Abstract
Helcococcus spp. are Gram-positive, catalase-negative, facultatively anaerobic cocci that are associated with wound and prosthetic joint infections as well bacteremia and empyema. Five Helcococcus spp. strains were isolated from our patient population, including 2 strains of Helcococcus kunzii from trauma-associated wounds, 2 Helcococcus sueciensis strains from blood and abscess, and a novel Helcococcus spp. strain from blood associated with urosepsis. Based on the phenotypic and phylogenetic evidence, we propose that the unknown bacterium be classified as Helcococcus seattlensis sp. nov. We found that all 5 tested Helcococcus strains grew as satellite colonies around Staphylococcus aureus and, interestingly, both H. kunzii strains were isolated together with S. aureus. In addition to 16S rRNA gene sequencing, conventional methods for leucine aminopeptidase (LAP) and pyrrolidonyl arylamidase (PYR) testing can be cost-effective and efficient for differentiation of Helcococcus spp. from Abiotrophia and Granulicatella species. Using nonstandard methods, we found that all tested Helcococcus spp. had high MICs of >4/76 μg/ml for trimethoprim-sulfamethoxazole, an antibiotic commonly used to treat urinary tract infections. High MICs for erythromycin, azithromycin, and clindamycin, and intermediate to high MICs for moxifloxacin, levofloxacin, and gentamicin were also observed among the Helcococcus strains.
INTRODUCTION
Three species of Helcococcus have been described by Collins et al.: Helcococcus kunzii, Helcococcus sueciensis, and Helcococcus ovis (1–3). H. kunzii has been isolated from blood, a prosthetic joint, wounds, abscesses, plantar phlegmon, and an implantable cardiac device (1, 4–9) and is thought to be a nonpathogenic skin colonizer (10). H. sueciensis has been isolated from a human wound (2), while H. ovis causes mastitis and urocystitis in animals (3). The susceptibility of H. kunzii to a few antibiotics has been described (11), but evaluation of a comprehensive antibiotics panel for Helcococcus spp. has been lacking. The identification of Helcococcus spp. remains challenging due to the slow growth of the organism and the inadequate databases for commercially available kits, such as API Rapid ID 32 Strep, API Rapid ID 32A, and API 20S Strep (bioMérieux) (1, 2, 8). The MicroSeq Full Gene database (v.0001a) and 500 database (v.0023b; Applied Biosystems) also lack any Helcococcus spp. Vitek 2 (bioMérieux) can only identify H. kunzii and no other Helcococcus spp. (8, 12). In the current study, we identified these isolates by 16S rRNA gene sequencing and aimed to develop a more efficient and affordable protocol to identify these bacteria of potential clinical significance. We also characterized a novel species of the genus Helcococcus, and we propose its classification as Helcococcus seattlensis sp. nov.
Case report of the novel Helcococcus sp.
An 80-year-old male with a history of type 2 diabetes, obesity, coronary disease with bypass surgery, Reiter's disease, and hypertension presented with a fever of 101.6°F. Elevated cardiac enzymes (troponin at 0.34 μg/liter and creatine kinase MB isoenzyme at 7.5 μg/liter) with a brain natriuretic peptide level of >30,000 indicated heart failure. In addition, the patient experienced painful urination, and the urinalysis results suggested a urinary tract infection (UTI): leukocyte esterase was positive (3+), nitrite was negative, white blood cell (WBC) and red blood cell counts were high (>182/high-power field [HPF] and 40/HPF, respectively). Many WBC clumps were seen in the urine. His blood test revealed an elevated WBC level (15.3 × 109/liter), the hemoglobin level was slightly decreased (12.7 g/dl), and the platelet count was normal (270 × 109/liter). Blood and urine cultures were ordered. With the BacT/Alert 3D blood culture system (bioMérieux), the anaerobic but not the aerobic bottle turned positive on day 4. Gram staining showed Gram-positive cocci in clusters with variable cell sizes (0.5 to 1.8 μm in diameter). On subculture, the organism showed negligible growth on Columbia blood agar (BA) and slightly better growth on chocolate agar (CA). The colonies were nonhemolytic, gray, and pinpoint after 48 h of incubation at 35°C in air enriched with 6 to 7% CO2. The results of 16S rRNA gene sequence analysis were compared to those for 4 closely related clinical strains that were previously recovered in our laboratory, as well as isolates from the GenBank database. The urine specimen collected at the same time as the blood was cultured on BA, CA, and MacConkey agar. Pinpoint colonies were observed on CA only, but they were not further identified because of a failure to appreciate the organism as a possible pathogen. However, Gram staining of the colonies on CA revealed Gram-positive cocci with the same morphology and sizes as the organism isolated from the blood culture. The fine lawn of colonies on CA resembled what was seen on the subculture of the blood isolate. The patient died the next day due to heart failure.
