Abstract
Johne's disease (JD) is a chronic enteric disease caused by Mycobacterium avium subsp. paratuberculosis that affects ruminants. Transmission occurs by the fecal-oral route. A commonly used antemortem diagnostic test for the detection of M. avium subsp. paratuberculosis in feces is liquid culture; however, a major constraint is the 2- to 3-month incubation period needed for this method. Rapid methods for the detection of M. avium subsp. paratuberculosis based on PCR have been reported, but comprehensive validation data are lacking. We describe here a new test, the high-throughput-Johnes (HT-J), to detect M. avium subsp. paratuberculosis in feces. Its diagnostic accuracy was compared with that of liquid radiometric (Bactec) fecal culture using samples from cattle (1,330 samples from 23 herds) and sheep (596 samples from 16 flocks). The multistage protocol involves the recovery of M. avium subsp. paratuberculosis cells from a fecal suspension, cell rupture by bead beating, extraction of DNA using magnetic beads, and IS900 quantitative PCR. The limit of detection of the assay was 0.0005 pg, and the limit of quantification was 0.005 pg M. avium subsp. paratuberculosis genomic DNA. Only M. avium subsp. paratuberculosis was detected from a panel of 51 mycobacterial isolates, including 10 with IS900-like sequences. Of the 549 culture-negative fecal samples from unexposed herds and flocks, 99% were negative in the HT-J test, while 60% of the bovine- and 84% of the ovine-culture-positive samples were positive in the HT-J test. As similar total numbers of samples from M. avium subsp. paratuberculosis-exposed animals were positive in culture and HT-J tests in both species, and as the results of a McNemar's test were not significant, these methods probably have similar sensitivities, but the true diagnostic sensitivities of these tests are unknown. These validation data meet the consensus-based reporting standards for diagnostic test accuracy studies for paratuberculosis and the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (S. A. Bustin et al., Clin. Chem. 55:611–622, 2009, doi:10.1373/clinchem.2008.112797). The HT-J assay has been approved for use in JD control programs in Australia and New Zealand.
INTRODUCTION
Johne's disease (JD) is a chronic enteric disease affecting cattle, sheep, and goats, as well as other ruminant species. It is caused by Mycobacterium avium subsp. paratuberculosis and is transmitted by the fecal-oral route. As it results in significant economic impacts on the livestock industry and because M. avium subsp. paratuberculosis has been implicated in human chronic enteric infection, control programs for JD have been implemented in many countries, and these require objective laboratory diagnostic tests. JD presents unique challenges in the design, conduct, validation, and reporting of diagnostic tests, as is the case with similar diseases, such as tuberculosis, due to the chronicity of the disease, variable disease progression in individuals, differential host immune response to infection, and intermittent shedding and shedding of low numbers of organisms during the subclinical stage of the disease (1–3).
Fecal culture in liquid medium is a commonly used noninvasive antemortem diagnostic test for the detection of M. avium subsp. paratuberculosis infection (4). However, fecal culture requires long incubation times (2 to 3 months), and some strains of M. avium subsp. paratuberculosis, particularly those known as S (sheep) and B (bison) strains, have failed to grow on some media (5–7). Therefore, the development of a rapid, sensitive, and robust test for the detection of M. avium subsp. paratuberculosis in feces has been a goal to facilitate the efforts of control programs for many years. There are many reports on direct fecal PCR assays for M. avium subsp. paratuberculosis, but meaningful estimates of diagnostic accuracy are lacking. One reason for this is sample sizes that are too low to be meaningful (1, 2). Another important consideration in test validation is selecting animals in various stages of disease to ensure that the results are representative of the full range of samples that the test is designed to assess and that they ultimately align with the purpose of the test (1). Furthermore, most of the assays have been compared to solid medium culture methods with lower sensitivities than that of Bactec liquid culture (8), leading to a potential overestimation of sensitivity. Unfortunately, due to the cryptic nature of the disease, estimates of the true diagnostic sensitivities and specificities for most tests for M. avium subsp. paratuberculosis infection are lacking.
Several technical factors interact to impede the development of reliable protocols for fecal PCR tests for M. avium subsp. paratuberculosis. Feces is generally considered to be a difficult sample type for molecular detection of microbial pathogens due to the presence of PCR inhibitors (9–11) and the large amount of nonspecific DNA that is extracted from other fecal flora species and host cells. M. avium subsp. paratuberculosis is present in very low numbers in feces in the majority of infected animals in a population, as the disease has a prolonged incubation period and only a minority of cases develop clinical disease with high-level fecal shedding (12). Finally, M. avium subsp. paratuberculosis is a robust organism and requires vigorous treatment to effect lysis/rupture of its cell wall and release of DNA for analysis (13).
In 2007, Kawaji et al. (14) reported a rapid test for the detection of M. avium subsp. paratuberculosis in ovine fecal samples that had a sensitivity and specificity similar to those of Bactec culture. The DNA extraction component was laborious and could not be scaled up for routine use in diagnostic laboratories, but the quantitative PCR (qPCR) component was robust. Here, we report the development of a practical new test (the high-throughput-Johnes [HT-J] test) based on this qPCR. The HT-J test has eliminated several labor-intensive aspects of DNA purification and replaced them with a semiautomated procedure in a 96-well plate format; not only does this reduce the time required to perform the test, but it significantly reduces the risk of cross-contamination due to manual handling. The purpose of the test is to identify M. avium subsp. paratuberculosis in fecal samples from cattle and sheep. Feces from cattle (1,330 samples from 23 herds) and sheep (596 samples from 16 flocks) were evaluated in the HT-J test in comparison with Bactec culture. We report here the analytical sensitivity and specificity of the HT-J test and provide estimates of M. avium subsp. paratuberculosis detectability relative to fecal culture to meet the recommended standards for reporting M. avium subsp. paratuberculosis and qPCR diagnostic test accuracy (2, 15).
MATERIALS AND METHODS
Mycobacterial strains.
A panel of 51 mycobacterial isolates that included defined C and S strains of M. avium subsp. paratuberculosis was used to assess the analytical specificity and sensitivity of the assay (Table 1). The two reference strains of M. avium subsp. paratuberculosis were Telford 9.2 and CM00/416. Telford 9.2 is a pure culture at passage level 5 (including its primary isolation from sheep feces) and is IS900 restriction fragment length polymorphism (RFLP) type S1, IS1311 PCR-restriction endonuclease analysis (REA) type S, pathogenic in sheep, and distinct from M. avium subsp. paratuberculosis strain K10 based on a whole-genome microarray comparison (16, 17). CM00/416 is a nonclonal culture at passage level 5 (including its primary isolation from cattle tissues) and is IS1311 PCR-REA type C, pathogenic for cattle (our unpublished data), and apparently identical to M. avium subsp. paratuberculosis K10 based on a whole-genome microarray comparison (17). Mycobacteria were reconstituted from lyophilized stock in 0.1 ml sterile water and inoculated into radiometric Bactec 12B culture vials containing PANTA (BD Biosciences). For all M. avium subsp. paratuberculosis strains, the radiometric culture vials were supplemented with egg yolk (obtained aseptically from fresh commercial eggs sold for human consumption) and mycobactin J (Allied Monitor, Inc., Fayette, MO), as previously described (18, 19). These were incubated at 37°C until they reached a growth index of 999. A 1-ml aliquot from each radiometric culture vial was stored at −80°C prior to DNA extraction using routine methods. DNA from Mycobacterium tuberculosis and Mycobacterium bovis strains were a kind gift from the National Institute of Animal Health, Japan. Purified genomic DNA was quantified using spectrophotometry (NanoDrop 1000; Thermo Scientific), diluted to a concentration of 2 ng/μl in nuclease-free water, and stored at −80°C.
TABLE 1.
Mycobacterial taxa and strains used in the specificity study
| Strain no. | Taxon | Strain/sourcea | IS900 resultb |
|---|---|---|---|
| 1 | M. avium subsp. paratuberculosis | 316v (C strain) | + |
| 2 | M. avium subsp. paratuberculosis | Telford (S strain) | + |
| 3 | M. avium subsp. aviumc | ST1 | − |
| 4 | M. avium subsp. aviumc | ST9 | − |
| 5 | M. avium subsp. aviumc | ST11 | − |
| 6 | M. avium subsp. aviumc | ST12 | − |
| 7 | M. avium subsp. aviumc | ST15 | − |
| 8 | M. avium subsp. aviumc | ST16 | − |
| 9 | M. avium subsp. aviumc | ST17 | − |
| 10 | M. avium subsp. aviumc | ST18 | − |
| 11 | M. avium subsp. aviumc | ST19 | − |
| 12 | M. avium subsp. aviumc | ST23 | − |
| 13 | M. avium subsp. aviumc | ST26 | − |
| 14 | M. avium subsp. aviumc | ST28 | − |
| 15 | M. avium subsp. aviumc | ST29 | − |
| 16 | M. avium subsp. aviumc | ST31 | − |
| 17 | M. avium subsp. aviumc | ST38 | − |
| 18 | M. avium subsp. aviumc | ST49 | − |
| 19 | M. avium subsp. aviumc | ST54 | − |
| 20 | M. avium subsp. aviumc | ST55 | − |
| 21 | M. avium subsp. aviumc | ST60 | − |
| 22 | M. avium subsp. aviumc | ST62 | − |
| 23 | M. avium subsp. avium | TMC715 | − |
| 24 | M. chitae | ATCC 19627 | − |
| 25 | M. flavescens | ATCC 14474 | − |
| 26 | M. gordonae | ATCC 14470 | − |
| 27 | M. intracellulare | ATCC 13950 | − |
| 28 | M. kansasii | ATCC 12478 | − |
| 29 | M. gordonae | ATCC 19530 | − |
| 30 | M. parafortuitum | ATCC 19686 | − |
| 31 | M. phlei | ATCC 11758 | − |
| 32 | M. scrofulaceum | ATCC 19981 | − |
| 33 | M. terrae | ATCC 15755 | − |
| 34 | M. thermoresistibile | ATCC 19527 | − |
| 35 | M. triviale | ATCC 23292 | − |
| 36 | M. vaccae | ATCC 15483 | − |
| 37 | M. xenopi | ATCC 10042 | − |
| 38 | M. tuberculosisd | Aoyama B | − |
| 39 | M. tuberculosisd | Tapir C-3 | − |
| 40 | M. bovis BCGd | Tokyo | − |
| 41 | M. bovisd | B-10 | − |
| 42 | Mycobacterium sp.e | 2333 IS900-like | − |
| 43 | Mycobacterium sp.f | AM1 IS900-likeg | − |
| 44 | Mycobacterium sp.f | AM3 IS900-likeg | − |
| 45 | Mycobacterium sp.f | AM4 IS900-likeg | − |
| 46 | Mycobacterium sp.f | AM5 IS900-likeg | − |
| 47 | Mycobacterium sp.f | IWGMT90236 IS900-likeg | − |
| 48 | Mycobacterium sp.h | JD00/266 IS900-likeg | − |
| 49 | Mycobacterium sp.f | P99-4609 IS900-likeg | − |
| 50 | M. scrofulaceum-likeh | Vic-1 IS900-likeg | − |
| 51 | M. scrofulaceum-likeh | Vic-2 IS900-likeg | − |
ST, sequence type.
