Abstract
The process of plate streaking has been automated to improve the culture readings, isolation quality, and workflow of microbiology laboratories. However, instruments have not been well evaluated under routine conditions. We aimed to evaluate the performance of the fully automated InoqulA instrument (BD Kiestra B.V., The Netherlands) in the automated seeding of liquid specimens and samples collected using swabs with transport medium. We compared manual and automated methods according to the (i) within-run reproducibility using Escherichia coli-calibrated suspensions, (ii) intersample contamination using a series of alternating sterile broths and broths with >105 CFU/ml of either E. coli or Proteus mirabilis, (iii) isolation quality with standardized mixed bacterial suspensions of diverse complexity and a 4-category standardized scale (very poor, poor, fair to good, or excellent), and (iv) agreement of the results obtained from 244 clinical specimens. By involving 15 technicians in the latter part of the comparative study, we estimated the variability in the culture quality at the level of the laboratory team. The instrument produced satisfactory reproducibility with no sample cross-contamination, and it performed better than the manual method, with more colony types recovered and isolated (up to 11% and 17%, respectively). Finally, we showed that the instrument did not shorten the seeding time over short periods of work compared to that for the manual method. Altogether, the instrument improved the quality and standardization of the isolation, thereby contributing to a better overall workflow, shortened the time to results, and provided more accurate results for polymicrobial specimens.
INTRODUCTION
In recent years, instruments for automated sample inoculation have become available to microbiology laboratories and represent an appealing means to complete a repetitive and tedious process (1). These instruments have been claimed to both improve the quality of colony separation and save technician time; however, the current scientific literature assessing the benefits of automation is scarce. In particular, the improvement in isolation quality has not been demonstrated or quantified in a peer-reviewed publication, as recently noted (2). Only three preliminary evaluation studies are currently available; these include a preliminary assessment of the MicroStreak instrument (bioMérieux, France) (3), a comparison of the WASP instrument (Copan, Italy) with the InocuLAB instrument (Dynacon Inc., Canada) (no longer commercially available) (4), and a study focusing on the microscopy performance associated with liquid swabs and the Previ Isola system (bioMérieux) (5). Moreover, no evaluation method for objectively quantifying the culture quality, the colony separation, or the instrument performance is available in the peer-reviewed literature.
In this study, we aimed to evaluate the performance of the fully automated (FA) InoqulA instrument (BD Kiestra B.V., The Netherlands) compared with manual inoculation. The recovery of bacterial strains, the quality of colony separation on agar plates and its variability, the overall reproducibility, and the total time required for inoculating the samples and streaking plates were evaluated for both mixed bacterial suspensions of various complexity (16 conditions with up to 4 bacterial species) and 244 clinical samples (including urine and various body specimens collected by swabbing).
MATERIALS AND METHODS
Instrument.
The FA InoqulA instrument (BD Kiestra B.V., The Netherlands) tested is a U-shaped automated system for pipetting and distributing liquid specimens onto agar plates and spreading bacteria over the agar surface with a magnetic rolling bead (1, 2). The liquid specimens used here included urine samples and other samples collected via swabs with liquid transport medium.
Bacterial suspensions and clinical samples. (i) Bacterial suspensions.
Combinations of bacteria, containing up to 4 species at ratios up to 1:1000, were designed to (a) ensure easy visual examination of the plates, (b) include species frequently recovered in mixed cultures from polymicrobial samples, and (c) cover a range of bacterial inocula commonly found during infectious processes. Each inoculum was measured with the DensiCHEK Plus instrument (bioMérieux, France). Monobacterial suspensions of Staphylococcus aureus, Escherichia coli, Pseudomonas aeruginosa, and Streptococcus agalactiae in sterile saline were adjusted to a 0.5 McFarland standard (108 CFU/ml). Then, 10-fold dilutions were prepared in sterile saline to obtain suspensions of 107, 106, 105, 104, and 103 CFU/ml. The monobacterial suspensions were mixed to obtain a total of 16 polymicrobial suspensions containing two to four species, with species ratios ranging from 1:1 to 1:1000 (Table 1). All monobacterial suspensions were plated (10 μl) on Mueller-Hinton agar to assess the number of colonies.