MATERIALS AND METHODS
Bacterial strains.
All 5 Helcococcus spp. strains were isolated during routine culture procedures at the VA Puget Sound Healthcare System. All isolates were stocked at −70°C using an alphanumeric system beginning with F for identification. Unless specified, all 5 clinical isolates, H. kunzii F5450, H. kunzii F7676, H. sueciensis F6341, H. sueciensis F6503, and the novel Helcococcus sp. strain F5780 were grown on CA (Remel, Lenexa, KS) at 35°C in air enriched with 6 to 7% CO2.
Growth characteristics.
The satellite test of all 5 strains was performed on BA (Remel, Lenexa, KS) with a streak of Staphylococcus aureus ATCC 25923 at 35°C in air enriched with 6 to 7% CO2 and in ambient air. In addition, novel Helcococcus strain F5780, H. kunzii F5450, and H. sueciensis F6503 were tested for satellite colony formation on Mueller-Hinton agar (MHA) at 35°C in 5% CO2–95% air. H. seattlensis F5780 was grown on CA at 3 temperatures under 4 atmospheric conditions: 30°C, 35°C, 42°C in ambient air, air enriched with 6 to 7% CO2, a microaerophilic atmosphere generated by a Pouch-MicroAero apparatus (Mitsubishi Gas Chemical America, Inc., New York), and anaerobic conditions generated by a Pack-Anaero instrument (Mitsubishi Gas Chemical America, Inc., New York).
Antibiotic susceptibility testing.
The antibiotic susceptibility testing of all 5 Helcococcus isolates was performed according to the manufacturer's instructions by using the Sensititre system with the Streptococcus panel (STP6F; Trek Diagnostic Systems, Cleveland, OH), except that the results were read on day 3 due to the slow growth of the organisms. Strains for which the MICs were greater than the highest dilution on the Sensititre panel were further tested with the Etest to determine precise MICs, except for trimethoprim-sulfamethoxazole (SXT). All Etest experiments were performed on CA with incubation at 35°C in air enriched with 6 to 7% CO2 for 2 days. Based on the facts that thymidine in sheep blood agar alters the action of sulfur drugs (13) and that Helcococcus strains showed negligible to no growth on MHA, an Etest of SXT could not be performed. The microbial resistance to SXT was subsequently confirmed by using the MicroScan system with a MICroSTREP plus panel (Siemens Healthcare Diagnostics, Deerfield, IL). Currently, no antimicrobial testing guideline from the Clinical and Laboratory Standards Institute (CLSI) is available for Helcococcus. The level of the MIC (high, intermediate, or low) determined in this study was in reference to that of S. aureus in CLSI guidelines (14).
Biochemical differentiation.
Biochemical tests were performed with the API 20S Strep (bioMérieux) and Vitek 2 (bioMérieux) systems with 2-day-old Helcococcus cultures grown on CA. Leucine aminopeptidase (LAP) and pyrrolidonyl arylamidase (PYR) results were confirmed by using conventional LAP and PYR disks (Remel, Lenexa, KS).
16S rRNA gene sequencing.
Strains were grown on CA for 48 h, and the DNA was extracted by using a Prepman apparatus (PE Applied Biosystems, Foster City, CA) following the manufacturer's instructions. 16S rRNA PCR and cycle sequencing of Helcococcus spp. were performed as previously described (15), with modifications. A 500-bp 16S rRNA gene sequence analysis was performed for H. kunzii F5450, H. kunzii F7676, H. sueciensis F6341, and H. sueciensis F6503, while 1,500-bp 16S 16S rRNA gene sequence analysis was performed for strain F5780. Three sets of primers (VB1 to VB6) were used to amplify the 1,500-bp 16S rRNA gene, and the first set (VB1 and VB2) was used for the 500-bp 16S rRNA sequencing. The primers were as follows: VB1 (TGGAGAGTTTGATCCTGGCTCAG), VB2 (TACCGCGGCTGCTGGCAC), VB3 (CCAGCAGCCGCGGTAATAC), VB4 (CGGGACTTAACCCAACATCTCAC), VB5 (GTGAGATGTTGGGTTAAGTCCCG), and VB6 (AAGGAGGTGATCCAGCCGCA) (15, 16). The sequences were analyzed and compared to information in the BIBI and GenBank databases (17). The phylogenetic tree was constructed with MUSCLE, a program for generating multiple alignments (18), with MEGA5.1 software (http://www.megasoftware.net/).
