Abstract
A total of 330 clinical Vibrio cholerae O1 serogroups from China dating between 1961 and 2010 were investigated. By phenotypic biotyping and genetic analysis, during the seventh pandemic of V. cholerae O1 in China, the isolates of hybrid biotype (mixed classical phenotypes) were present during the entire1961-2010 period, while El Tor genetic shifts appeared in 1992 and replaced the prototype El Tor from 2002 to 2010.
TEXT
Vibrio cholerae is the causative agent of the life-threatening diarrheal disease cholera (1). Seven distinct pandemics of cholera have been recorded since the first pandemic in 1817. The sixth pandemic, and presumably the earlier pandemics, were caused by V. cholerae O1 of the classical biotype. The current seventh pandemic, which originated in Indonesia in 1961, is the most extensive in geographic spread and duration, and the causative agent is V. cholerae O1 of the El Tor biotype. In 1992, an outbreak of O139 cholera emerged in the coastal areas of India and then spread to many countries in Asia (2, 3).
The classification of the classical and El Tor biotypes of V. cholerae O1 is based on several phenotypic and genetic traits. The phenotypic traits include chicken erythrocyte agglutination (CCA), Voges-Proskauer (VP) test results, susceptibility to polymyxin B (PB; 50 units), and biotype-specific phages (1). The genetic traits include the variants of the gene encoding the cholera toxin subunit B (ctxB). In addition, the repeat sequence transcriptional regulator (rstR) gene and the major toxin coregulated pilus gene (tcpA) possess classical and El Tor-specific alleles, while the repeat in the toxin gene (rtxC) is present in El Tor but absent in classical biotype isolates (4).
Several atypical or variant El Tor biotypes have recently been identified. The Matlab variant was the first atypical El Tor biotype. It was identified in Matlab, Bangladesh, between 1991 and 1994 (5). Another study (6) reported a hybrid CTXΦ isolate carrying El Tor rstR and classical ctxB that has completely replaced the El Tor biotype in Kolkata, India, since 1995. Other atypical El Tor isolates have been reported in other countries in Asia (7, 8) and Africa (9, 10), as well as in Mexico (11). Previously, we identified three novel El Tor variants from China in which the ctxB genotype was different from known genotypes (12). These results suggested that there were variants in China; however, the traits have not been investigated.
In this study, 330 V. cholerae O1 El Tor biotype isolates were characterized and compared; these isolates were collected over nearly 50 years (1961 to 2010) and were obtained from different provinces in China from 1961to 2010, either from outbreaks or sporadic cases. All of the bacterial isolates were screened for the oxidase reaction and were identified by a slide agglutination test using specific polyvalent antisera against V. cholerae O1 (Ogawa and Inaba; S&A Reagents Lab, Bangkok, Thailand). The serogroups of these isolates were reconfirmed by real-time PCR targeting the O1 rfb-specific O biosynthetic gene (13).
For phenotypic tests, polymyxin B (PB; 50 units) susceptibility test, CCA, and the VP reaction were performed using standard procedures (14) and a previous report (15). The V. cholerae reference classical isolate 569B and the reference El Tor isolate N16961 were included as controls. The phenotypic tests were repeated three times to ensure reliable results.
To complement the phenotypic characterization of the biotypes, PCR assays were carried out using conventional PCR amplification. The target genes included ctxB (16), El Tor and classical variants of rstR (17, 18) and tcpA (19), and the repeat in the toxin gene (rtxC) (4). Table 1 shows the sequences used for primer design and their origins. A commercial company (TaKaRa, Dalian, China) performed the sequencing of the PCR products of ctxB. Comparative analyses of the ctxB sequences were conducted using BioEdit. ClustalW was used to obtain multiple alignments of the nucleotide and predicted amino acid sequences of ctxB. The ctxB sequences of isolates N16961 and 569B were used as El Tor and classical references, respectively.
TABLE 1.