Results are presented as positive (+) or negative (−) based on the criteria of Tm range and DNA quantity used to determine a positive result for M. avium subsp. paratuberculosis in feces samples.
Collection of Queensland Department of Health.
Collection of National Institute of Animal Health, Japan.
Englund et al. (38).
Collection of Western Australian Department of Agriculture.
Cousins et al. (36).
Collection of Victorian Institute of Animal Science.
For fecal spiking experiments, M. avium subsp. paratuberculosis organisms were subcultured on modified Middlebrook 7H10 agar slopes supplemented with mycobactin J for 6 weeks, as previously described (18), harvested by gently scraping, and resuspended in phosphate-buffered saline (PBS) with 0.1% (wt/vol) Tween 20. A single-cell suspension was prepared by vortexing for 5 min and then passing the suspension through a 26-G needle and filtering through a sterile 8-μm filter. The integrity of the filter was checked before proceeding. A 1:100 dilution of the suspension was examined (×400 magnification) to confirm that the majority of cells were single. If clumping was evident, the 26-G needle aspiration and filter steps were repeated until a single-cell suspension was achieved. Enumeration of M. avium subsp. paratuberculosis was performed using a Helber counting chamber for visible count, and a most probable number (MPN) estimate was used to obtain a viable count (20).
16S rRNA PCR.
For specific experiments, mycobacterial 16S rRNA primers (forward, 5′-CCTGGTGTAGCGGTGGAATG-3′ and reverse, 5′-GCTCCTCAGCGTCAGTTACT-3′) were designed in the nonpolymorphic region, and qPCR was performed as previously described (21).
M. avium subsp. paratuberculosis fecal culture.
Liquid culture to detect M. avium subsp. paratuberculosis in fecal samples using radiometric Bactec 12B medium and PCR confirmation were performed as previously described (18, 19).
Spiking of feces with M. avium subsp. paratuberculosis.
To determine the analytical range of the HT-J test (described below), endpoint titrations of M. avium subsp. paratuberculosis S and C strain organisms were performed using 10-fold dilutions of suspensions to give final concentrations from 1 × 108 to 1 × 101 organisms/g feces, derived from healthy nonexposed cattle or sheep. The numbers of M. avium subsp. paratuberculosis organisms were based on Helber visible microscopic counts. Each spiked sample (n = 1/dilution) was evaluated using the HT-J test.
HT-J test validation.
All animal experiments were approved by the University of Sydney Animal Ethics Committee. For test development, fecal samples were tested from calves (<12 months of age) experimentally inoculated with M. avium subsp. paratuberculosis utilizing a previously validated infection model (22, 23). Clinical samples were collected on farm during a prospective study or retrieved from −80°C freezer laboratory archives for the purpose of HT-J test validation relative to the reference standard of Bactec culture (see below).
The sample collection, transport, and initial handling at the laboratory were identical to the handling requirements for M. avium subsp. paratuberculosis fecal culture. Briefly, fecal samples were collected from animals, put in labeled sterile sample containers, and stored at 4°C prior to and during transit to the laboratory, avoiding unnecessary delays in transit. Upon arrival, the samples were maintained at 4°C if they could be tested within 48 h; alternatively (i.e., for longer-term storage), they were stored at −80°C. All of the collected samples were tested. The interval between sample collection and testing with the HT-J test ranged from 0 to 8 months for cattle fecal samples and 3 months to 2.5 years for sheep fecal samples (other than archival samples as specified below), and between sample collection and testing in Bactec culture from 0 to 2 months for cattle fecal samples and 0 to 3 months for sheep fecal samples.
Both the HT-J and Bactec culture tests were performed at the University of Sydney laboratories at Camden by dedicated and highly experienced technical and postdoctoral staff. The samples were not tested blind, as the identities of the herds and flocks and regional JD status were known to some of the laboratory staff.
Bovine fecal samples.
A single fecal sample was tested from each of 1,330 beef cattle, which included both males and females, 1 to 12 years old, and represented 15 exposed herds (n = 870 cattle) and at least 8 unexposed herds (n = 460 cattle) (Table 2). The herds were selected opportunistically, and convenience samples were collected and tested between 6 January 2010 and 11 November 2011. The majority were extensively grazed cattle, with the exception of 189 unexposed fecal samples that were from animals in beef cattle feedlots. The samples from the exposed herds were from three geographically distinct regions (New South Wales, Tasmania, and Victoria). A diagnosis of M. avium subsp. paratuberculosis infection had been made in at least one animal from each herd using a blood serum antibody enzyme-linked immunosorbent assay (ELISA) screen with confirmation by pathological examination. All herds except one in Tasmania had a low prevalence of infection, most of which were not suspected to be infected prior to detection, which occurred during certification testing as part of a national market assurance program (24). Samples from unexposed herds were obtained from farms in regions of Australia where JD was not known to be present at the time of the study (Table 2); the samples obtained from Western Australia were collected at an abattoir without matching farm identification data, so the number of farms represented is uncertain.
TABLE 2.
Summary of sample numbers and results for sheep and cattle fecal samples from different regions of Australia
| Region | Locationa | Herd/flock Status | No. of farms | No. of samples tested | Culture-positive samples | HT-J test-positive results |
|||
|---|---|---|---|---|---|---|---|---|---|
| Low | Medium | High | Total | ||||||
| Cattle samples | |||||||||
| A | QLD | Unexposed | 7 | 243 | 0 | 1 | 1 | ||
| B | WA | Unexposed | 1 | 217 | 0 | 1 | 1 | ||
| A and B totals | 8 | 460 | 0 | 2 | |||||
| C | NSW | Exposed | 9 | 68 | 4 | 1 | 2 | 1 | 4 |
| D | VIC | Exposed | 4 | 260 | 0 | 8 | 4 | 1 | 13 |
| E | TAS | Exposed | 2 | 542 | 107 | 59 | 21 | 27 | 107 |
| C, D, and E totals | 15 | 870 | 111 | 124 | |||||
| Sheep samples | |||||||||
| A | NSW | Unexposed | 1 | 89 | 0 | 1 | 1 | ||
| B | NSWb | Exposed | 3 | 368 | 61 | 22 | 11 | 31 | 64 |
| C | NSW | Exposed | 11 | 132 | 44 | 12 | 20 | 14 | 46 |
| D | VIC | Exposed | 1 | 7 | 6 | 1 | 1 | 5 | 7 |
| B, C, and D totals | 15 | 507 | 111 | 117 | |||||
QLD, Queensland; WA, Western Australia; NSW, New South Wales; VIC, Victoria; TAS, Tasmania.
Sheep are from experimental infection/exposure trials run by the University of Sydney, as described in Materials and Methods.
Ovine fecal samples.
A total of 596 fecal samples were obtained from extensively grazed mixed-sex predominantly adult Merino sheep representing both M. avium subsp. paratuberculosis-exposed (507 samples from 15 flocks) and -unexposed flocks (89 samples from 1 flock) (Table 2). The flocks were selected opportunistically, and convenience samples were collected or retrieved from archives and tested between 27 October 2010 and 11 November 2011. Samples from M. avium subsp. paratuberculosis-exposed flocks included pooled fecal samples (n = 50 sheep/pool) sourced from 12 M. avium subsp. paratuberculosis-exposed/-infected commercial sheep flocks in New South Wales. Based on owner reports in 2002, 5 of the flocks were regarded as low prevalence, as they had had no clinical cases of JD, 2 had medium prevalence based on the occurrence of clinical cases but with <5% per annum (p.a.) mortality in adult sheep, and 5 were classified as high prevalence based on >5% p.a. mortality in adult sheep. An annual lamb vaccination program commenced in 2002 using the Gudair vaccine (Zoetis Australia), and by the time fecal samples for this study were collected from adult sheep (3 to 7 years of age) from 2009 to 2010, sheep in all age classes on each of the 12 properties had been vaccinated, with a drop in the prevalence of fecal shedders recorded compared to prevaccination prevalence (25).
In addition, individual fecal samples from sheep were tested from trials performed at the University of Sydney. These comprised: a trial in which Merino lambs (n = 20) were inoculated orally with live M. avium subsp. paratuberculosis and sampled at three time points (from 3 to 18 months of age) (22), and archived samples (stored at −80°C since 2004) from a trial in which unexposed sheep were cograzed with M. avium subsp. paratuberculosis-infected sheep (6 months to 5 years of age) (26). Due to the difficulty of locating unexposed sheep, fecal samples were obtained from only one unexposed source; on this farm, the sheep have consistently yielded negative results (with fecal culture and serological methods) in tests for JD over 10 years.
High-throughput Johnes test.