Nucleotide sequence accession numbers.
The nucleotide sequences of the 16S rRNA genes of the novel strain F5780, H. kunzii F7676, H. kunzii F5450, H. sueciensis F6341, and H. sueciensis F6503 have been deposited in the GenBank Data Library under accession numbers KF780874, KF780875, KF780876, KF780877, and KF780878, respectively.
RESULTS
The sites of isolation for all Helcococcus strains, patient demographics, and treatments are listed in Table 1. The phylogenetic and phenotypic characterizations of H. kunzii and H. sueciensis reported in previous studies (1, 2) matched our strains (Fig. 1; Table 2). The cell morphology of the novel strain F5780 is Gram-positive cocci in clusters of cells of variable sizes (0.5 to 1.8 μm in diameter). The organism grows best on CA (CA > BA) at 35°C (35°C > 30°C > 42°C) and equally well under different oxygen conditions (air, air enriched with 6 to 7% CO, microaerophilic, or anaerobic). The colonies are nonhemolytic, gray, and pinpoint on CA after 48 h of incubation at 35°C in air enriched with 6 to 7% CO2. The organism is catalase, PYR, LAP, and motility negative when tested using conventional methods. The API 20S Strep test failed to identify the organism, but the results of negative PYR and LAP concurred with those demonstrated with Remel disks. The organism utilizes ribose to produce acid.
TABLE 1.
Summary of helcococcal infections at the Veterans Administration Puget Sound Health Care System
| Case no. | Organism | Strain | Organism(s) co-recovered | Site of infection | Age (yrs)/gender of patient | Underlying disease(s) | Treatment for current episode | History | WBC result based on: |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Gram stain of specimen | Blood test (cells/liter) | |||||||||
| 1 | H. seattlensis sp. nov.a | F5780 | NAb | Blood | 80/M | CAD,c type 2 diabetes | NA | NA | NA | 15.3 × 109 |
| 2 | H. kunzii | F7676 | S. aureus | Toe abscess | 25/M | PTSDd | SXT, cephalexin | NA | Negative | NA |
| 3 | H. kunzii | F5450 | S. aureus | Inner thigh wound from trauma | 68/M | CAD, colonic polyps | Vancomycin and piperacillin-tazobactam inpatient; cephalexin outpatient | Previous episode of wound infection at same site within past mo., treated with SXT without improvement | Negative | 6.3 × 109 |
| 4 | H. sueciensis | F6341 | NA | Blood | 64/M | Colon carcinoma, CAD | Vancomycin, piperacillin-tazobactam | NA | NA | 8.7 × 109 |
| 5 | H. sueciensis | F6503 | Corynebacterium spp., two colony types | Surgical axillary wound | 67/M | Axillary squamous cell carcinoma | Vancomycin, piperacillin-tazobactam inpatient; clindamycin outpatient | NA | Moderate | 14 × 109 |
The name H. seattlensis is proposed in the current study.
NA, not available.
CAD, coronary artery disease.
PTSD, post-traumatic stress disorder.
FIG 1.
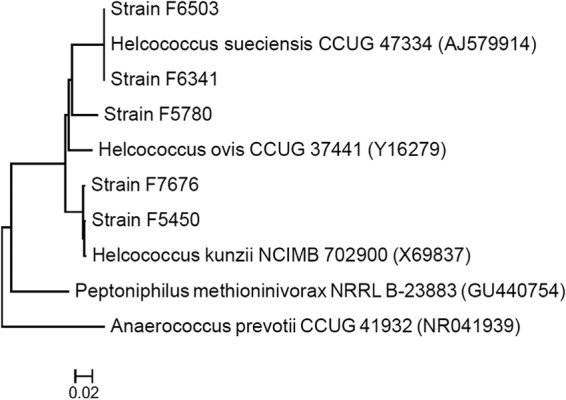
Unrooted tree showing the phylogenetic relationships of 5 Helcococcus spp. clinical strains to closely related species. The tree was inferred from 16S rRNA gene sequences (443 nucleotides) by using the neighbor-joining method, and the scale bar indicates the substitution rate per nucleotide. All names and accession numbers (in parentheses) are those cited in the GenBank database.