PCR primers used in this study
| Primer | Nucleotide sequence (5′ to 3′)a | Amplicon size (bp) | Use | Reference |
|---|---|---|---|---|
| O1-rfb | GCGTAAATATCTAAACGATTGCATTG, AAACTCAGTTTCGAAGCGATCAA | 83 | Real-time PCR | 13 |
| ctxB | GCCGGGTTGTGGGAATGCTCCAAG, CATGCGATTGCCGCAATTAGTATGGC | 536 | PCR, sequencing | 16 |
| rstRET | GAGCTAAAATACAGCAACCAATGC, ACTCACCTTGTATTCG | 487 | PCR | 17 |
| rstRC | TATTGGGATTGTAAACAGCTGTCC, ACTCACCTTGTATTCG | 480 | PCR | 18 |
| tcpA 72F | CACGATAAGAAAACCGGTCAAGAG | 19 | ||
| tcpA 477R | CGAAAGCACCTTCTTTCACGTTG | 451 (El Tor) | PCR | 19 |
| tcpA 647R | TTACCAAATGCAACGCCGAATG | 620 (classical) | PCR | 19 |
| rtxC | CGACGAAGATCATTGACGAC, CATCGTCGTTATGTGGTTGC | 265 | PCR | 4 |
In entries with two sequences, the sequence for the reverse primer follows that for the forward primer.
For classifying the biotypes of the variant V. cholerae O1 isolates, we referred to the literature (20). We designated “atypical El Tor” as all isolates with mixed classical and El Tor traits. Isolates having conventional phenotypic properties of both classical and El Tor biotypes were designated as having a “hybrid biotype,” and isolates similar to the El Tor biotype in conventional phenotypic traits but with the classical ctxB and/or rstR genotype were designated as being “El Tor variants.”
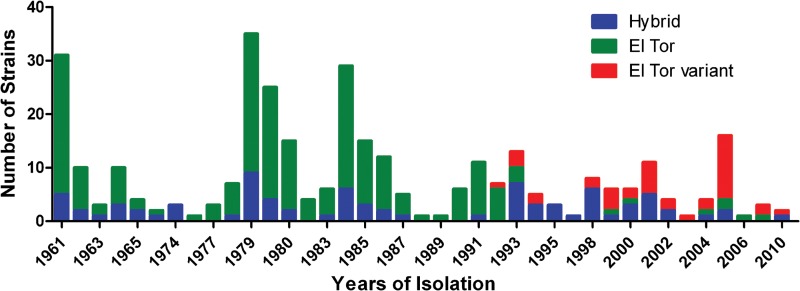
As shown in Table S1 in the supplemental material, among the 330 V. cholerae O1 isolates, 110 were identified as having the Inaba serotype and 220 were identified as having the Ogawa serotype. Phenotypic tests revealed that 250 isolates (75.8%) were typical El Tor prototypes (CCA+, VP+, and PB resistant [PBr]), identical to the El Tor reference isolate N16961. The other 80 isolates (24.2%) belonged to the classic phenotypes: classic phenotypes of CCA−, VP−, and PB susceptible [PBs] accounted for 30, 51, and 14 isolates, respectively. Specifically, the most common phenotype combinations were CCA+ VP− PBr (40 isolates) and then CCA− VP+ PBr (20 isolates), followed by CCA+ VP+ PBs (9 isolates), CCA− VP− PBr (6 isolates), CCA− VP− PBs (4 isolates), and CCA+ VP− PBs (1 isolate); no isolate had a phenotype combination of CCA− VP+ PBs. The classical phenotype isolates were present in the period from 1961 to 2010 (Fig. 1).
FIG 1.
Distribution of V. cholerae O1 El Tor isolates based on deduced biotypes, by year.
Genetic analysis showed that 278 isolates (84.2%) were positive for the ctxB gene. All isolates except four were positive for the rtxC gene, which verified that genetically, the majority of the isolates belonged to the El Tor biotype, with toxin-producing capacity and epidemic potential. The rstR PCR results showed that 304 (92.1%) isolates were positive for El Tor (rstRET), classical (rstRC), or El Tor and classical rstR genes (rstRET/C). In a detailed analysis of rstR-positive isolates, 254 (83.6%, 254/304) were positive for El Tor rstR only, 27 (8.9%) were positive for classical rstR only, and 23 (7.6%) were positive for El Tor and classical. During the period 1961 to 1991, all rstR-positive isolates were rstRET, except for two isolates from 1986 that were rstRC and rstRET/C, respectively. Between 1992 and 2010, the isolates carried either rstRET or rstRC or both rstRET and rstRC. Of the isolates in this study, 302 (91.5%) carried tcpAET, but tcpAC was not found (see Table S1 in the supplemental material).
