles of microbial systematics. Trends Microbiol. 16:200–207. 10.1016/j.tim.2008.02.005 [DOI] [PubMed] [Google Scholar]
- 476.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688–6719. 10.1093/nar/gkn668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Doolittle WF, Zhaxybayeva O. 2009. On the origin of prokaryotic species. Genome Res. 19:744–756. 10.1101/gr.086645.108 [DOI] [PubMed] [Google Scholar]
- 478.Poole AM. 2009. Horizontal gene transfer and the earliest stages of the evolution of life. Res. Microbiol. 160:473–480. 10.1016/j.resmic.2009.07.009 [DOI] [PubMed] [Google Scholar]
- 479.Brigulla M, Wackernagel W. 2010. Molecular aspects of gene transfer and foreign DNA acquisition in prokaryotes with regard to safety issues. Appl. Microbiol. Biotechnol. 86:1027–1041. 10.1007/s00253-010-2489-3 [DOI] [PubMed] [Google Scholar]
- 480.Case RJ, Boucher Y. 2011. Molecular musings in microbial ecology and evolution. Biol. Direct 6:58. 10.1186/1745-6150-6-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Popa O, Dagan T. 2011. Trends and barriers to lateral gene transfer in prokaryotes. Curr. Opin. Microbiol. 14:615–623. 10.1016/j.mib.2011.07.027 [DOI] [PubMed] [Google Scholar]
- 482.Vogan AA, Higgs PG. 2011. The advantages and disadvantages of horizontal gene transfer and the emergence of the first species. Biol. Direct 6:1. 10.1186/1745-6150-6-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Gao B, Gupta RS. 2012. Microbial systematics in the post-genomics era. Antonie Van Leeuwenhoek 101:45–54. 10.1007/s10482-011-9663-1 [DOI] [PubMed] [Google Scholar]
- 484.Koonin EV, Wolf YI. 2012. Evolution of microbes and viruses: a paradigm shift in evolutionary biology? Front. Cell. Infect. Microbiol. 2:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Berg OG, Kurland C. 2002. Evolution of microbial genomes: sequence acquisition and loss. Mol. Biol. Evol. 19:2265–2276. 10.1093/oxfordjournals.molbev.a004050 [DOI] [PubMed] [Google Scholar]
- 486.Hall BG, Yokoyama S, Calhoun DH. 1983. Role of cryptic genes in microbial evolution. Mol. Biol. Evol. 1:109–124 [DOI] [PubMed] [Google Scholar]
- 487.Baumdicker F, Hess WR, Pfaffelhuber P. 2012. The infinitely many genes model for the distributed genome of bacteria. Genome Biol. Evol. 4:443–456. 10.1093/gbe/evs016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Francis AR, Tanaka MM. 2012. Evolution of variation in presence and absence of genes in bacterial pathways. BMC Evol. Biol. 12:55. 10.1186/1471-2148-12-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Arber W. 2011. Molecular Darwinism: the contingency of spontaneous genetic variation. Genome Biol. Evol. 3:1090–1092. 10.1093/gbe/evr035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Solioz M, Yen HC, Marris B. 1975. Release and uptake of gene transfer agent by Rhodopseudomonas capsulata. J. Bacteriol. 123:651–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Lang AS, Beatty JT. 2007. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 15:54–62. 10.1016/j.tim.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 492.Stanton TB. 2007. Prophage-like gene transfer agents—novel mechanisms of gene exchange for Methanococcus, Desulfovibrio, Brachyspira, and Rhodobacter species. Anaerobe 13:43–49. 10.1016/j.anaerobe.2007.03.004 [DOI] [PubMed] [Google Scholar]
- 493.McDaniel LD, Young E, Delaney J, Ruhnau F, Ritchie KB, Paul JH. 2010. High frequency of horizontal gene transfer in the oceans. Science 330:50. 10.1126/science.1192243 [DOI] [PubMed] [Google Scholar]
- 494.Dubey GP, Ben-Yehuda S. 2011. Intercellular nanotubes mediate bacterial communication. Cell 144:590–600. 10.1016/j.cell.2011.01.015 [DOI] [PubMed] [Google Scholar]
- 495.Kolling GL, Matthews KR. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Yaron S, Kolling GL, Simon L, Matthews KR. 2000. Vesicle-mediated transfer of virulence genes from Escherichia coli O157:H7 to other enteric bacteria. Appl. Environ. Microbiol. 66:4414–4420. 10.1128/AEM.66.10.4414-4420.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 497.Mashburn-Warren LM, Whiteley M. 2006. Special delivery: vesicle trafficking in prokaryotes. Mol. Microbiol. 61:839–846. 10.1111/j.1365-2958.2006.05272.x [DOI] [PubMed] [Google Scholar]