The fecal DNA extraction when performed in tandem with the IS900 qPCR was termed the HT-J test.
(i) Fecal DNA extraction.
The extraction of DNA from feces involved: (a) the preparation of a fecal suspension and sedimentation to enrich for M. avium subsp. paratuberculosis organisms, (b) bead beating to release DNA from within the mycobacteria, and (c) automated magnetic bead purification of DNA. The controls in each batch test included: a positive-control fecal sample from an infected animal that was fecal culture positive; a negative-control fecal sample from a noninfected animal that was fecal culture negative; a process control (exposed to all buffers in the fecal suspension and DNA extraction process but with no feces), and an extraction plate control (DNA elution buffer from a well in the 96-well plate with all DNA extraction buffers but no fecal lysate). The protocol incorporated the BioSprint 96 one-for-all vet kit (Qiagen), with significant changes made to the sample input volume and processing steps to optimize M. avium subsp. paratuberculosis detection in feces.
(a) Preparation of fecal suspension. The fecal suspension was prepared by adding 1.2 g (dry weight) or 1.5 g (moist weight) of feces to a 15-ml sterile culture tube (11 cm by 1.6 cm diameter) containing 10 ml 0.85% (wt/vol) sterile saline. The tube was shaken vigorously to mix the suspension thoroughly and then allowed to settle for 30 min, with a gentle flick after 5 min to release air bubbles and dislodge floating debris. The top 3 to 5 ml of the supernatant was transferred to a 15-ml conical centrifuge tube using a sterile plastic transfer pipette and centrifuged at 1,231 × g for 30 min with low brake at room temperature. The supernatant was discarded without dislodging the pellet, and 600 μl of modified lysis/binding solution (BioSprint 96 one-for-all vet kit; Qiagen) (597.2 μl buffer RLT and 2.8 μl carrier RNA that had been reconstituted in buffer AVE, without isopropanol or MagAttract suspension G [Qiagen]) was added. The pellet was resuspended by repeated aspiration using a sterile plastic transfer pipette.
(b) Bead-beating step. The full volume of the resuspended pellet was transferred to a 2-ml conical-base screw-cap tube containing 0.3 g of zirconia/silica beads (BioSpec Products, Inc., Daintree Scientific) and disrupted using a mechanical cell disruptor/bead beater (6.5 m/s for 60s, twice, using a FastPrep-24 bead beater; MP Biomedicals). The bead tubes were centrifuged at 16,000 × g for 3 min, the supernatant (∼600 μl) was transferred to a new 1.5-ml tube, and the centrifugation step was repeated.
(c) Magnetic bead DNA purification. This was undertaken using a kit (BioSprint 96 one-for-all vet kit; Qiagen). The fecal supernatant in lysis/binding solution (400 μl) was transferred to the lysate plate (deep 96-well plate) along with 40 μl proteinase K and 300 μl magnetic bead mix (preparation per reaction, 300 μl isopropanol and 25 μl MagAttract suspension G vortexed thoroughly as recommended, which allowed for pipetting errors). Three additional deep 96-well (S-Block) plates and one standard 96-well plate were labeled and prepared: wash 1 (deep well, 700 μl buffer AW1/well), and washes 2 and 3 (2 × deep well, 500 μl buffer RPE/well), elution (standard 96-well, 75 μl buffer AVE/well), and these run on an automated magnetic particle processor (MagMAX Express-96; Life Technologies) using the BS96 Vet 100 instrument protocol (Qiagen). The contents of the individual wells of the elution plate were stored in 200-μl tubes at 4°C if qPCR was to be performed within 24 h; otherwise, they were stored at −20°C/−80°C
(ii) IS900 quantitative PCR.
qPCR for IS900 was performed using an Mx3000P real-time PCR instrument (Stratagene, Agilent). The reaction mixtures each contained 5 μl template DNA, forward and reverse primers (250 nM final concentration) (forward primer MP10-1, [5′-ATGCGCCACGACTTGCAGCCT-3′], and reverse primer MP11-1, [5′-GGCACGGCTCTTGTTGTAGTCG-3′]; Kawaji et al. [14]), and SensiMix SYBR Low-ROX qPCR mastermix (Bioline). Optimization of the primer concentration in the range of 100 to 600 nM was performed, with 250 nM forward and reverse primer found to be optimal (data not shown). A 5-step standard curve of M. avium subsp. paratuberculosis genomic DNA was included in every qPCR experiment (10-fold serial dilutions over the range of 10 to 0.001 pg/reaction). The limit of detection (LOD) was determined from the standard curves obtained from 12 experiments (Fig. 1A and B). No-template controls (all qPCR reagents with 5 μl purified sterile water) were included in every qPCR experiment. The cycling parameters were: initial denaturation at 95°C for 8 to 10 min, 40 cycles of denaturation at 95°C for 30 s, and annealing/extension at 68°C for 60 s with fluorescence acquisition at the end of the annealing/extension step, followed by a melt curve analysis from 65 to 95°C. The threshold was set in each run based on the M. avium subsp. paratuberculosis genomic DNA standard curve using the qPCR analysis software (MxPro; Stratagene) and then applied across the samples. The raw data were presented as the DNA quantities derived from the M. avium subsp. paratuberculosis genomic DNA standard curve to obtain normalization of the data between runs as distinct from quantification (15). Individual amplification curves with a threshold cycle (CT) were verified by visual examination to ensure that there was an exponential phase; test wells with CTs of >40 were disregarded (per MIQE guidelines) (15).
FIG 1.

Standard curve and Tm validation for the IS900 qPCR assay. (A and B) Results derived from 12 standard curves of duplicate reactions (n = 24 data points/concentration) with template concentrations from 0.0005 pg to 10 pg of M. avium subsp. paratuberculosis (MAP) genomic DNA/reaction. (A) Plots of the means ± SD of CT values detected for each DNA concentration. (B) Detection rate of individual replicates. The data are the percentage of individual qPCR replicates with positive results. The standards included in the standard curve in all qPCR experiments are shown (⧫). (C) Histogram of all Tm observations, including data from replicates for all samples tested but excluding sample values for which “No CT” was recorded. The mean Tm of each of the two major peaks is shown on the graph. The first peak, assumed to be a primer-dimer artifact, had a mean Tm of 82.3°C, while the M. avium subsp. paratuberculosis-specific IS900 sample results had a mean Tm of 89.1°C. The primer-dimer artifact was seen in the no-template controls (NTC) and samples with no specific template, and it was not seen to be competitive with the specific product.
The following criteria had to be met for the data from a qPCR experiment (run or plate) to be accepted: amplification efficiency of the M. avium subsp. paratuberculosis genomic DNA standard curve, including at least 4 of the 5 standards, between 90 and 110% (calculated using the inbuilt algorithm in the qPCR instrument software), at least one replicate of standard 5 (0.001 pg) giving a positive amplification, and no-template PCR control negative. A positive result for a single replicate, i.e., one reaction in a single well in a qPCR plate, required a fluorescence curve with an appropriate exponential phase confirmed by visual examination, DNA quantity exceeding the cut point, and a correct dissociation peak (Tm) in melt curve analysis. The DNA quantity cut point (≥0.001 pg M. avium subsp. paratuberculosis genomic DNA) was determined by an epidemiological approach using the complete validation data set. The acceptable Tm range was determined using data from all controls and all samples (Fig. 1C) to be 89.1 ± 1.5°C for the Mx3000 (Stratagene).
HT-J test acceptance criteria.
The results from an HT-J experiment were accepted when the negative fecal controls and extraction plate process controls gave negative results in both replicates of the DNA extract, positive fecal controls gave positive results in both replicates, and the IS900 qPCR data were accepted (see above).
Statistical analysis of HT-J test performance.
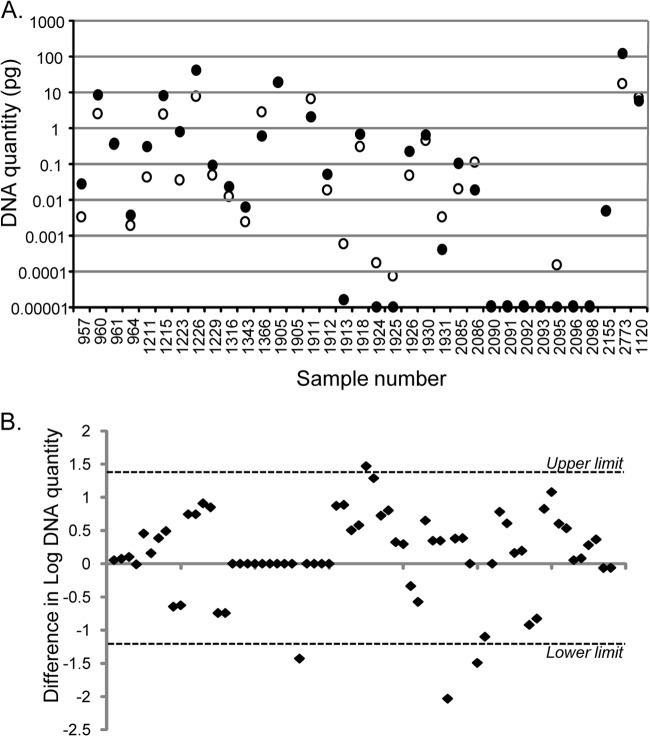
A restricted maximum likelihood linear mixed model (REML) was used to analyze the DNA quantity data from method optimization experiments using GenStat (release 12.1 [2009]; VSN International, Ltd.). Repeatability was assessed using a paired t test on DNA quantities of all individual qPCRs from two replicate HT-J experiments, and the 95% upper and lower limits of agreement for these were calculated from the difference in the log10 DNA quantity between the results for sample replicates using the formula d̄ ± 2 standard deviations (SD), where d̄ is the mean of the differences (27).
