Abstract
SUMMARY
Clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated (Cas) genes are present in many bacterial and archaeal genomes. Since the discovery of the typical CRISPR loci in the 1980s, well before their physiological role was revealed, their variable sequences have been used as a complementary typing tool in diagnostic, epidemiologic, and evolutionary analyses of prokaryotic strains. The discovery that CRISPR spacers are often identical to sequence fragments of mobile genetic elements was a major breakthrough that eventually led to the elucidation of CRISPR-Cas as an adaptive immunity system. Key elements of this unique prokaryotic defense system are small CRISPR RNAs that guide nucleases to complementary target nucleic acids of invading viruses and plasmids, generally followed by the degradation of the invader. In addition, several recent studies have pointed at direct links of CRISPR-Cas to regulation of a range of stress-related phenomena. An interesting example concerns a pathogenic bacterium that possesses a CRISPR-associated ribonucleoprotein complex that may play a dual role in defense and/or virulence. In this review, we describe recently reported cases of potential involvement of CRISPR-Cas systems in bacterial stress responses in general and bacterial virulence in particular.
INTRODUCTION
In 1987, a repetitive stretch of DNA was detected on the Escherichia coli K-12 chromosome, downstream of the alkaline phosphatase isozyme iap gene (1). Similarly organized repetitive elements were found on the chromosomes of Shigella dysenteriae and Salmonella enterica serovar Typhimurium (2). The physiological role of the repetitive DNA was not obvious at that time. In the subsequent decennium, repetitive sequences were frequently detected in the genomes of both bacteria and archaea (3). A typical feature was the fact that the repeats were interspaced by noncoding, nonrepetitive sequences of similar lengths (3). In 2002, Jansen et al. discovered that these repetitive loci were always accompanied by conserved sets of genes encoding nucleic acid processing enzymes, including nuclease or helicase proteins. The latter authors proposed the names clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated (Cas) genes/proteins (4), now referred to as CRISPR-Cas.
In 2005, three groups independently observed that some of the interspaced sequences were 100% identical to DNA sequences from viruses and plasmids; it was proposed that CRISPR-Cas could be a novel defense system (5–7). Comparative genomic analyses revealed that the CRISPRs and their associated (cas) genes were present in diverse bacterial phylogenetic groups, resulting in the classification of these genes into several protein families (8–10). In 2007, Barrangou et al. provided the first experimental evidence that the CRISPR-Cas system is an adaptive immune system that protects its host against invading viruses (11). A role for CRISPR-Cas in defense was further established in a range of subsequent studies on viral transfections and plasmid transformations (5, 12–20). In 2011, Makarova et al. (21) suggested the classification of the different CRISPR-Cas systems into the following three main types: type I CRISPR-Cas systems, based on the presence of the cas3 gene; type II CRISPR-Cas systems, based on the presence of the cas9 gene; and type III CRISPR-Cas systems, based on the presence of the cas10 gene. This has now become established nomenclature (21).
An overview of the CRISPR-Cas types in species that are covered in this review is provided in Table 1. A selection of well-studied bacteria, their distribution across the human body, and the diversity of cas gene expression is summarized in Fig. 1. In general, most strains of the same species appear to contain identical CRISPR-Cas types, with some exceptions (such as the rare occurrence of the type I-F system in E. coli strains and the type I-C system in some Streptococcus sanguis strains). At the genus level, the diversity of CRISPR-Cas systems is somewhat larger. One of the most striking examples is in the Campylobacter genus: while most species (Campylobacter concisus, Campylobacter curvus, Campylobacter fetus, Campylobacter hominis, and Campylobacter rectus) have been found to harbor a type I-B system, Campylobacter jejuni contains a type II-C system instead. Most bacterial species seem to contain either one or a combination of two CRISPR-Cas types, although some of the Streptococcus species harbor all three types of CRISPR-Cas system. Helicobacter species and at least two genera belonging to the Pasteurellaceae family (Haemophilus and Pasteurella) harbor either a type II-C system or a virulence-associated protein D (VapD) that exhibits homology to Cas2. In the sequenced genomes of Mycobacterium spp., mainly the type III-A system is observed; in some strains, only the VapD protein is detected. In the sequenced Clostridium species, type I-B, type II-B, type II-C, and type III-B CRISPR-Cas systems have been detected. In the sequenced genomes of Bacillus species, mainly type I-B and type I-C CRISPR-Cas systems have been observed, whereas in Bacillus cereus, only a large CRISPR array has been found, reminiscent of a degenerate CRISPR-Cas system. Also apparent is the complete absence of the type I-D system, which so far has been found in genomes of only a few pathogenic bacteria (21), in the species covered in this review.
TABLE 1.
CRISPR-Cas types in the species covered in this reviewa
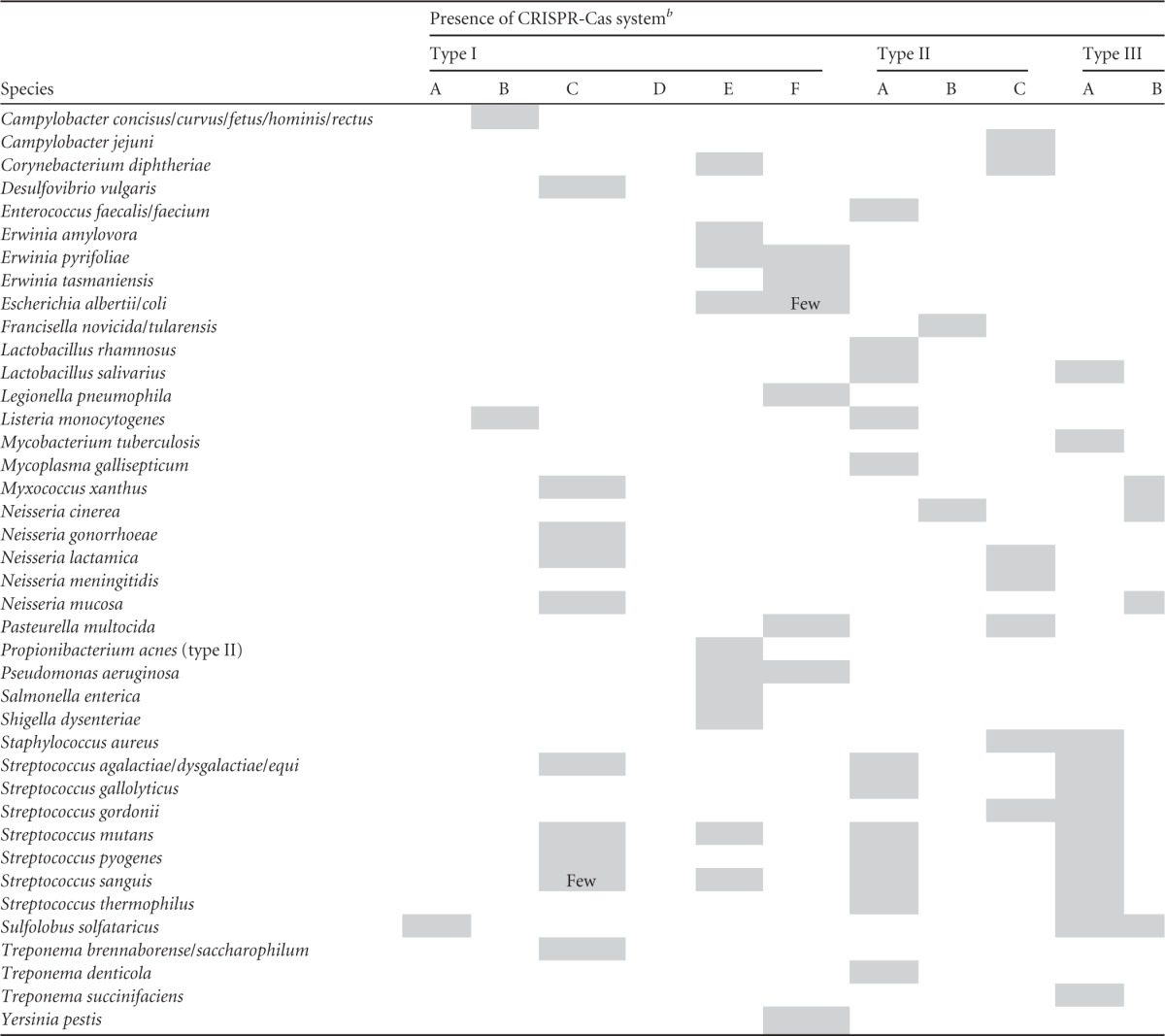
In general, most strains of the same species were found to contain identical CRISPR-Cas types.
Few, only a few species within the particular genus were found to contain the respective system.
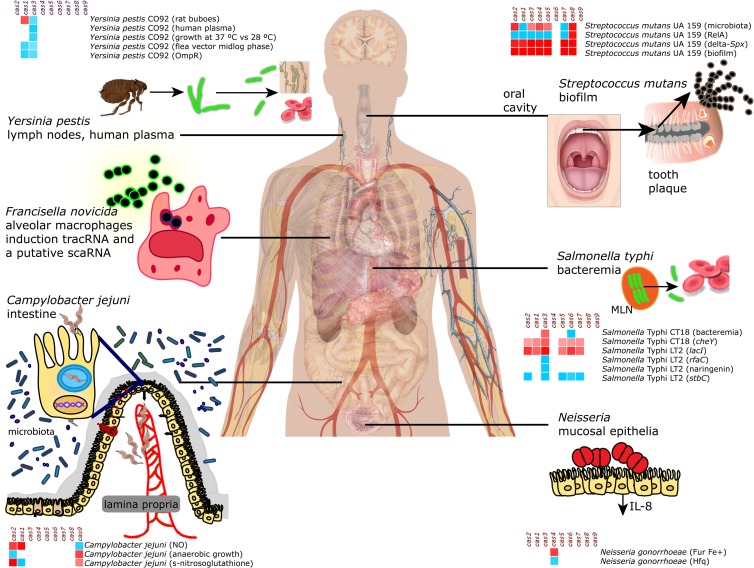
FIG 1.
Overview of expression of cas genes in human-associated bacteria that occupy different host niches. The heat maps indicate which cas genes are induced (shades of red) or repressed (shades of blue) during bacterial responses to changes in the environment. Details are given in the main text. The overview shows that modulation of cas gene expression occurs in diverse Gram-positive and Gram-negative bacteria that together occupy very diverse niches throughout the human body. For F. novicida, adaptation of gene expression in macrophages depends on Cas9, tracRNA, and possibly also scaRNA, which together inhibit expression of an immunogenic lipoprotein (shown in green) (22). All bacteria depicted in this figure possess Cas9, and scaRNA production has been predicted for F. novicida, C. jejuni, L. monocytogenes, and N. meningitidis (22), suggesting that a role of Cas9 in regulation of bacterial gene expression may be more widespread.