Based on amino acid residue substitutions at positions 39, 46, and 68, three ctxB genotypes have been identified among O1 V. cholerae isolates, genotypes 1, 2, and 3 (21). In our study, of all 330 V. cholerae O1 isolates, 278 (84.2%) were positive for ctxB, and 212 (64.2%) isolates were classified as genotype 3 on the basis of multiple sequence alignments with ctxB from the typical El Tor reference isolate N16961. The other 66 (20%) isolates were classified as genotype 1, carrying the classical trait of ctxB, typical of the classical reference isolate 569B (see Table S1 in the supplemental material).
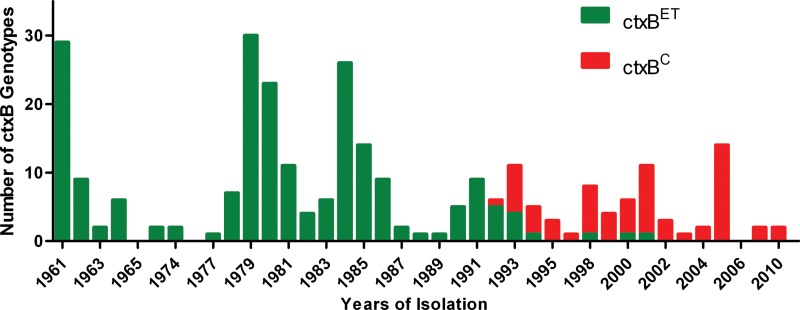
From 1961to 1991, only the El Tor allele of ctxB was present. The first classical biotype ctxB clinical isolate emerged in 1992, while the others were El Tor biotype ctxB. In 1993, isolates carrying classical ctxB were found in two other provinces, and classical ctxB isolates coexisted with the El Tor biotype of ctxB but gradually increased and became predominant by 2001. During the period 2002 to 2010, clinical isolates carrying classical biotype ctxB were completely replaced with the El Tor biotype ctxB allele (Fig. 2).
FIG 2.
Distribution of V. cholerae O1 El Tor isolates based on the ctxB subtypes (amino acid sequence alignment), by year.
This is the first study that describes the phenotypic traits of clinical V. cholerae O1 isolates in China over a long period of time (1961 to 2010). Although 75.8% of the isolates were typical El Tor, 24.2% were the classic phenotypes. It is noteworthy that the hybrid biotype isolates were present in all years from 1961 to 2010 (Fig. 1), whereas all hybrid biotype isolates except four harbored the rtxC gene, an El Tor biotype-specific genetic marker (22). This result indicates that the phenotypic changes in El Tor isolates occurred throughout the seventh pandemic in China. Although there is no other continual report of phenotypic changes in isolates from the first stage of the seventh pandemic, this hybrid biotype in the first stage of the seventh pandemic may be a universal phenomenon. We propose that there was a “phenotypic shifting period” before 1961, when classical and El Tor phenotypes coexisted among isolates, similar to the “genetic shifting period” of isolates between 1991 and 1994 in Matlab, Bangladesh (5), between 1990 and 1994 in Kolkata, India (6), and between 1992 and 2002 in China, although the mechanisms involved in the emergence of the hybrid biotype are not clear.
In addition to reports of atypical El Tor biotypes in Bangladesh and India and Mozambique variants in Africa, studies have also identified an altered variant that completely replaced the progenitor El Tor isolates in Thailand (7), Vietnam (8), and Angola (9), all around 1991. Taken together with our study in China, it is clear that the genetic shift in El Tor V. cholerae O1occurred around 1991 or even before, becoming the predominant isolate or replacing the progenitor El Tor isolate in many Asian and African countries.
In the present study, the biotypes of CTXΦ in China underwent the following shifts: a period of typical CTXΦET(rstRET ctxBET) (1961 to 1992); a period of coexistence of CTXΦET and CTXΦC (rstRC ctxBC) (1993 to 2001); and a period in which CTXΦC replaced CTXΦET (2002 to 2010). This process suggests that during the genetic shifting of El Tor V. cholerae O1, horizontal gene transfer of virulence genes, as well as genetic recombination and mutation, might have occurred (22).