Relative specificity was defined as the proportion of fecal culture-negative samples from flocks and herds not exposed to M. avium subsp. paratuberculosis that tested negative in the HT-J test. Relative sensitivity was defined as the proportion of fecal culture-positive samples that tested positive in the HT-J test. For the purposes of calculation of sensitivity and specificity relative to Bactec culture, a fecal sample was classified as HT-J test positive using a range of criteria for consideration of the qPCR data from replicate DNA extracts performed for each fecal sample (Table 3). The exact binomial confidence limits were calculated (Minitab Statistical Software). McNemar's test for paired observations was used for comparison of the fecal culture method and the HT-J method (28). The association between the HT-J-positive category based on an ordinal classification of normalized DNA quantity (high, medium, low) and the results of Bactec culture was assessed using a chi-square test, while the relationship between the weeks to peak growth index and the likelihood of a sample being positive in the HT-J test were determined using Spearman's rank correlation (GenStat 12th edition' VSN International, Ltd.).
TABLE 3.
Summary of sensitivity and specificity of the HT-J test relative to liquid radiometric (Bactec) culture with different scenarios for conduct of the HT-J test or test result reporting
| Scenario | Description | Sheep |
Cattle |
||
|---|---|---|---|---|---|
| Sensitivitya | Specificityb | Sensitivitya | Specificityb | ||
| 1c | DNA extracts tested in duplicate. Samples with one positive replicate and one negative replicate were classified as negative | 83.8 | 98.9 | 60.4 | 99.6 |
| 2 | DNA extracts tested in duplicate. Samples with one positive replicate and one negative replicate were classified as positive | 85.6 | 97.8 | 63.1 | 97.6 |
| 3 | DNA extracts tested in single wells | 84.7–85.7 | 97.8–98.9 | 61.3–62.2 | 98.5–98.7 |
| 4 | DNA extracts tested in duplicate. Average DNA quantity of the replicates used to determine result. | 85.6 | 98.9 | 62.2 | 98.7 |
Proportion (%) of culture-positive samples from exposed herds/flocks that tested positive in HT-J.
Proportion (%) of culture-negative samples from unexposed herds/flocks that tested negative in HT-J.
Scenario 1 was the format of test conduct and result reporting criteria that were applied for the HT-J test.
RESULTS
Optimization of DNA extraction method.
A range of methods were assessed for DNA extraction, including commercial column-based methods, but a magnetic bead isolation method utilizing the BioSprint 96 one-for-all vet kit (Qiagen) and a 96-well automated magnetic particle processor had the capacity for high-throughput processing and gave the highest analytical sensitivities (data not shown). Protocol variations were evaluated, including the initial storage temperature of feces (−20°C or −80°C), the fecal suspension method (amount of feces and total volume of the suspension), the ratio of fecal suspension to lysis buffer, and the volume of fecal lysate added to the extraction plate. Following the pilot experiments, a trial was conducted using Bactec culture-positive cattle fecal samples that were known to contain low levels of M. avium subsp. paratuberculosis and that had been stored as aliquots at −20°C or −80°C (Fig. 2). These were extracted with different method variants to evaluate suspensions of 0.3 g feces in 1 ml saline or 1.5 g feces in 10 ml saline and ratios of 1:4, 1:3, 1:2.5, or 1:1.7 of fecal suspension to lysis buffer. In each case, the DNA quantity detected by qPCR was greater for samples stored at −80°C than at −20°C (P < 0.05). The protocol that gave the highest analytical sensitivity was the suspension of 1.5 g moist feces (1.2 g dry feces) in 10 ml of saline, with a 1:3 ratio of fecal suspension to lysis solution, and storage of the fecal samples at −80°C. This was adopted as the recommended method for the HT-J test.
FIG 2.
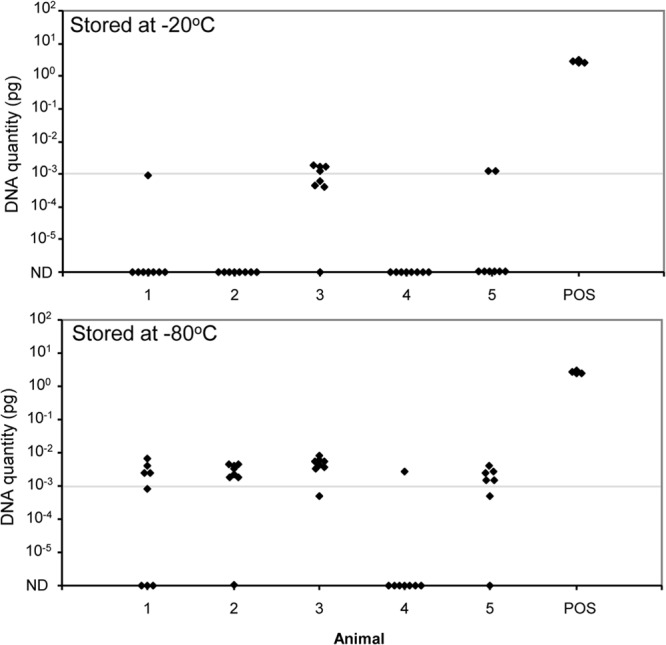
Effect of storage temperature on HT-J test results. Separate extractions were performed on fecal aliquots from 5 cattle and 1 positive-control fecal sample (POS), stored at −20°C or −80°C. The results shown are from 1.5 g feces in 10 ml saline and a ratio of 1:3 for suspension in lysis buffer. The data are the individual qPCR results (n = 8/animal; n = 4 positive control). The cut point for the determination of a positive result is shown as a horizontal line at 10−3 pg. ND, not detectable (no CT).
The controls included in the HT-J extraction method were a process control, a positive fecal control, a negative fecal control, and a buffer control on the 96-well extraction plate. The qPCR primer concentration was optimized (data not shown); although primer-dimer artifact was seen particularly in samples with no template (Fig. 1C), it was not determined to be competitive from an assessment of amplification and major Tm peaks for samples with concentrations of M. avium subsp. paratuberculosis genomic DNA approaching the LOD of the assay (0.001 pg). An experiment was conducted to confirm that the 96-well plate-based magnetic particle DNA purification method was robust and not subject to cross-contamination between the wells (see Fig. S1 in the supplemental material). All negative-control wells in this experiment gave no CT values, indicating that no cross-transfer of material occurred between the wells, even for wells located adjacent to samples with very high quantities of M. avium subsp. paratuberculosis DNA.
Analytical specificity.
The IS900 primers used in the HT-J assay (MP10-1 and MP11-1) were designed to be specific to M. avium subsp. paratuberculosis, with reference to mycobacterial isolates possessing known IS900-like sequences (14). BLASTn analysis conducted in July 2013 revealed M. avium subsp. paratuberculosis to be the only organism with a 100% match for these primers. Incomplete sequence alignments were identified for both the forward and reverse primers in Mycobacterium sp. strain 2333 (NCBI Nucleotide database accession no. AF455252.1), and an incomplete sequence was identified for the MP10-1 primer in Mycobacterium intracellulare (GenBank accession no. CP003324.1).
A panel of 51 mycobacterial isolates (Table 1), including environmental and pathogenic strains, as well as 10 strains previously identified as containing IS900-like elements in their genomes, were tested using the IS900 qPCR assay. The genomic DNA was isolated, quantified, and 10 ng was added to each qPCR. Only the S and C strain M. avium subsp. paratuberculosis isolates gave a positive result, with CT values of 19.0 ± 1.5 (Table 1). Importantly, neither of the mycobacterial strains with partial sequence identity to the primers nor any of the remaining mycobacterial isolates with IS900-like elements produced a positive result. However, there was amplification of all mycobacterial isolates in a PCR with nonpolymorphic 16S rRNA primers, with CT values of 19.7 ± 1.2 (mean ± SD).
Analytical sensitivity and range.
Analytical sensitivity was determined for the IS900 qPCR assay by performing replicate (n = 12) standard curves of M. avium subsp. paratuberculosis genomic DNA, with duplicate reactions from 10 pg to 0.0005 pg of input DNA (Fig. 1A and B). In addition to a 10-fold dilution series, intermediate DNA amounts were included. The limit of detection (LOD) was 0.0005 pg, defined as the DNA concentration at which at least 50% of the replicates were detected (29, 30). The limit of quantitation (LOQ), defined as the linear portion of the standard curve, was between 0.005 and 10 pg M. avium subsp. paratuberculosis genomic DNA (15).
The range of detection of M. avium subsp. paratuberculosis in feces was assessed using fecal samples spiked with known quantities of M. avium subsp. paratuberculosis cells. Both the C and S strains of M. avium subsp. paratuberculosis were included in this experiment. The analytical sensitivities of the HT-J test were similar for both animal species and strains, with the test being able to detect as few as 10 M. avium subsp. paratuberculosis organisms per gram of feces (Fig. 3).
FIG 3.
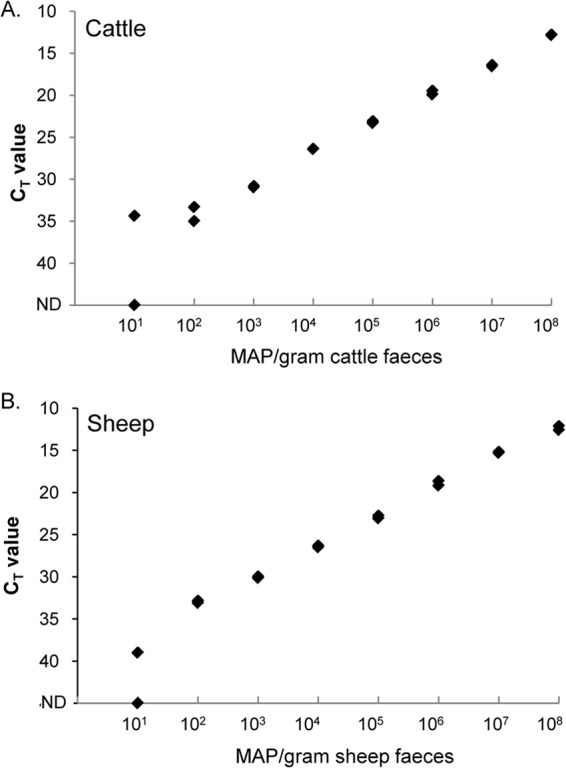
Endpoint titration of M. avium subsp. paratuberculosis organisms diluted in negative feces. (A) M. avium subsp. paratuberculosis C strain diluted in negative bovine feces. (B) M. avium subsp. paratuberculosis S strain diluted in negative ovine feces. Replicate qPCR results at each dilution are shown for individual fecal samples at each dilution of M. avium subsp. paratuberculosis; these results were reproducible in a replicate experiment. The number of M. avium subsp. paratuberculosis (MAP) organisms per gram of feces is shown on the x axis, and CT values are on the y axis. ND, not detectable (no CT).