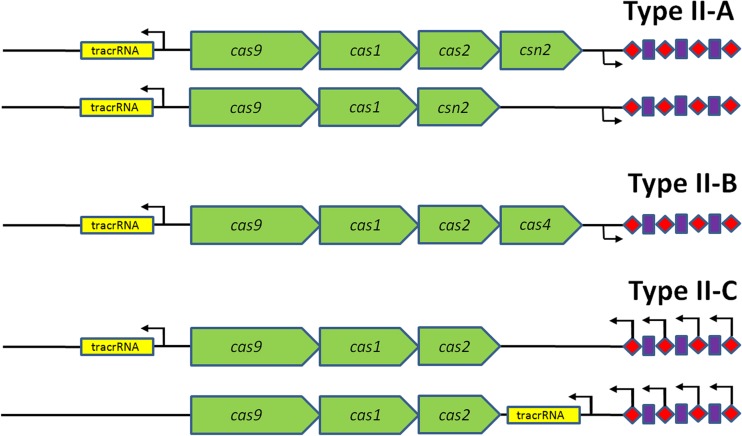
Whereas type I and type III CRISPR-Cas systems share some common features (21), the type II system is rather unique. Apart from a conserved set of cas genes (cas1, cas2, and cas9), three variant subtypes have been recognized: type II-A, with an additional csn2 gene; type II-B, with an additional cas4 gene; and type II-C, with no additional gene (21) (Fig. 2). The usual type II CRISPR-Cas system genomic arrangement is that the cas operons are adjacent to the CRISPR array, together with a DNA sequence encoding a trans-activating CRISPR RNA (tracrRNA). tracrRNA is partly complementary to the repeat part of the immature CRISPR-encoded RNA (crRNA): Watson-Crick base pairing results in an RNA duplex that, while associated with Cas9, is further processed by RNase III (20). Interestingly, type II CRISPR-Cas systems occur only in bacteria, not in archaea. Moreover, type II CRISPR-Cas systems are overrepresented in bacteria that use vertebrates as a host, including a wide variety of pathogens (8, 20, 22). Here we review recent insights into the role of CRISPR-Cas systems in the virulence of mainly pathogenic bacteria, but we also describe recently reported links between CRISPR-Cas and general stress responses in nonpathogenic bacteria.
FIG 2.
Overview of the three type II CRISPR-Cas subtypes. All three subtypes share a conserved set of cas genes: cas1, cas2, and cas9. Type II-A has an additional csn2 gene, and type II-B has an additional cas4 gene (21). Type II-C does not feature an additional cas gene beyond the three conserved cas genes (122). All subtypes feature a small trans-encoded RNA called trans-activating CRISPR RNA (tracrRNA); type II-C displays variation in the location of tracrRNA. cas9, cas1, and cas2 are indicated with green arrows, and tracrRNA is shown with yellow boxes. Transcription start sites are shown as black arrows upstream of the repeats (red diamonds) and spacers (purple squares) in type II-A (e.g., in Streptococcus spp.) and -B (e.g., in Legionella pneumophila) CRISPR loci, or within each spacer in the case of the minimal type II-C CRISPR systems of Neisseria meningitidis (upper) and Campylobacter jejuni (lower) (122).
CRISPR-Cas AS A TYPING TOOL
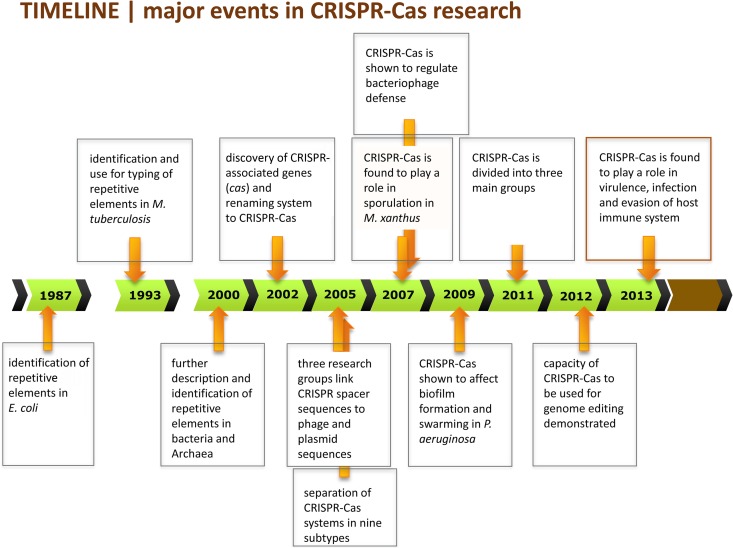
Major discoveries in CRISPR-Cas research, including the elucidation of its role in bacterial virulence, are highlighted in a timeline in Fig. 3. Initially, before an understanding of their physiological role, CRISPR-Cas systems were found to be a useful tool for typing bacterial diversity (23). In Mycobacterium tuberculosis (type III-A), for instance, CRISPR variability has been a gold standard for routine genotyping purposes and to study the epidemiology of M. tuberculosis (24–27). However, limitations exist for the use of CRISPR typing in evolutionary studies, because it is impossible to study which or how many evolutionary events are responsible for the loss of a cluster of neighboring spacers from the CRISPR (27). In 2003, CRISPR-Cas typing of C. jejuni was explored for the first time to reveal phylogenetic relationships between strains in population biology and epidemiology studies (28). For C. jejuni (type II-C), CRISPR typing alone appeared not to be useful, since the spacers were too diverse. However, a combination of CRISPR typing with amplified fragment length polymorphisms (AFLP) and multilocus sequence typing (MLST) techniques enhanced the discriminatory power, enabling subtyping of C. jejuni isolates (28). The CRISPR typing techniques for C. jejuni were further optimized, resulting in a technique called CRISPR high-resolution melt analysis (29). Six years later, exploration of sequence variation in the C. jejuni CRISPR-Cas locus established that (non)synonymous polymorphisms in the cas genes were linked to the presence of a gene found specifically in clinical C. jejuni isolates retrieved from Guillain-Barré syndrome (GBS) patients (30, 31). Louwen et al. thus established that not only the spacer variation of the CRISPR array but also single nucleotide polymorphisms (SNPs) of the cas genes in C. jejuni are useful for typing purposes (30). Likewise, in Corynebacterium diphtheriae, the CRISPR-Cas systems (type I-E and type II-C) were found to be useful for typing purposes (32, 33). Remarkably, spoligotyping, a technique making use of sequence information contained within the CRISPR spacers, was found to provide enhanced discriminatory power in C. diphtheriae compared to pulsed-field gel electrophoresis (PFGE), random amplification of polymorphic DNA (RAPD), and AFLP analyses (33). Furthermore, spoligotyping had discriminatory power for subtyping of Legionella pneumophila strains and enabled the identification of environmental sources that caused clinical outbreaks (34).
FIG 3.
Overview of the most important discoveries in CRISPR-Cas research. The original papers describing the major findings are discussed and cited in the main text.
As described above for the type II CRISPR-Cas system, the type I CRISPR-Cas system was found to be useful for typing of Yersinia pestis (type I-F) and Salmonella (types I-E and I-F) strains (35, 36). In Y. pestis, CRISPR typing was able to identify the origin of the ancestor strains that caused the black plague (36). Whereas it had been assumed that the black plague originated from Mongolia, CRISPR typing suggested that this region harbored a different, less virulent Yersinia species (Yersinia microtus clade) (37). A separate study had linked geographical sources to outbreaks of Y. pestis isolates based on sequence variation within the CRISPR array (38). From this study, it was concluded that the more virulent Y. pestis strains could have originated from China and Tajikistan (38). Interestingly, 59% of the 131 studied spacers harbored strong identity to a protospacer in a Y. pestis prophage (38), unfortunately without details addressing the correlation between the CRISPR genotype and virulence.
In Salmonella, established techniques such as combining MLST with virulence gene polymorphisms were not able to distinguish between individual outbreak strains, rendering subtyping of Salmonella isolates causing food-related infections during outbreaks impossible (39). The use of Salmonella CRISPR genotypes, either alone (39) or in combination with gene polymorphisms present in virulence-associated genes (35, 40), has strongly improved the ability to separate Salmonella strain collections into individual outbreak isolates. Thus, elevated acquisition and turnover of CRISPR spacers of type I-F and I-E CRISPR-Cas systems in Y. pestis and Salmonella spp., respectively, do allow for high-resolution typing. Indeed, a recent study of Salmonella enterica established that CRISPR typing in combination with a method exploiting the hypervariability of the virulence genes, CRISPR-MVLST, was useful for subtyping purposes (41).
In several other bacterial species, typing by making use of CRISPR array polymorphisms has shown some promise in delineating subgroups with biologically relevant characteristics. For the plant pathogen Erwinia amylovora, (type I-E) CRISPR typing enabled the separation of this bacterium into three major groups that reflected their geographic origins (42).
For Propionibacterium acnes, CRISPR typing revealed three lineages that had also been identified in previous typing studies (43). Remarkably, for this bacterium, there was a profound difference in virulence between the first lineage and the other two. Lineage I was associated with infections of sebaceous glands of the human skin and inflammatory acne, whereas lineages II and III caused more severe deep tissue infections (43). Lineage I was found to contain integrated phages and plasmid DNA, but it lacks a CRISPR-Cas system and harbors only the cas2 and cas1 genes (44). Lineage II has a complete type I-E CRISPR-Cas system with at least eight cas genes, and lineage III has four cas genes, possibly reflecting a degenerate type I-E system (44). The CRISPR-Cas-bearing lineages II and III are more invasive (44), which makes them interesting candidates for generating cas gene knockouts for each P. acnes lineage and investigating if the differential presence of CRISPR-Cas genes is a major genomic factor explaining their virulence. In 2012, Marinelli et al. revealed that some P. acnes strains harboring a complete type I-E CRISPR-Cas system comprising spacers with 100% identity against specific bacteriophage protospacers were resistant to these bacteriophages (45). This study also revealed that strains belonging to the more invasive lineages, but lacking such spacers or a complete CRISPR-Cas system, were lysed by these bacteriophages, suggesting that bacteriophage therapy might eventually be used to treat acne (45).
DIFFERENT EFFICIENCIES OF CRISPR-Cas TYPING
In enterococci, the presence or absence of CRISPR-Cas provides the ability to distinguish antibiotic-resistant species, with a wide variety of plasmids carrying antibiotic resistance genes, from less resistant species (46). van Schaik et al. sequenced seven Enterococcus faecium strains, including four clinical isolates and three fecal isolates. It was observed that all seven isolates contained a type II-A cas operon that did not include a cas1 gene. Since cas1 encodes a nuclease that is involved in spacer acquisition (47), this probably implies that in these isolates the CRISPR system had lost its potential for CRISPR adaptation (48). On the other hand, it cannot be ruled out that CRISPR interference is still effective despite the loss of cas1. Note that increased antibiotic resistance as well as the uptake of phages and pathogenicity islands by this bacterium was found to be associated with deletions of cas genes (48). Indeed, in a recent Canadian study, the emergence of ampicillin-resistant E. faecium isolates was associated with a total absence of CRISPR sequences (49). An anticorrelation between the presence/absence of a CRISPR-Cas system and the absence/presence of mobile elements providing antibiotic resistance was also observed in methicillin-resistant Staphylococcus aureus (MRSA) isolate ST779 (50). This isolate had acquired a composite island (CI) element (including methicillin resistance genes) that had integrated into the genome. In addition, this strain was found to harbor a type II-C CRISPR-Cas system, possibly resulting from a different integration event (50). Both the resistance genes and the type II-C CRISPR-Cas system were suggested to originate from coagulase-negative staphylococci (CoNS) (50), which are commensal bacteria that lack the ability to clot blood by the enzyme coagulase, a staphylococcal virulence factor. Links between the presence/absence of CRISPR systems and horizontal transmission of antibiotic resistance and virulence genes from CoNS to S. aureus are thought to be more prevalent than previously considered (51).