TABLE 1.
Characteristics of the 16 polymicrobial suspensions tested in the manual and automated inoculation and streaking
| Suspension no. | No. of strains | Initial suspension concn (CFU/ml) fora: |
|||
|---|---|---|---|---|---|
| S. agalactiae | P. aeruginosa | S. aureus | E. coli | ||
| 1 | 2 | 105 | 105 | ||
| 2 | 2 | 103 | 105 | ||
| 3 | 2 | 107 | 105 | ||
| 4 | 2 | 104 | 107 | ||
| 5 | 3 | 105 | 105 | 105 | |
| 6 | 3 | 103 | 105 | 105 | |
| 7 | 3 | 105 | 103 | 107 | |
| 8 | 3 | 107 | 105 | 103 | |
| 9 | 3 | 104 | 105 | 106 | |
| 10 | 3 | 106 | 104 | 105 | |
| 11 | 3 | 105 | 106 | 105 | |
| 12 | 4 | 105 | 105 | 105 | 105 |
| 13 | 4 | 107 | 106 | 105 | 104 |
| 14 | 4 | 106 | 107 | 104 | 105 |
| 15 | 4 | 105 | 104 | 106 | 107 |
| 16 | 4 | 104 | 105 | 105 | 106 |
Suspensions were mixed volume to volume, i.e., 1:1, 1:1:1, and 1:1:1:1, to produce final suspensions of two, three, and four species, respectively.
(ii) Specimen collection.
A total of 94 successive urine samples and 150 swabbed samples (1 swab/sample) from wounds (n = 40), rectums (n = 80), bedsores (n = 11), vaginas (n = 10), and other sites (n = 9) were collected from patients hospitalized in the Montpellier University Hospital (Montpellier, France). Soft polyurethane (PU) foam bud swabs (Sigma Transwabs MW176S; Medical Wire & Equipment Co. Ltd., UK) with liquid Amies medium were used for sample collection. Before inoculation, swabs were discharged into the Amies medium by vortexing the tube for 5 s.
Inoculation, streaking, and incubation procedures.
All suspensions and clinical samples were inoculated and streaked by both the manual and automated methods within a period of 15 min. The sampling procedures and culture conditions were as follows. (i) For bacterial suspensions, volumes of 100 μl were seeded onto Columbia agar with 5% sheep blood (Oxoid, Germany) using pipetting for the reproducibility study; volumes of 10 μl were seeded onto the same medium using a disposable loop. Suspensions were inoculated in triplicate using both the automated and manual methods, resulting in 48 inoculated plates per method. (ii) For urine samples, volumes of 10 μl were inoculated and streaked onto colistin aztreonam blood agar (CAP agar) (Oxoid) and MacConkey agar (bioMérieux) plates using a disposable loop for the manual method and an FA InoqulA pipetting device for the automated method. (iii) For PU swabs, volumes of 35 μl of transport medium were inoculated and streaked onto CAP agar, chocolate agar (Oxoid), and MacConkey agar plates by the InoqulA instrument, while 1 drop corresponding to an estimated volume of 35 μl was delivered with a disposable pipette, as currently performed in our routine practices, and streaked manually onto the same media by analysts.
Manual inoculations were performed by a microbiologist (bacterial suspensions) with 2 years of experience or by technicians (clinical samples) with experience of <2 years (n = 4), 2 to 5 years (n = 3), 5 to 10 years (n = 4), and >10 years (n = 4), using a quadrant isolation pattern in use in our daily practice.
Quality of bacterial isolation score.