Repeatability.
The repeatability of the IS900 qPCR for M. avium subsp. paratuberculosis genomic DNA standards is shown in Fig. 1. To assess the repeatability of the entire HT-J extraction and PCR method, a total of 34 exposed cattle (n = 15) and sheep (n = 19) samples were analyzed in two replicate experiments (Fig. 4). A paired t test of DNA quantities from all replicates indicated the results for the two tests were not significantly different. In terms of positive versus negative HT-J test outcomes, the results were identical for all 15 cattle samples (8 positive and 7 negative) and 18/19 sheep samples (15 positive and 3 negative). For the sheep sample for which the results of the two HT-J experiments were different, the “negative” test had detectable amounts of M. avium subsp. paratuberculosis DNA (0.0004 pg), but this was below the positive/negative cut point threshold of 0.001 pg. A plot of the variability in DNA quantity for the two independent HT-J tests showed that the majority of the replicate HT-J tests had DNA quantities within 1 log10 of each other (Fig. 4B).
FIG 4.
Repeatability of the HT-J assay. The results for replicate HT-J assays on 34 individual fecal samples are shown. (A) Mean DNA quantities for each sample in replicate HT-J assays (●, test 1; ○, test 2). (B) Plot of the difference between the log10 DNA quantities (DNA quantity from test 1 − DNA quantity from test 2) of all replicates for 34 individual fecal samples. The 95% upper and lower limits of agreement are shown as dotted lines. There was no bias in the results between the first and second test, as the values were scattered on both sides of the horizontal axis, confirmed by a paired t test (no significant difference).
Repeatability for samples containing a small quantity of M. avium subsp. paratuberculosis.
Thirty culture-positive samples from calves experimentally inoculated with M. avium subsp. paratuberculosis (<12 months of age, n = 26) and adult cattle from an exposed property (n = 4) were known to contain low numbers of M. avium subsp. paratuberculosis (Table 4) because the time to growth in Bactec liquid culture medium was ≥5 weeks (20). A DNA extract was prepared from each sample and tested in quadruplicate in two separate qPCR experiments. Replicates that were positive had low levels of M. avium subsp. paratuberculosis DNA ranging from 0.0008 pg to 0.02 pg. M. avium subsp. paratuberculosis was not detected in all replicates of a DNA extract. For example, samples 1 and 8 had a DNA quantity detectable in only 1 or 2 of the 8 replicate qPCRs (Table 4). These samples were at the LOD of the assay and demonstrate that for such samples, subsampling the same DNA extract will often lead to inconsistent detection of target DNA for stoichiometric reasons (15).
TABLE 4.
Pattern of results in repeated PCR experiments on individual fecal DNA extracts from 30 bovine fecal samples that were culture positive but contained low numbers of viable M. avium subsp. paratuberculosis based on time to peak growth in Bactec medium (20)
| Sample no. | Wks to GI of 999a | Experiment 1 |
Experiment 2 |
||
|---|---|---|---|---|---|
| Avg DNA quantity of positive replicates (pg)b | No. of positive replicatesc | Avg DNA quantity of positive replicates (pg)b | No. of positive replicatesc | ||
| 1 | 6 | 0.0059 | 2 | 0 | |
| 2 | 6 | 0 | 0 | ||
| 3 | 5 | 0 | 0 | ||
| 4 | 7 | 0.0037 | 4 | 0.0026 | 2 |
| 5 | 6 | 0.0023 | 3 | 0.0014 | 1 |
| 6 | 6 | 0.0032 | 4 | 0.0027 | 4 |
| 7 | 5 | 0.0062 | 4 | 0.0044 | 4 |
| 8 | 6 | 0 | 0.0042 | 1 | |
| 9 | 7 | 0.0022 | 4 | 0.0029 | 3 |
| 10 | 12 | 0.0041 | 3 | 0.0041 | 1 |
| 11 | 7 | 0.0034 | 3 | 0.0020 | 2 |
| 12 | 6 | 0.0009 | 2 | 0 | |
| 13 | 12 | 0.0051 | 4 | 0.0049 | 4 |
| 14 | 9 | 0.0043 | 4 | 0.0025 | 4 |
| 15 | 8 | 0.0041 | 2 | 0.0046 | 4 |
| 16 | 7 | 0.0048 | 4 | 0.0057 | 4 |
| 17 | 10 | 0 | 0 | ||
| 18 | 9 | 0.0028 | 3 | 0.0026 | 4 |
| 19 | 8 | 0.0040 | 4 | 0.0029 | 4 |
| 20 | 7 | 0 | 0 | ||
| 21 | 5 | 0.0011 | 4 | 0.0010 | 3 |
| 22 | 9 | 0 | 0 | ||
| 23 | 9 | 0.0026 | 4 | 0.0009 | 3 |
| 24 | 5 | 0.0019 | 4 | 0.0011 | 4 |
| 25 | 8 | 0.0025 | 4 | 0.0008 | 3 |
| 26 | 10 | 0.0016 | 3 | 0.0009 | 4 |
| 27 | 6 | 0.0214 | 4 | 0.0105 | 4 |
| 28 | 5 | 0.0051 | 4 | 0.0033 | 4 |
| 29 | 5 | 0.0062 | 4 | 0.0022 | 4 |
| 30 | 9 | 0.0022 | 4 | 0.0005 | 4 |
GI, growth index. Weeks of growth in radiometric liquid culture medium to reach a reading of 999.
No average DNA quantity value is given for sample extracts for which zero replicates were positive.
The number of replicates out of 4 that gave a positive result.
Detection of M. avium subsp. paratuberculosis in fecal samples relative to Bactec culture.
Overall, a total of 1,330 bovine fecal samples and 596 ovine fecal samples were tested. The samples tested included animals across a spectrum of disease from uninfected to early subclinical and clinical disease (see Materials and Methods).
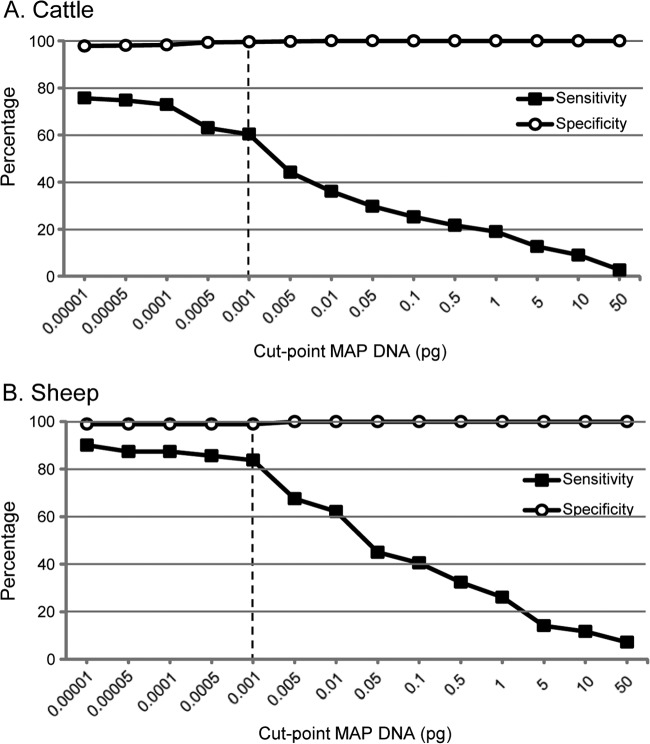
The sensitivity and specificity values for the HT-J assay relative to culture were dependent on the cut point imposed on the data to determine positive results, as shown in Fig. 5. A cut point was selected (0.001 pg M. avium subsp. paratuberculosis DNA) that led to high relative specificity values for both cattle and sheep while retaining relative sensitivity. A result was classified as positive only when both replicate qPCRs were within the correct Tm range (89.1 ± 1.5°C; Fig. 1C) and had a DNA quantity of ≥0.001 pg.
FIG 5.
The specificity and sensitivity of the HT-J test relative to liquid radiometric (Bactec) culture at different DNA quantity positive/negative cut points. The vertical dotted line shows the recommended cut point (0.001 pg).
Of the fecal samples that were collected from populations that were not exposed to M. avium subsp. paratuberculosis and that were uniformly culture negative, 99.6% from cattle and 98.9% from sheep were negative in the HT-J test (Table 5). These results suggest that both tests are highly specific. Of the fecal samples from M. avium subsp. paratuberculosis-exposed animals that were culture positive, 60.4% (from cattle) and 83.8% (from sheep) were positive in the HT-J test (Table 6). These outcomes were defined using the criteria whereby samples with one of two replicates being positive and the other one being negative were classified as qPCR negative; the performances of the test using alternate criteria are shown in Table 3. It is important to note that for both the bovine and ovine samples, the HT-J test identified slightly more positive samples from animals from M. avium subsp. paratuberculosis-exposed populations than did Bactec culture (Table 6). Many of the culture-positive samples that were not classified as HT-J positive had detectable levels of M. avium subsp. paratuberculosis DNA but did not meet the positive test criteria due to quantities of DNA below the positive/negative cut point and/or differing results for the two qPCR replicates (see Fig. S2 in the supplemental material). McNemar's test for paired data was used to confirm that the rates of detection of M. avium subsp. paratuberculosis in feces for the HT-J and Bactec culture tests were not significantly different (P > 0.05).