TABLE 2.
Important differences in phenotypic characteristics of Helcococcus, Abiotrophia, and Granulicatella isolates
| Characteristic | H. seattlensis sp. nov. (n = 1)a |
H. kunzii |
H. sueciensis |
Abiotrophia defectivad | Granulicatella elegansd | Granulicatella adiacensd | ||
|---|---|---|---|---|---|---|---|---|
| This study (n = 2) | Reported elsewhereb | This study (n = 2) | Reported elsewherec | |||||
| Catalase | − | 0/2 | − | 0/2 | − | − | − | − |
| Leucine aminopeptidase | − | 0/2 | − | 2/2 | NAe | + | + | + |
| Pyrrolidonyl arylamidase | − | 2/2 | + | 0/2 | − | + | + | + |
| Growth at 42°C | + | 1/2 | vf | 0/2 | NA | + | − | + |
| Acid produced from: | ||||||||
| Trehalose | − | 2/2 | + | 2/2 | + | + | − | − |
| Lactose | − | 2/2 | + | 2/2 | + | + | − | − |
| Glycogen | − | 2/2 | v | 1/2 | − | − | − | − |
| Ribose | + | 2/2 | − | 2/2 | − | NA | NA | NA |
| Insulin | − | 0/2 | − | 0/2 | − | − | − | + |
| Starch | − | 2/2 | v | 2/2 | + | + | − | − |
| Raffinose | − | 0/2 | − | 0/2 | − | + | + | − |
| Production of: | ||||||||
| β-Glucuronidase | + | 0/2 | − | 0/2 | − | − | − | + |
| β-Galactosidase | + | 2/2 | − | 2/2 | + | + | − | − |
The comparison of sequences to the database shown in Fig. 1 and 2 revealed that strain F5780 is most closely related to species of the genus Helcococcus. The species with the highest percentages of similarity in sequence to this organism were H. sueciensis (94%), H. ovis (94%), and H. kunzii (92%). Species of other related genera, such as Tissierella and Peptoniphilus, were related more distantly (≤87%). Based on the phylogenetic (Fig. 1 and 2) and phenotypic (Table 2) evidence, we suggest that strain F5780 is a new species of the genus Helcococcus, for which the name Helcococcus seattlensis sp. nov. is proposed.
FIG 2.

Unrooted tree showing the phylogenetic relationships of strain F5780 to closely related species and to more distant species that share similar morphological features and growth characteristics. The tree was inferred from 16S rRNA gene sequences (1,391 nucleotides) by using the neighbor-joining method, and the scale bar indicates the substitution rate per nucleotide. All names and accession numbers (in parentheses) are those cited in the GenBank database.
We performed a satellite test for all 5 clinical strains of Helcococcus on BA, and after 1 to 3 days of incubation at 35°C in air enriched with 6 to 7% CO2, we found that the colonies adjacent to S. aureus ATCC 25923 showed better growth. For Helcococcus seattlensis sp. nov. F5780, the mean colony size adjacent to and distant from S. aureus was 197 ± 81 μm and 81 ± 26 μm, respectively (means ± standard deviations; P < 0.001 by t test; n = 50). The satelliting phenomenon of H. kunzii F5450, H. sueciensis F6503, and H. seattlensis F5780 was further tested on MHA. The results revealed a more distinct satellite pattern, such that Helcococcus strains grew only as satellite colonies proximal to the streaking of S. aureus on MHA but not away from it. Strains F5450, F6503, and F5780 showed positive colony satellite formation on days 2, 4, and 6, respectively. With regard to the stimulating effect of S. aureus, it is interesting that our 2 H. kunzii strains (F7676 and F5450) were isolated from wounds together with S. aureus. Also, the veterinary H. ovis was isolated with a Staphylococcus sp. (3). These observations suggested that the presence of Staphylococcus spp. may enhance the growth of Helcococcus spp.
Helcococcus spp. and other Gram-positive, slow-growing genera that have specific nutrient requirements, such as Abiotrophia and Granulicatella, can be difficult to identify (19–21). The positive satellite test as well as the rapid and inexpensive LAP and PYR tests can help distinguish the organisms (Table 2). Only a small amount of bacterial inoculum is needed to perform the tests, thus favoring the identification of slow growers.