For enterohemorrhagic E. coli (EHEC) bacteria, (type I-E/I-F) CRISPR polymorphisms were found to correlate with the presence of two EHEC virulence genes, stx and eae, encoding the phage-delivered Shiga toxin and the intimin virulence factor, respectively (52). Interestingly, the CRISPR polymorphisms were found to provide a more specific typing profile than the established techniques, which were based on stx and eae gene polymorphisms alone or together with O:H serotypes (52). This suggests that a significant correlation exists between CRISPR genotypes and an isolate's virulence. Potential causality between CRISPR spacers and virulence is discussed below. In other E. coli strains, it is questionable whether the CRISPR is useful for diagnostic, epidemiology, or evolutionary studies. In some E. coli strains, it is observed that the CRISPR arrays not only are relatively small but also appear to have remained unaltered for evolutionarily relevant periods (hundreds of thousands of years), which argues against a role as an active bacterial immune system in these strains (53). This observation was corroborated by a study where the CRISPR-Cas system in E. coli was not found to provide strong resistance against the spread of antibiotic resistance plasmids (54). Although the E. coli CRISPR-Cas system has often been reported as static and small, a recent study suggests that this might be beneficial for separating commensal fecal E. coli isolates from more pathogenic variants (55). The observation that short or absent CRISPRs correspond with increased pathogenicity is in line with a recent study in C. jejuni, in which strains causing severe gastroenteritis and postinfectious complications also harbored short CRISPRs or completely lacked the CRISPR array (30).
Genome sequencing of a highly virulent Streptococcus pyogenes strain with a high transformation efficiency revealed the presence of a type II-A CRISPR-Cas system (56). In a subsequent study, 13 S. pyogenes strains were sequenced. Two distinct CRISPR loci with relatively small numbers of spacers compared to those in other streptococci were detected; five strains harbored a CRISPR-Cas system with cas gene deletions, and these strains contained larger numbers of prophages than the other isolates, which harbored a typical type II-A CRISPR-Cas system (57). Spacer analysis revealed that in four isolates, one spacer targeted a protospacer on its own chromosome, i.e., in a prophage-carried gene (57). Nozawa et al. concluded that the limited presence and activity of CRISPR-Cas systems in S. pyogenes have allowed the introduction of virulence genes by phages into S. pyogenes, thereby contributing to the strain-specific pathogenicity that is characteristic of this species (57).
There is some experimental evidence pointing at an interplay between CRISPR-Cas systems, mobile genetic elements, and host range. An association between a reduction of antibacteriophage CRISPR activity and bacterial virulence was found in Mycoplasma gallisepticum bacteria that can infect several bird species (58); these bacteria harbor a type II-A CRISPR-Cas system. During a host switch from poultry to a songbird, a strong reduction in CRISPR spacer diversity and a loss of all of the type II cas genes were observed (58). The authors concluded that the extremely rapid evolution of the bacterial genomes, including the CRISPR degradation following the host shift, pointed to an involvement of mobile genetic elements.
We can conclude that CRISPR typing, either alone or sometimes in combination with other markers, has been used successfully for strain typing. In some cases, CRISPR typing has enabled the characterization and identification of outbreak strains at the serotype and genomic subgroup levels in epidemiological or evolutionary contexts. For instance, variability in CRISPR repeat numbers and cas gene presence has allowed for clustering of clinical enterococcal isolates into subgroups of highly and lowly antibiotic-resistant isolates and for grouping of C. jejuni strains into isolates that induce either postinfectious complications or merely gastroenteritis. Phages and plasmids potentially play important roles in bacterial virulence, e.g., as delivery vehicles of antibiotic resistance and virulence genes (59, 60). Therefore, it is not surprising that grouping of bacterial isolates based on variations in antiphage CRISPR-Cas systems may yield groups of isolates that differ with respect to clinically relevant virulence features. In some peculiar cases, the persistent sequence conservation of the CRISPR-Cas system as observed in E. coli or the profound alterations in this system as observed in Legionella and C. jejuni can negatively affect the discriminatory power of variation in the CRISPR element for techniques such as spoligotyping. Remarkably, in contrast to the established CRISPR typing techniques, a strongly reduced or absent CRISPR in E. coli and C. jejuni enhanced the discriminatory power between pathogens and less virulent or commensal isolates belonging to the same species, respectively, whereas in enterococci, absence of the CRISPR array was associated with increased antibiotic resistance. In conclusion, the discriminatory power of CRISPR-Cas systems can be extremely high in diagnostic, epidemiology, evolutionary, virulence, and antibiotic resistance studies when they are used alone or in combination with other typing techniques, including MLST and PFGE.
FUNCTIONAL DIVERSITY OF CRISPR-Cas SYSTEMS IN BACTERIAL PATHOGENS
The increase in availability of sequenced bacterial, bacteriophage, and plasmid genomes has provided more detailed insights into CRISPR-Cas variation, as well as in the mobile elements that are targeted by CRISPR systems. In 2005, Mojica et al. suggested that on an evolutionary time scale, the pathogenicity of natural prokaryotic populations is largely controlled by bacteriophages and conjugative plasmids and that CRISPR spacers targeting these mobile elements might therefore affect bacterial evolution, including pathogenicity and virulence (5). As mentioned above, CRISPR activity may interfere with the uptake of bacteriophage DNA carrying virulence genes, including toxin and antibiotic resistance genes (51).
The early comparative analyses of CRISPR spacers revealed sequence homology not only to “nonself” DNA of mobile genetic elements but occasionally also to “self,” endogenous chromosomal DNA (5). A multigenome analysis (61) revealed that 1 in every 250 spacers is self-targeting and that such self-targeting occurs in 18% of all CRISPR-bearing organisms. Although the presence of self-spacers has been suggested to allow control of gene expression, the above-mentioned study (61) proposed that (at least in some cases) self-targeting is a form of autoimmunity. The complete lack of conservation of these self-spacers across species, combined with the cooccurrence of degraded repeats near self-targeting spacers, strongly suggests that the acquisition of these spacers is harmful to the stability of the host genome. Indeed, the incorporation of foreign chromosomal fragments with homology against endogenous genes as new CRISPR spacers has been demonstrated to occur frequently in the absence of an active interference system (47). When a complete, active CRISPR-Cas system is present, acquisition of self-targeting spacers is detrimental to genome integrity, and this autoimmunity issue may explain the abundance of degenerated CRISPR systems in prokaryotes with self-targeting spacers. Apart from that, the recent discovery of “CRISPR inhibitors” that reside in certain prophages may also explain the occurrence of self-targeting CRISPR spacers (62). The incidental incorporation of sequences as CRISPR spacers with high identity to endogenous genes thus suggests a role for self-targeting spacers in the regulation of endogenous gene expression in those cases where this type of autoimmunity is not lethal to the bacterium.
In a study on the virulence of Enterococcus faecalis isolates, a mouse urinary tract model (63) was used to analyze two strains: one with and one without a type II-A CRISPR-Cas system (64). Initially, the virulence of the type II CRISPR-Cas-harboring strain appeared to be lower (the 50% lethal dose [LD50] was higher); however, when equal inocula of both strains were used, the CRISPR-Cas-harboring strain induced a more rapid mortality in the mice (64). Histological examinations showed that the CRISPR-Cas-harboring strain had an increased capacity to form biofilms, and as such, it did colonize the organs of the mouse more efficiently than the isolate lacking the system (64). In conclusion, these Enterococcus studies suggest that CRISPR-Cas systems in addition to other genomic differences may influence bacterial pathogenicity via two non-mutually exclusive processes: on the one hand, defense by CRISPR-Cas may reduce the potential bacterial virulence when mobile elements could introduce foreign DNA carrying potential virulence factors (toxins or antibiotic resistance genes), whereas on the other hand, control of gene expression by CRISPR-Cas may enhance bacterial virulence, e.g., by promoting host colonization.
Kuenne et al. studied the genome sequences of 16 Listeria monocytogenes strains and divided the detected CRISPR-Cas systems into three different loci (65). CRISPR-Cas locus 1 was characterized by a single CRISPR array, CRISPR-Cas locus 2 belonged to type I-B, and CRISPR-Cas locus 3 was classified as type II-A (8, 21, 65). Interestingly, CRISPR-Cas locus 1 had previously been associated with the presence of a tracrRNA suggested to control virulence in L. monocytogenes strain 1/2a EGD-e during growth in macrophages (66), but it remained mechanistically unknown how this trans-acting noncoding RNA could regulate virulence. The suggestion that an antisense RNA interference system could form the basis for controlling bacterial pathogenicity in this L. monocytogenes isolate (66) was close to the actual molecular mechanism identified in a different bacterial species (see below).
To summarize, analyses of genomic sequences of diverse pathogenic bacteria and their virulence features suggest a role of CRISPR-Cas in processes other than defense, e.g., a potential involvement in regulation of endogenous gene expression (5, 67), including that of genes involved in virulence (57, 65). In the aforementioned examples, direct or indirect links between CRISPR-Cas and control of virulence were suggested but were not explored further. Below, we discuss recently reported studies in which more convincing evidence for the involvement of CRISPR-Cas in control of bacterial stress responses, including responses to host immunity, was obtained.
TRANSCRIPTIONAL INDUCTION OF THE CRISPR-Cas SYSTEM UPON STRESS
In Myxococcus xanthus, a gene operon, previously named dev, represents a type I-B CRISPR-Cas system in which the cas genes and CRISPR cassette are activated during stress (68). Expression of this operon contributes to the development of fruiting bodies from which bacterial spores are released (68). A few candidate CRISPR spacers might be involved in the regulation of this sporulation event in M. xanthus (68, 69), but no mechanistic connection has been established between these spacers and fruiting body development. An interesting hypothesis put forward was that mutations in one or more of the cas genes have led to exaptation of the M. xanthus CRISPR-Cas system into a regulatory system that controls the stress-dependent development of fruiting bodies (69).
In E. coli, stress on the cell envelope may result in an induction of cas gene expression (70). Apart from regulation by H-NS, LeuO, and Rcs/BglJ (71, 72), a two-component signal transduction system named BaeSR has been found to activate the expression of the E. coli cas genes (70). Perez-Rodriguez et al. established that the periplasmic expression of a green fluorescent protein (GFP) reporter (fused to the Tat-dependent excretion signal peptide of trimethylamine N-oxide reductase [ssTorA]) was diminished in the absence of the chaperone DnaK (70). Random mutagenesis of this E. coli DnaK mutant revealed that deletion of the type I-E cas operon restored the expression of ssTor-GFP, as did deletion of the BaeSR system (70). Hence, the CRISPR-Cas system appeared to be expressed by BaeSR upon cell envelope stress. Subsequently, upon its expression, the CRISPR-Cas system was found to target ssTorA-encoding sequences via partly complementary spacers, directly affecting protein transport across the bacterial membrane (70). Although the molecular details are not well understood, this study again suggests that the function of CRISPR-Cas goes beyond viral defense and plasmid conjugation.