- 498.Popa O, Hazkani-Covo E, Landan G, Martin W, Dagan T. 2011. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Res. 21:599–609. 10.1101/gr.115592.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Jain R, Rivera MC, Moore JE, Lake JA. 2003. Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20:1598–1602. 10.1093/molbev/msg154 [DOI] [PubMed] [Google Scholar]
- 500.Liu Y, Harrison PM, Kunin V, Gerstein M. 2004. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 5:R64. 10.1186/gb-2004-5-9-r64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Selman M, Corradi N. 2011. Microsporidia: horizontal gene transfers in vicious parasites. Mob. Genet. Elements 1:251–255. 10.4161/mge.18611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, McDonald L, Utterback TR, Malek JA, Linher KD, Garrett MM, Stewart AM, Cotton MD, Pratt MS, Phillips CA, Richardson D, Heidelberg J, Sutton GG, Fleischmann RD, Eisen JA, White O, Salzberg SL, Smith HO, Venter JC, Fraser CM. 1999. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399:323–329. 10.1038/20601 [DOI] [PubMed] [Google Scholar]
- 503.Navarre WW, McClelland M, Libby SJ, Fang FC. 2007. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 21:1456–1471. 10.1101/gad.1543107 [DOI] [PubMed] [Google Scholar]
- 504.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. 10.1038/nature02241 [DOI] [PubMed] [Google Scholar]
- 505.Hastings PJ, Rosenberg SM, Slack A. 2004. Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol. 12:401–404. 10.1016/j.tim.2004.07.003 [DOI] [PubMed] [Google Scholar]
- 506.Madsen JS, Burmølle M, Hansen LH, Sørensen SJ. 2012. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65:183–195. 10.1111/j.1574-695X.2012.00960.x [DOI] [PubMed] [Google Scholar]
- 507.Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- 508.Lawrence JG, Ochman H. 1998. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. U. S. A. 95:9413–9417. 10.1073/pnas.95.16.9413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Garcia-Vallve S, Romeu A, Palau J. 2000. Horizontal gene transfer in bacterial and archaeal complete genomes. Genome Res. 10:1719–1725. 10.1101/gr.130000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Kurland CG, Canback B, Berg OG. 2003. Horizontal gene transfer: a critical view. Proc. Natl. Acad. Sci. U. S. A. 100:9658–9662. 10.1073/pnas.1632870100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Kurland CG. 2005. What tangled web: barriers to rampant horizontal gene transfer. Bioessays 27:741–747. 10.1002/bies.20258 [DOI] [PubMed] [Google Scholar]
- 512.Dagan T, Artzy-Randrup Y, Martin W. 2008. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc. Natl. Acad. Sci. U. S. A. 105:10039–10044. 10.1073/pnas.0800679105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Dagan T, Martin W. 2007. Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution. Proc. Natl. Acad. Sci. U. S. A. 104:870–875. 10.1073/pnas.0606318104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Park C, Zhang J. 2012. High expression hampers horizontal gene transfer. Genome Biol. Evol. 4:523–532. 10.1093/gbe/evs030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Lerat E, Daubin V, Moran NA. 2003. From gene trees to organismal phylogeny in prokaryotes: the case of the gamma-Proteobacteria. PLoS Biol. 1:E19. 10.1371/journal.pbio.0000019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Bapteste E, Boucher Y, Leigh J, Doolittle WF. 2004. Phylogenetic reconstruction and lateral gene transfer. Trends Microbiol. 12:406–411. 10.1016/j.tim.2004.07.002 [DOI] [PubMed] [Google Scholar]
- 517.Cohen O, Gophna U, Pupko T. 2011. The complexity hypothesis revisited: connectivity rather than function constitutes a barrier to horizontal gene transfer. Mol. Biol. Evol. 28:1481–1489. 10.1093/molbev/msq333 [DOI] [PubMed] [Google Scholar]