The antibiotic susceptibilities of Helcococcus species were studied by employing the Sensititre, Etest, and the MicroScan systems (Table 3). The MICs of Helcococcus species were interpreted based on the standard for S. aureus in the guidelines. All the tested Helcococcus species revealed a high MIC for SXT (>4/76 μg/ml), an indication of drug resistance. Intermediate to high MICs for gentamicin were observed for all tested H. kunzii and H. sueciensis isolates, but not with H. seattlensis. High MICs for erythromycin, azithromycin, clindamycin, and intermediate to high MICs for moxifloxacin and levofloxacin were also observed among the Helcococcus spp. strains. There are no CLSI guidelines for performing and interpreting antibiotic susceptibility tests for Helcococcus. The microdilution and Etest results were read on day 3, because the organisms grew slowly.
TABLE 3.
Antibiotic susceptibilities of Helcococcus speciesa
| Antibiotic | MIC (μg/ml) |
||||
|---|---|---|---|---|---|
| H. seattlensis sp. nov. F5780 | H. kunzii F7676 | H. kunzii F5450 | H. sueciensis F6341 | H. sueciensis F6503 | |
| Penicillin | 0.06 | 0.25 | <0.03 | <0.03 | 0.06 |
| Cefuroxime | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| Ceftriaxone | <0.12 | 0.25 | <0.12 | <0.12 | <0.12 |
| Cefotaxime | <0.12 | <0.12 | <0.12 | <0.12 | <0.12 |
| Cefepime | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| Amoxicillin-clavulanic acid | <2/1 | <2/1 | <2/1 | <2/1 | <2/1 |
| Gentamicinb | 1 | 16 | 8 | 8 | 8 |
| Vancomycin | 1 | <0.5 | <0.5 | 4 | 1 |
| Linezolid | 2 | 4 | 2 | 2 | 2 |
| Azithromycinc | <0.25 | 1 | >2 | <0.25 | >2 |
| Erythromycin | 0.5 | 1 | >256 | <0.25 | >256 |
| Tetracycline | <1 | <1 | <1 | <1 | <1 |
| Tigecycline | <0.015 | 0.03 | <0.015 | <0.015 | 0.03 |
| Daptomycin | <0.06 | 0.5 | 0.5 | 0.12 | 0.12 |
| Clindamycin | 0.25 | 0.5 | 0.5 | <0.12 | >256 |
| Moxifloxacin | 4 | <1 | 2 | <1 | 8 |
| Levofloxacin | 16 | 1 | 1 | <0.5 | 8 |
| Trimethoprim-sulfamethoxazole | >4/76 | >4/76 | >4/76 | >4/76 | >4/76 |
| Meropenem | 0.5 | <0.25 | <0.25 | <0.25 | <0.25 |
| Ertapenem | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| Chloramphenicol | 8 | 16 | 8 | 4 | 4 |
An antibiotic susceptibility testing method for Helcococcus spp. has not been standardized by CLSI.
The susceptibility to gentamicin was measured by Etest.
Etest results for azithromycin were not available, and susceptibility was based solely on the result with the Sensititre system.
DISCUSSION
Strain F5780 resembled the species of the genus Helcococcus in cell morphology, microscopic appearance, growth rate (slow), and negative catalase reaction. 16S rRNA gene sequencing revealed that this organism is most closely related to H. sueciensis and H. ovis. The lack of PYR and LAP of F5780 was distinct from other medically significant bacteria that are similar based on Gram stain, catalase reaction and growth characteristics, such as other Helcococcus species, Abiotrophia, Granulicatella, and Streptococcus. Another important trait for all Helcococcus species was their high MIC for SXT, to which Abiotrophia and Granulicatella are also susceptible (19).
The facts that strain F5780 in our current study was recovered from a patient with urosepsis and that H. kunzii caused urocystitis in a sow suggest that Helcococcus species can cause UTI (12). Our finding that all tested Helcococcus species had high MICs (>4/76 μg/ml) for SXT is of medical importance. SXT is currently recommended by the Infectious Diseases Society of America (IDSA) to treat cystitis, pyelonephritis, and catheter-associated UTI (22, 23), while our data showed that SXT may not be effective against Helcococcus spp.