TRANSCRIPTOME ANALYSIS OF THE cas GENES
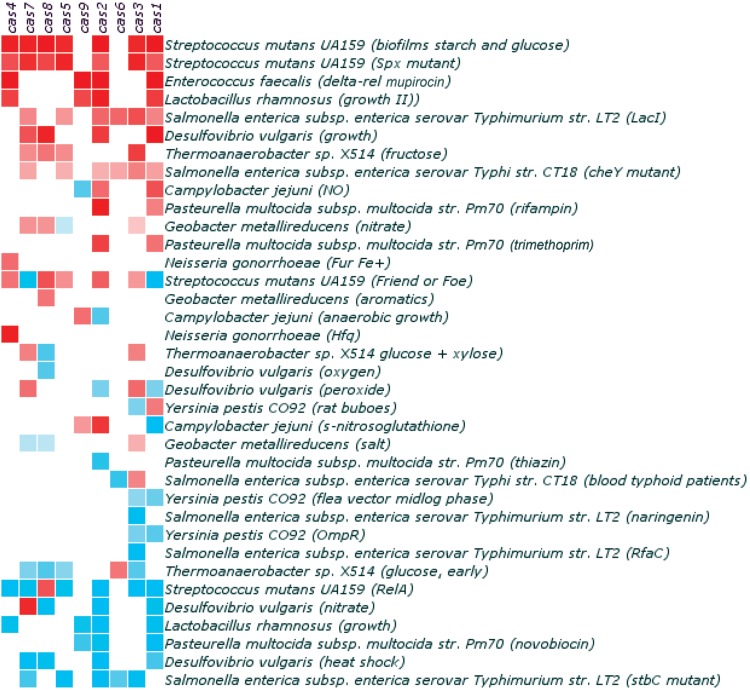
In several bacterial species, transcriptome studies have revealed that deletion of transcriptional regulators affects cas gene expression. For example, in Y. pestis CO92, the regulator OmpR was found to control the composition of the outer membrane and to be required for virulence and survival in macrophages (73). Deletion of OmpR in Y. pestis CO92 resulted in differential expression of 224 genes, including repression of cas1 transcription (Fig. 1 and 4) (73). The cas1 gene was also slightly downregulated during the preadaptation phase, when the Y. pestis CO92 strain adapted itself to the environment in fleas that parasitize rats (Fig. 1 and 4). In fleas, which present a low-temperature environment, Y. pestis forms a biofilm that promotes transmission when the fleas bite a rat host. Upon transmission to a rat, the temperature shift to 37°C induces production of Y. pestis virulence factors that confer resistance to innate immunity responses of the host. When the transcriptome of the Y. pestis CO92 wild-type strain that was used in the OmpR study was determined after passage in rats, cas1 transcription was induced (Fig. 1 and 4). Escape of Y. pestis from rat lymph nodes is characterized by systemic spread, leading to fatal sepsis (74). During dispersion, to escape the innate immune system, Y. pestis has evolved a strong adaptive response against nitric oxide (NO) (74), which is a free radical that is released in immune cells when bacteria are phagocytosed. NO molecules are highly toxic for bacteria by causing DNA damage (75). Escape from the innate immune response coincided with differential transcription of cas1 (Fig. 1 and 4), among other gene transcripts (73). It is noteworthy that Y. pestis OmpR and Cas1 are involved in the stress response (73), which is reminiscent of the involvement of transcriptional regulators and the CRISPR-Cas systems in the stress responses of E. coli and M. xanthus.
FIG 4.
Heat map showing expression of cas genes in human-associated bacteria that occupy different host niches. The heat map indicates which cas genes are induced (shades of red) or repressed (shades of blue) during bacterial responses to changes in the environment. All gene expression data displayed in this figure have been published, are publicly available in the MicrobesOnline (http://www.microbesonline.org/) and NCBI Entrez (http://www.ncbi.nlm.nih.gov/gene/) gene expression databases, and are further discussed in this review.
E. coli and S. enterica are closely related, and both harbor a type I-E CRISPR-Cas system (21, 76). The E. coli lac operon is important for maintaining fitness in the presence of lactose, and its expression is regulated by LacI. Both the lac operon and the LacI repressor are absent in Salmonella spp. (76). Eswarappa et al. addressed why the lac operon is absent from Salmonella and not from E. coli, whereas both E. coli and Salmonella are exposed to lactose in the mammalian gut (76). These authors showed that introducing the LacI repressor into S. enterica reduced virulence by affecting the transcription of genes on the SpI-1 and SpI-2 pathogenicity islands, while a lack of LacI enhanced the virulence of this bacterial species via SpI-2 (76). Introduction of the LacI repressor was also concomitant with reduced mouse serum resistance and a significantly induced transcription of the S. enterica cas genes (Fig. 1 and 4) (76). In a different study, transcriptomes were obtained for five clinical S. enterica serovar Typhi strains that were isolated from the blood circulation system; these transcriptomes were compared to those of in vitro-grown S. Typhi strains (77). The 331 transcripts that were altered also included multiple cas genes (Fig. 1 and 4), suggesting that the Salmonella CRISPR-Cas system is involved in infection in vivo (77). Also in C. jejuni, changes in cas gene expression were observed during intestinal passage in a mouse model (78).
E. faecalis is a bacterial species that can cause opportunistic infections of the intestine. The GTP pyrophosphokinase (RelA) of this bacterium, involved in (p)ppGpp biosynthesis during amino acid starvation, was shown to play an important role in stress adaptation and virulence (79). Notably, in E. faecalis, the stress response is controlled by the bifunctional synthetase/hydrolase RelA and the monofunctional guanosine pentaphosphate synthetase RelQ, by regulating the production of the effector molecule (p)ppGpp (80). When E. faecalis relA mutant and wild-type strains were treated with antibiotics, a strong downregulation of transcription was detected for a wide variety of genes (79), including the cas genes. In contrast, in a double relA relQ mutant, expression of the cas genes was induced (Fig. 4). In addition to E. faecalis, expression of RelA has been shown to occur in a context of reduced virulence in S. enterica serovar Typhimurium, M. tuberculosis, Vibrio cholerae, and L. monocytogenes (79). How RelA and RelQ control the regulation of cas gene expression is unknown; it appears that the cas genes function together with the RelAQ system during the stress response and the rapid adaptation to potentially unfavorable changes in the bacterial environment.
In Streptococcus mutans, the causative agent of tooth decay, mutations in virulence or global regulatory genes (including genes involved in stress responses) strongly affect gene transcription (81–85), including that of the type II cas genes (Fig. 1 and 4). In a protease gene deletion mutant of S. mutans, the cas genes were differentially transcribed compared to those of wild-type bacteria (Fig. 4). Likewise, deletion of genes from the spx operon, which is involved in the regulation of the stress response in survival and virulence features, led to an induction in transcription of the cas genes (Fig. 1 and 4). Five cas genes belonging to the type I-E and type II-A systems were induced in response to commensal bacteria or microbiota, and two cas genes, including cas1, were downregulated (Fig. 1 and 4). All of the cas genes in S. mutans were induced during biofilm formation in the presence of starch or sucrose (Fig. 1 and 4). In short, gene transcription data for S. mutans suggest that the cas genes of the two different CRISPR-Cas systems present in this bacterial species are activated during stress. Functional analysis of cas gene deletion mutants could shed light on the diverse involvement in stress responses of the cas genes of S. mutans.
Changes in cas gene expression in response to stress appear to be a general phenomenon. In Desulfovibrio vulgaris (86), Streptococcus sanguinis (87), Pasteurella multocida (88), Lactobacillus rhamnosus (89, 90), and C. jejuni (MicrobesOnline database [http://www.microbesonline.org]), it has been demonstrated that cas gene transcription is commonly altered in response to changes in growth, bile stress, reactive oxygen species (ROS) and nitrosative stress, antibiotics, and expression of genetic competence (Fig. 1 and 4). In addition, several studies support the idea that regulatory factors involved in stress and virulence, such as LacI in S. enterica, OmpR in Y. pestis, RelA and RelQ in S. mutans, and BaeSR/DnaK in E. coli, might somehow interact with the CRISPR-Cas systems of these bacteria. Virulence is a specific stress response of pathogenic bacteria during host infection, resulting in the coordinated expression of genes encoding virulence factors, including host colonization and survival factors. The findings listed above suggest that there might be a more general involvement of CRISPR-Cas in virulence than previously appreciated, although the evidence is circumstantial and descriptive, not providing clues for a molecular mechanism. For the remainder of this review, we discuss findings that have led to the unambiguous demonstration that at least in some bacteria, the CRISPR-Cas system does play an important role in regulating the expression of virulence genes.
CORRELATION OF CRISPR-Cas WITH BACTERIAL VIRULENCE
Legionella pneumophila strain 130b harbors a type II-B CRISPR-Cas system whose cas2 and cas1 genes were induced during intracellular growth in macrophages and aquatic amoebae (91). Analysis of knockout mutants of the type II-B CRISPR-Cas system showed that cas9, cas1, and cas4 were not essential for growth in macrophages and aquatic amoebae. In contrast, disruption of cas2 was found to affect intracellular survival and replication in amoebae (91). This study indicated that the type II CRISPR-Cas system of L. pneumophila plays an important role in withstanding stress encountered during intracellular growth in amoebae.
In 2009, Zegans et al. showed that in Pseudomonas aeruginosa, CRISPR-Cas was involved in biofilm formation and swarming, which are important characteristics of P. aeruginosa virulence (92). These authors observed that infection of P. aeruginosa isolate PA14 with the bacteriophage DMS3 blocked biofilm formation and swarming motility (92) and that these changes were dependent on the presence of a type I-F CRISPR-Cas system (92). A follow-up study demonstrated that a specific spacer present in the type I-F CRISPR array was required to inhibit biofilm formation (93). It was hypothesized that transcription of the specific spacer resulted in production of an antisense RNA that was involved in knockdown of the gene involved in P. aeruginosa biofilm formation (93). These findings and their associated hypotheses indicated that in addition to involvement in viral defense, CRISPR-Cas systems could indeed be involved in the regulation of expression of virulence genes.
Cas9 AS A REGULATOR OF BACTERIAL VIRULENCE
A wide variety of important pathogens of mammals bear a type II CRISPR-Cas system, including major pathogens such as L. monocytogenes, S. pyogenes, Streptococcus agalactiae, Neisseria meningitidis, C. jejuni, Haemophilus influenzae, and Helicobacter pylori (8, 20, 30, 94). These pathogens are able to cause acute or chronic damage to the host (95, 96) or are linked to (postinfectious) complications (97–107).