TABLE 5.
Results of the HT-J test relative to liquid radiometric (Bactec) culture for bovine and ovine fecal samples from M. avium subsp. paratuberculosis-unexposed herds/flocks
| HT-J test result by sample type | No. of Bactec culture results |
Relative specificity (% [95% CI])a | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Cattle samples | 99.6 (98.43–99.95) | |||
| Positive | 0 | 2 | 2 | |
| Negative | 0 | 458 | 458 | |
| Total | 0 | 460 | 460 | |
| Sheep samples | 98.9 (93.90–99.97) | |||
| Positive | 0 | 1 | 1 | |
| Negative | 0 | 88 | 88 | |
| Total | 0 | 89 | 89 | |
CI, confidence interval.
TABLE 6.
Results of the HT-J test relative to liquid radiometric (Bactec) culture for bovine and ovine fecal samples from M. avium subsp. paratuberculosis-exposed herds/flocks
| HT-J test result by sample type | No. of Bactec culture results |
Relative sensitivity (% [95% CI])a | ||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Cattle samples | 60.4 (50.63–69.52) | |||
| Positive | 67 | 57 | 124 | |
| Negative | 44 | 702 | 746 | |
| Total | 111 | 759 | 870 | |
| Sheep samples | 83.8 (75.59–90.10) | |||
| Positive | 93 | 24 | 117 | |
| Negative | 18 | 372 | 390 | |
| Total | 111 | 396 | 507 | |
CI, confidence interval.
The likelihood of a sample being culture positive was strongly related to the M. avium subsp. paratuberculosis DNA quantity detected in the sample (chi-square test, 50.1; P < 0.001) (Table 7). Almost all (97%) of the samples with a high quantity of M. avium subsp. paratuberculosis DNA were culture positive, while 25% of samples with a low quantity of M. avium subsp. paratuberculosis DNA quantity were culture positive. Similarly, the likelihood of obtaining a positive HT-J test result was correlated with the number of live M. avium subsp. paratuberculosis cells per gram of feces (Spearman's rank correlation, 0.96; P < 0.001) (Table 8).
TABLE 7.
Comparison of HT-J DNA quantity with the result for radiometric culture for samples from M. avium subsp. paratuberculosis-exposed cattle
| HT-J-positive categorya | DNA quantity (pg)b | BACTEC culture result |
||
|---|---|---|---|---|
| Positive | Negative | % Positive | ||
| High | >0.1 | 28 | 1 | 97 |
| Medium | 0.005–0.1 | 22 | 6 | 79 |
| Lowc | 0.001–0.005 | 17 | 50 | 25 |
The high, medium, and low categories are for grading of the HT-J-positive results, assessed according to the established positive/negative test criteria for the assay.
DNA quantity determined from the M. avium subsp. paratuberculosis genomic DNA standard curve included in the IS900 qPCR assay.
Both of the HT-J-positive results in the unexposed cohort had low levels of M. avium subsp. paratuberculosis genomic DNA.
TABLE 8.
Detection of radiometric culture-positive samples with various amounts of M. avium subsp. paratuberculosis using the HT-J test, based on samples from cattle
| Weeks to peak growth index | No. of live M. avium subsp. paratuberculosis cells per g of fecesa | Total no. of culture positives | % HT-J positive |
|---|---|---|---|
| 3 | ∼105 | 15 | 93.3 |
| 4 | ∼104 | 21 | 81.0 |
| 5 | ∼103 | 27 | 66.7 |
| 6 | ∼101 | 29 | 41.4 |
| 7 | ∼100 | 12 | 33.3 |
| 8 | ≤100 | 5 | 40.0 |
| 9 | ≤100 | 1 | 0.0 |
| 12 | ≤100 | 1 | 0.0 |
Estimate derived from L. Reddacliff et al. (20).
All exposed cattle herds that returned at least one culture-positive result were also identified as positive by the HT-J test. However, HT-J-positive samples were identified in an additional four exposed cattle herds in which none of the samples were culture positive. Similarly, culture-positive samples were identified in 13 sheep flocks, all of which were identified as positive by the HT-J test. Examples of the patterns of HT-J and fecal culture results from herds with various prevalences of M. avium subsp. paratuberculosis infection are shown in Fig. 6. Regardless of the region from which the sample originated, the majority of HT-J-positive results were in the low to medium DNA quantity ranges (Table 2).
FIG 6.
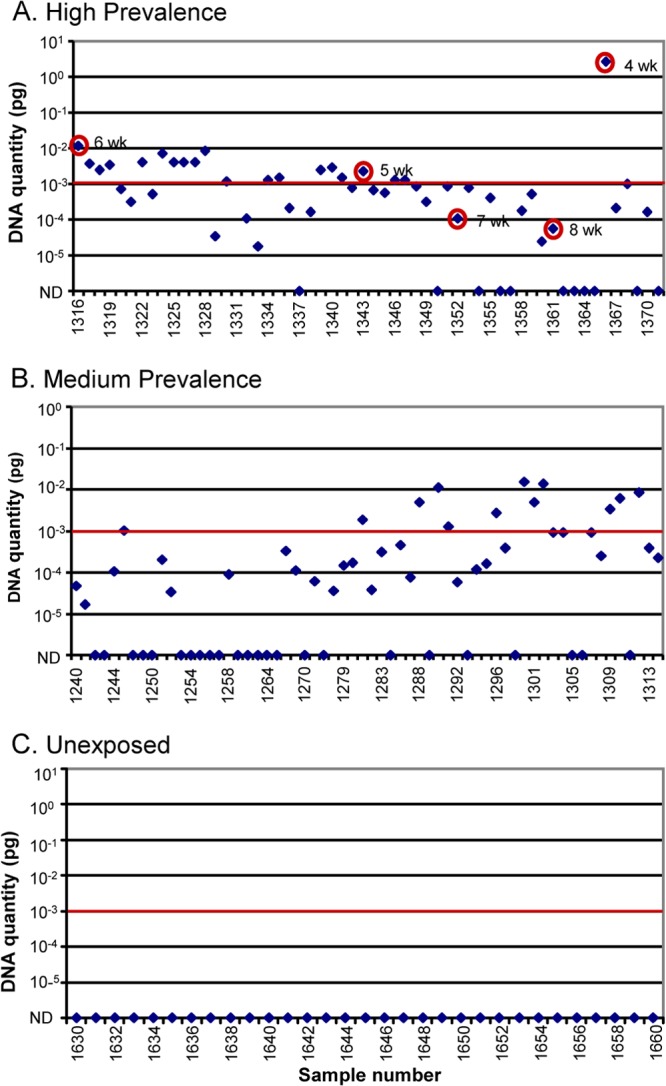
HT-J and culture results for samples from three cattle farms with differing disease prevalence: high prevalence (A), medium prevalence (B), and unexposed (C) to M. avium subsp. paratuberculosis. The quantity of DNA determined from the M. avium subsp. paratuberculosis DNA standard curve is shown on the y axis, and sample numbers (animal no.) are shown along the x axis. The PCR result of each individual animal is shown as a dot. The positive/negative cut point is shown as a horizontal line. (A) Culture-positive samples are circled with weeks of growth in radiometric culture indicated. ND, not detected.
DISCUSSION
We developed a new high-throughput fecal DNA extraction procedure coupled with a qPCR assay (termed the HT-J test) for the direct detection of M. avium subsp. paratuberculosis in fecal samples from cattle and sheep, and we provided an evaluation of its performance according to standards recently provided for M. avium subsp. paratuberculosis diagnostic test validation (2) and the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (15). The HT-J test enables the rapid reporting of results with a similar rate of detection of M. avium subsp. paratuberculosis-positive samples as that of fecal culture in liquid medium.
In this study, the optimization of fecal DNA extraction was undertaken, and critical steps affecting the sensitivity of the test were identified. The first was related to the handing of the fecal samples and identified that storage at −80°C rather than −20°C was required for greatest sensitivity. This is consistent with standard sample-handling protocols for the culture of M. avium subsp. paratuberculosis; however, the biological basis of the observed effects on sensitivity has yet to be established. The optimal fecal suspension method was also consistent with a method commonly used for fecal culture, with the volumes of feces and saline and principles being the same. Mechanical disruption of the M. avium subsp. paratuberculosis cell wall using a bead beater was previously shown to be critical for analytical sensitivity (14, 31). The use of a commercial DNA extraction procedure (Qiagen BioSprint 96 one-for-all vet kit) gave high sensitivity as well as high-throughput capacity in a diagnostic laboratory setting. Magnetic bead DNA purification methods generally lead to lower carryover of contaminants or potential PCR inhibitors from a sample, as the method involves the physical removal of the DNA from the remainder of the sample (31–34).
The critical components for the qPCR assay were identified by Kawaji et al. (14) as primer sequence and high annealing temperature (68°C). These were used in the qPCR for the HT-J test to ensure specificity, but the assay was modified to use a relatively inexpensive mastermix (SensiMix SYBR Low-ROX kit; Bioline). Primer location was based on gene regions where there were base mismatches with IS900-like sequences that have been identified in organisms other than M. avium subsp. paratuberculosis. Analytical specificity was reconfirmed in this study using the modified qPCR conditions across a panel of 51 mycobacterial isolates, including those with IS900-like sequences (14). IS900 is the most common molecular target sequence used to detect M. avium subsp. paratuberculosis. This is based on initial reports of its specificity for the M. avium subsp. paratuberculosis genome, and it being a multicopy gene, which increases the likelihood of detection compared to single-copy genes (35). However, there have been a number of reports of other mycobacterial species with IS900-like sequences (36–38), but no other microbial genera have been reported to have this insertion element. IS900 primers that do not discriminate between M. avium subsp. paratuberculosis and other mycobacteria with IS900-like sequences are still in general use (39). To overcome concerns with specificity, other elements, such as F57, have been applied in real-time PCR assays (40), but these generally have lower analytical sensitivity for the M. avium subsp. paratuberculosis genome than IS900 (41).