H. kunzii was isolated together with S. aureus from wounds (Table 1, cases 2 and 3) (1, 6). We speculate that helcococci may occur more commonly in clinical specimens but are not detected unless S. aureus is present to enhance the growth or unless particular culture conditions are used. Lacking a good database and a clear algorithm, Helcococcus might be neglected due to its slow growth and the difficulty in its identification, resulting in an inappropriate choice of antibiotics for the patient.
Description of Helcococcus seattlensis sp. nov.
Helcococcus seattlensis. se.at.tlen'sis. N.L. masc. adj. seattlensis of Seattle, the city in which the organism was isolated. Cells are Gram-positive cocci and occur singly or in clusters with variable cell sizes (0.5 to 1.8 μm in diameter). Cells are nonmotile and non-spore forming. The organism grows best on CA (CA > BA) at 35°C (35°C > 30°C > 42°C) and equally well under different oxygen conditions (air, air enriched with 6 to 7% CO2, microaerophilic, or anaerobic). The colonies are nonhemolytic, gray, and pinpoint on CA after 48 h of incubation at 35°C in air enriched with 6 to 7% CO2. It grows as satellite colonies adjacent to both clinical and ATCC 25923 strains of S. aureus, while the satellite phenomenon is more remarkable on Mueller-Hinton agar than on sheep blood agar. On blood agar, the mean colony sizes adjacent to and distant from S. aureus are 197 ± 81 μm and 81 ± 26 μm, respectively. On Mueller-Hinton agar, the organism only grows as satellite colonies proximal to the streaking of S. aureus. It takes 3 days and 6 days to grow as satellite colonies on blood agar and Mueller-Hinton agar, respectively. The organism is catalase, PYR, and LAP negative when tested by conventional methods. It produces β-glucuronidase and β-galactosidase. Acid is produced from ribose. The organism is resistant to trimethoprim-sulfamethoxazole.
The type strain of H. seattlensis sp. nov. is strain F5780, which was isolated as the sole organism from blood of a patient with urosepsis. Its 16S rRNA gene sequence has been deposited in the GenBank data library under accession number KF780874. The isolate has been accepted for deposition in the ATCC (ATCC number to be determined).
ACKNOWLEDGMENTS
We acknowledge the excellent support of personnel of the microbiology laboratory, especially Janelle Lee and Uyen Bui. We sincerely thank Jean P. Euzéby for his expertise in naming the novel organism.
This study was completed with the Puget Sound Veterans Administration Medical Center IRB approval (MIRB 01012).
Footnotes
Published ahead of print 26 December 2013
REFERENCES
- 1.Collins MD, Facklam RR, Rodrigues UM, Ruoff KL. 1993. Phylogenetic analysis of some Aerococcus-like organisms from clinical sources: description of Helcococcus kunzii gen. nov., sp. nov. Int. J. Syst. Bacteriol. 43:425–429 [DOI] [PubMed] [Google Scholar]
- 2.Collins MD, Falsen E, Brownlee K, Lawson PA. 2004. Helcococcus sueciensis sp. nov., isolated from a human wound. Int. J. Syst. Evol. Microbiol. 54:1557–1560. 10.1009/ijs.0.63077-0 [DOI] [PubMed] [Google Scholar]
- 3.Collins MD, Falsen E, Foster G, Monasterio LR, Dominguez L, Fernandez-Garazabal JF. 1999. Helcococcus ovis sp. nov., a Gram-positive organism from sheep. Int. J. Syst. Bacteriol. 49:1429–1432 [DOI] [PubMed] [Google Scholar]
- 4.Woo PC, Tse H, Wong SS, Tse CW, Fung AM, Tam DM, Lau SK, Yuen KY. 2005. Life-threatening invasive Helcococcus kunzii infections in intravenous-drug users and ermA-mediated erythromycin resistance. J. Clin. Microbiol. 43:6205–6208. 10.1128/JCM.43.12.6205-6208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Jorge C, Cordero J, Marin M, Esteban J. 2012. Prosthetic joint infection caused by Helcococcus kunzii. J. Clin. Microbiol. 50:528–530. 10.1128/JCM.01244-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riegel P, Lepargneur JP. 2003. Isolation of Helcococcus kunzii from a post-surgical foot abscess. Int. J. Med. Microbiol. 293:437–439 http://dx.doi.org/10.1078/1438-4221-00284 [DOI] [PubMed] [Google Scholar]