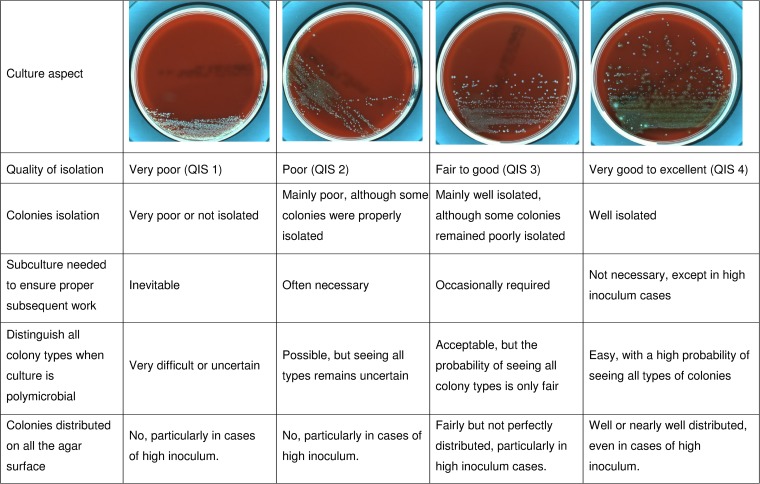
A standardized and mixed “reading culture scale,” here referred to as the quality of isolation score (QIS), was developed for evaluating each plate and is presented in Fig. 1. The plates were reviewed 24 h after incubation. The isolation quality was divided into 4 categories: very poor (QIS 1), poor (QIS 2), fair to good (QIS 3), and very good to excellent (QIS 4). This 4-category QIS was evaluated by two independent investigators through the blinded reading of 80 culture plates, which were randomly chosen from among the polymicrobial cultures taken from the routine analysis.
FIG 1.
Four-category scale used for evaluating the quality of bacterial isolation.
Definitions.
For the mean of the performance evaluation, the following definitions were used. A visible colony type was a type of microorganism that was recovered by culture from a sample or a suspension and was visible on the plates after growth. A nonrecovered microorganism was a microorganism that was known to be in a suspension but was not visible on plates after a culture step. A useable colony was an isolated (also named discrete) colony that may be used by a technician for subsequent analysis (e.g., bacterial identification or antimicrobial susceptibility testing) without requiring any subculture step.
Performance analysis.
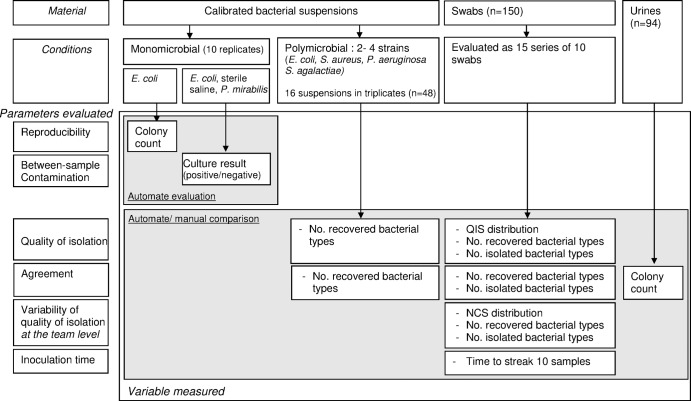
The flow chart of the study is shown in Fig. 2.
FIG 2.
Flow chart of the study showing the materials and conditions used and the variables measured according to the type of parameter evaluated.
(i) Instrument reproducibility.
The reproducibility of the instrument was assessed with colony counting of 10-μl and 100-μl samples taken from a 104 CFU/ml E. coli suspension that was distributed 10-fold. Cross-contamination between the samples was assessed by alternating samples inoculated with >105 CFU/ml E. coli or P. mirabilis and sterile saline.
(ii) Quality of isolation.
We evaluated the quality of isolation using both polymicrobial suspensions and a series of swabs. Plates from the clinical samples were described according to the 4-category QIS by a single investigator; the numbers of visible and discrete colony types were recorded for all plates.
(iii) Agreement between methods.
Agreement was evaluated on paired suspensions or clinical samples for (a) the number of recovered and discrete bacterial types (bacterial suspensions and swabs) and (b) the colony count (urine analysis).
(iv) Variability of the isolation quality within the laboratory.
We evaluated isolation quality using a series of swabs. Each series consisted of 10 swabs that were each streaked manually and by the instrument. A representative group of 15 investigators, consisting of recently trained and highly experienced bacteriology technicians, were randomly selected for this evaluation. For each series, a normalized cumulative score (NCS) was determined for the analyst and the instrument by summing the QIS of all the plates from the 10 swabs (30 plates) and normalizing the score to take into account the various proportions of sterile plates between each series. The resulting NCS represents a comprehensive score ranging from 0 (the poorest quality) to 100 (the best quality) to assess the colony isolation qualities between the series.