- 518.Abby SS, Tannier E, Gouy M, Daubin V. 2012. Lateral gene transfer as a support for the tree of life. Proc. Natl. Acad. Sci. U. S. A. 109:4962–4967. 10.1073/pnas.1116871109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Beiko RG, Harlow TJ, Ragan MA. 2005. Highways of gene sharing in prokaryotes. Proc. Natl. Acad. Sci. U. S. A. 102:14332–14337. 10.1073/pnas.0504068102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Homma K, Fukuchi S, Nakamura Y, Gojobori T, Nishikawa K. 2007. Gene cluster analysis method identifies horizontally transferred genes with high reliability and indicates that they provide the main mechanism of operon gain in 8 species of gamma-Proteobacteria. Mol. Biol. Evol. 24:805–813. 10.1093/molbev/msl206 [DOI] [PubMed] [Google Scholar]
- 521.Maslov S, Krishna S, Pang TY, Sneppen K. 2009. Toolbox model of evolution of prokaryotic metabolic networks and their regulation. Proc. Natl. Acad. Sci. U. S. A. 106:9743–9748. 10.1073/pnas.0903206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Bansal MS, Banay G, Gogarten JP, Shamir R. 2011. Detecting highways of horizontal gene transfer. J. Comput. Biol. 18:1087–1114. 10.1089/cmb.2011.0066 [DOI] [PubMed] [Google Scholar]
- 523.Andersson JO, Andersson SG. 1999. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9:664–671. 10.1016/S0959-437X(99)00024-6 [DOI] [PubMed] [Google Scholar]
- 524.Moran NA, Mira A. 2001. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2:RESEARCH0054–RESEARCH0054.12. 10.1186/gb-2001-2-12-research0054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Silva FJ, Latorre A, Moya A. 2001. Genome size reduction through multiple events of gene disintegration in Buchnera APS. Trends Genet. 17:615–618. 10.1016/S0168-9525(01)02483-0 [DOI] [PubMed] [Google Scholar]
- 526.Frank AC, Amiri H, Andersson SG. 2002. Genome deterioration: loss of repeated sequences and accumulation of junk DNA. Genetica 115:1–12. 10.1023/A:1016064511533 [DOI] [PubMed] [Google Scholar]
- 527.Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. 10.1016/S0092-8674(02)00665-7 [DOI] [PubMed] [Google Scholar]
- 528.Gomez-Valero L, Latorre A, Silva FJ. 2004. The evolutionary fate of nonfunctional DNA in the bacterial endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 21:2172–2181. 10.1093/molbev/msh232 [DOI] [PubMed] [Google Scholar]
- 529.Moran NA, Plague GR. 2004. Genomic changes following host restriction in bacteria. Curr. Opin. Genet. Dev. 14:627–633. 10.1016/j.gde.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 530.Bordenstein SR, Reznikoff WS. 2005. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 3:688–699. 10.1038/nrmicro1233 [DOI] [PubMed] [Google Scholar]
- 531.Wernegreen JJ. 2005. For better or worse: genomic consequences of intracellular mutualism and parasitism. Curr. Opin. Genet. Dev. 15:572–583. 10.1016/j.gde.2005.09.013 [DOI] [PubMed] [Google Scholar]
- 532.Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337. 10.1371/journal.pbio.0040337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190. 10.1146/annurev.genet.41.110306.130119 [DOI] [PubMed] [Google Scholar]
- 534.Moya A, Peretó J, Gil R, Latorre A. 2008. Learning how to live together: genomic insights into prokaryote-animal symbioses. Nat. Rev. Genet. 9:218–229. 10.1038/nrg2319 [DOI] [PubMed] [Google Scholar]
- 535.O'Fallon B. 2008. Population structure, levels of selection, and the evolution of intracellular symbionts. Evolution 62:361–373. 10.1111/j.1558-5646.2007.00289.x [DOI] [PubMed] [Google Scholar]
- 536.Plague GR, Dunbar HE, Tran PL, Moran NA. 2008. Extensive proliferation of transposable elements in heritable bacterial symbionts. J. Bacteriol. 190:777–779. 10.1128/JB.01082-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Feldhaar H, Gross R. 2009. Genome degeneration affects both extracellular and intracellular bacterial endosymbionts. J. Biol. 8:31. 10.1186/jbiol129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 7:2. 10.1186/1741-7007-7-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol. Direct 4:13. 10.1186/1745-6150-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Newton IL, Bordenstein SR. 2011. Correlations between bacterial ecology and mobile DNA. Curr. Microbiol. 62:198–208. 10.1007/s00284-010-9693-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10:13–26. 10.1038/nrmicro2670 [DOI] [PubMed] [Google Scholar]
- 542.Wernegreen JJ. 2012. Strategies of genomic integration within insect-bacterial mutualisms. Biol. Bull. 223:112–122 http://www.biolbull.org/content/223/1/112.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Moran NA, McLaughlin HJ, Sorek R. 2009. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science 323:379–382. 10.1126/science.1167140 [DOI] [PubMed] [Google Scholar]