- 7.Lemaitre N, Huvent D, Loiez C, Wallet F, Courcol RJ. 2008. Isolation of Helcococcus kunzii from plantar phlegmon in a vascular patient. J. Med. Microbiol. 57:907–908. 10.1099/jmm.0.2008/000471-0 [DOI] [PubMed] [Google Scholar]
- 8.Chagla AH, Borczyk AA, Facklam RR, Lovgren M. 1998. Breast abscess associated with Helcococcus kunzii. J. Clin. Microbiol. 36:2377–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McNicholas S, McAdam B, Flynn M, Humphreys H. 2011. The challenges of implantable cardiac device infection due to Helcococcus kunzii. J. Hosp. Infect. 78:337–338. 10.1016/j.jhin.2011.04.010 [DOI] [PubMed] [Google Scholar]
- 10.Haas J, Jernick SL, Scardina RJ, Teruya J, Caliendo AM, Ruoff KL. 1997. Colonization of skin by Helcococcus kunzii. J. Clin. Microbiol. 35:2759–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caliendo AM, Jordan CD, Ruoff KL. 1995. Helcococcus, a new genus of catalase-negative, gram-positive cocci isolated from clinical specimens. J. Clin. Microbiol. 33:1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grattarola C, Bellino C, Tursi M, Maggi E, D'Angelo A, Gianella P, Dondo A, Cagnasso A. 2010. Helcococcus kunzii isolated from a sow with purulent urocystitis. J. Clin. Microbiol. 48:3019–3020. 10.1128/JCM.00554-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Humphries RM, Lee C, Hindler JA. 2011. Aerococcus urinae and trimethoprim-sulfamethoxazole. J. Clin. Microbiol. 49:3934–3935. 10.1128/JCM.05535-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute 2013. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 15.Claassen SL, Reese JM, Mysliwiec V, Mahlen SD. 2011. Achromobacter xylosoxidans infection presenting as a pulmonary nodule mimicking cancer. J. Clin. Microbiol. 49:2751–2754. 10.1128/JCM.02571-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall L, Doerr KA, Wohlfiel SL, Roberts GD. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J. Clin. Microbiol. 41:1447–1453. 10.1128/JCM.41.4.1447-1453.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park KS, Ki CS, Kang CI, Kim YJ, Chung DR, Peck KR, Song JH, Lee NY. 2012. Evaluation of the GenBank, EzTaxon, and BIBI services for molecular identification of clinical blood culture isolates that were unidentifiable or misidentified by conventional methods. J. Clin. Microbiol. 50:1792–1795. 10.1128/JCM.00081-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo PC, Fung AM, Lau SK, Chan BY, Chiu SK, Teng JL, Que TL, Yung RW, Yuen KY. 2003. Granulicatella adiacens and Abiotrophia defectiva bacteraemia characterized by 16S rRNA gene sequencing. J. Med. Microbiol. 52:137–140. 10.1099/jmm.0.04950-0 [DOI] [PubMed] [Google Scholar]
- 20.Zheng X, Freeman AF, Villafranca J, Shortridge D, Beyer J, Kabat W, Dembkowski K, Shulman ST. 2004. Antimicrobial susceptibilities of invasive pediatric Abiotrophia and Granulicatella isolates. J. Clin. Microbiol. 42:4323–4326. 10.1128/JCM.42.9.4323-4326.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roggenkamp A, Abele-Horn M, Trebesius KH, Tretter U, Autenrieth IB, Heesemann J. 1998. Abiotrophia elegans sp. nov., a possible pathogen in patients with culture-negative endocarditis. J. Clin. Microbiol. 36:100–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE. 2011. International clinical practice guidelines for the treatment of acute uncomplicated cystitis and pyelonephritis in women: a 2010 update by the Infectious Diseases Society of America and the European Society for Microbiology and Infectious Diseases. Clin. Infect. Dis. 52:e103–e120. 10.1093/cid/ciq257 [DOI] [PubMed] [Google Scholar]
- 23.Hooton TM, Bradley SF, Cardenas DD, Colgan R, Geerlings SE, Rice JC, Saint S, Schaeffer AJ, Tambayh PA, Tenke P, Nicolle LE. 2010. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 International Clinical Practice Guidelines from the Infectious Diseases Society of America. Clin. Infect. Dis. 50:625–663. 10.1086/650482 [DOI] [PubMed] [Google Scholar]