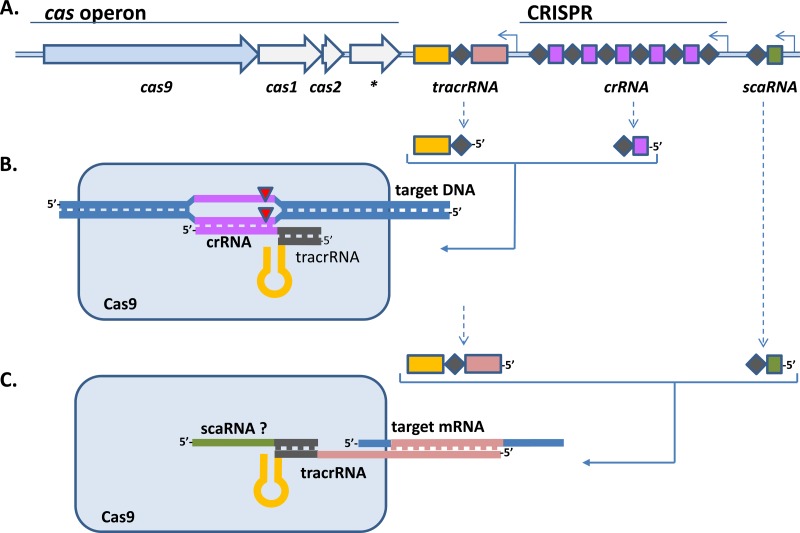
Which virulence factor(s) could be shared by all these diverse type II CRISPR-Cas-bearing pathogens? Type II CRISPR-Cas systems are represented by the CRISPR-Cas system that was originally found in the N. meningitidis isolate Z2491 (21) and later established by Barrangou et al. to be a functional viral defense system in Streptococcus thermophilus (11). Next, an important feature of N. meningitidis is the ability to express sialylated lipooligosaccharide (LOS) structures on its cell envelope (108). C. jejuni, H. influenzae, H. pylori, and P. multocida are also able to sialylate LOS by using species-specific sialyltransferase enzymes (108). C. diphtheriae, L. monocytogenes, M. gallisepticum, S. pyogenes, S. agalactiae, and S. mutans all produce sialidases to remove sialic acid from host glycoproteins in order to uncover host adhesion receptors (108). The enzymatically released sialic acids are then used either as an energy source or as building blocks for incorporation into the bacterial cell envelope (108–113), where they contribute to serum resistance (114). For N. meningitidis, H. influenzae, and C. jejuni, the sialylation of the cell envelope was found to be an important virulence factor that strongly contributed to the ability of these pathogens to adhere to, invade, and translocate across epithelial cells and to evade host immune responses (115–119). Sialylated cell envelopes produced by C. jejuni and H. influenzae have been linked to the induction of the postinfectious sequela Guillain-Barré syndrome (GBS) (102). In addition to a role in virulence, sialylated LOS produced by Gram-negative bacteria has been proposed to play a role in viral defense (120). Indeed, incorporation of sialic acid into LOS structures present on the cell envelope of C. jejuni can protect this bacterium from viral infections (30). Remarkably, deletion of cas9 in GBS-inducing C. jejuni isolates abolished the ability of these isolates to translocate across polarized intestinal epithelial cells, indicating that C. jejuni Cas9 not only protects against infection by viruses but also is crucial for virulence (30). An important observation is the fact that cas9 deletion mutants with sialylated LOS bind more strongly to human serum (30), indicating that Cas9 and sialylated LOS both might play important roles in avoiding immune recognition of C. jejuni. As proof of principle, supplementation of cas9 in an isolate lacking a CRISPR-Cas system led to a significant increase of virulence of this isolate, showing that Cas9 is indeed important for virulence of C. jejuni (30). The question remained: how could Cas9 influence virulence? An intriguing answer to this question has now been provided for a different Gram-negative pathogen, i.e., Francisella novicida.
In F. novicida, overproduction of a specific bacterial lipoprotein (BLP) in the bacterial membrane significantly decreased bacterial survival in host macrophages, and regulation of production of specific BLP strongly contributed to innate immune evasion (121). Sampson et al. demonstrated that predicted antisense base pairing between a 3′ extension of the tracrRNA, in a complex with Cas9, and a complementary BLP mRNA molecule forms a double-stranded RNA (dsRNA) complex that eventually leads to the degradation of BLP mRNA (22) (Fig. 5). Cas9-mediated degradation of BLP mRNA was shown to be activated after phagocytosis of bacteria by macrophages. tracrRNA-mediated silencing of BLP production led to avoidance of Toll-like receptor 2 (TLR2) signaling and was subsequently associated with intramacrophage survival of F. novicida bacteria (22). The importance of Cas9-mediated inhibition of BLP production could also be demonstrated experimentally in vivo. F. novicida knockout mutants of either cas9, tracrRNA, or an additional, putative small RNA containing a CRISPR repeat (scaRNA) were not able to cause lethal infections in a mouse model (22). This landmark paper therefore describes a molecular mechanism by which a CRISPR-Cas system is involved in bacterial pathogenicity, in this case, by repressing production of an immunogenic membrane protein via an antisense RNA-based silencing mechanism that uses two different RNA molecules and the Cas9 protein.
FIG 5.
Dual function of type II CRISPR-Cas systems. (A) Genomic locus of type II CRISPR-Cas system. The cas operon consists of at least three genes (cas9, cas1, and cas2). A fourth gene (*) is present in type II-A (csn2) and II-B (cas4) systems but not in type II-C systems (122). Adjacent to the cas operon, the CRISPR locus is present (dark purple diamonds indicate repeats, and bright purple squares indicate the spacers), as well as the trans-encoded CRISPR RNA (tracrRNA) gene and possibly the recently proposed scaRNA gene (22). The order and orientation of the CRISPR and the genes vary in different genomes. (B) A role in defense against DNAs of invading genetic elements is well established (11), in which processed crRNA and a short version of the tracrRNA (most likely resulting from processing of a longer tracrRNA transcript or transcription from a second promoter; see panel A) eventually are responsible for interaction with target DNA. Eventually, both DNA strands are cleaved at the active sites of Cas9 (red triangles) (20, 129, 130). (C) A distinct role of Cas9 in virulence has been suggested (30), and a molecular basis for how Cas9 can codetermine virulence has been revealed (22): a long version of the tracrRNA shares significant homology with a target transcript, resulting in silencing and probably degradation of this transcript. Involvement of another small CRISPR-associated RNA (scaRNA) has been proposed; if indeed important, this scaRNA may be involved in stabilizing the interaction of the tracrRNA in the Cas9 complex.
The requirement of different RNA molecules may differ between bacterial species, since in C. jejuni, a significant increase of virulence could be achieved by supplementation of cas9 in a natural strain that lacks CRISPR-Cas (30), an indication that other Cas9-dependent mechanisms that determine virulence may exist. Since cas9 is present in a wide variety of host-associated pathogenic and commensal bacteria, it is tempting to speculate that type II CRISPR-Cas is important not only in virulence but also in commensalism, as suggested by the presence of tracrRNA at or near CRISPR loci of different commensals (22, 122). In this respect, one might hypothesize that commensal bacteria use the type II CRISPR-Cas system to regulate their immune recognition.
The role of Cas9 in bacterial mRNA degradation could lead to confusion due to the established function of Cas9 as an endonuclease in targeting and cleavage of DNA instead of RNA. This apparent conflict can be reconciled by the observation that it is not Cas9 that digests the target dsRNA but, rather, RNase III (20). According to Deltcheva et al., Cas9 functions only as a stabilizer of dsRNA (20). Other RNases, including RNase A, RNase T1, and RNase H, might also be involved in the digestion of dsRNA molecules upon their stabilization by Cas9.
CONCLUDING REMARKS
Polymorphisms in bacterial CRISPR-Cas systems have been shown to be of use for typing purposes and to study evolution and epidemiology in some bacterial species. In several comparative genomic studies, it was noted that subgroups of bacteria with variant CRISPR-Cas systems also appeared to show substantial differences with respect to virulence. Independent studies of both commensal and pathogenic host-associated bacteria have suggested that there might be a role for type II CRISPR-Cas in virulence, perhaps based on an RNA-based mechanism. Indeed, in the bacterial pathogen F. novicida, a ribonucleoprotein complex of Cas9 and a short noncoding tracrRNA was found to suppress production of an immunogenic lipoprotein (BLP) by promoting its mRNA degradation, possibly involving a second short RNA molecule (scaRNA). Avoidance of BLP production is essential for full virulence and immune evasion of F. novicida. Such a mechanism might also be functional in N. meningitidis, C. jejuni, and other bacterial species harboring type II CRISPR-Cas systems, since in C. jejuni and N. meningitidis, cas9 knockout mutants also displayed a loss-of-virulence phenotype as observed for F. novicida.
These findings are of interest to those in both academia and companies and to society, since type II CRISPR-Cas-harboring pathogenic species belonging to the genera Neisseria, Campylobacter, Streptococcus, and others exert enormous pressure on the health care system and food industries. Vaccines are available for some of the type II CRISPR-Cas-harboring pathogens, but for most of these pathogens, vaccines still need to be developed or require continuous adaptation due to fast bacterial evolution. Genetic engineering of the type II CRISPR-Cas system might provide the opportunity to obtain suitably attenuated vaccine candidates, as suggested for F. novicida (22). Indications for a role of the other two CRISPR-Cas systems in pathogenesis have been reported but require more detailed investigations. Unraveling the mechanisms that lead to control of virulence via CRISPR-Cas systems will certainly provide deeper insight into the genetic regulation of gene expression and the ways that bacteria respond to sudden changes in their environment.
The discovery of CRISPR-type repetitive elements in the 1980s resulted in a fascinating research area of bacterial genetics, with several groundbreaking discoveries covering defense against invasion by genetic elements as well as regulation of gene expression. Potential applications of CRISPR-Cas systems range from protecting bacterial production systems against viral infections to multiplex genome editing of microbial and mammalian cells (123–128), potentially even contributing to future gene therapy approaches. The recent discovery that type II CRISPR-Cas is involved in bacterial pathogenesis is an exciting addition to the upsurge of papers describing CRISPR-Cas-associated functions. A picture now emerges that CRISPR-Cas systems may be involved in controlling virulence of very different pathogens that occupy different niches throughout the human body (Fig. 1). Progressive experience using CRISPR-Cas in epidemiology and evolutionary studies might also shed light on how bacterial epidemics and pandemics developed and evolved over time. In addition, it will be interesting to see if CRISPR-Cas systems codetermine commensal and pathogenic lifestyles and transitions between these two lifestyles in pathobionts. More discoveries are expected in the near future concerning CRISPR-Cas control of bacterial commensalism, pathogenicity, innate immune evasion, and virulence.
ACKNOWLEDGMENTS
We thank E. Charpentier (Helmholtz Centre for Infection Research, Braunschweig, Germany) and D. Weiss (Department of Microbiology and Immunology, Emory University, Atlanta, GA) for their advice on the development of the figure that discusses the dual function of type II CRISPR-Cas systems.
We declare that there are no competing financial interests.
Biographies

Rogier Louwen is a postdoctoral researcher who was trained at the Department of Medical Microbiology and Infectious Diseases at the Erasmus Medical Centre (Erasmus MC) Rotterdam (Netherlands). He received his Ph.D. in microbiology in 2012, for research on the role of sialylated lipooligosaccharide structures and the type II CRISPR-Cas system in Campylobacter jejuni pathogenesis and bacteriophage defense. He is currently working as a Junior Scientist with the same research group, trying to elucidate how type II CRISPR-Cas regulates the expression of sialylated LOS structures and lipoproteins on the cell envelope of C. jejuni.