- 544.Lopez-Sanchez MJ, Neef A, Patino-Navarrete R, Navarro L, Jimenez R, Latorre A, Moya A. 2008. Blattabacteria, the endosymbionts of cockroaches, have small genome sizes and high genome copy numbers. Environ. Microbiol. 10:3417–3422. 10.1111/j.1462-2920.2008.01776.x [DOI] [PubMed] [Google Scholar]
- 545.Mendonca AG, Alves RJ, Pereira-Leal JB. 2011. Loss of genetic redundancy in reductive genome evolution. PLoS Comput. Biol. 7:e1001082. 10.1371/journal.pcbi.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Kuo CH, Ochman H. 2009. Deletional bias across the three domains of life. Genome Biol. Evol. 1:145–152. 10.1093/gbe/evp016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P, Berry K, Young MB, Utterback T, Weidman J, Nierman WC, Paulsen IT, Nelson KE, Tettelin H, O'Neill SL, Eisen JA. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. 10.1371/journal.pbio.0020069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Bordenstein SR, Wernegreen JJ. 2004. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21:1981–1991. 10.1093/molbev/msh211 [DOI] [PubMed] [Google Scholar]
- 549.Moliner C, Fournier PE, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol. Rev. 34:281–294. 10.1111/j.1574-6976.2009.00209.x [DOI] [PubMed] [Google Scholar]
- 550.Bertelli C, Greub G. 2012. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front. Cell. Infect. Microbiol. 2:110. 10.3389/fcimb.2012.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Lamrabet O, Merhej V, Pontarotti P, Raoult D, Drancourt M. 2012. The genealogic tree of mycobacteria reveals a long-standing sympatric life into free-living protozoa. PLoS One 7:e34754. 10.1371/journal.pone.0034754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603. 10.1038/284601a0 [DOI] [PubMed] [Google Scholar]
- 553.Orgel LE, Crick FH. 1980. Selfish DNA: the ultimate parasite. Nature 284:604–607. 10.1038/284604a0 [DOI] [PubMed] [Google Scholar]
- 554.Kobayashi I. 1998. Selfishness and death: raison d'etre of restriction, recombination and mitochondria. Trends Genet. 14:368–374. 10.1016/S0168-9525(98)01532-7 [DOI] [PubMed] [Google Scholar]
- 555.Kidwell MG, Lisch DR. 2001. Perspective: transposable elements, parasitic DNA, and genome evolution. Evolution 55:1–24. 10.1111/j.0014-3820.2001.tb01268.x [DOI] [PubMed] [Google Scholar]
- 556.Lan R, Reeves PR. 1996. Gene transfer is a major factor in bacterial evolution. Mol. Biol. Evol. 13:47–55. 10.1093/oxfordjournals.molbev.a025569 [DOI] [PubMed] [Google Scholar]
- 557.Doolittle WF. 1999. Phylogenetic classification and the universal tree. Science 284:2124–2129. 10.1126/science.284.5423.2124 [DOI] [PubMed] [Google Scholar]
- 558.Spratt BG, Hanage WP, Feil EJ. 2001. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Curr. Opin. Microbiol. 4:602–606. 10.1016/S1369-5274(00)00257-5 [DOI] [PubMed] [Google Scholar]
- 559.Mira A, Klasson L, Andersson SG. 2002. Microbial genome evolution: sources of variability. Curr. Opin. Microbiol. 5:506–512. 10.1016/S1369-5274(02)00358-2 [DOI] [PubMed] [Google Scholar]
- 560.Kunin V, Ouzounis CA. 2003. The balance of driving forces during genome evolution in prokaryotes. Genome Res. 13:1589–1594. 10.1101/gr.1092603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Lerat E, Daubin V, Ochman H, Moran NA. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3:e130. 10.1371/journal.pbio.0030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Lopez P, Bapteste E. 2009. Molecular phylogeny: reconstructing the forest. C. R. Biol. 332:171–182. 10.1016/j.crvi.2008.07.003 [DOI] [PubMed] [Google Scholar]
- 563.Olendzenski L, Gogarten JP. 2009. Evolution of genes and organisms: the tree/web of life in light of horizontal gene transfer. Ann. N. Y. Acad. Sci. 1178:137–145. 10.1111/j.1749-6632.2009.04998.x [DOI] [PubMed] [Google Scholar]
- 564.Raoult D. 2010. The post-Darwinist rhizome of life. Lancet 375:104–105. 10.1016/S0140-6736(09)61958-9 [DOI] [PubMed] [Google Scholar]