(v) Time for streaking.
The streaking time was directly measured during the swab series study.
Statistical analysis.
Statistical analyses were accomplished using the R project software (http://www.r-project.org). The levels of interrater agreement between the results of two independent investigators and between the automated and manual methods were evaluated with kappa statistics using the irr package (http://cran.r-project.org/web/packages/irr/index.html). The magnitude of kappa was considered to be poor (<0.40), fair to good (0.4 to 0.75), or excellent (>0.75) (6). Differences between the groups were assessed using the Wilcoxon signed-rank test and the Kruskal-Wallis test as appropriate. A P value of ≤0.05 was considered significant.
RESULTS
Quality of isolation score validation.
An 83% agreement (κ = 0.73) was observed between the two independent investigators, allowing the validation of the QIS for the assessment of the isolation quality.
Reproducibility and cross contamination in the automated method.
The instrument produced highly reproducible results. With 10 and 100 μl of a 104 CFU/ml E. coli suspension, the numbers of colonies varied between 8 and 18 (mean = 12.6; SD = 3.8) and between 95 and 136 (mean = 110; SD = 13.6), respectively. No cross-contamination was observed.
Quality of isolation.
Higher QIS values were obtained using the automated method than with the manual method (Table 2; see also Fig. S1 in the supplemental material); 81% of the positive cultures spiked by the manual method were attributed to QIS 2 and 3, while 87% of the positive cultures spiked by the instrument were attributed to QIS 3 and 4 (P = 1.4 × 10−33). The frequencies of plates attributed to QIS 1 and 3 were similar using both methods (see Fig. S1A in the supplemental material). The QIS distribution differed according to the mono- or polymicrobial characteristics of the plates for both the automated and manual methods (P = 0.0006 and P = 0.00002, respectively) (see Fig. S1B and C in the supplemental material).
TABLE 2.
Comparative culture results according to the type of sample and method of sample inoculation
| Sample type, parameter | No. of plates with indicated result |
Statistical analysis |
|||
|---|---|---|---|---|---|
| In agreement | Discrepant (greater) |
κ | P | ||
| Manual | Instrument | ||||
| Urine, colony countb | 156 | 26c | 6 | 0.71d | 0.0002 |
| Negative | 113 | ||||
| <103 CFU/ml | 13 | 2 | 4 | ||
| 103 CFU/ml | 13 | 3 | 2 | ||
| 104 CFU/ml | 5 | 1 | |||
| 105 CFU/ml | 3 | 6 | |||
| 106 CFU/ml | 0 | 6 | |||
| 107 CFU/ml | 8 | 7 | |||
| >107 CFU/ml | 1 | 1 | |||
| Swabs, QISe | |||||
| Total | 224 | 16f | 210g | 1.4 × 10−33 | |
| Very poor, 1 | 4 | ||||
| Poor, 2 | 37 | ||||
| Fair to good, 3 | 57 | 4 | 88 | ||
| Very good to excellent, 4 | 50 | 12 | 122 | ||
| Negative | 76 | ||||
| Plate culture, no. of colony types visible | |||||
| Total | 376 | 18h | 56i | 0.74 | 2.9 × 10−6 |
| 0 | 52 | ||||
| 1 | 31 | 3 | 2 | ||
| 2 | 80 | 1 | 7 | ||
| 3 | 137 | 10 | 25 | ||
| 4 | 64 | 4 | 12 | ||
| 5 | 12 | 8 | |||
| 6 | 2 | ||||
| Plate culture, no. of colony types with discrete colonies | |||||
| Total | 306 | 31j | 113k | 0.57 | 4.3 × 10−11 |
| 0 | 60 | ||||
| 1 | 45 | 3 | 2 | ||
| 2 | 87 | 4 | 21 | ||
| 3 | 78 | 19 | 53 | ||
| 4 | 31 | 4 | 30 | ||
| 5 | 5 | 1 | 7 | ||
Urine: data show agreement in colony count and frequency of discrepancies according to the inoculum level (from <103 to >107 CFU/ml). Swabs: data show (i) quality of isolation score (QIS) agreement and number of discrepancies according to the QIS category, (ii) agreement and discrepancies in the numbers of colony types visible on plates according to the total numbers of colony types visible on plates, and (iii) agreement and discrepancies in the numbers of colony types with discrete colonies on plates according to the total numbers of discrete colony types on the plates.