Raymond Staals worked as a Ph.D. student in the department of Biomolecular Chemistry in the Nijmegen Centre for Molecular Life Sciences (NCMLS) at the Radboud University Nijmegen (Netherlands). There, he worked on a multifunctional protein complex which is involved in many stages of human RNA metabolism: the RNA exosome. Over the last 3 years, he has worked as a researcher for the bacterial genetics research group (in the Laboratory of Microbiology, Wageningen University, Netherlands) led by John van der Oost, where he characterized several fundamental aspects of CRISPR-Cas-based defense, with a special focus on the type III-B CRISPR-Cas system of Thermus thermophilus.

Hubert Endtz obtained his M.D. from Leiden University in 1989 and his Ph.D. from the same university in 1993. In 1990, he joined Erasmus MC and was Deputy Head of the Department of Medical Microbiology and Head R&D until 2013. He occupies the Chair of Tropical Bacteriology at Erasmus MC. In 2009, this chair was created as part of a strategic collaboration between the International Centre for Diarrheal Research in Bangladesh and Erasmus MC. Since 2013, he has been Scientific Director of the Foundation Merieux in Lyon, France, a family foundation dedicated to fighting infectious diseases in low-income developing countries. His key interests are infectious diseases in low-income developing countries, diarrheal diseases, respiratory diseases, antimicrobial resistance, Guillain-Barré syndrome and other postinfectious neuropathies, and innovative diagnostics.

Peter van Baarlen obtained his Ph.D. in genetics at Wageningen University in 2001. Since then, he has worked for a private company (2001) and as a postdoctoral researcher in the phytopathology group of Wageningen University (Netherlands), studying the population genetics of and host responses to infection by pathogenic fungi (2002 to 2006). From 2006 to 2008, he worked as a postdoctoral researcher at the Radboud University Medical Centre (Nijmegen, Netherlands), where he studied human responses to lactic acid bacteria by using transcriptomics. In 2008, he joined the host-microbe interactomics (HMI) group at Wageningen University, where he studied interactions between hosts (humans and mice) and pathogenic and commensal bacteria by using functional genomic approaches. Currently, he is assistant professor in the HMI group and performs and supervises research in population genetics, functional genomics, pathway analysis, network biology, and host-microbe interactions.

John van der Oost received his Ph.D. in molecular microbiology at the Free University of Amsterdam (Netherlands) in 1989. He then took Ph.D. positions at Helsinki University (Finland) and at EMBL in Heidelberg (Germany). After 3 years, he returned to the Free University of Amsterdam, on a fellowship of the Netherlands Academy of Science (KNAW). Since 1995, he has been leader of the bacterial genetics research group within the Laboratory of Microbiology at Wageningen University (Netherlands). Since 2005, Dr. van der Oost has been a Full Professor with a Personal Chair in Molecular Microbiology and Biochemistry at Wageningen University. Current research topics include (i) protein optimization, (ii) pathway engineering, and (iii) unraveling of microbial host-virus interactions, including the CRISPR-Cas system.
REFERENCES
- 1.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. 1987. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 169:5429–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakata A, Amemura M, Makino K. 1989. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J. Bacteriol. 171:3553–3556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mojica FJ, Diez-Villasenor C, Soria E, Juez G. 2000. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 36:244–246. 10.1046/j.1365-2958.2000.01838.x [DOI] [PubMed] [Google Scholar]
- 4.Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 43:1565–1575. 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- 5.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60:174–182. 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- 6.Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663. 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- 7.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151:2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- 8.Haft DH, Selengut J, Mongodin EF, Nelson KE. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR-Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. 10.1371/journal.pcbi.0010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lillestol RK, Redder P, Garrett RA, Brugger K. 2006. A putative viral defence mechanism in archaeal cells. Archaea 2:59–72. 10.1155/2006/542818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 1:7. 10.1186/1745-6150-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 12.Westra ER, Staals RH, Gort G, Hogh S, Neumann S, de la Cruz F, Fineran PC, Brouns SJ. 2013. CRISPR-Cas systems preferentially target the leading regions of MOBF conjugative plasmids. RNA Biol. 10:749–761. 10.4161/rna.24202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pougach K, Semenova E, Bogdanova E, Datsenko KA, Djordjevic M, Wanner BL, Severinov K. 2010. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 77:1367–1379. 10.1111/j.1365-2958.2010.07265.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deveau H, Garneau JE, Moineau S. 2010. CRISPR-Cas system and its role in phage-bacteria interactions. Annu. Rev. Microbiol. 64:475–493. 10.1146/annurev.micro.112408.134123 [DOI] [PubMed] [Google Scholar]
- 16.Marraffini LA, Sontheimer EJ. 2010. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 11:181–190. 10.1038/nrg2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrangou R, Horvath P. 2012. CRISPR: new horizons in phage resistance and strain identification. Annu. Rev. Food Sci. Technol. 3:143–162. 10.1146/annurev-food-022811-101134 [DOI] [PubMed] [Google Scholar]
- 18.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U. S. A. 108:10098–10103. 10.1073/pnas.1104144108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. 10.1126/science.1165771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. 2011. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471:602–607. 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. 2011. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9:467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. 2013. A CRISPR-Cas system mediates bacterial innate immune evasion and virulence. Nature 497:254–257. 10.1038/nature12048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grissa I, Bouchon P, Pourcel C, Vergnaud G. 2008. On-line resources for bacterial micro-evolution studies using MLVA or CRISPR typing. Biochimie 90:660–668. 10.1016/j.biochi.2007.07.014 [DOI] [PubMed] [Google Scholar]
- 24.Abadia E, Zhang J, dos Vultos T, Ritacco V, Kremer K, Aktas E, Matsumoto T, Refregier G, van Soolingen D, Gicquel B, Sola C. 2010. Resolving lineage assignation on Mycobacterium tuberculosis clinical isolates classified by spoligotyping with a new high-throughput 3R SNPs based method. Infect. Genet. Evol. 10:1066–1074. 10.1016/j.meegid.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 25.Gomgnimbou MK, Abadia E, Zhang J, Refregier G, Panaiotov S, Bachiyska E, Sola C. 2012. “Spoligoriftyping,” a dual-priming-oligonucleotide-based direct-hybridization assay for tuberculosis control with a multianalyte microbead-based hybridization system. J. Clin. Microbiol. 50:3172–3179. 10.1128/JCM.00976-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comas I, Homolka S, Niemann S, Gagneux S. 2009. Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. 10.1371/journal.pone.0007815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schouls LM, Reulen S, Duim B, Wagenaar JA, Willems RJ, Dingle KE, Colles FM, Van Embden JD. 2003. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J. Clin. Microbiol. 41:15–26. 10.1128/JCM.41.1.15-26.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price EP, Smith H, Huygens F, Giffard PM. 2007. High-resolution DNA melt curve analysis of the clustered, regularly interspaced short-palindromic-repeat locus of Campylobacter jejuni. Appl. Environ. Microbiol. 73:3431–3436. 10.1128/AEM.02702-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Louwen R, Horst-Kreft D, de Boer AG, van der Graaf L, de Knegt G, Hamersma M, Heikema AP, Timms AR, Jacobs BC, Wagenaar JA, Endtz HP, van der Oost J, Wells JM, Nieuwenhuis EE, van Vliet AH, Willemsen PT, van Baarlen P, van Belkum A. 2013. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barré syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 32:207–226. 10.1007/s10096-012-1733-4 [DOI] [PubMed] [Google Scholar]
- 31.van Belkum A, van den Braak N, Godschalk P, Ang W, Jacobs B, Gilbert M, Wakarchuk W, Verbrugh H, Endtz H. 2001. A Campylobacter jejuni gene associated with immune-mediated neuropathy. Nat. Med. 7:752–753. 10.1038/89831 [DOI] [PubMed] [Google Scholar]
- 32.Mokrousov I, Limeschenko E, Vyazovaya A, Narvskaya O. 2007. Corynebacterium diphtheriae spoligotyping based on combined use of two CRISPR loci. Biotechnol. J. 2:901–906. 10.1002/biot.200700035 [DOI] [PubMed] [Google Scholar]
- 33.Mokrousov I, Vyazovaya A, Kolodkina V, Limeschenko E, Titov L, Narvskaya O. 2009. Novel macroarray-based method of Corynebacterium diphtheriae genotyping: evaluation in a field study in Belarus. Eur. J. Clin. Microbiol. Infect. Dis. 28:701–703. 10.1007/s10096-008-0674-4 [DOI] [PubMed] [Google Scholar]
- 34.Ginevra C, Jacotin N, Diancourt L, Guigon G, Arquilliere R, Meugnier H, Descours G, Vandenesch F, Etienne J, Lina G, Caro V, Jarraud S. 2012. Legionella pneumophila sequence type 1/Paris pulsotype subtyping by spoligotyping. J. Clin. Microbiol. 50:696–701. 10.1128/JCM.06180-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, de Romans S, Lim C, Roux C, Passet V, Diancourt L, Guibourdenche M, Issenhuth-Jeanjean S, Achtman M, Brisse S, Sola C, Weill FX. 2012. CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One 7:e36995. 10.1371/journal.pone.0036995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vergnaud G, Li Y, Gorge O, Cui Y, Song Y, Zhou D, Grissa I, Dentovskaya SV, Platonov ME, Rakin A, Balakhonov SV, Neubauer H, Pourcel C, Anisimov AP, Yang R. 2007. Analysis of the three Yersinia pestis CRISPR loci provides new tools for phylogenetic studies and possibly for the investigation of ancient DNA. Adv. Exp. Med. Biol. 603:327–338. 10.1007/978-0-387-72124-8_30 [DOI] [PubMed] [Google Scholar]