- 565.Woese C. 1998. The universal ancestor. Proc. Natl. Acad. Sci. U. S. A. 95:6854–6859. 10.1073/pnas.95.12.6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Vetsigian K, Woese C, Goldenfeld N. 2006. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. U. S. A. 103:10696–10701. 10.1073/pnas.0603780103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Lawrence JG, Roth JR. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Ballouz S, Francis AR, Lan R, Tanaka MM. 2010. Conditions for the evolution of gene clusters in bacterial genomes. PLoS Comput. Biol. 6:e1000672. 10.1371/journal.pcbi.1000672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Kolesov G, Wunderlich Z, Laikova ON, Gelfand MS, Mirny LA. 2007. How gene order is influenced by the biophysics of transcription regulation. Proc. Natl. Acad. Sci. U. S. A. 104:13948–13953. 10.1073/pnas.0700672104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Fang G, Rocha EP, Danchin A. 2008. Persistence drives gene clustering in bacterial genomes. BMC Genomics 9:4. 10.1186/1471-2164-9-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Price MN, Huang KH, Arkin AP, Alm EJ. 2005. Operon formation is driven by co-regulation and not by horizontal gene transfer. Genome Res. 15:809–819. 10.1101/gr.3368805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Pal C, Hurst LD. 2004. Evidence against the selfish operon theory. Trends Genet. 20:232–234. 10.1016/j.tig.2004.04.001 [DOI] [PubMed] [Google Scholar]
- 573.Davids W, Zhang Z. 2008. The impact of horizontal gene transfer in shaping operons and protein interaction networks—direct evidence of preferential attachment. BMC Evol. Biol. 8:23. 10.1186/1471-2148-8-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Feil EJ, Spratt BG. 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55:561–590. 10.1146/annurev.micro.55.1.561 [DOI] [PubMed] [Google Scholar]
- 575.Rivera MC, Lake JA. 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431:152–155. 10.1038/nature02848 [DOI] [PubMed] [Google Scholar]
- 576.Kunin V, Goldovsky L, Darzentas N, Ouzounis CA. 2005. The net of life: reconstructing the microbial phylogenetic network. Genome Res. 15:954–959. 10.1101/gr.3666505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Zhaxybayeva O, Lapierre P, Gogarten JP. 2005. Ancient gene duplications and the root(s) of the tree of life. Protoplasma 227:53–64. 10.1007/s00709-005-0135-1 [DOI] [PubMed] [Google Scholar]
- 578.Dagan T, Martin W. 2006. The tree of one percent. Genome Biol. 7:118. 10.1186/gb-2006-7-10-118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Koonin EV. 2009. Darwinian evolution in the light of genomics. Nucleic Acids Res. 37:1011–1034. 10.1093/nar/gkp089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Beauregard-Racine J, Bicep C, Schliep K, Lopez P, Lapointe FJ, Bapteste E. 2011. Of woods and webs: possible alternatives to the tree of life for studying genomic fluidity in E. coli. Biol. Direct 6:39. 10.1186/1745-6150-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Williams D, Fournier GP, Lapierre P, Swithers KS, Green AG, Andam CP, Gogarten JP. 2011. A rooted net of life. Biol. Direct 6:45. 10.1186/1745-6150-6-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Snel B, Bork P, Huynen MA. 1999. Genome phylogeny based on gene content. Nat. Genet. 21:108–110. 10.1038/5052 [DOI] [PubMed] [Google Scholar]
- 583.Brochier C, Bapteste E, Moreira D, Philippe H. 2002. Eubacterial phylogeny based on translational apparatus proteins. Trends Genet. 18:1–5. 10.1016/S0168-9525(01)02522-7 [DOI] [PubMed] [Google Scholar]
- 584.Daubin V, Gouy M, Perriere G. 2002. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 12:1080–1090. 10.1101/gr.187002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Wolf YI, Rogozin IB, Grishin NV, Koonin EV. 2002. Genome trees and the Tree of Life. Trends Genet. 18:472–479. 10.1016/S0168-9525(02)02744-0 [DOI] [PubMed] [Google Scholar]
- 586.Daubin V, Moran NA, Ochman H. 2003. Phylogenetics and the cohesion of bacterial genomes. Science 301:829–832. 10.1126/science.1086568 [DOI] [PubMed] [Google Scholar]
- 587.Gu X, Zhang H. 2004. Genome phylogenetic analysis based on extended gene contents. Mol. Biol. Evol. 21:1401–1408. 10.1093/molbev/msh138 [DOI] [PubMed] [Google Scholar]