Urine samples: total of 188 plates (2 plates per urine sample, 94 samples).
All cases correspond with a higher colony count of 1 log10.
The magnitude of the kappa value indicates the level of agreement between two tests as follows: <0.40, poor agreement; 0.40 to 0.75, fair to good agreement; and >0.75, excellent agreement (6).
Swabs: total of 450 plates (3 plates per swab; 150 swabs).
Increased QIS by 1 (n = 14, 88% of the 16 cases) or 2 (n = 2) classes.
Increased QIS by 1 (n = 146, 69% of the 210 cases), 2 (n = 62), or 3 classes (n = 2).
One more type of colony in all of the 18 cases.
One (n = 47) or two (n = 9) more types of colonies (one more type in 84% of the 56 cases).
One (n = 26, 84% of the 31 cases), two (n = 4), or three (n = 1) more types of colonies.
One (n = 86, 84% of the 56 cases), two (n = 23), or three (n = 4) more types of colonies.
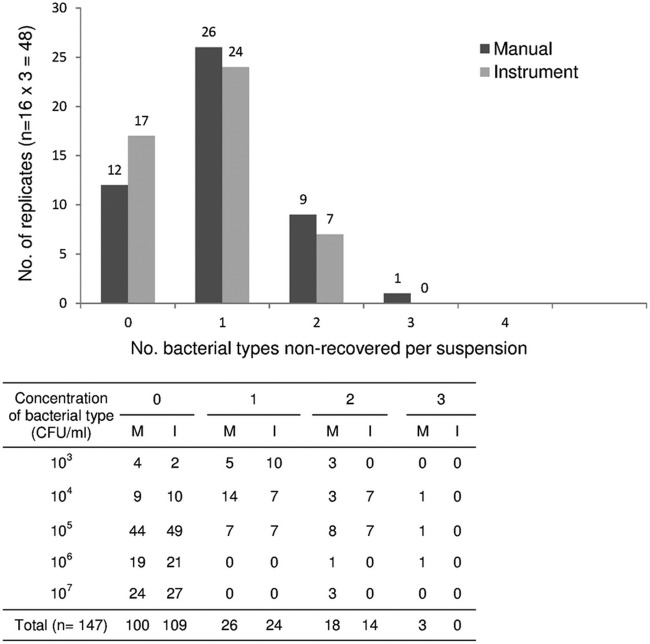
Neither the manual nor the automated method was able to recover all of the bacterial types from all of the suspensions. However, the instrument recovered the microorganisms with greater frequency (Fig. 3). Of the 49 bacterial types from the 16 mixed suspensions, 9 and 14 were not recovered from any of the triplicate cultures streaked by the instrument or manually, respectively. Thus, the instrument produced recovery improvements of 10% of the bacterial types present in the suspensions and 14% (5 of 35) compared with those for the manual method performance. The triplicate cultures analyzed as independent tests showed that of the total 147 microorganisms from the 48 replicate cultures, 38 and 47 of the organisms were not recovered with the automated and manual methods, respectively, leading to recovery rate defaults of 25.8% and 32%, respectively, and a 24% recovery improvement with the instrument. The nonrecovered microorganisms were frequently associated with an inoculum of ≤104 CFU/ml for the two methods (24 of 38 [63%] with the instrument and 26 of 47 [55%] with the manual method). Briefly, with concentrations ≤104 CFU/ml, the main differences between the 2 methods concerned S. agalactiae that was not recovered with the manual method in 14 of the 15 evaluated cases (compared with 9 nonrecovered cases with the instrument) and P. aeruginosa that was not recovered by the instrument in 6 of the 9 cases, compared with 3 nonrecovered cases with the manual method. Additionally, the manual method failed to recover 6 cases of the 18 evaluated cases with a 105 CFU/ml S. aureus inoculum compared with only 2 cases with the instrument. The manual method failed to recover 5 E. coli of the 42 microorganisms (11.9%) with a ≥106 CFU/ml inoculum. Finally, 14 and 21 of the cultures were associated with at least two nonrecovered microorganisms using the instrument and manual method, respectively (Fig. 3).