In conclusion, a retrospective assay of the phenotypic and genetic characteristics of clinical isolates from the seventh pandemic V. cholerae O1 in China was undertaken. The El Tor variants have replaced the prototype seventh pandemic El Tor variant in China, which is consistent with the shift in most countries in Asia and Africa. Recently, V. cholerae O1 El Tor isolates producing Haitian variant cholera toxin (HCT) and showing reduced susceptibility to ciprofloxacin caused a cholera outbreak associated with a high case fatality rate in India (23). HCT-secreting strains have been responsible for severe cholera epidemics in western Africa (24) and Haiti (25). The role of new variants in cholera epidemics and pathogenicity should be noted, and additional surveillance is required to understand the epidemiology and the pathogenic and molecular evolution of atypical El Tor isolates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NSFC:30872260), the State Key Laboratory for Infectious Diseases Prevention and Control (2011SKLID201), and a Special Grant for Prevention and Treatment of Infectious Diseases (2008ZX10004-012).
Footnotes
Published ahead of print 18 December 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03121-13.
REFERENCES
- 1.Kaper JB, Morris JG, Jr, Levine MM. 1995. Cholera. Clin. Microbiol. Rev. 8:48–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert MJ, Siddique AK, Islam MS, Faruque AS, Ansaruzzaman M, Faruque SM, Sack RB. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. 10.1016/0140-6736(93)90481-U [DOI] [PubMed] [Google Scholar]
- 3.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703–704. 10.1016/0140-6736(93)90480-5 [DOI] [PubMed] [Google Scholar]
- 4.Chow KH, Ng TK, Yuen KY, Yam WC. 2001. Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39:2594–2597. 10.1128/JCM.39.7.2594-2597.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair GB, Faruque SM, Bhuiyan NA, Kamruzzaman M, Siddique AK, Sack DA. 2002. New variants of Vibrio cholerae O1 biotype El Tor with attributes of the classical biotype from hospitalized patients with acute diarrhea in Bangladesh. J. Clin. Microbiol. 40:3296–3299. 10.1128/JCM.40.9.3296-3299.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raychoudhuri A, Patra T, Ghosh K, Ramamurthy T, Nandy RK, Takeda Y, Balakrish-Nair G, Mukhopadhyay AK. 2009. Classical ctxB in Vibrio cholerae O1, Kolkata, India. Emerg. Infect. Dis. 15:131–132. 10.3201/eid1501.080543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Na-Ubol M, Srimanote P, Chongsa-Nguan M, Indrawattana N, Sookrung N, Tapchaisri P, Yamazaki S, Bodhidatta L, Eampokalap B, Kurazono H, Hayashi H, Nair GB, Takeda Y, Chaicumpa W. 2011. Hybrid & El Tor variant biotypes of Vibrio cholerae O1 in Thailand. Indian J. Med. Res. 133:387–394 [PMC free article] [PubMed] [Google Scholar]
- 8.Tran HD, Alam M, Trung NV, Kinh NV, Nguyen HH, Pham VC, Ansaruzzaman M, Rashed SM, Bhuiyan NA, Dao TT, Endtz HP, Wertheim HF. 2012. Multi-drug resistant Vibrio cholerae O1 variant El Tor isolated in northern Vietnam between 2007 and 2010. J. Med. Microbiol. 61:431–437. 10.1099/jmm.0.034744-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceccarelli D, Spagnoletti M, Bacciu D, Cappuccinelli P, Colombo MM. 2011. New V. cholerae atypical El Tor variant emerged during the 2006 epidemic outbreak in Angola. BMC Microbiol. 11:130. 10.1186/1471-2180-11-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naha A, Chowdhury G, Ghosh-Banerjee J, Senoh M, Takahashi T, Ley B, Thriemer K, Deen J, Seidlein LV, Ali SM, Khatib A, Ramamurthy T, Nandy RK, Nair GB, Takeda Y, Mukhopadhyay AK. 2013. Molecular characterization of high-level-cholera-toxin-producing El Tor variant Vibrio cholerae strains in the Zanzibar Archipelago of Tanzania. J. Clin. Microbiol. 