- 588.Lienau EK, DeSalle R, Rosenfeld JA, Planet PJ. 2006. Reciprocal illumination in the gene content tree of life. Syst. Biol. 55:441–453. 10.1080/10635150600697416 [DOI] [PubMed] [Google Scholar]
- 589.House CH. 2009. The Tree of Life viewed through the contents of genomes. Methods Mol. Biol. 532:141–161. 10.1007/978-1-60327-853-9_8 [DOI] [PubMed] [Google Scholar]
- 590.Horiike T, Miyata D, Tateno Y, Minai R. 2011. HGT-Gen: a tool for generating a phylogenetic tree with horizontal gene transfer. Bioinformation 7:211–213. 10.6026/97320630007211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Forterre P. 2012. Darwin's goldmine is still open: variation and selection run the world. Front. Cell. Infect. Microbiol. 2:106. 10.3389/fcimb.2012.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Fox GE, Wisotzkey JD, Jurtshuk P., Jr 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166–170. 10.1099/00207713-42-1-166 [DOI] [PubMed] [Google Scholar]
- 593.Cohan FM. 2002. What are bacterial species? Annu. Rev. Microbiol. 56:457–487. 10.1146/annurev.micro.56.012302.160634 [DOI] [PubMed] [Google Scholar]
- 594.Doolittle WF, Papke RT. 2006. Genomics and the bacterial species problem. Genome Biol. 7:116. 10.1186/gb-2006-7-9-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Konstantinidis KT, Ramette A, Tiedje JM. 2006. The bacterial species definition in the genomic era. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:1929–1940. 10.1098/rstb.2006.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Franklin L. 2007. Bacteria, sex, and systematics. Philos. Sci. 74:69–95. 10.1086/519476 [DOI] [Google Scholar]
- 597.Caro-Quintero A, Konstantinidis KT. 2012. Bacterial species may exist, metagenomics reveal. Environ. Microbiol. 14:347–355. 10.1111/j.1462-2920.2011.02668.x [DOI] [PubMed] [Google Scholar]
- 598.Burke C, Steinberg P, Rusch D, Kjelleberg S, Thomas T. 2011. Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. U. S. A. 108:14288–14293. 10.1073/pnas.1101591108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Koonin EV, Wolf YI. 2009. Is evolution Darwinian or/and Lamarckian? Biol. Direct 4:42. 10.1186/1745-6150-4-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Haerter JO, Sneppen K. 2012. Spatial structure and Lamarckian adaptation explain extreme genetic diversity at CRISPR locus. mBio 3(4):e00126–12. 10.1128/mBio.00126-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Podglajen I, Breuil J, Collatz E. 1994. Insertion of a novel DNA sequence, IS1186, upstream of the silent carbapenemase gene cfiA, promotes expression of carbapenem resistance in clinical isolates of Bacteroides fragilis. Mol. Microbiol. 12:105–114. 10.1111/j.1365-2958.1994.tb00999.x [DOI] [PubMed] [Google Scholar]
- 602.Coyne S, Courvalin P, Galimand M. 2010. Acquisition of multidrug resistance transposon Tn6061 and IS6100-mediated large chromosomal inversions in Pseudomonas aeruginosa clinical isolates. Microbiology 156:1448–1458. 10.1099/mic.0.033639-0 [DOI] [PubMed] [Google Scholar]
- 603.Martin C, Gomez-Lus R, Ortiz JM, Garcia-Lobo JM. 1987. Structure and mobilization of an ampicillin and gentamicin resistance determinant. Antimicrob. Agents Chemother. 31:1266–1270. 10.1128/AAC.31.8.1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Franke AE, Clewell DB. 1981. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of “conjugal” transfer in the absence of a conjugative plasmid. J. Bacteriol. 145:494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Ulycznyj PI, Salmon KA, Douillard H, DuBow MS. 1995. Characterization of the Pseudomonas aeruginosa transposable bacteriophage D3112 A and B genes. Biochim. Biophys. Acta 1264:249–253. 10.1016/0167-4781(95)00186-7 [DOI] [PubMed] [Google Scholar]
- 606.Pattee PA. 1981. Distribution of Tn551 insertion sites responsible for auxotrophy on the Staphylococcus aureus chromosome. J. Bacteriol. 145:479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Reddy SP, Rasmussen WG, Baseman JB. 1996. Isolation and characterization of transposon Tn4001-generated, cytadherence-deficient transformants of Mycoplasma pneumoniae and Mycoplasma genitalium. FEMS Immunol. Med. Microbiol. 15:199–211. 10.1111/j.1574-695X.1996.tb00086.x [DOI] [PubMed] [Google Scholar]