FIG 3.
Distribution of recovered and nonrecovered microorganisms from the polymicrobial suspensions by the manual and automated methods. The histogram presents the numbers of bacterial types recovered (0) and nonrecovered (1, 2, or 3 bacterial types per suspension) for the 48 replicates (M, manual method; I, instrument). The table details the bacterial concentration for the recovered (0) and nonrecovered bacterial types. A total of 49 bacterial types from the 16 polymicrobial suspensions with 2 to 4 distinct bacterial types were tested in triplicate, corresponding to a total of 147 distinct bacterial types to be recovered from the 48 plate cultures.
Similar results were obtained from the 150-swab study characterized by a high rate of polymicrobial cultures; 110 of the swabs (73.3%) and 348 of the inoculated plates (77.3%) had polymicrobial cultures (mean number of cultivated microorganisms of 2.3, with a range of 0 to 5). Microorganism recovery was higher with the instrument both in terms of the numbers of visible colony types (1,142 versus 1,095, P = 6.4 × 10−11) and the numbers of discrete colony types (1,012 versus 905, P = 4.9 × 10−13). Eighteen (1.5%) of the 1,160 colony types were recovered using only the manual method, while 65 (5.6%) were recovered using the instrument only. Regarding the 144 discrepant plates obtained for 96 of the swabs (64%), the instrument isolated more colony types in 113 of the cases (78.5%), with up to 3 additional colony types isolated (Table 2).
Manual and instrument agreement. (i) Complex polymicrobial suspensions.
Agreement between the numbers of bacterial types recovered with the two methods was low (52% [25 of 48 conditions], κ = 0.21), related to the greater number of colonies recovered with the instrument.
(ii) Swabs.
The numbers of recovered microorganism types were very consistent between the two methods (κ = 0.74), while the numbers of discrete colony types only fairly agreed (κ = 0.57), because the manual method isolated a lower number of colonies.
(iii) Urine samples.
Forty-six of the 94 specimens yielded positive cultures, with colony counts varying from <103 to >107 CFU/ml (Table 2). The colony counts from the manual and automated inoculations were consistent (83%, κ = 0.71). All discrepant counts, which involved 32 plates from 19 samples, varied by 1 log10, and of these discrepant counts, higher colony counts were observed more frequently with the manual method than with the instrument (26 versus 6 plates, respectively, P = 0.0002). Of note, most of the higher bacterial counts observed with the manual method involved counts ≥105 CFU/ml (20 of 26) (Table 2). The 6 cases with higher colony counts observed with the InoqulA corresponded to counts ≤103 CFU/ml.
Variation of the isolation quality according to the inoculation method used.
In addition to the lower QIS obtained with the instrument for the 450 inoculated plates for the 150 swabs (see Fig. S1A in the supplemental material), the distribution of the normalized cumulated score (NCS), presented in Fig. 4A, showed that the NCS per technician (10 swabs) varied from 48% to 90% (median of 61%), corresponding to an interinvestigator NCS variation of 42% (see Table S1 in the supplemental material). Importantly, the use of the FA InoqulA decreased the NCS variation to only 14%, with values ranging from 74% to 88% (median of 83%). Differences in the NCS between the automated system and the technicians were ≥10 with the instrument for all but two of the series, and only one technician matched the instrument (range, 1 to 29%, median of 21%) (see Table S1 in the supplemental material). This result demonstrates that the instrument consistently performed isolations of better and more standardized quality (P = 0.0007).
FIG 4.