- 37.Riehm JM, Vergnaud G, Kiefer D, Damdindorj T, Dashdavaa O, Khurelsukh T, Zoller L, Wolfel R, Le Fleche P, Scholz HC. 2012. Yersinia pestis lineages in Mongolia. PLoS One 7:e30624. 10.1371/journal.pone.0030624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui Y, Li Y, Gorge O, Platonov ME, Yan Y, Guo Z, Pourcel C, Dentovskaya SV, Balakhonov SV, Wang X, Song Y, Anisimov AP, Vergnaud G, Yang R. 2008. Insight into microevolution of Yersinia pestis by clustered regularly interspaced short palindromic repeats. PLoS One 3:e2652. 10.1371/journal.pone.0002652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F, Kariyawasam S, Jayarao BM, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Subtyping Salmonella enterica serovar Enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl. Environ. Microbiol. 77:4520–4526. 10.1128/AEM.00468-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao G, Meng J, Strain E, Stones R, Pettengill J, Zhao S, McDermott P, Brown E, Allard M. 2013. Phylogenetics and differentiation of Salmonella newport lineages by whole genome sequencing. PLoS One 8:e55687. 10.1371/journal.pone.0055687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shariat N, Kirchner MK, Sandt CH, Trees E, Barrangou R, Dudley EG. 2013. Subtyping of Salmonella enterica serovar Newport outbreak isolates by CRISPR-MVLST and determination of the relationship between CRISPR-MVLST and PFGE results. J. Clin. Microbiol. 51:2328–2336. 10.1128/JCM.00608-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rezzonico F, Smits TH, Duffy B. 2011. Diversity, evolution, and functionality of clustered regularly interspaced short palindromic repeat (CRISPR) regions in the fire blight pathogen Erwinia amylovora. Appl. Environ. Microbiol. 77:3819–3829. 10.1128/AEM.00177-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruggemann H, Lomholt HB, Tettelin H, Kilian M. 2012. CRISPR-Cas loci of type II Propionibacterium acnes confer immunity against acquisition of mobile elements present in type I P. acnes. PLoS One 7:e34171. 10.1371/journal.pone.0034171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruggemann H, Lomholt HB, Kilian M. 2012. The flexible gene pool of Propionibacterium acnes. Mob. Genet. Elements 2:145–148. 10.4161/mge.21204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marinelli LJ, Fitz-Gibbon S, Hayes C, Bowman C, Inkeles M, Loncaric A, Russell DA, Jacobs-Sera D, Cokus S, Pellegrini M, Kim J, Miller JF, Hatfull GF, Modlin RL. 2012. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. mBio 3:e00279–12. 10.1128/mBio.00279-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer KL, Gilmore MS. 2010. Multidrug-resistant enterococci lack CRISPR-Cas. mBio 1:e00227-10. 10.1128/mBio.00227-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yosef I, Goren MG, Qimron U. 2012. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 40:5569–5576. 10.1093/nar/gks216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H, Willems RJ. 2010. Pyrosequencing-based comparative genome analysis of the nosocomial pathogen Enterococcus faecium and identification of a large transferable pathogenicity island. BMC Genomics 11:239. 10.1186/1471-2164-11-239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tremblay CL, Charlebois A, Masson L, Archambault M. 2013. Characterization of hospital-associated lineages of ampicillin-resistant Enterococcus faecium from clinical cases in dogs and humans. Front. Microbiol. 4:245. 10.3389/fmicb.2013.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinnevey PM, Shore AC, Brennan GI, Sullivan DJ, Ehricht R, Monecke S, Slickers P, Coleman DC. 2013. Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals. Antimicrob. Agents Chemother. 57:524–531. 10.1128/AAC.01689-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Otto M. 2013. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection: staphylococcal commensal species such as Staphylococcus epidermidis are being recognized as important sources of genes promoting MRSA colonization and virulence. Bioessays 35:4–11. 10.1002/bies.201200112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delannoy S, Beutin L, Fach P. 2012. Use of clustered regularly interspaced short palindromic repeat sequence polymorphisms for specific detection of enterohemorrhagic Escherichia coli strains of serotypes O26:H11, O45:H2, O103:H2, O111:H8, O121:H19, O145:H28, and O157:H7 by real-time PCR. J. Clin. Microbiol. 50:4035–4040. 10.1128/JCM.02097-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Touchon M, Charpentier S, Clermont O, Rocha EP, Denamur E, Branger C. 2011. CRISPR distribution within the Escherichia coli species is not suggestive of immunity-associated diversifying selection. J. Bacteriol. 193:2460–2467. 10.1128/JB.01307-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Touchon M, Charpentier S, Pognard D, Picard B, Arlet G, Rocha EP, Denamur E, Branger C. 2012. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology 158:2997–3004. 10.1099/mic.0.060814-0 [DOI] [PubMed] [Google Scholar]
- 55.Dang TN, Zhang L, Zollner S, Srinivasan U, Abbas K, Marrs CF, Foxman B. 2013. Uropathogenic Escherichia coli are less likely than paired fecal E. coli to have CRISPR loci. Infect. Genet. Evol. 19:212–218. 10.1016/j.meegid.2013.07.017 [DOI] [PubMed] [Google Scholar]
- 56.McShan WM, Ferretti JJ, Karasawa T, Suvorov AN, Lin S, Qin B, Jia H, Kenton S, Najar F, Wu H, Scott J, Roe BA, Savic DJ. 2008. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190:7773–7785. 10.1128/JB.00672-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nozawa T, Furukawa N, Aikawa C, Watanabe T, Haobam B, Kurokawa K, Maruyama F, Nakagawa I. 2011. CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS One 6:e19543. 10.1371/journal.pone.0019543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Delaney NF, Balenger S, Bonneaud C, Marx CJ, Hill GE, Ferguson-Noel N, Tsai P, Rodrigo A, Edwards SV. 2012. Ultrafast evolution and loss of CRISPRs following a host shift in a novel wildlife pathogen, Mycoplasma gallisepticum. PLoS Genet. 8:e1002511. 10.1371/journal.pgen.1002511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hacker J, Blum-Oehler G, Hochhut B, Dobrindt U. 2003. The molecular basis of infectious diseases: pathogenicity islands and other mobile genetic elements. A review. Acta Microbiol. Immunol. Hung. 50:321–330. 10.1556/AMicr.50.2003.4.1 [DOI] [PubMed] [Google Scholar]
- 60.Cheetham BF, Katz ME. 1995. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol. Microbiol. 18:201–208. 10.1111/j.1365-2958.1995.mmi_18020201.x [DOI] [PubMed] [Google Scholar]
- 61.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 26:335–340. 10.1016/j.tig.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bondy-Denomy J, Pawluk A, Maxwell KL, Davidson AR. 2013. Bacteriophage genes that inactivate the CRISPR-Cas bacterial immune system. Nature 493:429–432. 10.1038/nature11723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frimodt-Moller N. 1993. The mouse peritonitis model: present and future use. J. Antimicrob. Chemother. 31(Suppl D):55–60. 10.1093/jac/31.suppl_D.55 [DOI] [PubMed] [Google Scholar]
- 64.Bourgogne A, Garsin DA, Qin X, Singh KV, Sillanpaa J, Yerrapragada S, Ding Y, Dugan-Rocha S, Buhay C, Shen H, Chen G, Williams G, Muzny D, Maadani A, Fox KA, Gioia J, Chen L, Shang Y, Arias CA, Nallapareddy SR, Zhao M, Prakash VP, Chowdhury S, Jiang H, Gibbs RA, Murray BE, Highlander SK, Weinstock GM. 2008. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol. 9:R110. 10.1186/gb-2008-9-7-r110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuenne C, Billion A, Mraheil MA, Strittmatter A, Daniel R, Goesmann A, Barbuddhe S, Hain T, Chakraborty T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics 14:47. 10.1186/1471-2164-14-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, Hain T. 2011. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 39:4235–4248. 10.1093/nar/gkr033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimomura Y, Okumura K, Murayama SY, Yagi J, Ubukata K, Kirikae T, Miyoshi-Akiyama T. 2011. Complete genome sequencing and analysis of a Lancefield group G Streptococcus dysgalactiae subsp. equisimilis strain causing streptococcal toxic shock syndrome (STSS). BMC Genomics 12:17. 10.1186/1471-2164-12-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Viswanathan P, Murphy K, Julien B, Garza AG, Kroos L. 2007. Regulation of dev, an operon that includes genes essential for Myxococcus xanthus development and CRISPR-associated genes and repeats. J. Bacteriol. 189:3738–3750. 10.1128/JB.00187-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrangou R, van der Oost J. 2013. CRISPR-Cas systems: RNA-mediated adaptive immunity in bacteria and archaea. Springer, Berlin, Germany. [Google Scholar]
- 70.Perez-Rodriguez R, Haitjema C, Huang Q, Nam KH, Bernardis S, Ke A, DeLisa MP. 2011. Envelope stress is a trigger of CRISPR RNA-mediated DNA silencing in Escherichia coli. Mol. Microbiol. 79:584–599. 10.1111/j.1365-2958.2010.07482.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Westra ER, Pul U, Heidrich N, Jore MM, Lundgren M, Stratmann T, Wurm R, Raine A, Mescher M, Van Heereveld L, Mastop M, Wagner EG, Schnetz K, Van Der Oost J, Wagner R, Brouns SJ. 2010. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol. Microbiol. 77:1380–1393. 10.1111/j.1365-2958.2010.07315.x [DOI] [PubMed] [Google Scholar]
- 72.Arslan Z, Stratmann T, Wurm R, Wagner R, Schnetz K, Pul U. 2013. RcsB-BglJ-mediated activation of Cascade operon does not induce the maturation of CRISPR RNAs in E. coli K12. RNA Biol. 10:708–715. 10.4161/rna.23765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao H, Zhang Y, Han Y, Yang L, Liu X, Guo Z, Tan Y, Huang X, Zhou D, Yang R. 2011. Phenotypic and transcriptional analysis of the osmotic regulator OmpR in Yersinia pestis. BMC Microbiol. 11:39. 10.1186/1471-2180-11-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sebbane F, Lemaitre N, Sturdevant DE, Rebeil R, Virtaneva K, Porcella SF, Hinnebusch BJ. 2006. Adaptive response of Yersinia pestis to extracellular effectors of innate immunity during bubonic plague. Proc. Natl. Acad. Sci. U. S. A. 103:11766–11771. 10.1073/pnas.0601182103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wink DA, Kasprzak KS, Maragos CM, Elespuru RK, Misra M, Dunams TM, Cebula TA, Koch WH, Andrews AW, Allen JS, Keefer LK. 1991. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science 254:1001–1003. 10.1126/science.1948068 [DOI] [PubMed] [Google Scholar]
- 76.Eswarappa SM, Karnam G, Nagarajan AG, Chakraborty S, Chakravortty D. 2009. Lac repressor is an antivirulence factor of Salmonella enterica: its role in the evolution of virulence in Salmonella. PLoS One 4:e5789. 10.1371/journal.pone.0005789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sheikh A, Charles RC, Sharmeen N, Rollins SM, Harris JB, Bhuiyan MS, Arifuzzaman M, Khanam F, Bukka A, Kalsy A, Porwollik S, Leung DT, Brooks WA, LaRocque RC, Hohmann EL, Cravioto A, Logvinenko T, Calderwood SB, McClelland M, Graham JE, Qadri F, Ryan ET. 2011. In vivo expression of Salmonella enterica serotype Typhi genes in the blood of patients with typhoid fever in Bangladesh. PLoS Negl. Trop. Dis. 5:e1419. 10.1371/journal.pntd.0001419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jerome JP, Bell JA, Plovanich-Jones AE, Barrick JE, Brown CT, Mansfield LS. 2011. Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS One 6:e16399. 10.1371/journal.pone.0016399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan X, Zhao C, Budin-Verneuil A, Hartke A, Rince A, Gilmore MS, Auffray Y, Pichereau V. 2009. The (p)ppGpp synthetase RelA contributes to stress adaptation and virulence in Enterococcus faecalis V583. Microbiology 155:3226–3237. 10.1099/mic.0.026146-0 [DOI] [PubMed] [Google Scholar]