- 608.Briolat V, Reysset G. 2002. Identification of the Clostridium perfringens genes involved in the adaptive response to oxidative stress. J. Bacteriol. 184:2333–2343. 10.1128/JB.184.9.2333-2343.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Roberts AP, Mullany P. 2009. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 17:251–258. 10.1016/j.tim.2009.03.002 [DOI] [PubMed] [Google Scholar]
- 610.Nurk A, Tamm A, Horak R, Kivisaar M. 1993. In-vivo-generated fusion promoters in Pseudomonas putida. Gene 127:23–29. 10.1016/0378-1119(93)90612-7 [DOI] [PubMed] [Google Scholar]
- 611.Lers A, Bitoun R, Zamir A. 1989. Outreading promoters are located at both ends of the gamma-delta transposon. Mol. Gen. Genet. 216:138–143. 10.1007/BF00332242 [DOI] [PubMed] [Google Scholar]
- 612.Ciampi MS, Schmid MB, Roth JR. 1982. Transposon Tn10 provides a promoter for transcription of adjacent sequences. Proc. Natl. Acad. Sci. U. S. A. 79:5016–5020. 10.1073/pnas.79.16.5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Lin TP, Lai EM, To KY, Chang YS, Liu ST. 1994. Transcriptional activation of flanking sequences by Tn1000 insertion. Mol. Gen. Genet. 245:417–423. 10.1007/BF00302253 [DOI] [PubMed] [Google Scholar]
- 614.Ou X, Zhang B, Zhang L, Zhao G, Ding X. 2009. Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl. Environ. Microbiol. 75:2158–2165. 10.1128/AEM.02209-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 615.Rakin A, Noelting C, Schubert S, Heesemann J. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265–5274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 616.Rouquette-Loughlin CE, Balthazar JT, Hill SA, Shafer WM. 2004. Modulation of the mtrCDE-encoded efflux pump gene complex of Neisseria meningitidis due to a Correia element insertion sequence. Mol. Microbiol. 54:731–741. 10.1111/j.1365-2958.2004.04299.x [DOI] [PubMed] [Google Scholar]
- 617.Teras R, Hõrak R, Kivisaar M. 2000. Transcription from fusion promoters generated during transposition of transposon Tn4652 is positively affected by integration host factor in Pseudomonas putida. J. Bacteriol. 182:589–598. 10.1128/JB.182.3.589-598.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.De Gregorio E, Silvestro G, Petrillo M, Carlomagno MS, Di Nocera PP. 2005. Enterobacterial repetitive intergenic consensus sequence repeats in yersiniae: genomic organization and functional properties. J. Bacteriol. 187:7945–7954. 10.1128/JB.187.23.7945-7954.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Shimoji Y, Mori Y, Sekizaki T, Shibahara T, Yokomizo Y. 1998. Construction and vaccine potential of acapsular mutants of Erysipelothrix rhusiopathiae: use of excision of Tn916 to inactivate a target gene. Infect. Immun. 66:3250–3254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Foster TJ, Lundblad V, Hanley-Way S, Halling SM, Kleckner N. 1981. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell 23:215–227. 10.1016/0092-8674(81)90286-5 [DOI] [PubMed] [Google Scholar]
- 621.Swartley JS, McAllister CF, Hajjeh RA, Heinrich DW, Stephens DS. 1993. Deletions of Tn916-like transposons are implicated in tetM-mediated resistance in pathogenic Neisseria. Mol. Microbiol. 10:299–310. 10.1111/j.1365-2958.1993.tb01956.x [DOI] [PubMed] [Google Scholar]
- 622.Merrick M, Filser M, Dixon R, Elmerich C, Sibold L, Houmard J. 1980. The use of translocatable genetic elements to construct a fine-structure map of the Klebsiella pneumoniae nitrogen fixation (nif) gene cluster. J. Gen. Microbiol. 117:509–520 [DOI] [PubMed] [Google Scholar]
- 623.Nisen PD, Kopecko DJ, Chou J, Cohen SN. 1977. Site-specific DNA deletions occurring adjacent to the termini of a transposable ampicillin resistance element (Tn3). J. Mol. Biol. 117:975–978. 10.1016/S0022-2836(77)80008-9 [DOI] [PubMed] [Google Scholar]