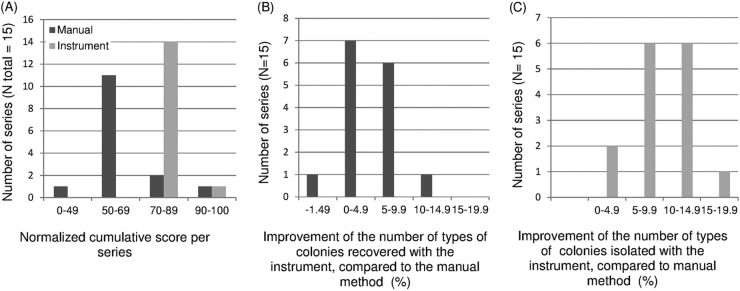
Variability of the quality of isolation at the laboratory team level (15 technicians) and improvement obtained with the instrument over each technician. (A) Distribution of the normalized cumulative score (NCS), which corresponds with the following equation: [cumulative QIS (technician) − minimum QIS possible) × 100/(cumulative QISmax − cumulative QISmin). (B and C) Distribution of the rate improvement with the instrument compared with manual isolation at the team level in terms of recovered (B) and isolated (C) colony types. Each evaluation included a series of 10 swabs (30 cumulated plates).
The levels of improvement varied between series in relation to technician variability from 0 to 11% (median of 4.3%) and 0 to 17% (median of 9%) for the visible and usable colony types, respectively (Fig. 4B and C). The instrument recovered fewer colony types than the technician in only one series, with a default of 1.49%.
Streaking rate and inoculation time estimations.
The time per technician for inoculating 10 swabs varied from 9 min 34 s to 17 min 3 s, with a median time of 11 min 52 s. The instrument can inoculate 10 swabs with a median of 14 min 10 s.
DISCUSSION
Evaluation of the colony isolation quality.
Colony isolation is a critical step in sample processing for generating fast, accurate, and relevant results. However, the colony isolation quality remains difficult to objectively quantify. Some authors have evaluated instruments using the number of isolated colonies (3–5). Although this is an attractive approach for demonstrating the absolute quality of an automated isolation, it only theoretically demonstrates the real impact of automation on the quality of final, routine results. Indeed, isolating an average of 36 rather than 26 colonies (3) is better, but as only one or two isolated colonies are sufficient to perform the analyses, the real improvement is lower than estimated and, in this example, is nothing. In this study, we chose the number of recovered colony types and number of usable colony types, and a 4-category scale as the main parameters for evaluating the real impact of automation on the quality of our results. We observed satisfactory levels of interrater agreement. The main confounding factor was the inoculum load. Not surprisingly, the probability of proper colony separation increased according to the low inoculum load and monomicrobial nature of the culture, as the QIS is artificially maximized in these situations. Despite this limitation, the QIS is easy to use for assessing the quality of colony separation without enumerating colonies, allowing easy evaluations of paired series by the technicians and instrument.
We deliberately evaluated the manual and automated streaking with a test panel that included a high frequency of polymicrobial cultures because this most closely approximates the events of routine diagnosis. Comprehensively, our results suggest that the streaking quality evaluation is highly dependent on the rate of polymicrobial plates, due to the combination of the frequency of polymicrobial samples and selective media used (data not shown). Further evaluations of automated inoculation instruments should systematically identify these factors.
Instrument performances.
To our knowledge, this is the first peer-reviewed study to evaluate the performance of the FA InoqulA instrument. Overall, our results showed that the instrument recovered more microorganisms and performed higher and more standardized quality isolations than the manual method, independently of the experience level of the analyst.
The number of suspensions (11.9%) that had a nonrecovered isolate with a high inoculum using the manual method was disappointing, suggesting a significant impact on the diagnostic results. The 24% improvement in the quality of isolation observed with the instrument should improve the quality of the final results. By improving the numbers of recovered and isolated bacterial types, we can reasonably expect more reliable and accurate results for a median frequency of 5 to 10% of samples (range, 0 to 25%) with an exhaustive microorganism recovery, a lesser need for subculture, a better workflow, and a shortened time to results. Further evaluations are necessary to confirm these results by direct assessment.
We noticed a 1-log disagreement in the bacterial count for 17% of the urine plate cultures between the instrument and manual disposable loop-based method, which was not associated with differences in patient management. Most of the cases (20 of the 26 observed) corresponded to a 1-log higher colony count with the manual method. The disposable loop supplier claimed a 30% precision but provided no information on the accuracy of the volume dispensed. Given that the instrument sampling is based on a Hamilton pipettor with a precision of 3%, the bacterial count was likely more reliable with the instrument. A similar phenomenon was observed with the MicroStreak instrument (bioMérieux), although the frequency of inconsistencies was much lower (3 of 500 cultures versus 26 of 188 plate cultures in our study) (3).