The analytical sensitivity of the qPCR test based on the M. avium subsp. paratuberculosis genomic DNA standard curve was comparable to that reported by Kawaji et al. (14), as was the analytical sensitivity of the HT-J test for the detection of M. avium subsp. paratuberculosis in spiked fecal samples. The inclusion of a M. avium subsp. paratuberculosis genomic DNA standard curve allowed for the normalization of results between experiments and will facilitate interlaboratory standardization of test results, as comparisons of raw CT values are subject to run-to-run variability (15). Furthermore, the criteria developed for the determination of HT-J-positive test results depend on the inclusion of the DNA standard curve in every run. To obtain the maximum potential specificity for use in control programs in Australia, the criteria for a positive HT-J test result required that both qPCR replicates be positive, while samples with one positive and one negative qPCR replicate were considered negative. Alternative criteria, including those that consider test results with a single positive replicate to be positive, were assessed with respect to their effects on relative diagnostic specificity and sensitivity (Table 3). Although all options had high apparent specificities (≥97.6%), the criteria that gave the highest apparent specificity across both animal species were adopted for the purposes of the analyses shown in Tables 5 and 6.
The HT-J test, like other qPCR-based diagnostic assays, provides results on a continuous scale, making it possible to set different positive/negative cut points (Fig. 5) (30). In this study, both Tm and DNA quantity were used to define a cut point. The Tm was specified as a range and needed to be determined for other mastermixes and qPCR machine platforms (average Tm of M. avium subsp. paratuberculosis genomic DNA standards ± 1.5°C). The selection of the 0.001-pg cut point was determined using a rigorous epidemiological approach, based on the probability of misclassification of the disease status while prioritizing high specificity (30), and it was within the scope of MIQE guidelines, relating to the establishment of assay limits of detection (15). This DNA quantity is equivalent to approximately one-fifth of a M. avium subsp. paratuberculosis genome; however, the target gene (IS900) is a multicopy gene present in 15 to 20 copies per genome (35). Depending on the purpose of the test, there is an opportunity to use different cut points for interpretation of test result data. A high/medium/low test categorization scale was developed in this study that closely aligned with the likelihood of a sample returning a positive-culture result (Table 6).
In Australia, a test cannot be used in a national animal disease control program unless it has been fully validated and assessed (see www.scahls.org.au/). The sampling strategy incorporated regional areas of Australia where the test is likely to be applied. A spectrum of exposure/infection was included to avoid skewed sensitivity estimates that might have occurred by selecting samples from animals based on the results of other tests, for example, serum antibody ELISA. In addition, a large sample size was used. The strategy was to include fecal samples likely to have a range of concentrations of viable M. avium subsp. paratuberculosis cells, and it was not biased by inclusion only of ELISA reactors or fecal samples only with high numbers of M. avium subsp. paratuberculosis, which are relatively easy to detect. This is reflected by the range of culture incubation times to peak growth index (GI) in radiometric culture (3 to 12 weeks), for which the number of weeks to peak GI is inversely proportional to the number of viable M. avium subsp. paratuberculosis in the sample (20). These design elements are consistent with recommendations for M. avium subsp. paratuberculosis diagnostic test validation (1, 2). Despite these design considerations, we acknowledge that the samples evaluated in this study are not truly representative of paratuberculosis prevalence. In particular, as paratuberculosis is endemic and cryptic, it is not possible to obtain a reliable or representative sample from nonexposed noninfected individuals from within the M. avium subsp. paratuberculosis-exposed region of Australia. Therefore, we used samples from regions of Australia where at the time it was thought that exposure to M. avium subsp. paratuberculosis did not occur. This is no longer the case due to the detection in 2013 of JD in beef cattle in Queensland (24). However, we believe the probability of the supposedly nonexposed herds from which samples were collected being exposed to M. avium subsp. paratuberculosis is low, based on verbal advice from Chief Veterinary Officers in each region. For this reason, valid estimates of diagnostic specificity are not available, notwithstanding the fact that both fecal culture and HT-J appeared to be specific tests when applied to the samples in this study. With respect to diagnostic sensitivity, the limited number of flocks and herds examined and the lack of reliable independent classifications of individual animals as infected or noninfected make it impossible to determine the true diagnostic sensitivity for either the HT-J test or fecal culture.
Infectious animals shedding large numbers of M. avium subsp. paratuberculosis (so-called heavy or super shedders) are likely to be detected by most protocols for direct fecal PCR (42) and a range of other antemortem tests (43). However, it is epidemiologically and diagnostically important to be able to detect very low numbers of M. avium subsp. paratuberculosis, as the majority of infected animals in a flock/herd are subclinical cases (12). For this reason, during optimization of the HT-J test, extensive use was made of a panel of fecal samples that were known to contain very low numbers of M. avium subsp. paratuberculosis cells. The results from this study show that there was variable detection between the replicate qPCRs. It is important to be aware that when an analyte (in this case, M. avium subsp. paratuberculosis genomic DNA in the fecal DNA extract) is present at a very low concentration in a sample, it is a matter of chance whether a small aliquot removed from the sample will actually contain the analyte (15). Stochastic variation therefore applies to such analyses and is the reason why some replicates of a DNA extraction produce a positive result and others do not. This also applies to the comparison between HT-J and radiometric culture, as the culture was performed on a separate aliquot of feces to that used in the HT-J testing. The agreement between the culture and HT-J test results decreased with decreasing numbers of M. avium subsp. paratuberculosis in the original sample because of the lower likelihood that a positive test result will be obtained from all subsamples of the original.
Although sensitivity estimates for HT-J are provided here relative to those of Bactec culture, it is clear from other studies that no antemortem test for M. avium subsp. paratuberculosis infection is capable of detecting all infected cattle or sheep due to the epidemiology and pathogenesis of JD (12). Our results show that the HT-J test detects a set of animals that may overlap but is not equivalent to those detected by fecal culture. A comparison of the HT-J test with fecal culture, intestinal tissue culture, histopathology, cell-mediated immune assays, and antibody ELISA is required to better understand the nature of HT-J-positive results from fecal culture-negative samples. All fecal tests for M. avium subsp. paratuberculosis work more reliably at the herd/flock level. Consequently, the HT-J test has been approved by the regulatory authority in Australia for herd/flock testing in the national JD program (see www.scahls.org.au/).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Meat and Livestock Australia and by the Cattle Council of Australia, Sheepmeat Council of Australia, and WoolProducers Australia through Animal Health Australia.
We thank Craig Kristo, Nobel Toribio, and James Dalton for animal husbandry support, Adelyn Bolithon and Nicole Carter for technical support, and the many veterinarians and livestock producers who assisted in this study.
Footnotes
Published ahead of print 18 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03233-13.
REFERENCES
- 1.Nielsen SS, Toft N, Gardner IA. 2011. Structured approach to design of diagnostic test evaluation studies for chronic progressive infections in animals. Vet. Microbiol. 150:115–125. 10.1016/j.vetmic.2011.01.019 [DOI] [PubMed] [Google Scholar]
- 2.Gardner IA, Nielsen SS, Whittington RJ, Collins MT, Bakker D, Harris B, Sreevatsan S, Lombard JE, Sweeney R, Smith DR, Gavalchin J, Eda S. 2011. Consensus-based reporting standards for diagnostic test accuracy studies for paratuberculosis in ruminants. Prev. Vet. Med. 101:18–34. 10.1016/j.prevetmed.2011.04.002 [DOI] [PubMed] [Google Scholar]
- 3.Whittington RJ, Begg DJ, de Silva K, Plain KM, Purdie AC. 2012. Comparative immunological and microbiological aspects of paratuberculosis as a model mycobacterial infection. Vet. Immunol. Immunopathol. 148:29–47. 10.1016/j.vetimm.2011.03.003 [DOI] [PubMed] [Google Scholar]
- 4.Whittington RJ, Whittington AM, Waldron A, Begg DJ, de Silva K, Purdie AC, Plain KM. 2013. Development and validation of a liquid medium (M7H9C) for routine culture of Mycobacterium avium subsp. paratuberculosis to replace modified Bactec 12B medium. J. Clin. Microbiol. 51:3993–4000. 10.1128/JCM.01373-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sibley JA, Woodbury MR, Appleyard GD, Elkin B. 2007. Mycobacterium avium subspecies paratuberculosis in bison (Bison bison) from northern Canada. J. Wildl. Dis. 43:775–779. 10.7589/0090-3558-43.4.775 [DOI] [PubMed] [Google Scholar]