- 624.Csonka LN, Clark AJ. 1979. Deletions generated by the transposon Tn10 in the srl recA region of the Escherichia coli K-12 chromosome. Genetics 93:321–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Faelen M, Toussaint A. 1978. Stimulation of deletions in the Escherichia coli chromosome by partially induced Mucts62 prophages. J. Bacteriol. 136:477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Song H, Hwang J, Yi H, Ulrich RL, Yu Y, Nierman WC, Kim HS. 2010. The early stage of bacterial genome-reductive evolution in the host. PLoS Pathog. 6:e1000922. 10.1371/journal.ppat.1000922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Claus H, Elias J, Meinhardt C, Frosch M, Vogel U. 2007. Deletion of the meningococcal fetA gene used for antigen sequence typing of invasive and commensal isolates from Germany: frequencies and mechanisms. J. Clin. Microbiol. 45:2960–2964. 10.1128/JCM.00696-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 628.Weber PC, Levine M, Glorioso JC. 1988. Simple assay for quantitation of Tn5 inversion events in Escherichia coli and use of the assay in determination of plasmid copy number. J. Bacteriol. 170:4972–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Rice LB, Carias LL, Marshall S, Rudin SD, Hutton-Thomas R. 2005. Tn5386, a novel Tn916-like mobile element in Enterococcus faecium D344R that interacts with Tn916 to yield a large genomic deletion. J. Bacteriol. 187:6668–6677. 10.1128/JB.187.19.6668-6677.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Cabezón T, Faelen M, De Wilde M, Bollen A, Thomas R. 1975. Expression of ribosomal protein genes in Escherichia coli. Mol. Gen. Genet. 137:125–129. 10.1007/BF00341678 [DOI] [PubMed] [Google Scholar]
- 631.Haack KR, Roth JR. 1995. Recombination between chromosomal IS200 elements supports frequent duplication formation in Salmonella typhimurium. Genetics 141:1245–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Sinclair MI, Holloway BW. 1991. Chromosomal insertion of TOL transposons in Pseudomonas aeruginosa PAO. J. Gen. Microbiol. 137:1111–1120. 10.1099/00221287-137-5-1111 [DOI] [PubMed] [Google Scholar]
- 633.Chumley FG, Roth JR. 1980. Rearrangement of the bacterial chromosome using Tn10 as a region of homology. Genetics 94:1–147399262 [Google Scholar]
- 634.Daveran-Mingot ML, Campo N, Ritzenthaler P, Le Bourgeois P. 1998. A natural large chromosomal inversion in Lactococcus lactis is mediated by homologous recombination between two insertion sequences. J. Bacteriol. 180:4834–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Chiang SJ, Clowes RC. 1980. Intramolecular transposition and inversion in plasmid R6K. J. Bacteriol. 142:668–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Scott AE, Timms AR, Connerton PL, Loc Carrillo C, Adzfa Radzum K, Connerton IF. 2007. Genome dynamics of Campylobacter jejuni in response to bacteriophage predation. PLoS Pathog. 3:e119. 10.1371/journal.ppat.0030119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Weightman AJ, Topping AW, Hill KE, Lee LL, Sakai K, Slater JH, Thomas AW. 2002. Transposition of DEH, a broad-host-range transposon flanked by ISPpu12, in Pseudomonas putida is associated with genomic rearrangements and dehalogenase gene silencing. J. Bacteriol. 184:6581–6591. 10.1128/JB.184.23.6581-6591.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Desmet L, Faelen M, Lefèbvre N, Résibois A, Toussaint A, van Gijsegem F. 1981. Genetic study of Mu transposition and Mu-mediated chromosomal rearrangements. Cold Spring Harb. Symp. Quant. Biol. 45(Part 1):355–363 [DOI] [PubMed] [Google Scholar]
- 639.Reimmann C, Haas D. 1987. Mode of replicon fusion mediated by the duplicated insertion sequence IS21 in Escherichia coli. Genetics 115:619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Mahillon J, Lereclus D. 1988. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 7:1515–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Robillard NJ, Tally FP, Malamy MH. 1985. Tn4400, a compound transposon isolated from Bacteroides fragilis, functions in Escherichia coli. J. Bacteriol. 164:1248–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Shoemaker NB, Li L-Y, Salyers AA. 1994. An unusual type of cointegrate formation between a Bacteroides plasmid and the excised circular form of an integrated element (NBU1). Plasmid 32:312–317. 10.1006/plas.1994.1070 [DOI] [PubMed] [Google Scholar]
- 643.Faelen M, Toussaint A. 1980. Chromosomal rearrangements induced by temperate bacteriophage D108. J. Bacteriol. 143:1029–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]