51:1040–1045. 10.1128/JCM.03162-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alam M, Islam MT, Rashed SM, Johura FT, Bhuiyan NA, Delgado G, Morales R, Mendez JL, Navarro A, Watanabe H, Hasan NA, Colwell RR, Cravioto A. 2012. Vibrio cholerae classical biotype strains reveal distinct signatures in Mexico. J. Clin. Microbiol. 50:2212–2216. 10.1128/JCM.00189-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang P, Zhou H, Kan B, Wang D. 2013. Novel ctxB variants of Vibrio cholerae O1 isolates, China. Infect. Genet. Evol. 20:48–53. 10.1016/j.meegid.2013.08.004 [DOI] [PubMed] [Google Scholar]
- 13.Wang XM, Wang DC, Tan HL, Zhong HJ, Chen JD, Li BS, Ke CW, Yan MY, Zhang J, Kan B. 2007. Development and application of real-time polymerase chain reaction to detect Vibrio cholerae O1 and O139 in river water. Zhonghua Liu Xing Bing Xue Za Zhi. 28:768–771 (In Chinese.) [PubMed] [Google Scholar]
- 14.WHO 1987. Manual for laboratory investigations of acute enteric infections. World Health Organization, Geneva, Switzerland [Google Scholar]
- 15.Son MS, Megli CJ, Kovacikova G, Qadri F, Taylor RK. 2011. Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes. J. Clin. Microbiol. 49:3739–3749. 10.1128/JCM.01286-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Jain M, Goel AK, Bhadauria S, Sharma SK, Kamboj DV, Singh L, Ramamurthy T, Nair BG. 2009. A large cholera outbreak due to a new cholera toxin variant of the Vibrio cholerae O1 El Tor biotype in Orissa, Eastern India. J. Med. Microbiol. 58:234–238. 10.1099/jmm.0.002089-0 [DOI] [PubMed] [Google Scholar]
- 17.Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910–1914. 10.1126/science.272.5270.1910 [DOI] [PubMed] [Google Scholar]
- 18.Mukhopadhyay AK, Chakraborty S, Takeda Y, Nair GB, Berg DE. 2001. Characterization of VPI pathogenicity island and CTXphi prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737–4746. 10.1128/JB.183.16.4737-4746.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera IN, Chun J, Huq A, Sack RB, Colwell RR. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421–2429. 10.1128/AEM.67.6.2421-2429.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raychoudhuri A, Mukhopadhyay AK, Ramamurthy T, Nandy RK, Takeda Y, Nair GB. 2008. Biotyping of Vibrio cholerae O1: time to redefine the scheme. Indian J. Med. Res. 128:695–698 [PubMed] [Google Scholar]
- 21.Olsvik O, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth IK, Fields PI. 1993. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J. Clin. Microbiol. 31:22–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Safa A, Nair GB, Kong RY. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol. 18:46–54. 10.1016/j.tim.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 23.Kumar P, Mishra DK, Deshmukh DG, Jain M, Zade AM, Ingole KV, Yadava P. 18 September 2013. Haitian variant ctxB producing Vibrio cholerae O1 with reduced susceptibility to ciprofloxacin is persistent in Yavatmal, Maharashtra, India after causing a cholera outbreak. Clin. Microbiol. Infect. 10.1111/1469-0691.12393 [DOI] [PubMed] [Google Scholar]
- 24.Quilici ML, Massenet D, Gake B, Bwalki B, Olson DM. 2010. Vibrio cholerae O1 variant with reduced susceptibility to ciprofloxacin, Western Africa. Emerg. Infect. Dis. 16:1804–1805. 10.3201/eid1611.100568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjolund-Karlsson M, Reimer A, Folster JP, Walker M, Dahourou GA, Batra DG, Martin I, Joyce K, Parsons MB, Boncy J, Whichard JM, Gilmour MW. 2011. Drug-resistance mechanisms in Vibrio cholerae O1 outbreak strain, Haiti, 2010. Emerg. Infect. Dis. 17:2151–2154. 10.3201/eid1711.110720 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.