- 6.Forde T, De Buck J, Elkin B, Kutz S, van der Meer F, Orsel K. 2013. Contrasting results of culture-dependent and molecular analyses of Mycobacterium avium subsp. paratuberculosis from wood bison. Appl. Environ. Microbiol. 79:4448–4454. 10.1128/AEM.00995-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittington RJ. 2009. Factors affecting isolation and identification of Mycobacterium avium subsp. paratuberculosis from fecal and tissue samples in a liquid culture system. J. Clin. Microbiol. 47:614–622. 10.1128/JCM.01986-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whittington RJ. 2010. Cultivation of Mycobacterium avium subsp. paratuberculosis, p 244–266 In Behr MA, Collins DM. (ed), Paratuberculosis: organism, disease, control. CABI, Wallingford, United Kingdom [Google Scholar]
- 9.Monteiro L, Bonnemaison D, Vekris A, Petry KG, Bonnet J, Vidal R, Cabrita J, Mégraud F. 1997. Complex polysaccharides as PCR inhibitors in feces: Helicobacter pylori model. J. Clin. Microbiol. 35:995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thornton CG, Passen S. 2004. Inhibition of PCR amplification by phytic acid, and treatment of bovine fecal specimens with phytase to reduce inhibition. J. Microbiol. Methods 59:43–52. 10.1016/j.mimet.2004.06.001 [DOI] [PubMed] [Google Scholar]
- 11.Kreader CA. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittington RJ, Sergeant ES. 2001. Progress towards understanding the spread, detection and control of Mycobacterium avium subsp. paratuberculosis in animal populations. Aust. Vet. J. 79:267–278. 10.1111/j.1751-0813.2001.tb11980.x [DOI] [PubMed] [Google Scholar]
- 13.Bull TJ, McMinn EJ, Sidi-Boumedine K, Skull A, Durkin D, Neild P, Rhodes G, Pickup R, Hermon-Taylor J. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915–2923. 10.1128/JCM.41.7.2915-2923.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaji S, Taylor DL, Mori Y, Whittington RJ. 2007. Detection of Mycobacterium avium subsp. paratuberculosis in ovine faeces by direct quantitative PCR has similar or greater sensitivity compared to radiometric culture. Vet. Microbiol. 125:36–48. 10.1016/j.vetmic.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 15.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- 16.Begg D, Whittington R. 2010. Paratuberculosis in sheep, p 157–168 In Behr MA, Collins DM. (ed), Paratuberculosis: organism, disease, control. CABI Wallingford, United Kingdom [Google Scholar]
- 17.Marsh IB, Bannantine JP, Paustian ML, Tizard ML, Kapur V, Whittington RJ. 2006. Genomic comparison of Mycobacterium avium subsp. paratuberculosis sheep and cattle strains by microarray hybridization. J. Bacteriol. 188:2290–2293. 10.1128/JB.188.6.2290-2293.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whittington RJ, Marsh I, McAllister S, Turner MJ, Marshall DJ, Fraser CA. 1999. Evaluation of modified Bactec 12B radiometric medium and solid media for culture of Mycobacterium avium subsp. paratuberculosis from sheep. J. Clin. Microbiol. 37:1077–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whittington RJ, Marsh I, Turner MJ, McAllister S, Choy E, Eamens GJ, Marshall DJ, Ottaway S. 1998. Rapid detection of Mycobacterium paratuberculosis in clinical samples from ruminants and in spiked environmental samples by modified Bactec 12B radiometric culture and direct confirmation by IS900 PCR. J. Clin. Microbiol. 36:701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddacliff LA, Nicholls PJ, Vadali A, Whittington RJ. 2003. Use of growth indices from radiometric culture for quantification of sheep strains of Mycobacterium avium subsp. paratuberculosis. Appl. Environ. Microbiol. 69:3510–3516. 10.1128/AEM.69.6.3510-3516.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plain KM, Purdie AC, Begg DJ, de Silva K, Whittington RJ. 2010. Toll-like receptor (TLR)6 and TLR1 differentiation in gene expression studies of Johne's disease. Vet. Immunol. Immunopathol. 137:142–148. 10.1016/j.vetimm.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 22.Begg DJ, de Silva K, Di Fiore L, Taylor DL, Bower K, Zhong L, Kawaji S, Emery D, Whittington RJ. 2010. Experimental infection model for Johne's disease using a lyophilised, pure culture, seedstock of Mycobacterium avium subspecies paratuberculosis. Vet. Microbiol. 141:301–311. 10.1016/j.vetmic.2009.09.007 [DOI] [PubMed] [Google Scholar]
- 23.Purdie AC, Plain KM, Begg DJ, de Silva K, Whittington RJ. 2012. Expression of genes associated with the antigen presentation and processing pathway are consistently regulated in early Mycobacterium avium subsp. paratuberculosis infection. Comp. Immunol. Microbiol. Infect. Dis. 35:151–162. 10.1016/j.cimid.2011.12.007 [DOI] [PubMed] [Google Scholar]
- 24.Animal Health Australia 2013. Bovine Johne's disease in Australia. Animal Health Australia, Deakin, Australian Capital Territory, Australia: http://www.animalhealthaustralia.com.au/programs/johnes-disease/bovine-johnes-disease-in-australia/ [Google Scholar]
- 25.Dhand NK, Johnson WO, Eppleston J, Whittington RJ, Windsor PA. 2013. Comparison of pre- and post-vaccination ovine Johne's disease prevalence using a Bayesian approach. Prev. Vet. Med. 111:81–91. 10.1016/j.prevetmed.2013.03.003 [DOI] [PubMed] [Google Scholar]
- 26.McGregor H, Dhand NK, Dhungyel OP, Whittington RJ. 2012. Transmission of Mycobacterium avium subsp. paratuberculosis: dose-response and age-based susceptibility in a sheep model. Prev. Vet. Med. 107:76–84. 10.1016/j.prevetmed.2012.05.014 [DOI] [PubMed] [Google Scholar]
- 27.Petrie A, Watson P. 2006. Statistics for veterinary and animal science, 2nd ed. Blackwell Publishing, Oxford, United Kingdom [Google Scholar]
- 28.Motulsky H. 1995. Intuitive biostatistics: a nonmathematical guide to statistical thinking. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 29.World Organisation for Animal Health (OIE) 2013. Chapter 1.1.5: principles and methods of validation of diagnostic assays for infectious diseases. In Manual of diagnostic tests and vaccines for terrestrial animals. OIE, Paris, France [Google Scholar]
- 30.Caraguel CG, Stryhn H, Gagné N, Dohoo IR, Hammell KL. 2011. Selection of a cutoff value for real-time polymerase chain reaction results to fit a diagnostic purpose: analytical and epidemiologic approaches. J. Vet. Diagn. Invest. 23:2–15. 10.1177/104063871102300102 [DOI] [PubMed] [Google Scholar]
- 31.Leite FL, Stokes KD, Robbe-Austerman S, Stabel JR. 2013. Comparison of fecal DNA extraction kits for the detection of Mycobacterium avium subsp. paratuberculosis by polymerase chain reaction. J. Vet. Diagn. Invest. 25:27–34. 10.1177/1040638712466395 [DOI] [PubMed] [Google Scholar]
- 32.Petrich A, Mahony J, Chong S, Broukhanski G, Gharabaghi F, Johnson G, Louie L, Luinstra K, Willey B, Akhaven P, Chui L, Jamieson F, Louie M, Mazzulli T, Tellier R, Smieja M, Cai W, Chernesky M, Richardson SE, Ontario Laboratory Working Group for the Rapid Diagnosis of Emerging Infections 2006. Multicenter comparison of nucleic acid extraction methods for detection of severe acute respiratory syndrome coronavirus RNA in stool specimens. J. Clin. Microbiol. 44:2681–2688. 10.1128/JCM.02460-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fang X, Willis RC, Burrell A, Evans K, Hoang Q, Xu W, Bounpheng M. 2007. Automation of nucleic acid isolation on KingFisher magnetic particle processors. J. Lab. Autom. 12:195–201. 10.1016/j.jala.2007.05.001 [DOI] [Google Scholar]
- 34.Kim S, Park SJ, Namgoong S, Sung H, Kim MN. 2009. Comparative evaluation of two automated systems for nucleic acid extraction of BK virus: NucliSens easyMAG versus BioRobot MDx. J. Virol. Methods 162:208–212. 10.1016/j.jviromet.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 35.Green EP, Tizard ML, Moss MT, Thompson J, Winterbourne DJ, McFadden JJ, Hermon-Taylor J. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063–9073. 10.1093/nar/17.22.9063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cousins DV, Whittington R, Marsh I, Masters A, Evans RJ, Kluver P. 1999. Mycobacteria distenct [sic] from Mycobacterium avium subsp. paratuberculosis isolated from the faeces of ruminants possess IS900-like sequences detectable by IS900 polymerase chain reaction: Implications for diagnosis. Mol. Cell Probes 13:431–442. 10.1006/mcpr.1999.0275 [DOI] [PubMed] [Google Scholar]
- 37.Taddei R, Barbieri I, Pacciarini ML, Fallacara F, Belletti GL, Arrigoni N. 2008. Mycobacterium porcinum strains isolated from bovine bulk milk: implications for Mycobacterium avium subsp. paratuberculosis detection by PCR and culture. Vet. Microbiol. 130:338–347. 10.1016/j.vetmic.2008.02.007 [DOI] [PubMed] [Google Scholar]
- 38.Englund S, Bölske G, Johansson KE. 2002. An IS900-like sequence found in a Mycobacterium sp. other than Mycobacterium avium subsp. paratuberculosis. FEMS Microbiol. Lett. 209:267–271. 10.1111/j.1574-6968.2002.tb11142.x [DOI] [PubMed] [Google Scholar]
- 39.Cook KL, Britt JS. 2007. Optimization of methods for detecting Mycobacterium avium subsp. paratuberculosis in environmental samples using quantitative, real-time PCR. J. Microbiol. Methods 69:154–160. 10.1016/j.mimet.2006.12.017 [DOI] [PubMed] [Google Scholar]
- 40.Möbius P, Hotzel H, Rassbach A, Köhler H. 2008. Comparison of 13 single-round and nested PCR assays targeting IS900, ISMav2, f57 and locus 255 for detection of Mycobacterium avium subsp. paratuberculosis. Vet. Microbiol. 126:324–333. 10.1016/j.vetmic.2007.07.016 [DOI] [PubMed] [Google Scholar]
- 41.Castellanos E, de Juan L, Domínguez L, Aranaz A. 2011. Progress in molecular typing of Mycobacterium avium subspecies paratuberculosis. Res. Vet. Sci. 92:169–179. 10.1016/j.rvsc.2011.05.017 [DOI] [PubMed] [Google Scholar]
- 42.Wells SJ, Collins MT, Faaberg KS, Wees C, Tavornpanich S, Petrini KR, Collins JE, Cernicchiaro N, Whitlock RH. 2006. Evaluation of a rapid fecal PCR test for detection of Mycobacterium avium subsp. paratuberculosis in dairy cattle. Clin. Vaccine Immunol. 13:1125–1130. 10.1128/CVI.00236-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen SS, Toft N. 2008. Ante mortem diagnosis of paratuberculosis: a review of accuracies of ELISA, interferon-gamma assay and faecal culture techniques. Vet. Microbiol. 129:217–235. 10.1016/j.vetmic.2007.12.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.