The estimated time for streaking with the instrument was similar to that of the manual method over short periods of sample inoculation. The yield of the manual method over sustained periods of time was not estimated here, which is a limitation of our study.
Although the instrument supplier claimed a throughput of up to 250 plates/h with the FA module, the rate observed in routine conditions was lower, ranging from 120 to 150 plates/h. Although unrealistic, the only means to maximize this rate is to increase the number of streaked plates to 5/sample in relation to the total number of positions at the streaking station. A more useful feature is to increase the number of distinct samples that can be streaked simultaneously; the supplier should consider improving this capability. Despite this lower than advertised yield, automated plating was associated with an improved workflow with a constant rate and an expanded range of work and a more efficient process because the analyst working with the instrument can simultaneously accomplish additional tasks. Finally, analysts working on subsequent cultures performed fewer subcultures.
Comprehensive evaluation at the team level.
By taking into account the variability of manual streaking between technicians, this study is the first to estimate the variability in culture quality between a laboratory team and an automated instrument and to assess the overall impact of automated streaking on the standardization of culture quality. Although we included only 15 of the 28 technicians of our team (53%), the variability described here is a valuable estimation because it is based on a large number of randomly selected analysts. With an improvement in the number of usable colonies ranging from 0 to 25% (median 10%), the instrument produced results at least equivalent to those of technicians and optimally produced significant improvements in the quality of sample processing.
In summary, our results showed a better recovery of microorganisms with the instrument than with the manual procedure and identified sample characteristics (e.g., the number of bacterial types, the bacterial load, etc.) that should be specified in further evaluations. Specimen processing automation represents an attractive method, given the growing shortage of human and material resources allocated to hospital laboratories, the growing demand for improved quality and traceability, the reductions in hospital inpatient times, and the overall increases in testing volumes driven by the aging population and growing infection control demands. The FA InoqulA instrument was effective for addressing these changes. It was flexible and enabled the use of wide panels of distinct media and specimen containers. Although we did not observe a 50% reduction in the time spent by full-time technicians for specimen processing, as hypothesized elsewhere (1, 2), automation allowed the consolidation of plate-reading quality and improvements in the overall workflow. Overall, our study highlights the lack of such instrument evaluation and shows the importance of the sample strategy (rate of polymicrobial samples) in the evaluation of results and the impact and range of the team variability on quality results.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Centre Méditerranéen d'Epidémiologie (CME) and the Centre Hospitalier Universitaire de Montpellier (Equipe Leader).
We thank C. Carrière and M. Brun and gratefully acknowledge C. Paolini, C. Kieffer, and C. Hate for technical assistance, and we thank E. Barbotte for assistance in statistical analysis.
We declare no conflict of interest.
Footnotes
Published ahead of print 18 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02341-13.
REFERENCES
- 1.Greub G, Prod'hom G. 2011. Automation in clinical bacteriology: what system to choose? Clin. Microbiol. Infect. 17:655–660. 10.1111/j.1469-0691.2011.03513.x [DOI] [PubMed] [Google Scholar]
- 2.Bourbeau PP, Ledeboer NA. 2013. Automation in clinical microbiology. J. Clin. Microbiol. 51:1658–1665. 10.1128/JCM.00301-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glasson JH, Guthrie LH, Nielsen DJ, Bethell FA. 2008. Evaluation of an automated instrument for inoculating and spreading samples onto agar plates. J. Clin. Microbiol. 46:1281–1284. 10.1128/JCM.01687-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourbeau PP, Swartz BL. 2009. First evaluation of the WASP, a new automated microbiology plating instrument. J. Clin. Microbiol. 47:1101–1106. 10.1128/JCM.01963-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mischnik A, Mieth M, Busch CJ, Hofer S, Zimmermann S. 2012. First evaluation of automated specimen inoculation for wound swab samples by use of the Previ Isola system compared to manual inoculation in a routine laboratory: finding a cost-effective and accurate approach. J. Clin. Microbiol. 50:2732–2736. 10.1128/JCM.05501-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleiss JL. 1981. Statistical methods for rates and proportions. John Wiley & Sons, New York, NY [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.