- 80.Gaca AO, Abranches J, Kajfasz JK, Lemos JA. 2012. Global transcriptional analysis of the stringent response in Enterococcus faecalis. Microbiology 158:1994–2004. 10.1099/mic.0.060236-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie Z, Okinaga T, Niu G, Qi F, Merritt J. 2010. Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Mol. Microbiol. 78:1431–1447. 10.1111/j.1365-2958.2010.07417.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 192:2546–2556. 10.1128/JB.00028-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kajfasz JK, Abranches J, Lemos JA. 2011. Transcriptome analysis reveals that ClpXP proteolysis controls key virulence properties of Streptococcus mutans. Microbiology 157:2880–2890. 10.1099/mic.0.052407-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J, Wu C, Huang IH, Merritt J, Qi F. 2011. Differential response of Streptococcus mutans towards friend and foe in mixed-species cultures. Microbiology 157:2433–2444. 10.1099/mic.0.048314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ahn SJ, Wen ZT, Burne RA. 2007. Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 189:8519–8527. 10.1128/JB.01180-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mukhopadhyay A, Redding AM, Joachimiak MP, Arkin AP, Borglin SE, Dehal PS, Chakraborty R, Geller JT, Hazen TC, He Q, Joyner DC, Martin VJ, Wall JD, Yang ZK, Zhou J, Keasling JD. 2007. Cell-wide responses to low-oxygen exposure in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 189:5996–6010. 10.1128/JB.00368-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodriguez AM, Callahan JE, Fawcett P, Ge X, Xu P, Kitten T. 2011. Physiological and molecular characterization of genetic competence in Streptococcus sanguinis. Mol. Oral Microbiol. 26:99–116. 10.1111/j.2041-1014.2011.00606.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Melnikow E, Schoenfeld C, Spehr V, Warrass R, Gunkel N, Duszenko M, Selzer PM, Ullrich HJ. 2008. A compendium of antibiotic-induced transcription profiles reveals broad regulation of Pasteurella multocida virulence genes. Vet. Microbiol. 131:277–292. 10.1016/j.vetmic.2008.03.007 [DOI] [PubMed] [Google Scholar]
- 89.Koskenniemi K, Laakso K, Koponen J, Kankainen M, Greco D, Auvinen P, Savijoki K, Nyman TA, Surakka A, Salusjarvi T, de Vos WM, Tynkkynen S, Kalkkinen N, Varmanen P. 2011. Proteomics and transcriptomics characterization of bile stress response in probiotic Lactobacillus rhamnosus GG. Mol. Cell. Proteomics 10:M110.002741. 10.1074/mcp.M110.002741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Laakso K, Koskenniemi K, Koponen J, Kankainen M, Surakka A, Salusjarvi T, Auvinen P, Savijoki K, Nyman TA, Kalkkinen N, Tynkkynen S, Varmanen P. 2011. Growth phase-associated changes in the proteome and transcriptome of Lactobacillus rhamnosus GG in industrial-type whey medium. Microb. Biotechnol. 4:746–766. 10.1111/j.1751-7915.2011.00275.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gunderson FF, Cianciotto NP. 2013. The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio 4:e00074–13. 10.1128/mBio.00074-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zegans ME, Wagner JC, Cady KC, Murphy DM, Hammond JH, O'Toole GA. 2009. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J. Bacteriol. 191:210–219. 10.1128/JB.00797-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Palmer KL, Whiteley M. 2011. DMS3-42: the secret to CRISPR-dependent biofilm inhibition in Pseudomonas aeruginosa. J. Bacteriol. 193:3431–3432. 10.1128/JB.05066-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539–547. 10.1038/41483 [DOI] [PubMed] [Google Scholar]
- 95.Casadevall A, Pirofski LA. 2009. Virulence factors and their mechanisms of action: the view from a damage-response framework. J. Water Health 7(Suppl 1):S2–S18. 10.2166/wh.2009.036 [DOI] [PubMed] [Google Scholar]
- 96.Pirofski LA, Casadevall A. 2008. The damage-response framework of microbial pathogenesis and infectious diseases. Adv. Exp. Med. Biol. 635:135–146. 10.1007/978-0-387-09550-9_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hamon MA, Ribet D, Stavru F, Cossart P. 2012. Listeriolysin O: the Swiss army knife of Listeria. Trends Microbiol. 20:360–368. 10.1016/j.tim.2012.04.006 [DOI] [PubMed] [Google Scholar]
- 98.Mori M, Kuwabara S, Miyake M, Noda M, Kuroki H, Kanno H, Ogawara K, Hattori T. 2000. Haemophilus influenzae infection and Guillain-Barré syndrome. Brain 123:2171–2178. 10.1093/brain/123.10.2171 [DOI] [PubMed] [Google Scholar]
- 99.Harrison LH, Simonsen V, Waldman EA. 2008. Emergence and disappearance of a virulent clone of Haemophilus influenzae biogroup aegyptius, cause of Brazilian purpuric fever. Clin. Microbiol. Rev. 21:594–605. 10.1128/CMR.00020-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bennetto LP, Lyons P. 2004. Miller Fisher syndrome associated with Pasteurella multocida infection. J. Neurol. Neurosurg. Psychiatry 75:1786–1787. 10.1136/jnnp.2004.044032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Polk DB, Peek RM., Jr 2010. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer 10:403–414. 10.1038/nrc2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van Doorn PA, Ruts L, Jacobs BC. 2008. Clinical features, pathogenesis, and treatment of Guillain-Barré syndrome. Lancet Neurol. 7:939–950. 10.1016/S1474-4422(08)70215-1 [DOI] [PubMed] [Google Scholar]
- 103.Brigham KS, Sandora TJ. 2009. Neisseria meningitidis: epidemiology, treatment and prevention in adolescents. Curr. Opin. Pediatr. 21:437–443. 10.1097/MOP.0b013e32832c9668 [DOI] [PubMed] [Google Scholar]
- 104.Galazka AM, Robertson SE. 1995. Diphtheria: changing patterns in the developing world and the industrialized world. Eur. J. Epidemiol. 11:107–117. 10.1007/BF01719955 [DOI] [PubMed] [Google Scholar]
- 105.Ginsburg I, Ward PA, Varani J. 1999. Can we learn from the pathogenetic strategies of group A hemolytic streptococci how tissues are injured and organs fail in post-infectious and inflammatory sequelae? FEMS Immunol. Med. Microbiol. 25:325–338. 10.1111/j.1574-695X.1999.tb01357.x [DOI] [PubMed] [Google Scholar]
- 106.Marteau P, Chaput U. 2011. Bacteria as trigger for chronic gastrointestinal disorders. Dig. Dis. 29:166–171. 10.1159/000323879 [DOI] [PubMed] [Google Scholar]
- 107.Chen GY, Nunez G. 2011. Inflammasomes in intestinal inflammation and cancer. Gastroenterology 141:1986–1999. 10.1053/j.gastro.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Severi E, Hood DW, Thomas GH. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 153:2817–2822. 10.1099/mic.0.2007/009480-0 [DOI] [PubMed] [Google Scholar]
- 109.Davis L, Baig MM, Ayoub EM. 1979. Properties of extracellular neuraminidase produced by group A streptococcus. Infect. Immun. 24:780–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim S, Oh DB, Kwon O, Kang HA. 2010. Identification and functional characterization of the NanH extracellular sialidase from Corynebacterium diphtheriae. J. Biochem. 147:523–533. 10.1093/jb/mvp198 [DOI] [PubMed] [Google Scholar]
- 111.Villanueva MS, Beckers CJ, Pamer EG. 1994. Infection with Listeria monocytogenes impairs sialic acid addition to host cell glycoproteins. J. Exp. Med. 180:2137–2145. 10.1084/jem.180.6.2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beighton D, Whiley RA. 1990. Sialidase activity of the “Streptococcus milleri group” and other viridans group streptococci. J. Clin. Microbiol. 28:1431–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bercic RL, Slavec B, Lavric M, Narat M, Zorman-Rojs O, Dovc P, Bencina D. 2008. A survey of avian Mycoplasma species for neuraminidase enzymatic activity. Vet. Microbiol. 130:391–397. 10.1016/j.vetmic.2008.02.004 [DOI] [PubMed] [Google Scholar]
- 114.Fox KL, Cox AD, Gilbert M, Wakarchuk WW, Li J, Makepeace K, Richards JC, Moxon ER, Hood DW. 2006. Identification of a bifunctional lipopolysaccharide sialyltransferase in Haemophilus influenzae: incorporation of disialic acid. J. Biol. Chem. 281:40024–40032. 10.1074/jbc.M602314200 [DOI] [PubMed] [Google Scholar]
- 115.Harvey HA, Swords WE, Apicella MA. 2001. The mimicry of human glycolipids and glycosphingolipids by the lipooligosaccharides of pathogenic Neisseria and Haemophilus. J. Autoimmun. 16:257–262. 10.1006/jaut.2000.0477 [DOI] [PubMed] [Google Scholar]
- 116.Unkmeir A, Kammerer U, Stade A, Hubner C, Haller S, Kolb-Maurer A, Frosch M, Dietrich G. 2002. Lipooligosaccharide and polysaccharide capsule: virulence factors of Neisseria meningitidis that determine meningococcal interaction with human dendritic cells. Infect. Immun. 70:2454–2462. 10.1128/IAI.70.5.2454-2462.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Louwen R, Heikema A, van Belkum A, Ott A, Gilbert M, Ang W, Endtz HP, Bergman MP, Nieuwenhuis EE. 2008. The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect. Immun. 76:4431–4438. 10.1128/IAI.00321-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Louwen R, Nieuwenhuis EE, van Marrewijk L, Horst-Kreft D, de Ruiter L, Heikema AP, van Wamel WJ, Wagenaar JA, Endtz HP, Samsom J, van Baarlen P, Akhmanova A, van Belkum A. 2012. Campylobacter jejuni translocation across intestinal epithelial cells is facilitated by ganglioside-like lipooligosaccharide structures. Infect. Immun. 80:3307–3318. 10.1128/IAI.06270-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guerry P, Ewing CP, Hickey TE, Prendergast MM, Moran AP. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656–6662. 10.1128/IAI.68.12.6656-6662.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Angata T, Varki A. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439–469. 10.1021/cr000407m [DOI] [PubMed] [Google Scholar]
- 121.Jones CL, Sampson TR, Nakaya HI, Pulendran B, Weiss DS. 2012. Repression of bacterial lipoprotein production by Francisella novicida facilitates evasion of innate immune recognition. Cell. Microbiol. 14:1531–1543. 10.1111/j.1462-5822.2012.01816.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chylinski K, Le Rhun A, Charpentier E. 2013. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 10:726–737. 10.4161/rna.24321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Brouns SJ. 2012. Molecular biology. A Swiss army knife of immunity. Science 337:808–809. 10.1126/science.1227253 [DOI] [PubMed] [Google Scholar]
- 124.Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. 2013. Genome editing with RNA-guided Cas9 nuclease in zebrafish embryos. Cell Res. 23:465–472. 10.1038/cr.2013.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barrangou R. 2013. CRISPR-Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA 4:267–278. 10.1002/wrna.1159 [DOI] [PubMed] [Google Scholar]
- 126.Horvath P, Barrangou R. 2013. RNA-guided genome editing a la carte. Cell Res. 23:733–734. 10.1038/cr.2013.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cho SW, Kim S, Kim JM, Kim JS. 2013. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31:230–232. 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- 128.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. 2013. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 31:233–239. 10.1038/nbt.2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. 2012. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gasiunas G, Barrangou R, Horvath P, Siksnys V. 2012. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]